O-Linked glycosylation is an evolutionarily conserved protein modification found across species, such as mammals, worms, insects, protozoa and certain types of fungi. Whereas, N-linked glycosylation occurs in eukaryotes and widely in archaea, but very rarely in bacteria. Recent studies of cancer immunotherapy are based on immunogenicity of truncated O-glycan antigens [1,2]. Despite the importance of N-linked glycans for normal cells to develop tumor cells, immunotherapy that targets N-linked glycans has not yet been developed mainly due to the lack of specificity of N-linked glycans between normal and malignant cells. O- or N-glycan chains are synthesized by the sequential action of glycosyl transferases in the Golgi apparatus. It is a daunting task to discover drug-like inhibitors of glycosyl transferases that block the synthesis of specific branching processes in cancer cells, killing tumor cells selectively. It has long been known that N-glycan biosynthesis can be inhibited by disruption of the first committed enzyme, DPAGT1 [3,4]. Selective DPAGT1 inhibitors have the promising therapeutic potential for certain solid cancers that require increased branching of N-linked glycans in their growth progressions. Recently, two groups successfully co-crystalized DPAGT1 with UDP-GlcNAc or tunicamycin [4,5]. This structural information clarifies the binding domains of the enzymatic substrates and an inhibitor molecule, and provides valuable insight into a catalytic mechanism of DPAGT1, allowing us to design de novo DPAGT1 inhibitors.
N-glycosylation & DPAGT1
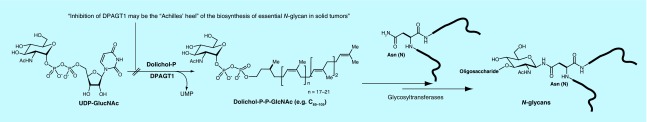
N-linked glycans play a key role in transformation-associated glycosylation changes for normal cells to develop tumor cells. Increased branching of N-linked glycans has been observed in certain solid cancer cells [6]. Abnormal glycosylation of N-linked glycans in cancers is typically associated with upregulation of β1,6-N-acetylyglucosaminyltransferase-5, enhancing β1,6-branching. An antibiotic, tunicamycin is known to be an inhibitor of two different bacterial phosphotransferases (MraY and WecA). Tunicamycin also interferes with the first committed enzyme of N-glycan biosynthesis, DPAGT1 (Figure 1) [7–10]. The recent studies have demonstrated that selective DPAGT1 inhibitors have the promising therapeutic potential for certain solid cancers that require increased branching of N-linked glycans in their growth progressions [11].
Figure 1. . DPAGT1 in N-glycan biosynthesis.
DPAGT1 is an integral membrane protein localized in the endoplasmic reticulum (ER) that catalyzes the transformation from UDP-GlcNAc to N-acetyl-D-glucosaminyl-diphosphodolichol. Anchored N-acetyl-D-glucosaminyl-diphosphodolichol in the ER membrane is modified by sequential glycosyltransferases to form dolichol-linked oligosaccharide precursors that are transferred to selected asparagine residues of polypeptide chains by oligosaccharyltransferase. DPAGT1 catalyzes N-glycosylation of β-catenin and E-cadherin. The mutations of β-catenin are found in a variety of cancers. A high level of β-catenin observed in the cytoplasm increases translocation to the nucleus, and drives transcription of the target genes including Wnt genes. Numerous studies concluded that the Wnt/β-catenin signaling pathway regulates the metabolic pathway of protein N-glycosylation by targeting DPAGT1 expression [12]. Based on these observed biological processes, inhibition of DPAGT1 could induce the loss of cell–cell adhesion and metathesis, and trigger an apoptotic pathway [13].
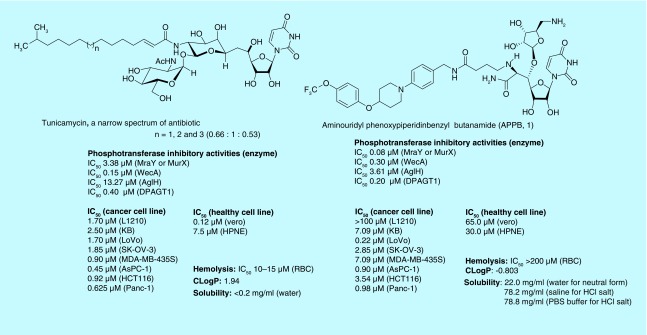
DPAGT1 inhibitors
A majority of in vitro studies associated with N-linked glycosylations through DPAGT1 and protein misfolding by DPAGT1 inhibition have been studied with tunicamycin. Tunicamycin is a nucleoside antibiotic that inhibits MraY and WecA. These enzymes are responsible for biosynthesis of peptidoglycan and cell wall in bacteria [14]. Tunicamycin shows greater than 20-times stronger inhibitory activity against WecA than that of MraY. It also inhibits DPAGT1 [15]. The cytotoxicity studies of tunicamycin against a series of cancer and healthy cells concluded that tunicamycin kills all cell lines with the IC50 values between 0.1 and 7.50 μM (Figure 2). Tunicamycin displays a strong hemolytic activity, causing anemia. Thus, one of the mechanisms of cytotoxicity of tunicamycin results from its perturbation ability of cell membrane structure (Mitachi et al.) [15]. These cytotoxicity statistics suggest that tunicamycin is not a therapeutic agent that is tolerated for in vivo studies. As of today, over 100 scientific articles have been produced about anticancer effects of tunicamycin. Among the widespread application of tunicamycin, quite a few studies have addressed its anticancer activity that is linked to the N-linked glycan biosynthesis. At 0.3–1.2 μM concentrations, tunicamycin can suppress migration and invasion of a human colon cancer cell line by an anchorage-dependent colony formation [16]. Their studies may demonstrate that inhibition of N-linked glycosylation by tunicamycin prevents the migration and adhesion of the colon cancer cells.
Figure 2. . Phosphotransferase enzyme and bacterial growth inhibiotors, cytotoxicities and physicochemical properties of tunicamycin, and a new DPAGT1 inhibitor, APPB (1).
In our screening of ribosamino-uridine derivatives against DPAGT1, we discovered that APPB (1) showed a strong DPAGT1 inhibitory activity [17]. A sharp difference between APPB and tunicamycin was that the hemolytic activity of APPB is significantly attenuated. A novel DPAGT1 inhibitor APPB selectively inhibits growth of the solid tumors at low micromolar concentrations, but does not inhibit growth of a leukemia cell and the healthy cells at these concentrations [19]. DPAGT1 expression levels of the cancer tissues summarized in the Human Protein Atlas revealed that several renal cancers and lymphomas show very low expression. However, several carcinomas of breast; melanomas and hepatocellular; adenocarcinomas of lung; pancreatic and ovarian cancers display very high expression. These data may imply that growth inhibition of APPB against the solid cancers is correlated with the expression level of DPAGT1.
Co-crystal structure of DPAGT1
Two groups have recently reported co-crystal structures of DAPGT1 with tunicamycin or UDP-GlcNAc [4,5]. These groups are primarily interested in mitigation of cytotoxicity of bacterial phosphotransferase inhibitor antibacterial agents.
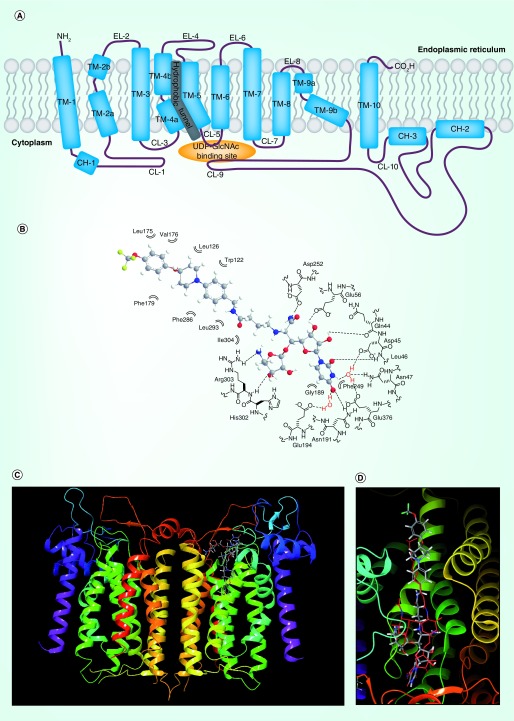
DPAGT1 comprises ten transmembrane segments (TM1–10). Three loops on the ER side and five loops on the cytoplasmic side connect the transmembrane segments, where TM-9, TM-4, CL-9, CL-7, CL-5 and CL-1 form the UDP-GlcNAc-binding domain. Dolychol-phosphate (Dol-P) is predicated to bind the ‘hydrophobic tunnel’ created by CL-9, TM-5 and TM-4 within the lipid bilayer (Figure 3A). In solution, DPAGT1 exists predominantly as a dimer. The uridine moiety of UDP-GlcNAc was coordinated with eight hydrogen bonds between the one backbone amide (Leu46), seven sidechains of the amino acids and two water molecules. The uridine moiety of tunicamycin occupies the identical binding sites of UDP-GlcNAc [5]. In the DPAGT1–tunicamycin co-crystal structure, it was realized that an Mg2+ ion is not involved in the binding mode; Arg301, Asn185 and Asp252 residues directly form hydrogen bonding with the C4-OH, C2-NH and C3-OH groups of the GalNAc moiety [4,6]. The fatty acid chain of tunicamycin is occupied in the hydrophobic tunnel. A majority of our antibacterial uridine-nucleoside derivatives that do not possess a hydrophobic sidechain did not display strong DPAGT1 inhibitory activity [15,17]. These results may suggest that introduction of pharmacologically benign hydrophobic groups which occupy the proposed Dol-P binding site is essential to exhibit DPAGT1 inhibitory activity.
Figure 3. . Two-dimensional representation of DPAGT1 (A) and docking studies of APPB (B, C & D).
(B) Schematic representation of interactions between APPB andits DPAGT1 binding domain. (C) The superimposed structures of DPAGT1-tunicamycin and DPAGT1-APPBa. (D) Close-up view of the superimposed structures of tunicamycin and APPBa. (C & D) aAPPB (1) was dockedwith DPAGT1 in complex with tunicamycin (Protein Data Bank ID: 6BW6) using Glide on Maestro from Schrodinger. The carbons of 1 are shown in grey; the hydrogens are white; the nitrogens are blue; the fluorines are green; the oxygens are red. Tunicamycin is shown in red. The docking score was -12.57 (Schrödinger, LLC, OR, USA). These images were generated with Maestro 11.5. DPAGT1 shows a homodimerict structure.
CH: Cytoplasmic helix segment; CL: Cytoplasmic loop; EL: ER loop; TM: Transmembrane segment.
Significant conformational changes are observed in the C-terminal end of TM-9, CL-9 and CL-1 in DPAGT1-ligand bound structures, however, the other portions of DPAGT1 are subtly changed upon binding [5]. Thus, a flexible ligand-rigid receptor docking method could provide meaningful guidance for evaluation of designed DPAGT1 inhibitors. Molecular docking techniques for structure-based drug discovery have been applied in a number of computer-aided drug design projects, and their basic theories include sampling algorithms, scoring functions and available docking software are summarized in review articles. APPB is an analog of uridine-nucleoside antibiotics, FR-900493 and the muraymycins [18,19]. Therefore, binding conformation of the uridine moiety of APPB and its binding site can be predicted. The uridine moiety of APPB should be buried in the cleft formed by the CL-5 and CL-7. APPB was manually docked with DPAGT1 (Protein Data Bank code: 6BW6) using Glide on Maestro from Schrödinger. Figure 3B illustrates interactions between APPB and its DPAGT1 binding domain, and Figure 3C illustrates the superimposed structures of DAPGT1-APPB and DPAGT1-tunicamycin. Docking analysis of a predicted binding conformer of APPB revealed that the ribosamino-uridine moiety occupies the tunicamycin GlcNAc-binding domain, and the hydrophobic portion of APPB is conformationally well organized to fit in the hydrophobic binding site as observed in the lipid-sidechain tunicamycin (Figures 3B & D). These docking studies of a biochemically validated DPAGT1 inhibitor, APPB, imply that the co-crystal structures of DPAGT1-tunicamycin are very powerful assets to develop novel DPAGT1 inhibitors.
Conclusion
Due to its interactions with human phosphotransferases enzyme(s), inhibition of eukaryotic phosphotransferase activities is believed to be attributable to the cytotoxicity of tunicamycin. Tunicamycin has a membrane disrupting activity that induces cytotoxic activity against a wide range of mammalian cells in a nonselective fashion. DPAGT1 is overexpressed in certain solid cancers. APPB (1) inhibits growth of solid cancer cell lines at low concentrations, but requires high concentrations to kill lymphomas and healthy cells. Thus, it was demonstrated that a selective DPAGT1 inhibitor can inhibit growth of solid tumors without deleterious consequences against normal cells. Recent co-crystal structures of DPAGT1 revealed the binding site of tunicamycin and the hydrophobic tunnel of DPAGT1. A case study summarized in Figure 3 proved that the flexible ligand-rigid enzyme docking method (a classic molecular docking) can provide critical insight into binding mode of newly designed DPAGT1 inhibitors. APPB is the only DPAGT1 inhibitor that has therapeutic potential, and is also an ideal seed molecule to initiate structure-based DPAGT1 inhibitor development toward new anticancer drugs.
Footnotes
Financial & competing interests disclosure
The NIH is greatly acknowledged for financial support of this work (grant number GM114611). I also thank University of Tennessee Health Science Center for generous financial support (CORNET award and UTRF Innovation Award). The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Vankemmelbeke M, Chua JX, Durrant LG. Cancer cell associated glycans as targets for immunotherapy. Oncoimmunology. 2016;5(1):e1061177. doi: 10.1080/2162402X.2015.1061177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belaya K, Finlayson S, Slater CR, et al. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am. J. Human Genetics. 2012;91(1):193–201. doi: 10.1016/j.ajhg.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou H, Sun H, Lu P, et al. Tunicamycin potentiates cisplatin anticancer efficacy through the DPAGT1/Akt/ABCG2 pathway in mouse xenograft models of human hepatocellular carcinoma. Mol. Cancer Ther. 2013;12(12):2874–2884. doi: 10.1158/1535-7163.MCT-13-0201. [DOI] [PubMed] [Google Scholar]
- 4.Yoo J, Mashalidis EH, Kuk A, et al. GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat. Struct. Mol. Biol. 2018;25(3):217–224. doi: 10.1038/s41594-018-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong YY, Wang H, Pike AC, et al. Structures of DPAGT1 explain glycosylation disease mechanisms and advance TB antibiotic design. BioRobix. 2018;175(4):1045–1058. doi: 10.1016/j.cell.2018.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27(55):6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sengupta PK, Bouchie MP, Kukuruzinska MA. N-glycosylation gene DPAGT1 is a target of the Wnt/β-catenin signaling pathway. J. Biol. Chem. 2010;285(41):31164–31173. doi: 10.1074/jbc.M110.149195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang SH, Wang DI, Zhang SH, et al. Tunicamycin potentiates paclitaxel-induced apoptosis through inhibition of PI3K/AKT and MAPK pathways in breast cancer. Cancer Chemother. Pharmacol. 2017;80(4):685–696. doi: 10.1007/s00280-017-3393-7. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee A, Martinez J, Longas M, et al. N-acetylglucosaminyl 1-phosphate transferase: an excellent target for developing new generation breast cancer therapeutic. Advances Exper. Med. Biol. 2015;842:355–374. doi: 10.1007/978-3-319-11280-0_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nami B, Donmez H, Kocak N. Tunicamycin-induced endoplasmic reticulum stress reduces in vitro subpopulation and invasion of CD44+/CD24-phenotype breast cancer stem. Cells. 2016;68(7):419–426. doi: 10.1016/j.etp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Kartha VK, Alamoud KA, Sadykov K, et al. Functional and genomic analyses reveal therapeutic potential of targeting β-catenin/CBP activity in head and neck cancer. Genome Med. 2018;10(54):1–18. doi: 10.1186/s13073-018-0569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nita-Lazar M, Rebustini I, Walker J, et al. Hypoglycosylated E-cadherin promotes the assembly of tight junctions through the recruitment of PP2A to adherens junctions. Experimental Cell Res. 2010;316(11):1871–1884. doi: 10.1016/j.yexcr.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim E, Jin H, Jeonghoon K, et al. Tunicamycin promotes apoptosis in leukemia cells through ROS generation and downregulation of surviving expression. Apoptosis. 2015;20(8):1087–1098. doi: 10.1007/s10495-015-1135-z. [DOI] [PubMed] [Google Scholar]
- 14.Katsuhiko M, Siricilla S, Yang D, et al. Fluorescence-based assay for polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) and identification of novel antimycobacterial WecA inhibitors. Anal. Biochem. 2016;512:78–90. doi: 10.1016/j.ab.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsuhiko M, Yun HG, Kurosu SM, et al. Novel FR-900493 analogs that inhibit outgrowth of Clostridium difficile spores. ACS Omega. 2018;3(2):1726–1739. doi: 10.1021/acsomega.7b01740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de-Freitas-Junior JCM, Bastos LG, Freire-Neto CA, et al. N-Glycan biosynthesis inhibitors induce in vitro anticancer activity in colorectal cancer cells. J. Cellular Biochem. 2012;113(9):2957–2966. doi: 10.1002/jcb.24173. [DOI] [PubMed] [Google Scholar]
- 17.Kurosu M. Inhibition of N-glycosylation towards novel anti-cancer chemotherapeutics. J. Mol Pharm. Org. Process Res. 2018;6(1):141–143. [PMC free article] [PubMed] [Google Scholar]
- 18.Katsuhiko M, Eslamimehr S, Kurosu S, et al. Boston, MA, USA: 2018. N-glycosylation inhibitors towards novel anti-cancer chemotherapeutics. Abstracts of Papers, 256th ACS National Meeting Exposition. 19–23 August. [Google Scholar]
- 19.Mitachi K, Kurosu S, Eslamimehr S, et al. A Semisynthesis of an anticancer DPAGT1 inhibitor from a muraymycin biosynthetic intermediate. Org. Lett. 2019;21(4):876–879. doi: 10.1021/acs.orglett.8b03716. [DOI] [PMC free article] [PubMed] [Google Scholar]