Abstract
A zwitterionic dimer model constructed of inter-molecular –N–H⋯O bonding has been proposed for the solid sample of DL-3-Aminoisobutyric acid consistent with IR absorption and Raman spectral features measured in the 3500–400/50 cm−1. This zwitterionic dimer model in water as solvent has been computed at B3LYP/6–311++G(d,p) and B3LYP-D3/6–311++G(d,p) levels including Grimme's dispersion correction associated with the –N–H⋯O interaction and SCRF-SMD method. Of the several possible monomer and dimer conformational structures, the most stable dimer constructed of two zwitterion monomer units has produced vibrational modes due to the –NH3+ cation and –CO2‾ anion involved in the –N–H⋯O bonding in fair agreement with the observed broad but composite IR modal features near the 3500–2000 cm−1. Except for the frequency of asymmetric stretching mode of the –NH3+ cation, its symmetric and bending modes agree with the observed values. As for the –CO2‾ anion, the frequencies of all of its modes are in good agreement with the experiment. Natural bond orbital (NBO), molecular electrostatic potential (MEP), atoms-in-molecules (AIM) and non-covalent interaction (NCI) analyses have been used to understand electronic characterization of the –N–H⋯O bonding.
Keywords: Physical chemistry, Theoretical chemistry, Molecular physics, DL-3-Aminoisobutyric acid, IR, Raman, DFT, AIM, N–H⋯O bonding
1. Introduction
Amino acids are the building blocks of polypeptides and proteins with diverse structural and functional properties. They have been and still are of fundamental importance because of their vital role in the vast biological landscape. Lately non-proteinogenic amino acids, also called unnatural amino acids, have also aroused interest for similar reasons including pharmaceutical applications [1, 2]. Biochemical activities in biological species primarily involve non-covalent interactions, say, among amino acids and other molecular fragments, for example, in bases in DNA and RNA; the interactions include H-bonding and stacking. The common H-bonds are O–H⋯O, N–H⋯O, C–H⋯O, and so on [3]. Invariably the amino acids or unnatural amino acids have the neutral structures (NE) in the gas phase and zwitterionic structure (ZW) in the condensed phase. They are categorized as α, β, γ and δ-amino acids depending on the position of amino group (−NH2) in the carbon chain [4, 5, 6]. Although β-amino acids are less abundant than their α-analogues, they occur in nature both in free and peptide-bound species [7, 8]. The oligomers containing β-amino acids have emerged as promising tools in medicinal chemistry with higher stability toward common peptides as they possess the potential to fold into secondary structures consisting of helices, turns and pleated sheets [7, 8, 9, 10]. The title molecule, DL-3-Aminoisobutyric acid (for short 3AIBA) being synonymous with α-Methyl-β-alanine, is a β-amino acid which occurs naturally in all living organisms. In humans and animals, 3AIBA can be formed from the nucleotide thymine and also from the amino acid valine [11, 12]. Since 3AIBA is structurally similar to β-alanine, it can also be called as α-methyl-β-alanine having one additional methyl group on α-carbon atom. In their free form, β-alanine and 3AIBA are found as metabolites in the mammals and also as moieties in several pharmacologically active compounds such as antibiotics, antitumor agents and antifungal agents [7, 8, 10, 13].
Among the non-covalent interactions, namely, H-bonding, π-stacking, van der Waals and electrostatic types, the landscape for H-bonding interactions is vast with implications across diverse supramolecular systems, nanostructures and nano-biomaterials [14, 15]. The nearest competitor to H-bonding is π-stacking interaction. Both H-bonding and π-stacking interactions are proved determinants in DNA-protein interactions for molecular recognition. The van der Waals and hydrophobic interactions, of course, play their role as well but as secondary determinants [16]. In a combined theoretical and experimental study, Krylov et al. have shown that H-bonding, among non-covalent interactions, influence ionization energies (IEs) of the dimers of nucleic acid bases: adenine-adenine, thymine-thymine and adenine-thymine dimers [17]. In a detailed theoretical and experimental study on the nine possible 2-pyridone-n-fluorobenzene dimer species, Leutwyler et al. have shown that H-bonding and π-stacking compete with small energy differences, both of which being accounted satisfactorily by the coupled-cluster model comprising single and double excitations with a perturbative corrections for triple excitations (CCSD(T)) [18]. In constructing 2D supramolecular networks, the role of multiple H-bonds has been ascertained in the structural polymorphism at the solid-liquid interface [19, 20, 21, 22, 23, 24]. In particular, the role of multiple H-bonds in inducing chirally different morphologies in a series of self-assembled nanostructures of 2-hydroxyl-7-pentadecyloxy-fluorenone has been demonstrated by Wenli Deng et al [25]. Some experimental vibrational studies and electronic structure calculations have also been reported for NE and ZW species of amino acids and aminophosphates [26, 27, 28].
Experimental and theoretical vibrational spectroscopy are proved techniques for investigating structural and/or conformational aspects of H-bonded molecular systems and their complexation with other molecules with potential implications for understanding the role of H-bonding in diverse material and biological phenomena [29, 30, 31, 32]. In the present work we study the role of inter-molecular –N–H⋯O bonding in building dimer species of 3AIBA and characterize dimerization on the basis of the structural, spectral and electronic properties. H-bond-induced dimer structures are of interest in decoding molecular vibrational structures since it is reasonable to assume the dimer species as basic units of intermolecular association in condensed phases [33, 34]. It is profitable to correlate structural, electronic and vibrational characterization with observed vibrational IR and Raman spectral features. Electronic structure calculations from density functional theory (DFT) codes provide an opportunity to model electronic and vibrational properties corresponding to different stable NE conformers in gas phase and ZW conformers in solvent media which are otherwise experimentally difficult to realize. Experimental IR spectrum of 3AIBA shows a very broad composite band structure across 3500–2000 cm−1, making assignments of the bands untenable. It calls for the construction of possible ZW dimer species defined by H-bonding between –NH3+and –CO2‾ groups and such dimer species would yield vibration modes for the satisfactory characterization of the observed IR and Raman spectral features and provide enhanced understanding of the involvement of electronic molecular orbital in –N–H⋯O bonding. We have carried out this proposed work using DFT modeling and NBO analysis. The NBO provides a satisfactory understanding of the stability of the H-bonded dimer species in terms of perturbative interaction energies resulting from the overlap integral between the lone-pair orbital of the donor and the anti-bonding orbital of the acceptor in the –N–H⋯O bonding. All these aspects constitute the principal aim of the present work.
Experimental and computational studies of NE form of β-alanine crystals and its ZW form in the aqueous media have been reported [35, 36]. The XRD structural studies of H-bonding in a series of amino-substituted carboxylic acids have been investigated [37, 38, 39]. The first among the series is γ–aminobutyric acid (GABA). The room and low temperatures structural studies of monoclinic phase of GABA have been carried out by different workers [40, 41, 42]. In monoclinic phase, the molecule is observed as partially folded zwitterion in gauche conformation. The ZW form of GABA is again observed in the tetragonal phase [37]. The three hydrogen atoms of –NH3+ form three strong H-bonds as in the monoclinic phase, but there is additional fourth weaker H-bond resulting in a two-centre bifurcated bond. Also the evidence suggests that there is an intramolecular –N–H⋯O bridge bond. The three strong H-bonds are similar in both monoclinic and tetragonal phases. There are infinite chains of H-bonded molecules running in one dimension that are cross-linked by the remaining H-bonds to form a three dimensional network. The major difference in both phases is that monoclinic structure shows gauche conformation while the tetragonal structure shows trans conformation of CH2–CH2 groups. Subsequent XRD study on another member of the series, 8-Aminocaprylic acid (8ACA) has been shown to crystallize inthe centrosymmetric space group P21/n in the extended ZW form upon dissolving in water [38]. It forms five intermolecular H-bonds, two of which are bifurcated. Each molecule is linked to six neighbouring molecules by a total of ten H-bonds in the monoclinic phase. DL-3-Aminoisobutyric acid monohydrate (DL-3-AIBA·H2O) whose structure is comparable with the afore-mentioned molecules crystallizes in the orthorhombic with centrosymmetric space group Pbca as a partially folded ZW. With this structure each acid molecule and its associated water molecule are directly H-bonded to five acid molecules and two water molecules. The three hydrogens in the –NH3+ group form inter-molecular H-bonding with the two –CO2‾ groups of the neighboring molecules and one water molecule. The N⋯O distances range from 2.758 to 2.809 Å and –N–H⋯O angles from 149 to 171° [39]. It is therefore reasonable to assume that the 3AIBA has also ZW form with affinity for inter-molecular –N–H⋯O bonding as is true for DL-3-Aminoisobutyric acid monohydrate. This is borne out by the fact that in the present work, some strong IR absorptions in the region 1675-1400 cm−1 are presumably produced by the –NH3+ and –CO2‾ groups as characteristic vibrational modes and a medium weak but highly characteristic band at 2139 cm−1 is the signature of the sum of the frequencies of asymmetric bending and torsional modes of –NH3+ group. This sum mode is the most reliable characteristic band for the identification of the –NH3+ cation in all the amino acids with ZW structure [43, 44]. In our previous study, it was satisfactorily explained the observed vibrational spectral features of L-β-Homoserine in terms of ZW dimer structures characterized by inter-molecular –N–H⋯O and –O–H⋯O bonding [45]. To account for the vibrational structure of 3AIBA, we computed seven –N–H⋯O bonded ZW dimer structures, of which the most stable one has produced a molecular structure and vibrational bands, namely, due to the –NH3+, –CO2‾ groups and –N–H⋯O bonding, in fair agreement with the experimental IR and Raman spectral features. Overall, the computed structural parameters of this dimer species are in fair agreement with the experimental crystal structure of 3AIBA monohydrate within 4%. A detailed NBO analysis has provided such properties as molecular orbital occupancies, charges on the atoms and the second order perturbative energies which support the strong –N–H⋯O bonding in the most stable dimer species (D1) referred to above. Further, topological AIM and NCI analyses have been performed to characterize the H-bonding in D1 and its MEP surface has been plotted to understand the reactivity of the molecule.
2. Experimental
2.1. Experimental measurements
The solid sample of DL-3-Aminoisobutyric acid was purchased from Sigma Aldrich Chemical Company and used without further purification. FT-IR spectra of the pellets made up of solid sample were recorded on a Nicolet 6700 FT-IR spectrometer that uses an Alum standard ETC Ever-Glo IR source, Deuterated Triglycine Sulphate (DTGS) detector equipped with KBr window. Pellets were prepared by taking the sample and KBr in 1:100 ratio and the spectra were recorded with 4 cm−1 resolution in the region 4000 − 400 cm−1 for 50 scans. Raman spectra of solid sample were recorded without sample preparation on the Nicolet NXR-FT-Raman Module spectrometer that uses a Nd:YVO4 (1064 nm) laser as a source of excitation, CaF2 beam splitter and liquid nitrogen cooled Ge detector. The spectra were recorded with 4 cm−1 resolution in the region 4000–100 cm−1 for 500 scans.
2.2. Computational modeling
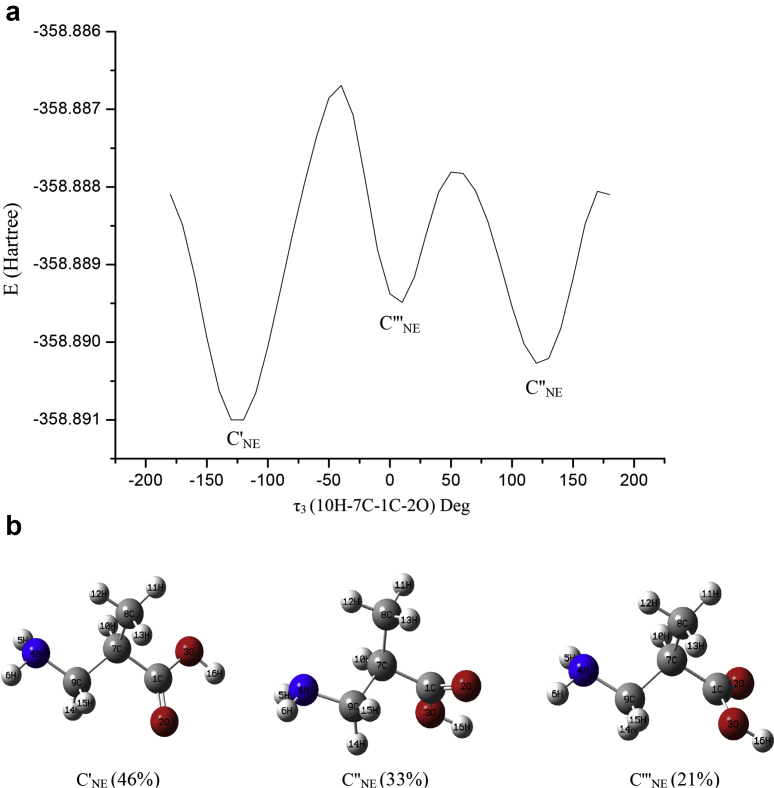
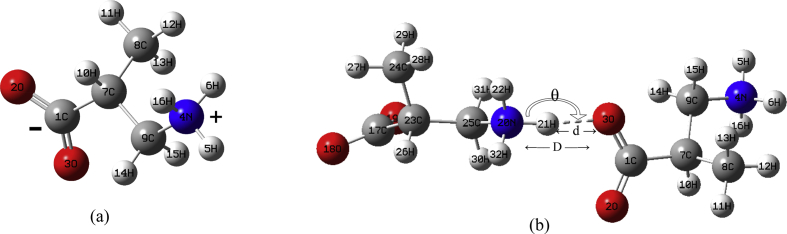
The electronic structure calculations have been performed using the Gaussian 09W and GaussView5 suite of programs [46, 47]. In order to minimize the computational cost, we did two jobs in tandem, firstly Restricted Hartree-Fock (RHF)/3-21G level of calculation followed by a second calculation at B3LYP/6–311++G(d,p) level [48]. To reduce the computational cost, the most stable NE conformers in gas phase were searched by a relaxed potential energy surface (PES) scan at the RHF/3-21G level. Initially, the dihedral angles, τ1 (16H–3O–1C–2O) and τ2 (14H–9C–4N–6H) were varied simultaneously with 10° interval from -180° to 180° and from 0° to 360° (refer Fig. 1 for τ1, τ2, τ3 and 2(b) for atom numbering). During the relaxed scan, all the internal coordinates were relaxed while the dihedral angles τ1 and τ2 were fixed at the specified values. The PES for this scan shows three minimum energy structures (Fig. 2(a)). The full optimization of these structures followed by frequency calculation was performed at B3LYP/6–311++G(d,p) level. Among the optimized structures, the one with lowest energy is considered to be more stable. This stable structure in NE form is shown in Fig. 2(b). In order to fix the orientation of carboxylic group for this minimum energy structure, we further scanned the dihedral angle τ3 (10H–7C–1C–2O) from -180° to 180°. This relaxed PES scan resulted in three minima (Fig. 3(a)). Further optimization of these structures followed by frequency calculations at B3LYP/6–311++G(d,p) level yielded three true structures with no imaginary harmonic frequencies. These three NE conformers we call C'NE, C″NE, C‴NE are presented in Fig. 3(b) and their optimized dihedral angles, geometrical parameters, Gibbs free energies and relative Boltzmann populations are presented in Tables 1 and 2. Further optimization of the conformers to search for ZW forms was performed using the Self-Consistent Reaction Field implicit Solvation Model based on Charge Density (SCRF-SMD) in water medium [35, 49, 50]. The optimization yielded only one stable ZW monomer, say, CZW, shown in Fig. 4 and its geometrical parameters are given in Table 2. Next we considered building possible dimer structures as a combination of C'NE, C″NE, C‴NE and CZW, inter-linked by –N–H⋯O bonding. We ran calculations at RHF/3-21G level, giving rise to seven ZW dimer structures, say, D1, D2,….,D7, that are collected in Fig. 5. The Gibbs free energies and Boltzmann populations of all the dimers are presented in Table 3. Consecutive differences in Boltzmann populations of D1 to D7 show that structurally and energetically D1 is the most stable structure. Further, the five dimer species, D1–D5, are close to one another with respect to their energies and vibrational frequencies and all of their Boltzmann populations add up to 98%. Accordingly, as a reasonable approximation, we chose D1 as a representative species for further optimization, harmonic frequency calculation and NBO analysis at B3LYP/6–311++G(d,p) level and its results will be used in the forthcoming discussion. The resultant geometrical structure of D1 is shown in Fig. 4 and H-bonding parameters are presented in Table 2. In order to include Grimme's dispersion correction associated with the –N–H⋯O interaction, we ran calculation at B3LYP-D3/6–311++G(d,p) level. We found that both the optimized dispersion-corrected energies and frequencies hardly differed from the results without dispersion correction [51, 52]. We have based vibrational mode analysis on potential energy distributions (PED) computed from VEDA program [54]. The AIM analysis has been carried out using Multiwfn software and the isosurfaces are visualized using VMD software package [55, 56].
Fig. 1.

Molecular structure of DL-3-Aminoisobutyric acid showing the dihedral angles τ1, τ2, τ3 used for PES scan.
Fig. 2.
(a). PES scan curve of DL-3-Aminoisobutyric acid showing three minima for the dihedral angles τ1 = 16H– 3O–1C–2O and τ2 = 14H–9C–4N–6H at RHF/3-21G level. (b). Optimized structure of DL-3-Aminoisobutyric acid obtained by PES scan for τ1 and τ2 in NE form at B3LYP/6–311++G(d, p) level.
Fig. 3.
(a). PES curve for the dihedral angle τ3 = 10H–7C–1C–2O yielding the stable conformers C'NE, C″NE and C‴NE of DL-3-Aminoisobutyric acid at RHF/3-21G level. (b). Optimized structures (C'NE, C″NE and C‴NE) of DL-3-Aminoisobutyric acid in NE form at B3LYP/6–311++G(d, p) level (values given in paranthesis are their Boltzmann populations).
Table 1.
Optimized dihedral angles, Gibbs free and relative energies, Boltzmann population of the NE monomers of DL-3-Aminoisobutyric acid in gas phase.
| Monomers | Dihedral angles (in deg)∗ |
Gibbs free energy G (Hartree) |
ΔG (kcal/mol) | % Population | ||
|---|---|---|---|---|---|---|
| τ1 | τ2 | τ3 | ||||
| C'NE | -1.3 | 72.3 | -145.3 | -363.0746 | 0 | 46 |
| C″NE | 0.8 | 73.5 | 157.0 | -363.0743 | 0.201 | 33 |
| C‴NE | 0.3 | 71.8 | 4.8 | -363.0738 | 0.483 | 21 |
τ1, τ2, τ3 as shown in Fig. 1.
Table 2.
Optimized geometrical parameters of NE monomers (C'NE ₋ C‴NE), ZW monomer (CZW) and dimer (D1) of DL-3-Aminoisobutyric acid.
| Parameters | C'NE | C″NE | C‴NE | CZW | D1 | Expta |
|---|---|---|---|---|---|---|
| Bond length (Å) | ||||||
| 1C–2O | 1.206 | 1.206 | 1.206 | 1.264 | 1.256 | 1.247 |
| 1C–3O/17C–19O∗ | 1.358 | 1.359 | 1.360 | 1.264 | 1.272/1.263∗ | 1.251 |
| 1C–7C | 1.514 | 1.516 | 1.515 | 1.544 | 1.540 | 1.520 |
| 3O–16H | 0.969 | 0.969 | 0.969 | -- | -- | -- |
| 9C–4N/25C–20N∗ | 1.467 | 1.465 | 1.465 | 1.507 | 1.507/1.499∗ | 1.484 |
| 9C–7C | 1.538 | 1.550 | 1.544 | 1.527 | 1.527 | 1.522 |
| 9C–14H | 1.098 | 1.099 | 1.100 | 1.088 | 1.088 | 1.000 |
| 9C–15H | 1.092 | 1.093 | 1.092 | 1.088 | 1.088 | 1.020 |
| 4N–5H/20N–21H∗ | 1.015 | 1.014 | 1.015 | 1.023 | 1.022/1.063∗ | 0.960 |
| 4N–6H/20N–22H∗ | 1.013 | 1.013 | 1.013 | 1.023 | 1.023/1.022∗ | 1.050 |
| 4N–16H/20N–32H∗ | -- | -- | -- | 1.023 | 1.023/1.022∗ | 0.970 |
| 8C–7C | 1.542 | 1.530 | 1.538 | 1.539 | 1.539 | 1.528 |
| 8C–11H | 1.091 | 1.092 | 1.092 | 1.092 | 1.092 | 0.970 |
| 8C–12H | 1.090 | 1.090 | 1.090 | 1.094 | 1.093 | 1.060 |
| 8C–13H | 1.093 | 1.092 | 1.092 | 1.093 | 1.093 | 1.010 |
| 7C–10H | 1.095 | 1.094 | 1.093 | 1.094 | 1.094 | 0.980 |
| RMSD | 0.0700 | 0.0704 | 0.0704 | 0.0619 | 0.0649 | |
| Bond angle (in deg) | ||||||
| 2O–1C–3O | 122.0 | 121.9 | 121.8 | 125.2 | 124.8 | 124.3 |
| 2O–1C–7C | 125.9 | 125.9 | 125.8 | 118.0 | 117.6 | 118.0 |
| 3O–1C–7C | 111.9 | 111.9 | 112.2 | 116.7 | 117.4 | 117.6 |
| 1C–3O–16H | 107.0 | 107.0 | 107.0 | -- | -- | -- |
| 4N–9C–7C/20N–25C–23C∗ | 110.3 | 110.3 | 110.1 | 111.3 | 111.3/111.9∗ | 112.3 |
| 4N–9C–14H | 113.4 | 113.2 | 113.2 | 105.9 | 106.0 | 104.5 |
| 4N–9C–15H | 108.3 | 108.2 | 108.2 | 106.5 | 106.5 | 106.5 |
| 7C–9C–14H | 109.0 | 109.1 | 109.0 | 111.4 | 111.3 | 112.5 |
| 7C–9C–15H | 108.8 | 108.4 | 108.9 | 112.2 | 112.2 | 111.0 |
| 15H–9C–14H | 106.5 | 107.2 | 107.0 | 108.9 | 108.9 | 109.0 |
| 9C–4N–5H | 111.1 | 111.1 | 111.1 | 110.9 | 110.5 | 108.0 |
| 9C–4N–6H | 110.9 | 111.1 | 111.0 | 110.5 | 111.6 | 106.7 |
| 9C–4N–16H | -- | -- | -- | 111.7 | 111.0 | 112.6 |
| 5H–4N–6H | 107.1 | 107.4 | 107.3 | 107.9 | 107.6 | 106.0 |
| 5H–4N–16H | -- | -- | -- | 107.7 | 107.9 | 111.0 |
| 6H–4N–16H | -- | -- | -- | 107.7 | 107.9 | 110.0 |
| 7C–8C–11H | 111.2 | 111.2 | 111.1 | 110.2 | 110.3 | 110.0 |
| 7C–8C–12H | 109.0 | 109.3 | 109.1 | 111.9 | 111.6 | 112.0 |
| 7C–8C–13H | 110.5 | 110.4 | 110.8 | 110.8 | 110.8 | 109.0 |
| 11H–8C–12H | 109.1 | 108.8 | 108.7 | 107.1 | 107.1 | 107.0 |
| 11H–8C–13H | 108.5 | 107.9 | 108.3 | 108.3 | 108.4 | 110.0 |
| 12H–8C–13H | 108.2 | 108.9 | 108.5 | 108.1 | 108.2 | 106.0 |
| 1C–7C–9C | 110.4 | 108.1 | 110.7 | 110.4 | 110.5 | 113.2 |
| 1C–7C–8C | 109.3 | 111.4 | 111.1 | 109.0 | 109.1 | 108.6 |
| 1C–7C–10H | 107.2 | 107.2 | 104.9 | 107.1 | 106.7 | 109.4 |
| 9C–7C–8C | 112.3 | 112.1 | 112.2 | 112.8 | 112.9 | 109.6 |
| 9C–7C–10H | 109.1 | 107.4 | 108.3 | 108.4 | 108.5 | 108.1 |
| 8C–7C–10H | 108.2 | 110.2 | 109.1 | 108.7 | 108.6 | 109.8 |
| RMSD | 3.4560 | 3.5830 | 3.5062 | 1.7860 | 1.8596 | |
| Dihedral angle (in deg) | ||||||
| 16H–3O–1C–2O | -1.3 | 0.8 | 0.3 | -- | -- | -- |
| 14H–9C–4N–6H | 72.3 | 73.5 | 71.8 | -60.1 | -60.84 | -- |
| 10H–7C–1C–2O | -145.3 | 157.0 | 4.8 | -149.6 | -146.5 | -- |
| Hydrogen bonding parameters | ||||||
| 20N–21H (r) | -- | -- | -- | -- | 1.063 | 1.050 |
| 21H⋯3O (d) | -- | -- | -- | -- | 1.663 | 1.730 |
| 20N⋯3O (D) | -- | -- | -- | -- | 2.721 | 2.758 |
| 20N–21H⋯3O (θ) | -- | -- | -- | -- | 172.9 | 166.0 |
XRD data of DL-3-Aminoisobutyric acid monohydrate [39].
Fig. 4.
Optimized (a) CZW, (b) D1 structures of DL-3-Aminoisobutyric acid showing the hydrogen bonding parameters d, D, θ (parameter values are given in Table 2).
Fig. 5.
Optimized structures of zwitterionic dimers D1 to D7 computed at RHF/3-21G level.
Table 3.
Gibbs free and relative energies of seven ZW dimers with their Boltzmann population.
| Dimer | Gibbs free energy G(Hartree) | Relative Energy ΔG (kcal/mol) |
Population (%) |
|---|---|---|---|
| D1 | -717.601533 | 0 | 35.56 |
| D2 | -717.600996 | 0.337 | 20.20 |
| D3 | -717.600917 | 0.387 | 18.59 |
| D4 | -717.600594 | 0.589 | 13.23 |
| D5 | -717.600318 | 0.762 | 9.89 |
| D6 | -717.598533 | 1.883 | 1.51 |
| D7 | -717.598126 | 2.138 | 0.98 |
3. Results and discussion
3.1. Structural analysis
The computed CZW and D1 species with the –N–H⋯O bonding are shown in Fig. 4. The optimized geometrical parameters of NE monomers C'NE, C″NE, C‴NE, ZW monomer CZW and dimer D1 compared with the XRD data of DL-3-Aminoisobutyric acid monohydrate by means of root-mean-square-deviation (RMSD) values are presented in Table 2. The low RMSD values of CZW and D1 for bond lengths (0.0163 and 0.0231 Å) and bond angles (0.8093 and 0.3905°) show the good agreement with XRD data than NE monomers (0.0487–0.0498 Å for bond lengths and 5.0272–5.1152° for bond angles). From Table 2, we note that the –N–H bond lengths are 1.023 Å in CZW as well as in D1 where the –N–H bonds are free. But the r = N–H bond length at the H-bonded region is 1.063 Å showing the elongation of the bond up to 4%. The bond angle Ɵ = N–H⋯O is 173°. The d = H⋯O distance is 1.663 Å, being shorter than the sum of the van der Waals radii of H and O, D = 2.721 Å and the –N⋯O distance is 2.726 Å, apparently indicating rather a strong H-bonding [3, 57]. Elongation in r causes decrease in both force constant and stretching frequency in D1. For example, the 20N–21H bond vibrates at the frequency, 2612 cm−1, which is lower than the frequencies, 3399 and 3349 cm−1 of the 20N–22H and 20N–32H bonds. The bonds adjacent to the –N–H⋯O, namely, the –C=O corresponding to the 1C–3O bond and the –C–N corresponding to the 25C–20N bond have respectively also shown a small increase and decrease in their values from CZW to D1. These changes are consistent with NBO analysis to be discussed in the following sections.
3.2. NBO analysis
NBO analysis has originated as a method for studying hybridization and covalency effects in polyatomic wave functions associated with, among other factors, intermolecular associations [58, 59, 60, 61, 62]. NBO analysis was performed for the monomer Czw and dimer D1 of 3AIBA using NBO Version 3.1 implemented in the Gaussian 09 package at B3LYP/6–311++G(d,p) level and the results are presented in Tables 4 and 5. The Tables show the occupancies, description of NBOs, polarization coefficients cA2, cB2 (in %) and the contribution of s character (in %) on each atom forming a bond in the monomer Czw and dimer D1. The following features are observed from the NBO results:
-
(i)
The σ(1C–2O), π(1C–2O) and σ(1C–3O) are strongly polarized towards the oxygen atom in both the Czw and D1. The occupancies of these bond orbitals are less in the D1 compared to those in the Czw.
-
(ii)
Again in both Czw and D1, the σ*(1C–2O) and σ*(1C–3O) are polarized towards the carbon atom while π*(1C–2O) is still more polarized towards the carbon atom. Occupancies of σ* and π* orbitals of the 1C–2O bond are less in the D1 whereas that of σ*(1C–3O) is large.
-
(iii)
In both Czw and D1 for σ(1C–2O), σ(1C–3O), σ*(1C–2O) and σ*(1C–3O), natural hybrids on the carbon and oxygen atoms are sp2 hybridized as expected with sufficiently large p character on both the atoms compared to the s character (in Tables 4 and 5, only s contributions on the atoms are given). The hybrid orbitals of π and π* of 1C–2O exhibit p character.
-
(iv)
The σ(N–H) bond orbitals are polarized more towards the nitrogen atom and σ*(N–H) are polarized towards hydrogen. In σ*(N–H) orbitals, the NHO on N are sp3 hybridized and NHO on H is completely of s character.
-
(v)
The nonbonding type of NBOs which are due to lone-electron pair orbitals, n1(2O), n1(3O) are of sp0.65, sp0.61 hybridized in CZW and sp0.64, sp0.82 in D1. The remaining lone pair orbitals, n2(2O), n2(3O) and n3(3O) exhibit pure pcharacter both in CZW and D1 thus favoring the π delocalization at –CO2‾ site.
Table 4.
Computed NBO parameters for some of the important natural bond orbitals in CZW.
| NBOs Ω/Ω∗ (A-B) | Occupancy q | Polarization coefficients (%) |
Description of NBO | Contribution of s character (%) |
ΔqΩΩ* | ||
|---|---|---|---|---|---|---|---|
| cA2 | cB2 | A | B | ||||
| σ(1C–2O) | 1.99624 | 34.31 | 65.69 | 0.5858 (sp2.10) C1 + 0.8105 (sp1.55)O2 | 32.23 | 39.15 | 1.94647 |
| σ∗(1C–2O) | 0.04977 | 65.69 | 34.31 | 0.8105 (sp2.10)C1 - 0.5858 (sp1.55)O2 | |||
| π(1C–2O) | 1.99334 | 22.75 | 77.25 | 0.4769 (p)C1 + 0.8789 (p)O2 | 0.45 | 0.15 | 1.61977 |
| π∗(1C–2O) | 0.37357 | 77.25 | 22.75 | 0.8789 (p)C1 - 0.4769 (p)O2 | |||
| σ(1C–3O) | 1.99484 | 34.15 | 65.85 | 0.5843 (sp2.14)C1 + 0.8115 (sp1.69)O3 | 31.74 | 37.09 | 1.93103 |
| σ∗(1C–3O) | 0.06381 | 65.85 | 34.15 | 0.8115 (sp2.14) C1 - 0.5843 (sp1.69) O3 | |||
| σ(4N–5H) | 1.99316 | 72.96 | 27.04 | 0.8542 (sp3.38)N4 + 0.5200 (s)H5 | 22.83 | 99.93 | 1.98787 |
| σ∗(4N–5H) | 0.00529 | 27.04 | 72.96 | 0.5200 (sp3.38) N4 - 0.8542 (s)H5 | |||
| σ(4N–6H) | 1.99371 | 72.82 | 27.18 | 0.8533 (sp3.31)N4 + 0.5214 (s)H6 | 23.20 | 99.93 | 1.98535 |
| σ∗(4N–6H) | 0.00836 | 27.18 | 72.82 | 0.5214 (sp3.31)N4 - 0.8533 (s)H6 | |||
| σ(4N–16H) | 1.99380 | 72.81 | 27.19 | 0.8533 (sp3.34)N4 + 0.5214 (s)H16 | 23.03 | 99.93 | 1.98549 |
| σ∗(4N–16H) | 0.00831 | 27.19 | 72.81 | 0.5214 (sp3.34)N4 - 0.8533 (s)H16 | |||
| n1(2O) | 1.97923 | sp0.65 | 60.73 | ||||
| n2(2O) | 1.90320 | p | 0.01 | ||||
| n1(3O) | 1.98025 | sp0.61 | 61.95 | ||||
| n2(3O) | 1.90938 | p | 0.10 | ||||
| n3(3O) | 1.62882 | p | 0.92 | ||||
Note: ΔqΩΩ* = Difference in occupancies qΩ – qΩ*. Where Ω - bonding and Ω∗ - antibonding orbitals on A and B.
Table 5.
Computed NBO parameters for some of the important natural bond orbitals in the H-bonding region in D1.
| NBOs Ω/Ω∗ (A-B) |
Occupancy q | Polarization coefficients (%) |
Description of NBO | Contribution of s character (%) |
ΔqΩΩ* | ||
|---|---|---|---|---|---|---|---|
| cA2 | cB2 | A | B | ||||
| σ(1C–2O) | 1.99575 | 34.37 | 65.63 | 0.5862 (sp2.15)C1 + 0.8101 (sp1.57)O2 | 31.64 | 38.86 | 1.94797 |
| σ∗(1C–2O) | 0.04778 | 65.63 | 34.37 | 0.8101 (sp2.15)C1 - 0.5862 (sp1.57)O2 | |||
| π(1C–2O) | 1.98812 | 23.06 | 76.94 | 0.4802 (p)C1 + 0.8772 (p)O2 | 1.45 | 0.25 | 1.63194 |
| π∗(1C–2O) | 0.35618 | 76.94 | 23.06 | 0.8772 (p)C1 - 0.4802 (p)O2 | |||
| σ(1C–3O) | 1.98748 | 32.45 | 67.55 | 0.5697 (sp2.20)C1 + 0.8219 (sp2.05)O3 | 31.15 | 32.77 | 1.88137 |
| σ∗(1C–3O) | 0.10611 | 67.55 | 32.45 | 0.8219 (sp2.20)C1 - 0.5697 (sp2.05)O3 | |||
| σ(20N–21H) | 1.99045 | 76.22 | 23.78 | 0.8730 (sp2.84)N20 + 0.4877 (s)H21 | 26.03 | 99.89 | 1.90924 |
| σ∗(20N–21H) | 0.08121 | 23.78 | 76.22 | 0.4877 (sp2.84)N20 - 0.8730 (s)H21 | |||
| σ(20N–22H) | 1.99275 | 72.06 | 27.94 | 0.8489 (sp3.50)N20 + 0.5286 (s)H22 | 22.19 | 99.94 | 1.98324 |
| σ∗(20N–22H) | 0.00951 | 27.94 | 72.06 | 0.5286 (sp3.50)N20 - 0.8489 (s)H22 | |||
| σ(20N–32H) | 1.99277 | 72.07 | 27.93 | 0.8489 (sp3.55)N20 + 0.5285 (s)H32 | 21.96 | 99.94 | 1.98385 |
| σ∗(20N–32H) | 0.00892 | 27.93 | 72.07 | 0.5285 (sp3.55)N20 - 0.8489 (s)H32 | |||
| n1(2O) | 1.97908 | sp0.64 | 60.93 | ||||
| n2(2O) | 1.90125 | p | 0.01 | ||||
| n1(3O) | 1.96243 | sp0.82 | 54.98 | ||||
| n2(3O) | 1.86544 | p | 7.70 | ||||
| n3(3O) | 1.61485 | p | 4.56 | ||||
Note: ΔqΩΩ* = Difference in occupancies qΩ – qΩ*. Where Ω - bonding and Ω∗ - antibonding orbitals on A and B.
According to the NBO procedure, the strength of concerned bonds is computed by considering the difference in occupancies of bonding and antibonding NBOs. From Tables 4 and 5, it is evident that in D1, the 1C–2O bond is strong and in the Czw stronger bond is 1C–3O. The difference in occupancies of bonding and antibonding σ and π components of 1C–2O are 1.94647 and 1.61977 in CZW, 1.94797 and 1.63194 in D1. The σ component of 1C–2O has slightly more occupancy in the D1 than in Czw. The total occupancies (σ + π) are 3.566 in CZW and 3.579 in D1 showing that 1C–2O carbonyl bond is stronger in the dimer than in the monomer.
The atomic charges are useful in evaluating the electronic structure of bonds [63]. The charge distribution among the atoms in CZW and D1 has also been calculated by the natural population analysis (NPA) and is presented in Table 6. It is noted from Table 6 that, the charges on the atoms 21H, 22H and 32H in D1 are 0.460, 0.435 and 0.435 respectively while on the 2O and 3O are −0.825 suggesting the strong electrostatic interaction between 21H and 3O. Charge transfer between the proton donor –NH3+ group and the proton acceptor –CO2‾ group elongate the –N–H bond length and hence weaken the –N–H bond. The difference in natural atomic charges between 5H in Czw and 21H in D1 and proton acceptor 3O are 0.004 and −0.035 respectively, suggesting that the dimer is stabilized by the red-shifted H-bonding. This is consistent with the increase in occupancy (see Tables 4 and 5) from 0.00529 in the monomer (σ*(4N–5H)) to 0.08121 in dimer (σ*(20N–21H)).
Table 6.
Natural atomic charges of CZW and D1 computed by NBO analysis.
| CZW |
D1 |
||||
|---|---|---|---|---|---|
| Atom | Charge (a.u) | Atom | Charge (a.u) | Atom | Charge (a.u) |
| 1C | 0.796 | 1C | 0.815 | 17C | 0.795 |
| 2O | -0.842 | 2O | -0.825 | 18O | -0.855 |
| 3O | -0.860 | 3O | -0.825 | 19O | -0.850 |
| 4N | -0.673 | 4N | -0.672 | 20N | -0.707 |
| 5H | 0.456 | 5H | 0.456 | 21H | 0.460 |
| 6H | 0.450 | 6H | 0.451 | 22H | 0.435 |
| 7C | -0.332 | 7C | -0.329 | 23C | -0.329 |
| 8C | -0.582 | 8C | -0.582 | 24C | -0.582 |
| 9C | -0.167 | 9C | -0.167 | 25C | -0.171 |
| 10H | 0.222 | 10H | 0.225 | 26H | 0.219 |
| 11H | 0.216 | 11H | 0.217 | 27H | 0.215 |
| 12H | 0.206 | 12H | 0.208 | 28H | 0.204 |
| 13H | 0.207 | 13H | 0.208 | 29H | 0.206 |
| 14H | 0.227 | 14H | 0.227 | 30H | 0.221 |
| 15H | 0.224 | 15H | 0.225 | 31H | 0.217 |
| 16H | 0.450 | 16H | 0.451 | 32H | 0.435 |
The second order perturbation energies (E(2)) were computed at B3LYP/6–311++G(d,p) level. The energy associated with each donor and acceptor is estimated using second order perturbation theory as,
| (1) |
where n is the population of the donor orbital, Fij is the Fock matrix element between the i and j NBOs, and, where and are donor and acceptor NBOs.
The mixing coefficients can be estimated as, and the occupancy transfer (charge transfer) from as, .
Larger the value, more intensive is the interaction between donor and acceptor. Fig. 6 shows the interaction of the filled orbital with the unfilled non-Lewis orbital [64]. The E(2) values corresponding to –N–H⋯O interaction in the dimers D1, D2, D3, D4, D5, D6 and D7, are 35.01, 23.70, 27.42, 20.85, 2.53, 19.08 and 27.13 kcal/mol respectively, suggesting that more charge transfer interactions exist in the case of D1. These intermolecular –N–H⋯O bonds are formed by the orbital overlap between lone pair (n) of oxygen (O) and antibonding orbital (σ*) of N–H. Lone pair of oxygen and antibonding orbitals of N–H are identified as electron donor and acceptor NBO species. The H-bonding between n(O) and σ∗(N–H) results in charge transfer causing stabilization of the dimer species. This –N–H⋯O intermolecular bonding increases the electron density which leads to the elongation of the N–H bond lengths and hence lowering the corresponding stretching vibrational frequencies in D1. For the D1, the intermolecular interactions between some of the important donor-acceptor NBO pairs such as n(3O) and σ*(20N–21H) give rise to the energies 28.46, 6.10 and 0.45 kcal/mol which are presented in Table 7 and the graphical electron density maps of NBOs corresponding to these interactions are presented in Fig. 7.
Fig. 6.
Second order perturbative donor-acceptor interaction involving a filled orbital i and an unfilled orbital j.
Table 7.
Computed mixing coefficients (λ), charge transfer (Q) giving NBOs with stabilization energy corresponding to –N–H⋯O interaction in D1.
| NBOs Ωi→Ω*j |
Mixing coefficient λi→j |
Charge transfer Qi→j |
E(2)a kcal/mol |
|---|---|---|---|
| n1(3O)→σ*(20N₋21H) | 0.1853 | 0.03433 | 28.46 |
| n2(3O)→σ*(20N–21H) | 0.0696 | 0.00969 | 6.10 |
| n3(3O)→σ*(20N–21H) | 0.0266 | 0.001411 | 0.45 |
| CR(3O)→σ*(20N–21H) | 0.0043 | 0.000037 | 0.43 |
| σ(1C–3O)→σ*(20N–21H) | 0.0156 | 0.000488 | 0.37 |
Note: Ωi, Ωj∗ = donor, acceptor NBOs, σ∗ = antibonding orbital, n = lone pair on oxygen atom, CR = core.
Energy of hyperconjugative interaction (stabilization energy).
Fig. 7.
Electron density maps of selected NBOs involved in –N–H⋯O bonding in D1. Donor (lone pair orbital of 3O) and acceptor (antibonding orbital of 20N–21H) NBOs are shown separately in left columns and their overlapping due to H-bonding is shown in right columns. Green and yellow colours of the orbitals correspond to positive and negative signs respectively.
From this analysis, we can conclude the following. The decreased occupancy of σ component of 1C–3O bond from Czw to D1 is consistent with the increase in the 1C–3O bond length value from 1.264 Å in Czw to 1.272 Å in D1. Similarly, the difference in atomic charges 5H in Czw, 21H in D1 and 3O suggest that the dimer is stabilized by the red-shifted H-bonding. This is in agreement with the increase in occupancy from 0.00529 in the CZW (σ*(4N–5H)) to 0.08121 in D1 (σ*(20N–21H)). The computed second order perturbative energies also suggest that the strong H-bonding exist in D1.
3.3. AIM analysis
The –N–H⋯O bonding in D1 species of 3AIBA has been studied through AIM analysis. According to AIM theory, a chemical bond including hydrogen bond is characterised by the presence of a bond critical point (BCP). The topological parameters like electron density ρ(r) and their Laplacian 2ρ(r) computed at BCP are useful tools to characterize the strength of a bond between donor and acceptor atoms [65, 66, 67, 68, 69]. The important topological parameter values obtained for D1 species include the electron density (0.0519 a.u), its corresponding Laplacian (0.1326 a.u), potential energy density (V(r) = -0.0486 a.u). Interaction energy (Eint = 15.2 kcal/mol) of the –N–H⋯O bond has been calculated at BCP using the relation Eint = V(r)/2. The low value of ρ(r) and positive value of 2ρ(r) and the value of interaction energy satisfy the criteria for existence of medium H-bond according to Rozas et al [70]. With all these values we can conclude that, a medium strong –N–H⋯O inter-molecular bond exist in D1.
Non-covalent interactions (NCI) in the molecular systems can be visualized using reduced density gradient (RDG) [71, 72]. The RDG is given by:
| (2) |
where ρ(r) is the electron density and ρ(r) is its gradient. NCI isosurfaces illustrate the nature of the interactions in real space through colour codes. Blue, green, red colours are used to represent attractive hydrogen bond, weak van der Waals and steric interactions respectively. The NCI isosurface for the D1 species is shown in Fig. 8. A blue patch between the hydrogen atom of –NH3+ group from one ZW monomer and oxygen atom of –CO2‾ group from another ZW monomer unit in Fig. 8 is attributed to –N–H⋯O bonding. Green and red patches between –NH3+, CH3 and –CO2‾, CH2 groups within the monomer units represent the van der Waals and repulsive steric interactions respectively.
Fig. 8.
Reduced density gradient isosurface for D1 of DL-3-Aminoisobutyric acid. Blue, green, red colours are used to represent attractive hydrogen bond, weak van der Waals and steric interactions respectively.
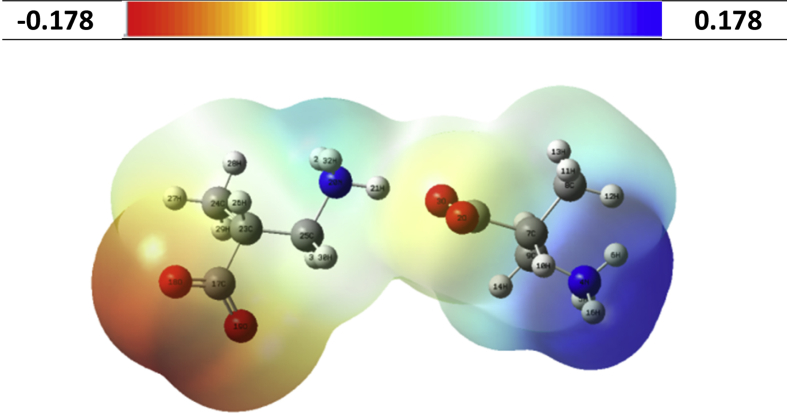
3.4. MEP analysis
The molecular electrostatic potential (MEP) serves as a useful quantity to explain H-bonding and reactivity of molecules [73]. The MEP surface of the D1 species was calculated from the optimized molecular structure discussed in section 2.2 and shown in Fig. 9. The different values of electrostatic potentials at the MEP surface are represented by different colors: red, blue and green represent the regions of most negative, most positive and zero electrostatic potential respectively. The negative region is localized over –CO2‾ group indicating the most reactive site for electrophillic attack and positive region is localized over –NH3+ group which is reactive site for nucleophillic attack. The loss of red and blue colors on –CO2‾ and –NH3+ groups in the bonding region of the D1 species is attributed to –N–H⋯O donor-acceptor interaction.
Fig. 9.
Molecular electrostatic potential surface for D1 of DL-3-Aminoisobutyric acid (in the range between -0.178 to 0.178 a.u).
3.5. Vibrational analysis
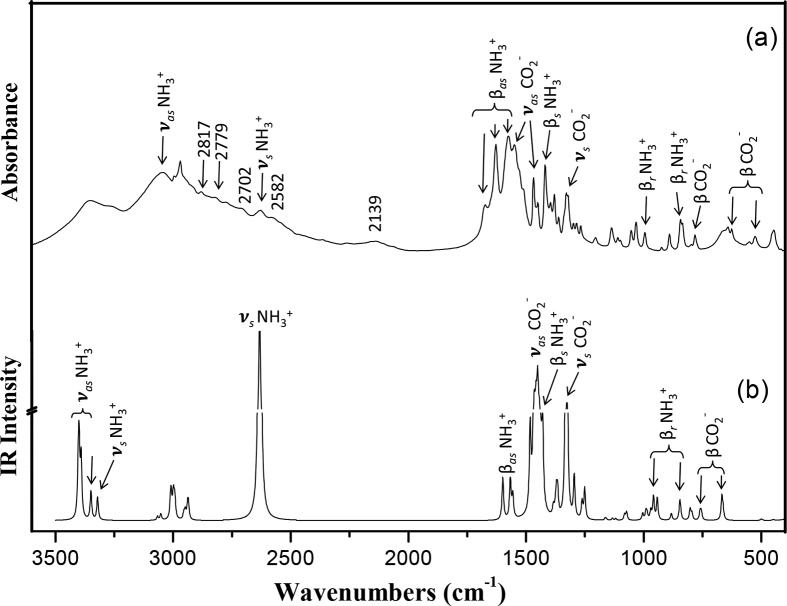
The experimental IR and Raman spectra of 3AIBA and their comparison with the simulated spectra of D1 are shown in Figs. 10 and 11. The IR spectrum marked by a broad composite band structure in the region 3500 − 2000 cm−1 with FWHM 700 cm−1 is indicative of inter-molecular interactions of which we assume H-bonding. A series of multiple peaks on this broad band and, medium to strong bands in the 1675 − 1400 cm−1 are characteristic modes of –NH3+ and –CO2− groups, strongly suggesting the ZW nature of the 3AIBA. However, the Raman spectrum has not shown these bands since the Raman modes are generally weak for ionic group vibrations. The strong bands it has shown are due to –CH, –CH2 and –CH3 groups in the 3000 − 2850 cm−1. In other regions, the 1675 − 1400 cm−1 is marked by very sharp IR bands readily assigned to the stretching and bending modes of –CO2− and –NH3+ groups. The Raman bands are weak in this region. We now present a detailed vibrational analysis of the observed and computed bands of the CZW monomer and D1 dimer species. We refer to modes arising from H-bonded vibrations as ‘bonded modes’; otherwise, they will be referred to as ‘free modes’.
Fig. 10.
(a) Experimental and (b) simulated dimer (D1) IR spectra of DL-3-Aminoisobutyric acid. Frequency values corresponding to vibrational modes are given in Table 8. Only the stretching, bending vibrations of –NH3+, –CO2− and their combinations are assigned in the above spectra. Ordinate in Dimer spectrum (b) is broken to adjust the bands from overshooting because of their largest intensity.
Fig. 11.
(a) Experimental and (b) simulated dimer (D1) Raman spectra of DL-3-Aminoisobutyric acid. Frequency values corresponding to vibrational modes are given in Table 8. Only the stretching, bending vibrations of –NH3+, ₋COO¯ and their combinations are assigned in the above spectra. Ordinate in Dimer spectrum (b) is broken to adjust the bands from overshooting because of their largest intensity.
3.5.1. NH3+ modes
In the IR spectrum no absorption is observed in the usual N–H stretching region, 3500 − 3300 cm−1; instead a broad absorption of medium intensity at 3044 cm−1 is identified as asymmetric stretching mode of –NH3+ as against the predicted band at 3391 cm−1 in CZW and at 3399 cm−1 in D1 [74,75]. Second asymmetric stretching mode has not been observed near 3383 cm−1 but is computed in CZW and it is at 3390 cm−1 in D1 (bonded mode is at 3349 cm−1). The symmetric stretching mode is not seen in the Raman spectrum since this band is apparently obscured by the more intense –C–H stretching band, but is predicted at 3321 cm−1 [76]. As for the bonded mode we identify a medium weak absorption at 2629 cm−1, which is correlated to a predicted band at 2612 cm−1 [77]. Bonded asymmetric bending mode identified as a medium weak absorption at 1675 cm−1 with its Raman band at 1689 cm−1 is correlated to the predicted band at 1599 cm−1; the free mode is predicted at 1568 cm−1. The second bonded mode is assigned to a very strong absorption at 1627 cm−1 (Raman band at 1616 cm−1) is correlated to a predicted band at 1566 cm−1, the free mode is predicted at 1556 cm−1. Another asymmetric mode appearing as a very strong absorption at 1574 cm−1 with its weak Raman band at 1584 cm−1 is correlated to a predicted band at 1482 cm−1. The symmetric bending mode observed as a medium weak absorption at 1400 cm−1 is correlated to a predicted band at 1431 cm−1. As for the rocking modes, weak and medium weak absorptions observed at 995 and 844 cm−1 are correlated to the predicted bands at 970 and 843 cm−1 respectively.
3.5.2. CO2− modes
As for the –CO2− modes, the asymmetric stretching bonded mode is identified as a very strong absorption at 1546 cm−1 with its Raman mode at 1544 cm−1 and is computed at 1458 cm−1; the free mode is predicted at 1465 cm−1. The second asymmetric mode predicted at 1450 cm−1 is correlated to a strong absorption and medium strong Raman band near 1468 cm−1. Symmetric stretching mode identified as a medium strong absorption at 1329 cm−1 is computed at 1332 cm−1. Another symmetric stretching mode predicted at 1325 cm−1 is correlated to the medium strong Raman band at 1312 cm−1. Weak to medium absorptions at 782, 642 and 526 cm−1 are identified as deformation modes of –CO2− and are predicted at 761, 668 and 502 cm−1 respectively.
3.5.3. Combination bands
Further, some bands appeared only in the IR spectrum in the region 2700 − 2000 cm−1 as a substructure on the broad absorption are assigned to a combination of asymmetric bending of –NH3+ with –C–N stretching or torsional modes of –NH3+ [10, 57]. The weak to medium absorptions at 2702 and 2629 cm−1 may arise due to the –N–H⋯O bonding, and it is reasonable to assign them to a combination of –NH3+ asymmetric stretch and deformation of the –C–N bond [78]. The band at 2139 cm−1 being a combination of –NH3+ asymmetric stretch and –C–N torsional mode is identified as a marker band for the identification of cation –NH3+ moiety in amino acids [10, 11]. Similarly, the weak absorptions at 2817, 2779 and 2582 cm−1 are combinations of –NH3+ asymmetric stretch with –C–N deformation and stretching vibrations.
3.5.4. C–H, C–C and C–N bands
All the other bands, which are not influenced by the H-bonding, have appeared in their usual region. As for methyl vibrations, the Raman spectra shows distinct, intense bands assigned to asymmetric and symmetric stretching vibrations in the region 3000 − 2850 cm−1 whereas the IR bands in this region appeared as weak bands or sometimes as shoulder to the other bands. The scissoring and deformation modes appear in the region 1450 − 1360 cm−1. The –C–N bending modes are assignedto the coupled vibrations with−C–C bending and stretching vibrations in the region 1300 − 800 cm−1. Medium weak Raman bands at 1033 and 891 cm−1 are identified as –C–C skeletal vibrations suggesting that the carbon chain is not perturbed due to the H-bonding. A detailed assignments of all the modes compared with the assignments of β-alanine are presented in Table 8.
Table 8.
Experimental and computed frequencies (cm−1) of CZW and D1 structures with assignments for DL-3-Aminoisobutyric acid.
| Observed Frequenciesd |
β-alaninee |
Computed Frequenciesa |
Assignmentsc | |||
|---|---|---|---|---|---|---|
| IR | Raman | IR | Raman | CZW | D1 | |
| 3044 s | -- | 3391 | 3399 | Vas (NH3+, 90) | ||
| -- | -- | 3383 | 3390 (3349)b | Vas (NH3+, 98) | ||
| 2629 mw | -- | 3313 | 3321 (2612) | Vs (NH3+, 100) | ||
| 2988 sh | 2991 vs | 3017 | 3016 | 3066 | 3066 | Vas (CH2, 99) |
| 2969 s | -- | -- | 2936 | 3012 | 3011 | Vas (CH2, 99) |
| -- | 2949 vs | 3011 | 3009 | Vas (CH3, 96) | ||
| -- | 2908 vvs | 2993 | 2993 | Vas (CH3, 95) | ||
| -- | 2889 sh | 2945 | 2948 | V(CH, 96) | ||
| 2879 mw | -- | 2936 | 2935 | Vs (CH3, 99) | ||
| 2817 w | -- | -- | -- | 1675 + 1136 | ||
| 2779 w | -- | -- | -- | 1627 + 1136 | ||
| 2702 w | -- | -- | -- | 1627 + 1053 | ||
| 2629 mw | -- | -- | -- | 1574 + 1053 | ||
| 2582 w | -- | -- | -- | 1574 + 923 | ||
| 2139 w | -- | -- | -- | 1675 + 447 | ||
| 1675 mw | 1689 w | 1653 | 1686 | 1561 | 1568 (1599) | δas (NH3+, 86) |
| 1627 vs | 1616 w | 1554 | 1556 (1566) | δas (NH3+, 90) | ||
| 1574 vvs | 1584 w | 1573 | 1556 | -- | 1482 | δas (NH3+, 71) |
| 1546 vs | 1544 w | 1633 | 1630 | 1463 | 1465 (1458) | Vas (CO2‾, 64) |
| 1468 s | 1468 ms | 1452 | 1450 | Vas (CO2‾, 18), δs (CH3, 26) | ||
| 1452 mw | 1445 mw | 1466 | 1473 | 1441 | 1441 | δsc (CH2, 73) |
| 1419 s | -- | 1436 | 1435 | δas (CH3, 63) | ||
| 1400 w | 1399 w | 1423 | 1431 | δs (NH3+, 56), δas (CH3, 27) | ||
| 1379 mw | 1386 w | 1447 | 1434 | 1383 | 1384 | Ω (CH2, 46) |
| 1360 w | 1365 w | 1371 | 1371 | δ (CH3, 34), δ (CH2, 14) | ||
| 1329 ms | 1331 mw | 1403 | 1395 | 1330 | 1332 | V (CO2‾, 17), δ (CN, 34), δt (CH2, 19) |
| -- | 1312 ms | 1320 | 1325 | V (CO2‾, 36), δ (CN, 28) | ||
| 1299 w | -- | 1290 | 1294 | δ (CC, 39), δ (CH, 15) | ||
| 1272 w | 1267 s | 1248 | 1251 | δ (CH, 49), (CO2‾, 12) | ||
| 1136 mw | 1134 w | 1160 | 1159 | δ (CC, 30), δ (CN, 10) | ||
| 1108 w | 1107 mw | 1121 | 1119 | V (CC, 33), δ (CN, 14) | ||
| 1053 mw | 1051 mw | 1072 | 1072 | δ (CC, 13), δ (CN, 11), δ (CO, 11) | ||
| 1032 mw | 1033 mw | 990 | 991 | Δ (CC, 36), (CC, 28) | ||
| 995 w | 1003 w | 1061 | 1068 | 965 | 970 | ρ (NH3+, 30), δ (CC, 10), δ (CO, 10) |
| 923 vw | 921 w | 937 | 943 | V (CN, 64), (CC, 11) | ||
| 891 w | 891 w | 877 | 882 | V (CC, 33), (CN, 17) | ||
| 844 mw | 837 ms | 991 | 998 | 838 | 843 | Ρ (NH3+, 15), δ (CC, 24) |
| 799 vw | -- | 794 | 794 | V (CC, 38), δ (CO2‾, 14) | ||
| 782 w | 773 w | 753 | 761 | δ (CO2‾, 59), δ (CC, 10) | ||
| 642 mw | 632 w | 654 | 659 | 654 | 668 | δ (CO2‾, 35) |
| 526 w | 539 w | 538 | 541 | 490 | 502 | Δ (CO2‾, 37), (CC, 10) |
| 447 w | 445 w | -- | 450 | Δ (N–H⋯O, 27), δ (CN, 40) | ||
| -- | -- | 357 | 358 | Δ (CN, 27), τ (CC, 11), τ (OH, 12) | ||
| -- | -- | 345 | 346 | Τ (CO, 12), τ (CC, 34) | ||
| -- | 296 w | 327 | 326 | 321 | Τ (CC, 43), τ (CN, 20) | |
| -- | 252 vw | 224 | 250 | 275 | Τ (CC, 36), τ (CN, 11) | |
| -- | -- | 218 | 227 | Τ (CC, 35), (O…H, 10) | ||
| -- | 170 w | 167 | 197 | 202 | Τ (CC, 78) | |
| -- | 126 mw | 112 | 119 | 119 | Τ (CCN, 77) | |
| -- | -- | -- | 107 | τ (CN, 11), τ (CO, 11), τ (O⋯H, 45) | ||
| -- | -- | -- | 76 | Δ (H...OC, 41), τ (CN, 19) | ||
| -- | -- | 53 | 46 | τ (CN, 23), τ (NCO, 10), τ (CO, 22) | ||
ν = stretching; δ = bending; ω = wagging; ρ = rocking, δt = twisting, τ = torsional vibration below 400 cm−1; as = antisymmetric; s = symmetric.
Frequencies are scaled by 0.9688 [53].
A dimer frequency shown in bracket belongs to bonded mode.
The quantities given in brackets are PED contribution (in %).
Our experimental results.
Ref [4].
4. Conclusions
A satisfactory zwitterionic dimer model at B3LYP/6–311++G(d,p) level and SCRF-SMD method, constructed of inter-molecular –N–H⋯O bonding between –NH3+ group of one zwitterionic monomer unit and –CO2− group of another identical zwitterionic monomer unit, has been proposed for DL-3-Aminoisobutyric acid. The vibration modal features predicted by the model are in fair agreement with the IR and Raman modes in 3040–3000 cm−1 and 1680–1460 cm−1. The Grimme's dispersion correction associated with the –N–H⋯O interaction computed at B3LYP-D3/6–311++G(d,p) level which yielded both the optimized dispersion-corrected energies and frequencies hardly differed from the results without dispersion correction. As for the NBO characterization of the –N–H⋯O bonding, it has been shown that it is formed by the orbital overlap between lone pair (n) of oxygen (O) as electron donor and antibonding orbital (σ*) of N–H bond as acceptor. The stabilization of the dimer species has been shown to arise from the hyperconjugation interaction between n(O) and σ∗(N–H) that has explained both structural aspects of DL-3-Aminoisobutyric acid and its empirical IR red spectral shift. The AIM analysis has shown the intermolecular –N–H⋯O bondingin D1 of 3AIBA. The values of ρ(r) and 2ρ(r) and hydrogen bond energy at bond critical points indicate a strong intermolecular –N–H⋯O bonding. The same result has been deduced from the NCI analysis as well. Overall all the DFT, NBO, AIM and NCI calculations mutually support each other. It may be said that the stretching modes of –NH3+ and –CO2− groups might have been predicted more accurately perhaps for a trimer model.
Declarations
Author contribution statement
Shashikala Yalagi: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Jagdish Tonannavar: Conceived and designed the experiments; Analyzed and interpreted the data.
Jayashree Tonannavar: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the Department of Physics under the Centre of Advanced Study at Level II (Grant no: No.F.530/9/CAS-II/2015(SAP-I)) funded by University Grants Commission, New Delhi. Jayashree Tonannavar was supported by the University Grants Commission, New Delhi by the Major Research Programme grant (F.37–243/2009(SR)).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank the Director, USIC, Karnatak University, Dharwad for FT IRand Raman spectrometer facilities sponsored under DST-funded PURSE Program.
References
- 1.http://www.sigmaaldrich.com/.
- 2.Nsangou M. DFT study of geometrical and vibrational features of small amino acids with polar side chains in hydrated media: L-Threonine and L-serine. Computational and Theoretical Chemistry. 2011;966:364–374. [Google Scholar]
- 3.Desiraju G.R., Steiner T. Oxford University Press; New York: 1999. The Weak Hydrogen Bond. [Google Scholar]
- 4.Rosado M.T.S., Duarte M.L.R.S. RuiFausto, vibrational spectra (FT-IR, Raman and MI-IR) of α- and β-alanine. J. Mol. Struct. 1997;410–411:343–348. [Google Scholar]
- 5.Galeno A., Alvarez-Idaboy J.R. Ab initio study of β-alanine conformers in the gas phase. ARKIVOC vi. 2005:7–18. [Google Scholar]
- 6.Yenagi J., Yalagi S., Tonannavar J. A study of vibrational spectra of zwitterionic 3-Aminobutanoic acid, as supported by density functional calculations. Asian J. Phys. 2015;24:703–711. [Google Scholar]
- 7.Lelais G., Seebach D. β2-Amino acids-syntheses, occurrence in natural products, and components of β-peptides1,2. Biopolymers. 2004;76:206–243. doi: 10.1002/bip.20088. [DOI] [PubMed] [Google Scholar]
- 8.Seebach D., Matthews J.L., Meden A., Wessels T., Baerlocher C., McCusker L.B. Cyclo-β-peptides: structure and tubular stacking of cyclic tetramers of 3-aminobutanoic acid as determined from powder diffraction data. Helv. Chim. Acta. 1997;80:173–182. [Google Scholar]
- 9.Kudo F., Miyanaga A., Eguchi T. Biosynthesis of natural products containing β-amino acids. Nat. Prod. Rep. 2014;31:1056–1073. doi: 10.1039/c4np00007b. [DOI] [PubMed] [Google Scholar]
- 10.Dobrowolski J.Cz., Jamoz M.H., Kolos R., Rode J.E., Sadlej J. IR low-temperature matrix and ab initio study on β-alanine conformers. ChemPhysChem. 2008;9:2042–2051. doi: 10.1002/cphc.200800383. [DOI] [PubMed] [Google Scholar]
- 11.Fink K., Henderson R.B., Fink R.M. Beta-aminoisobutyric acid, a possible factor in pyrimidine metabolism. Pro. Soc. Exp. Biol Med. 1951;78(1):135–141. doi: 10.3181/00379727-78-19000. Oct. [DOI] [PubMed] [Google Scholar]
- 12.Kupiecki F.P., Coon M.J. The enzymatic synthesis of β-aminoisobutyrate. A product of valine metabolism, and of β-alanine, a product of β-hydroxy-propionate metabolism. J. Biol. Chem. 1957;229(2):743–754. Jan 12. [PubMed] [Google Scholar]
- 13.Griffith O.W. β-Amino acids: mammalian metabolism and utility as α-amino acid analogues. Annu. Rev. Biochem. 1986;55:855–878. doi: 10.1146/annurev.bi.55.070186.004231. [DOI] [PubMed] [Google Scholar]
- 14.Toksoz S., Acar H., Guler M.O. Self-assembled one-dimensional soft nanostructures. Soft Matter. 2010;6:5839–5849. [Google Scholar]
- 15.Gao P., Wu Y., Wu L. Co-assembly of polyoxometalates and peptides towards biological applications. Soft Matter. 2016;12:8464–8479. doi: 10.1039/c6sm01433j. [DOI] [PubMed] [Google Scholar]
- 16.Cysewski P. A post-SCF complete basis set study on the recognition patterns of uracil and cytosine by aromatic and π-aromatic stacking interactions with amino acid residues. Phys. Chem. Chem. Phys. 2008;10:2636–2645. doi: 10.1039/b718394a. [DOI] [PubMed] [Google Scholar]
- 17.Bravaya K.B., Kostko O., Ahmed M., Krylov A.I. The effect of π-stacking, H-bonding, and electrostatic interactions on the ionization energies of nucleic acid bases: adenine-adenine, thymine-thymine and adenine-thymine dimers. Phys. Chem. Chem. Phys. 2010;12:2292–2307. doi: 10.1039/b919930f. [DOI] [PubMed] [Google Scholar]
- 18.Bachorz R.A., Bischoff F.A., Höfener S., Klopper W., Ottiger P., Leist R., Frey J.A., Leutwyler S. Scope and limitations of the SCS-MP2 method for stacking and hydrogen bonding interactions. Phys. Chem. Chem. Phys. 2008;10:2758–2766. doi: 10.1039/b718494h. [DOI] [PubMed] [Google Scholar]
- 19.Wu S., Xu S., Geng Y., Liu Z., Nie H., Shu L., Deng K., Zeng Q., Wang C. Scanning tunneling microscopy study on self-assembly behavior of hexylaniline derivatives spaced with diynes. J. Phys. Chem. C. 2016;120:12618–12625. [Google Scholar]
- 20.Pividori M., Dri C., Orselli M.E., Berti F., Peressi M., Comelli G. Spontaneous symmetry breaking on ordered, racemic monolayers of achiral theophylline: formation of unichiral stripes on Au(111) Nanoscale. 2016;8:19302–19313. doi: 10.1039/c6nr05301g. [DOI] [PubMed] [Google Scholar]
- 21.Cai L., Sun Q., Bao M., Ma H., Yuan C., Xu W. Competition between hydrogen bonds and coordination bonds steered by the surface molecular coverage. ACS Nano. 2017;11:3727–3732. doi: 10.1021/acsnano.6b08374. [DOI] [PubMed] [Google Scholar]
- 22.Xu L., Miao X., Cui L., Liu P., Miao K., Chen X., Deng W. Chiral transition of the supramolecular assembly by concentration modulation at the liquid/solid interface. J. Phys. Chem. C. 2015;119:17920–17929. [Google Scholar]
- 23.Miao K., Hu Y., Zha B., Xu L., Miao X., Deng W. Hydroxyl versus carbonyl substituent: effects of competitive and cooperative multiple hydrogen bonds on concentration-controlled assembly. J. Phys. Chem. C. 2016;120:14187–14197. [Google Scholar]
- 24.Miao K., Hu Y., Zha B., Xu L., Dong M., Miao X., Deng W. Polymorphic self-assemblies of 2,7-Bis(decyloxy)-9-fluorenone at the solid/gas interface: role of C-H···O=C hydrogen bond. J. Phys. Chem. C. 2017;121:3947–3957. [Google Scholar]
- 25.Miao K., Hu Y., Xu L., Dong M., Wu J., Miao X., Deng W. Chiral polymorphism in the self-assemblies of achiral molecules induced by multiple hydrogen bonds. Phys. Chem. Chem. Phys. 2018;20:11160–11173. doi: 10.1039/c8cp00591e. [DOI] [PubMed] [Google Scholar]
- 26.Guzzetti K.A., Brizuela A.B., Romano E., Brandan S.A. Structural and vibrational study on zwitterions of L-threonine in aqueous phase using the FT-Raman and SCRFcalculations. J. Mol. Struct. 2013;1045:171–179. [Google Scholar]
- 27.Leyton P., Brunet J., Silva V., Paipa C., Castillo M.V., Brandan S.A. An experimental and theoretical study of L-tryptophan in an aqueous solution combining two-layered ONIOM and SCRF calculations. Spectrochim. Acta, Part A. 2012;88:162–170. doi: 10.1016/j.saa.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Roldan M.L., Ledesma A.E., Raschi A.B., Castillo M.V., Romano E., Brandan S.A. A new experimental and theoretical investigation on the structures of aminoethyl phosphonic acid in aqueous medium based on the vibrational spectra and DFT calculations. J. Mol. Struct. 2013;1041:73–81. [Google Scholar]
- 29.Vener M.V., Medvedev A.G., Churakov A.V., Prikhadchenko P.V., Tripol’skaya T.A., Lev O. H-bond network in amino acid cocrystals with H2O or H2O2. The study of serine-H2O and serine-H2O2. J. Phys. Chem. A. 2011;115:13657–13663. doi: 10.1021/jp207899z. [DOI] [PubMed] [Google Scholar]
- 30.Parker S.F. Assignment of the vibrational spectrum of L-cysteine. Chem. Phys. 2013;424:75–79. [Google Scholar]
- 31.Janus S.O., Szymborska K., Komorowska M., Lipinski J. Conformational changes of L-phenylalanine – near infrared-induced mechanism of dimerization: B3LYP studies. J. Mol. Struct.: THEOCHEM. 2009;911:1–7. [Google Scholar]
- 32.Fisher G., Cao X., Cox N., Francis M. The FT-IR spectra of glycine and glycylglycine zwitterions isolated in alkali halide matrices. Chem. Phys. 2005;313:39–49. [Google Scholar]
- 33.Tonannavar J., Chavan Y.B., Yenagi J. (R)-(−)-2-Pyrrolidinemethanol: a combined experimental and DFT vibrational analysis of monomers, dimers and hydrogen bonding. Spectrochim. Acta, Part A. 2015;149:860–868. doi: 10.1016/j.saa.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Tonannavar J., Chavan Y.B., Yenagi J. A study of hydrogen bonded vibrational spectra of (R)-(+)-Methylsuccinic acid, as aided by DFT dimer analysis. Spectrochim. Acta, Part A. 2016;160:19–25. doi: 10.1016/j.saa.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen P.A., Norrby P.O., Liljefors T., Rega N., Barone V. Quantum mechanical conformational analysis of β-alanine zwitterion in aqueous solution. J. Am. Chem. Soc. 2000;122:3151–3155. [Google Scholar]
- 36.Berezhinsky L.I., Dovbeshko G.I., Lisitsa M.P., Litvinov G.S. Vibrational spectra of crystalline β-alanine. Spectrochim. Acta, Part A. 1998;54:349–358. [Google Scholar]
- 37.Dobson A.J., Gerkin R.E. γ-Aminobutyric acid: a novel tetragonal phase. Acta Crystallogr. 1996;C52:3075–3078. doi: 10.1107/s0108270196010001. [DOI] [PubMed] [Google Scholar]
- 38.Dobson A.J., Gerkin R.E. 8-Aminocaprylic acid. Acta Crystallogr. 1998;C54:969–972. doi: 10.1107/s0108270198001309. [DOI] [PubMed] [Google Scholar]
- 39.Dobson A.J., Gerkin R.E. DL-3-Aminoisobutyric acid monohydrate. Acta Crystallogr. 1998;C54:972–974. doi: 10.1107/s0108270198001310. [DOI] [PubMed] [Google Scholar]
- 40.Tomita K., Higashi H., Fujiwara T. Crystal and molecular structure of ω-amino sulfonic acids and their derivatives. IV. The crystal and molecular structure of γ-aminobutyric acid (GABA), a nervous inhibitory transmitter. Bull. Chem. Soc. Jpn. 1973;46:2199–2204. [Google Scholar]
- 41.Steward E.G., Player R.B., Warner D. The crystal and molecular structure of γ-aminobutyric acid determined at low temperature. Acta Crystallogr. 1973;B29:2038–2040. [Google Scholar]
- 42.Weber H.P., Craven B.M., McMullan R.K. The neutron structure of and thermal motion in γ-aminobutyric acid (GABA) at 122 k. Acta Crystallogr. 1983;B39:360–366. [Google Scholar]
- 43.Krishnan R.S., Sankaranarayanan V.N., Krishnan K. Raman and infrared spectra of amino acids. J. Indian Inst. Sci. 1973;55:66–116. [Google Scholar]
- 44.Jarmelo S., Reva I., Carey P.R., Fausto R. Infrared and Raman spectroscopic characterization of the hydrogen-bonding network in L-serine crystal. Vib. Spectrosc. 2007;43:395–404. [Google Scholar]
- 45.Yalagi S., Tonannavar J., Yenagi J. Experimental and DFT dimer modeling studies of the H-bond induced-vibration modes of L-β-Homoserine. Spectrochim. Acta, Part A. 2017;181:109–115. doi: 10.1016/j.saa.2017.03.041. [DOI] [PubMed] [Google Scholar]
- 46.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., aricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Jr., Montgomery J.A., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas Ö., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian Inc.; Wallingford CT: 2009. Gaussian 09, Revision A.1. [Google Scholar]
- 47.Dennington R., Keith T., Millam J. Semichem Inc.; 2009. GaussView 5.0. [Google Scholar]
- 48.Gangopadhyay D., Sharma P., Singh R.K. Temperature dependent Raman and DFT study of creatine. Spectrochim. Acta, Part A. 2015;150:9–14. doi: 10.1016/j.saa.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Tomasi J., Mennucci B., Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 50.Yao G., Zhang J., Huang Q. Conformational and vibrational analyses of meta-tyrosine: an experimental and theoretical study. Spectrochim. Acta, Part A. 2015;151:111–123. doi: 10.1016/j.saa.2015.06.073. [DOI] [PubMed] [Google Scholar]
- 51.Moellmann J., Grimme S. Importance of London dispersion effects for the packing of molecular crystals: a case study for intramolecular stacking in a bis-thiophene derivative. Phys. Chem. Chem. Phys. 2010;12:8500–8504. doi: 10.1039/c003432k. [DOI] [PubMed] [Google Scholar]
- 52.Grimme S., Antony J., Ehrlich S., Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132:154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 53.Merrick J.P., Moran D., Radom L. An evaluation of harmonic vibrational frequency scale factors. J. Phys. Chem. A. 2007;111:11683–11700. doi: 10.1021/jp073974n. [DOI] [PubMed] [Google Scholar]
- 54.Jamroz M.H. Vibrational energy distribution analysis (VEDA): scopes and limitations. Spectrochim. Acta, Part A. 2013;114:220–230. doi: 10.1016/j.saa.2013.05.096. [DOI] [PubMed] [Google Scholar]
- 55.Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 56.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 57.Colthup N.B., Daly L.H., Wiberley S.E. Academic Press Inc; New York and London: 1964. Introduction to Infrared and Raman Spectroscopy. [Google Scholar]
- 58.Glendening E.D., Landis C.R., Weinhold F. Natural bond orbital methods. Comput Mol Sci. 2012;2:1–42. [Google Scholar]
- 59.Reed A.E., Curtiss L.A., Weinhold F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988;88:899–926. [Google Scholar]
- 60.Foster J.P., Weinhold F. Natural hybrid orbitals. J. Am. Chem. Soc. 1980;102:7211–7218. [Google Scholar]
- 61.Pauling L. The nature of the chemical bond. Application of results obtained from the quantum mechanics and from a theory of paramagnetic susceptibility to the structure of molecules. J. Am. Chem. Soc. 1931;53:1367–1400. [Google Scholar]
- 62.Slater J.C. Directed valence in polyatomic molecules. Phys. Rev. 1931;37:481–489. [Google Scholar]
- 63.Irshaidat T. Modulating the electronic structure of amino acids: interaction of ModelLewis acids with anthranilic acid. Quim. Nova. 2014;37(9):1446–1452. [Google Scholar]
- 64.Alabugin I.V., Manoharan M., Peabody S., Weinhold F. Electronic basis of improper hydrogen bonding: a subtle balance of hyperconjugation and rehybridization. J. Am. Chem. Soc. 2003;125:5973–5987. doi: 10.1021/ja034656e. [DOI] [PubMed] [Google Scholar]
- 65.Ghalla H., Issaoui N., Bardak F., Atac A. Intermolecular interactions and molecular docking investigations on 4-methoxybenzaldehyde. Comput. Mater. Sci. 2018;149:291–300. [Google Scholar]
- 66.Gatfaoui S., Issaoui N., Menzi A., Bardak F., Roisnel T., Atac A., Marouani H. Synthesis, structural and spectroscopic features, and investigation of bioactive nature of a novel organic-inorganic hybrid material 1H-1,2,4-triazole-4-ium trioxonitrate. J. Mol. Struct. 2017;1150:242–257. [Google Scholar]
- 67.Trabelsi S., Issaoui N., Brandan S.A., Bardak F., Roisnel T., Atac A., Marouani H. Synthesis and physic-chemical properties of a novel chromate compound with potentialbiological applications, bis(2-phenylethylammonium) chromate(VI) J. Mol. Struct. 2019;1185:168–182. [Google Scholar]
- 68.Issa T.B., Sayari F., Ghalla H., Benhamada L. Synthesis, crystal structure,DFT calculations and molecular docking of L-pyroglutamic acid. J. Mol. Struct. 2019;1178:436–449. [Google Scholar]
- 69.Issa T.B., Ghalla H., Marzougui S., Benhamada L. Crystal structure and theoretical studies on quinoline phosphate. J. Mol. Struct. 2017;1150:127–134. [Google Scholar]
- 70.Rozas I., Alkorta I., Elguero J. Behavior of ylides containing N, O, and C atoms as hydrogen bond acceptors. J. Am. Chem. Soc. 2000;122:11154–11161. [Google Scholar]
- 71.Malaganvi S.S., Yenagi J.T., Tonannavar J. Spectroscopic and electronic structure characterization of hydrogen bonding in 2-Bromohydroquinone. J. Mol. Struct. 2019;1181:71–82. [Google Scholar]
- 72.Venkataramanan N.S., Suvitha A. Nature of bonding and cooperativity in linear DMSO clusters: a DFT, AIM and NCI analysis. J. Mol. Graph. Model. 2018;81:50–59. doi: 10.1016/j.jmgm.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 73.Sorocco E., Tomasi J. Electronic molecular structure, reactivity and intermolecular forces: an euristic interpretation by means of electrostatic molecular potentials. Adv. Quant. Chem. 1978;11:115–193. [Google Scholar]
- 74.Bellamy L.J. third ed. Chapman and Hall; London: 1975. The Infra-red Spectra of Complex Molecules. [Google Scholar]
- 75.Rao C.N.R. Academic Press Inc; New York and London: 1963. Chemical Applications of Infrared Spectroscopy. [Google Scholar]
- 76.Dollish F.R., Fateley W.G., Bentley F.F. John Wiley and Sons; Chichester, New York, Brisbane, Toronto: 1974. Characteristic Raman Frequencies of Organic Compounds. [Google Scholar]
- 77.Socrates G. John Wiley and Sons; New York, London, Sydney, Toronto: 1980. Infrared Characteristic Group Frequencies. [Google Scholar]
- 78.Krishnan R.S., Katiyar R.S. Raman and infrared spectra of β-alanine. Bull. Chem. Soc. Jpn. 1968;42:2098–2101. [Google Scholar]