Abstract
N6-Methyladenosine (m6A) is the most prevalent internal modification in mammalian mRNAs. Although m6A is important in many biological processes, its roles in the placenta are unclear.
Methods: Levels of global mRNA m6A methylation and ALKBH5 expression in recurrent miscarriage (RM) patients were determined using quantitative reverse transcription-PCR (qRT-PCR), m6A RNA methylation quantification, and immunohistochemical methods. Using ALKBH5 overexpression and knockdown methods, we determined the role of ALKBH5 in trophoblast invasion at the maternal interface through trophoblasts and an extravillous explant culture experiments. Furthermore, the regulation of CYR61 by ALKBH5 was explored by RNA-sequencing coupled with methylated RNA immunoprecipitation.
Results: We found that the level of global mRNA m6A methylation was significantly decreased in placental villous tissue from RM patients, while ALKBH5 expression was specifically unregulated. Furthermore, we demonstrated that ALKBH5 knockdown in human trophoblast promoted trophoblast invasion. Conversely, overexpression of ALKBH5 inhibited cell invasion. ALKBH5 knockdown promoted trophoblast invasion in villous explant culture experiments, while overexpression of ALKBH5 repressed these effects. Furthermore, we clarified that ALKBH5 inhibited trophoblast invasion by regulating CYR61 mRNA stability, and this RNA regulation is m6A dependent. Mechanistic analyses showed that decreased ALKBH5 in trophoblast increased the half-life of CYR61 mRNA and promoted steady-state CYR61 mRNA expression levels.
Conclusions: We elucidated the functional roles of ALKBH5 and mRNA m6A methylation in trophoblast and identified a novel RNA regulatory mechanism, providing a basis for further exploration of broad RNA epigenetic regulatory patterns in RM diseases.
Keywords: N6-methyladenosine, ALKBH5, CYR61, trophoblast invasion, recurrent miscarriage.
Introduction
Among the more than 100 types of post-transcriptional modifications identified in RNAs thus far, N6-methyladenosine (m6A) RNA methylation is the most abundant internal mRNA modification 1. This modification is present transcriptome-wide in at least one-fourth of all RNAs, accounting for approximately 50% of the total methylated ribonucleotides and 0.1-0.4% of all adenosines in total cellular RNAs 2. M6A RNA methylation is a reversible nucleotide modification detected in at least 20% of mouse and human cellular mRNAs in diverse cell types 3 and is typically located in a consensus motif RRACH (R = A or G; methylatedadenosine residue is underscored) and enriched near the translation termination codon in 3'-untranslated regions (3'-UTRs) 4-6. In addition, previous study indicates that m6A modification has been most strongly linked to increased mRNA degradation 7. In mammal, m6A is deposited cotranscriptionally by a methyltransferase complex consisting of METTL3, METTL14, WTAP, KIAA1429, and RBM15/RBM15B 8-10. In turn, it can be removed by two m6A demethylases, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) 11-13. Recently, a relationship between FTO and obesity was established in animal research. Study found that FTO regulates adipogenesis and energy homeostasis, and FTO overexpression results in increased food intake and obesity in mice. While FTO deficiency protected against obesity 14, 15. By contrast, ALKBH5 is localized in the nucleus, which is most highly expressed in the testes but has low expression in the heart and brain, and the m6A mRNA level significantly decreased in ALKBH5-overexpressing cells 16. ALKBH5 strictly selects a substrate in catalytic progression, and the loss of ALKBH5 impairs RNA metabolism, mRNA export and assembly 16. Interestingly, ALKBH5 has been shown to be involved in glioblastoma and breast cancer stem cell 17, 18. In addition, ALBKH5 has been reported to play a key role in spermatogenesis. Male mice ALKBH5 knockout are completely infertile due to severe germ cell apoptosis and affects meiotic metaphase-stage spermatocytes 16, 19. Thus, these studies clearly indicate that changes in the expression of key genes, which are sensitive to the function of m6A modulator, can cause g significant phenotype changes. However, to date, very few studies have focused on how ALKBH5 is dysregulated in the pathogenesis of recurrent miscarriage (RM).
Cysteine-rich angiogenic inducer 61 (CYR61), a matricellular protein, is a member of the CCN family of secreted matricellular proteins that includes connective tissue growth factor (CCN2), NOV (CCN3), WISP-1 (CCN4), WISP-2 (CCN5), and WISP-3 (CCN6) 20. This protein consists of 4 conserved domains: insulin-like growth factor-binding protein (IGFBP), von Willebrand factor type C repeat (VWC), thrombospondin type 1 (TSP-1) repeat and a carboxyl-terminal (CT) domain containing a cysteine knot motif. CYR61 was originally shown to act as a growth factor‑inducible immediate‑early gene, present in serum and platelet‑derived growth factor‑stimulated mouse BALB/c 3T3 cells 21. The biological properties of CYR61 in the regulation of cell survival, proliferation, differentiation, migration, adhesion and synthesis of ECM have been demonstrated to be important in the progression of embryogenesis 22. In the human placenta, CYR61 is expressed in endothelial cells of placental vessels, stromal cells, and interstitial extravillous trophoblast (EVT) giant cells, and the expression levels increase during pregnancy 23. Kipkeew et al. reprted that CYR61 could enhance the migratory capability of SGHPL-5 trophoblast cells through the activation of the FAK signaling pathway 24. Moreover, clinical evidence has also shown the downregulation of CYR61 in the placenta in women with preeclampsia compared with normal matched controls 25. These studies indicate that the dysfunctional expression of CYR61 might be responsible for the impaired trophoblast cell invasion in pregnancy- associated diseases.
Thus, our study demonstrated that the m6A modification is crucial for the control of trophoblast invasion and that this regulation is disrupted in RM diseases. RM patients had significantly higher expression of the m6A demethylase ALKBH5 in chorionic villous tissue than normal controls; this result is consistent with the finding that ALKBH5 overexpression inhibits trophoblast invasion and decreases RNA m6A methylation levels. Furthermore, using RNA-sequencing experiments, we identified the impact of ALKBH5 on the expression of CYR61 in trophoblast. These results suggested that ALKBH5 plays a key role in the pathogenesis of RM by regulating trophoblast invasion.
Methods and materials
Patient Characteristics
Between July 2016 and April 2018, 69 patients with RM (23-38 years old; mean age, 30.6 ± 7.4 years) who had been treated at the Department of Obstetrics and Gynaecology at Shanghai First Maternity and Infant Hospital at the Tongji University School of Medicine, were included in this study. Patients with the following features were excluded: (1) presence of uterine abnormalities or cervical incompetence on pelvic examination and ultrasound, (2) abnormal karyotype analysis of the parents or abortus, (3) comprehensive hormonal status assessment to rule out luteal phase defects, hyperprolactinaemia and hyperandrogenaemia, and (4) symptoms of endocrine or metabolic diseases (e.g., diabetes, hyperthyroidism, and hypothyroidism).
Additionally, 65 women aged 22-38 years (mean age, 30.3 ± 7.9 years) with normal early pregnancies were recruited as healthy controls. All women have had previous pregnancies without any history of spontaneous abortion, preterm labor, or preeclampsia. All women recruited to the control group had undergone artificial abortions to terminate their unwanted pregnancies at 6-10 weeks of gestation, and villous tissue samples were collected from these patients, and the samples treated for isolation of primary trophoblast or stored in liquid nitrogen until analysis. The study protocol was approved by the Medical Ethics Committee of the Shanghai First Maternity and Infant Hospital, Shanghai. Written informed consents were obtained from all the participants before enrolment.
Cell Culture
Primary trophoblast were isolated by trypsin-DNase I digestion (Sigma-Aldrich, St. Louis, MO) and discontinuous Percoll (GE Healthcare, Amersham Place, UK) gradient centrifugation from pooled villi obtained from 3-5 patients, as previously described 26. The resultant trophoblast cell culture had a purity of approximately 95%, which was determined by flow cytometry for cytokeratin-7 positive, HLA-G positive, and vimentin negative cells. Purified trophoblast were seeded in the wells of 12-well plates at a concentration of 4 × 105 cells/mL and cultured in Dulbecco's modified Eagle's Medium (DMEM)/F12 (GE Healthcare) plus 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) for further experiments.
The HTR-8/SVneo cell line 27, which is derived from human invasive EVTs, was a kind gift from Dr. PK Lala (University of Western Ontario, London, Ontario, Canada). The cells were cultured in DMEM/F12 with 10% fetal bovine serum (FBS) plus Penicillin/ Streptomycin antibiotics (Invitrogen, Carlsbad, CA, USA). JEG-3 cells, a human choriocarcinoma cell line, were obtained from the cell bank at the Chinese Academy of Sciences (Shanghai, China) with the original source being the American Type Culture Collection (ATCC) (Manassas, VA, USA) and cultured in 1640 complete medium supplemented with 10% FBS plus Penicillin/ Streptomycin antibiotics in 5% CO2 at 37 °C.
Overexpression of ALKBH5
The pLV-EGFP-T2A-Puro-hALKBH5 plasmid and the control vector were purchased from Cyagen company, and GV141-CYR61 plasmid and the control vector were purchased from Shanghai Genechem company, the plasmids were purified using an Endofree Plasmid kit (Qiagen, Hilden, Germany) and transfected into the cells using a jetPRIME® kit (Polyplus, Illkirch, France). For lentivirus construction, the pLV-EGFP-T2A-Puro-hALKBH5 plasmid co-transfected with VSVG and ∆8.9 plasmids into HEK293 cells for 48 h to produce lentivirus overexpressing ALKBH5.
Knockdown of ALKBH5 and CYR61
ALKBH5 or CYR61 knockdown was performed using specific small interfering RNA specific for ALKBH5 (siALKBH5 or siCYR61). Unless otherwise indicated, all oligonucleotides were purchased from GenePharma Inc., and transfected into the cells at a final concentration of 100 nM/L using Oligofectamine regent (Invitrogen, Carlsbad, CA, USA).
Explant Culture
Small 2-3 mm tissue sections were obtained from the tips of first-trimester human placental villi (6-10 weeks), dissected, and explanted in 24-well culture dishes pre-coated with phenol red-free Matrigel® substrate (Corning Life Sciences, New York, NY), as previously described 28. Coated inserts were placed into 24-well culture dishes (Costar, Cambridge, MA). Villi explants were cultured in DMEM/F12 media containing 10% FBS. Placental villi that successfully anchored on Matrigel matrix and initiated outgrowth were used for the subsequent experiments, and are referred to as 24 h samples. EVT sprouting and migration from the distal end of the villous tips were recorded daily for up to 3 days. The extent of migration was measured via using ImageJ Pro 6.0 software. To test the effect of ALKBH5 on the migration of EVTs, 250 nM siRNA specifically targeting ALKBH5 or an equal concentration of control siRNA was introduced into two wells of extravillous explants from HCs or RM patients. The images were obtained after 24 h and 72 h of in vitro culture under a light microscope. Extravillous explants from HCs were incubated with lenti-ctrl or lenti-ALKBH5 lentiviral, and images after 24 h and 72 h of in vitro culture were taken under a light microscope. All explant experiments with cultured villi were repeated three times.
ALKBH5 knockdown Transcriptome sequencing
A total amount of 3 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. The PFKM was assessed and log2 transformed, the data was further calculated and displayed as heatmaps to help visualize differential expression. Details are provided in the Supplementary materials and methods.
Quantitative Real-time PCR
Total RNA was extracted from cultured cells or primary cells using the TRIzol reagent (Life Technologies, Grand Island, NY), according to the manufacturer's instructions, and used to generate cDNA with a 5 × All-In-One RT MasterMix from Applied Biological Materials Inc (Richmond, BC, Canada). Realtime-PCR (qRT-PCR) was performed using SYBR Green kit (Qiagen, Hilden, Germany). For in vitro experiments, relative expression was calculated using the 2-ΔΔCt method and normalized to the internal control gene GAPDH mRNA (human). For clinical data, relative expression was calculated using the 2-ΔCt method and normalized against GAPDH mRNA values. The primers used are shown in Supplementary Primers.
MeRIP-qPCR
For quantification of m6A-modified CYR61 levels, methylated RNA immunoprecipitation was performed. Total RNA was isolated from HTR-8 cells by Trizol. 3 μg of anti-m6A antibody (Millipore, ABE572) or anti-IgG (Cell Signaling Technology) was conjugated to protein A/G magnetic beads in IP buffer (20 mM Tris pH 7.5, 140 mM NaCl, 1% NP-40, 2 mM EDTA) for overnight at 4 oC. A 100 μg aliquot of total RNA was then incubated with the antibody in IP buffer supplemented with RNase inhibitor and protease inhibitor. RNA was eluted from the beads by incubating with 200 μl 0.5 mg/mL N6-methyladenosine 5-monophosphate sodium salt (Sigma-Aldrich) for 1 h at 4 ºC. Total RNA was eluted with elution buffer, purified through phenol-chloroform extraction. For further qRT-PCR assay, 10 ng of eluate or input total RNA was reverse-transcribed using Superscript III with random hexamers, and enrichment of m6A-containing transcripts. Fold enrichment was calculated by calculating the 2-ΔCt of eluate relative to the input sample. The primers used for PCR were as follows: CYR61 primer for MeRIP-qPCR #1 F: 5'-GAATGCAGCAAGACCAAGAAAT-3', R: 5'-ACGCAGTACTTGGGCCGGTAT-3'; CYR61 primer for MeRIP-qPCR #2 F: 5'-TTTCCAAGAACGTCATGATGAT-3', R: 5'-CCTGGAAACC- CAGGTAGCAT-3'.
Statistical Analysis
All statistical values were calculated using SPSS 22.0 (Chicago, IL, USA). Experimental studies were analyzed by independent sample t-test for the comparison between 2 groups, and by one-way ANOVA followed by post-hoc Tukey's test for the comparison among multiple groups. For clinical studies, the nonparametric Mann-Whitney test was used to compare ALKBH5 and CYR61 expression. Correlations were analyzed using the Spearman's rank correlation test. Data are presented as mean ± SD. All P values are two-sided. A P value of < 0.05 was considered statistically significant.
Results
RM is associated with high levels of m6A mRNA methylation
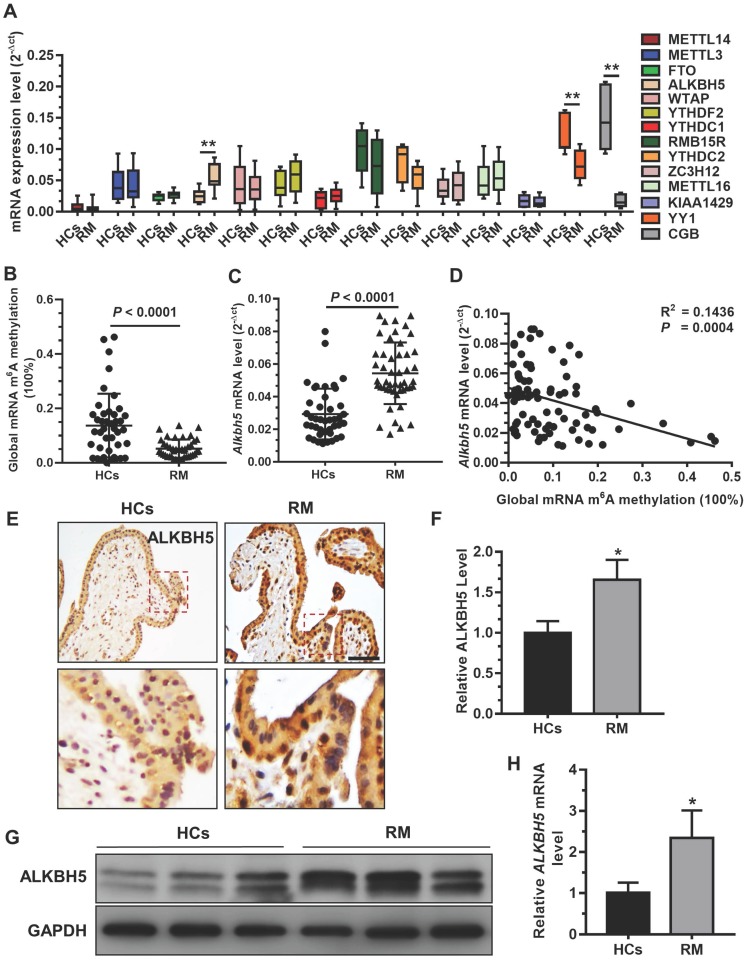
A previous study demonstrated that insufficient proliferation and invasion of cytotrophoblasts (CTBs) is associated with early or late RM 29. To explore whether mRNA m6A methylation is involved in the pathogenesis of RM, we analyzed the expression profiles of m6A 'writer', 'eraser', and 'reader' associated genes in chorionic villous tissues derived from RM patients (n = 12) and HCs (n = 12) using qRT-PCR. Our results showed that ALKBH5 mRNA expression was significantly increased in the chorionic villi of RM patients compared to HCs (Figure 1A). In contrast, level of YY1 and CGB mRNA expression, which was reported in our previous paper 26, was significantly decreased in the chorionic villi of RM patients compared to HCs. We further examined the expression levels of m6A and ALKBH5 mRNA in chorionic villi from RM patients (n = 65) and HCs (n = 57). Consistently, we found a significantly higher ALKBH5 mRNA level and a significantly lower m6A level in the chorionic villi from the RM group compared to the HC group (Figures 1B-C). Linear correlation analysis showed that the ALKBH5 mRNA level was negatively correlated with the mRNA m6A level in villous tissue (Figure 1D). These results suggest that ALKBH5 inhibits RNA m6A methylation in trophoblasts from RM patients. Furthermore, immunohistochemical analysis of paraffin-embedded first-trimester chorionic villous tissues was performed to investigate the localization of ALKBH5 in chorionic villous tissue. ALKBH5 expression in normal chorionic villous tissue from HCs was primarily observed in the nucleus of cytotrophoblasts (CTBs). A stronger positive signal for ALKBH5 was detected in chorionic villous tissue from the RM group compared to the HC group (Figures 1E-F). These findings were confirmed by western blotting and qRT-PCR analysis, which showed that ALKBH5 was expressed at a higher level in primary trophoblast of RM patients than HCs (Figure 1G-H). Collectively, these results indicate that ALKBH5 expression is increased in trophoblast in RM patients and suggest that this increase may be correlated with trophoblast function.
Figure 1.
ALKBH5 is overexpressed in first-trimester placental trophoblast from recurrent miscarriages patients. (A) Expression of RNA m6A methylation-regulated associated genes was determined in first-trimester human villous tissues from patients with RM or in healthy controls (HCs) using quantitative RT-PCR (qRT-PCR). (B) Global RNA m6A methylation of villous tissues from patients with RM or in HCs was assessed using the EpiQuik m6A RNA methylation quantification ELISA kit. (C) The ALKBH5 mRNA expression levels in the villous tissues of patients with RM (n = 65) and HCs (n = 57) were determined by qRT-PCR. (D) Global RNA m6A methylation level was measured in the villous tissues of patients with RM and HCs by EpiQuik m6A RNA methylation quantification ELISA kit and was correlated to level of ALKBH5 mRNA expression. (E and F) Single staining of maternal villi (cytotrophoblasts and syncytiotrophoblasts) using anti-IgG (rabbit) or anti-ALKBH5 antibody (rabbit), which was visualized with a labeled streptavidin biotin horseradish peroxidase (HRP) kit. Sections were counterstained with hematoxylin, and positive cells were quantified using Image-Pro Plus 6.0 software (n = 19); Right scale bar = 100 μm. (G and H) Primary trophoblasts were isolated from first-trimester villi of RM patients and HCs, and ALKBH5 levels were determined by western blotting and qRT-PCR.
ALKBH5 regulates trophoblast invasion at the maternal-fetal face
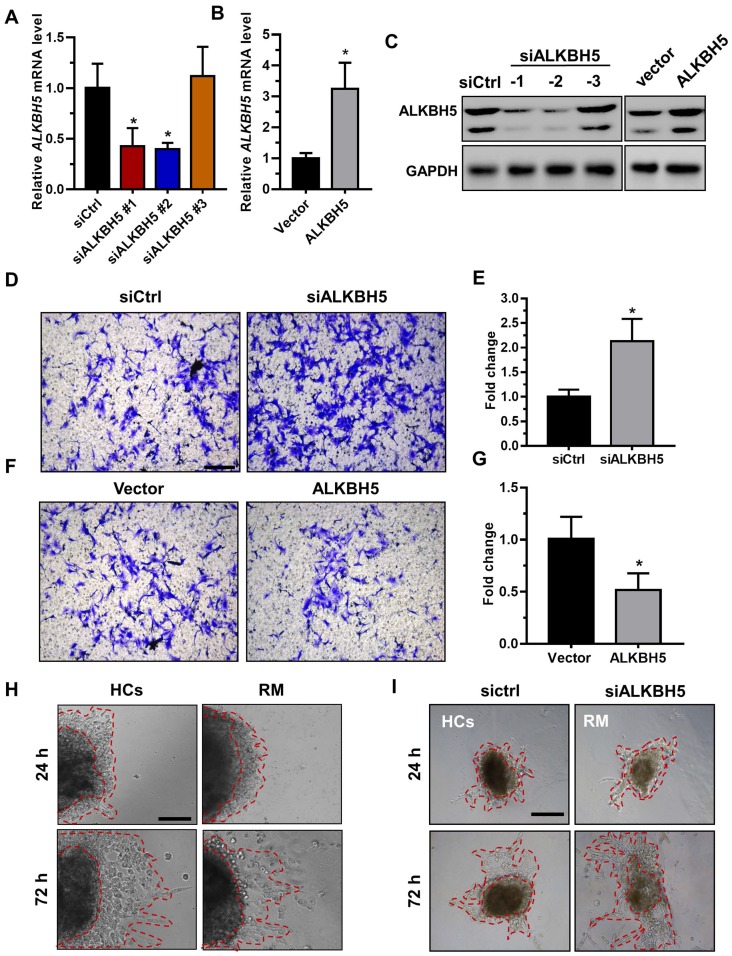
To directly address the role of ALKBH5 in human trophoblast, the HTR-8/SVneo (HTR-8) cell line, a first-trimester human extravillous trophoblast-derived cell line, was transfected with siALKBH5 oligonucleotides or with the ALKBH5-expressing vector. ALKBH5 expression was reduced after siALKBH5 transfection and was increased after transfection with the ALKBH5 expression vector (Figures 2A-C). Further, Matrigel® invasion assays revealed that overexpression of ALKBH5 significantly inhibited the invasive ability of HTR-8 cells, whereas knockdown of ALKBH5 significantly promoted cell invasion (Figures 2D-G).
Figure 2.
ALKBH5 inhibits trophoblast invasive ability in vitro and trophoblast outgrowth in a villous explant culture model. (A and B) QRT-PCR and western blotting analyses were performed to determine the levels of ALKBH5 in HTR-8 cells transfected with siCtrl, siALKBH5-1, siALKBH5-2, siALKBH5-3, control vector, or ALKBH5-overexpressing vector after 48 h. (D and E) ALKBH5 knockdown in HTR-8 cells increased trophoblast invasion compared to that of the control cell line. The invasive ability of the cells was assessed by Image-Pro Plus 6.0 software. (Panel: original magnification × 200 magnification). (F and G) ALKBH5 overexpression significantly reduced cell invasion compared to that of the vector control cell line. The invasive ability of the cells was assessed by Image-Pro Plus 6.0 software. (Panel: original magnification × 200 magnification). (H) Extravillous explants were obtained from HCs and RM patients at 6-10 weeks of gestation and cultured on Matrigel. Serial pictures of the explants were taken under a light microscope after 24 h and 72 h of culture in vitro. (I) Extravillous explants from RM at 6-10 weeks of gestation were maintained in culture on Matrigel. Serial pictures of the explants transfected with siALKBH5 or siCtrl were taken under a light microscope after 24 h and 72 h of culture in vitro.
Extravillous explants from first-trimester villi from the RM and age- and gestational week-matched control groups were cultured on Matrigel-coated dishes. The distance of outgrowth on the Matrigel surface was measured at 24 h and 72 h. Following 24 h of culture, the explants were anchored into the Matrigel and started to exhibit outgrowth. No significant difference was observed between the control and RM groups at this point. At 72 h of in vitro culture, the RM explants had migrated shorter distances than the control explants (Figure 2H and Figure S1A). To further confirm the role of ALKBH5 in trophoblast invasion, we cultured explants freshly obtained from the RM villous tissue samples (6-10 weeks of gestation) in 24-well dishes for 24 h and then treated them with siCtrl and siALKBH5 oligonucleotides. The results showed that the explants treated with siALKBH5 migrated significantly farther than the siCtrl-treated explants (Figure 2I and Figure S1B). Therefore, these results clearly suggest that ALKBH5 plays an important role in the regulation of trophoblast invasion.
The m6A modification is enriched in CYR61
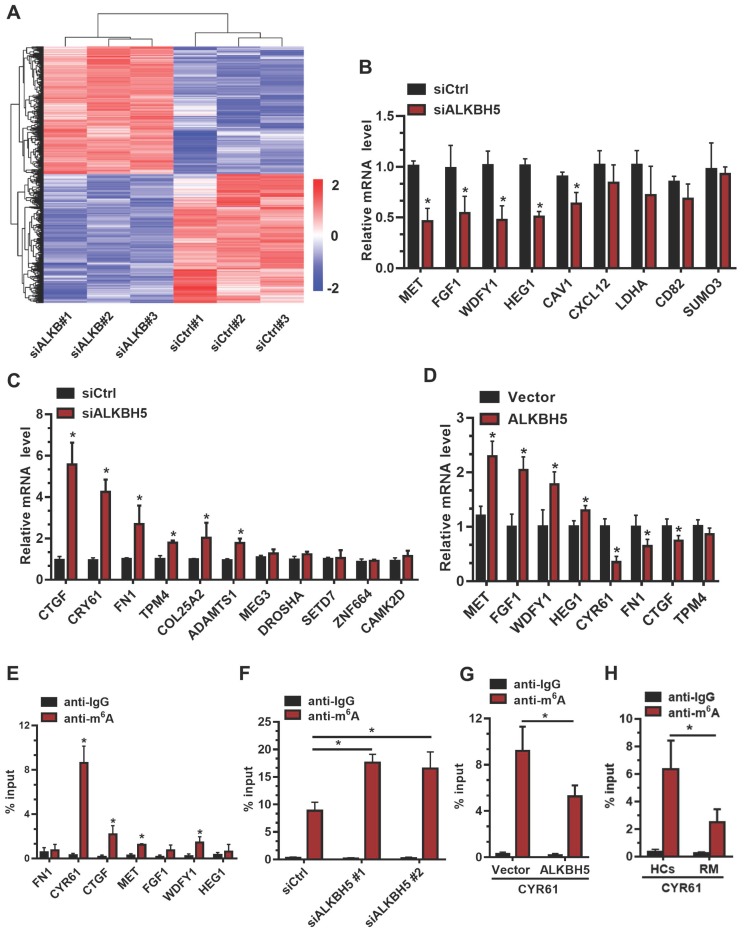
To further elucidate the molecular mechanism underlying ALKBH5 regulation of trophoblast invasion, we transfected HTR-8 cells with siALKBH5 oligos for 48 h. Transcriptome RNA sequencing was employed to identify genes dysregulated by ALKBH5 knockdown in HTR-8 cells. A total of 6891 genes were found to be dysregulated after ALKBH5 knockdown. Of these, 3383 were upregulated and 3408 were downregulated (Figure 3A), the majority of which had a fold change value (siALKBH5/siCtrl) between 0.5 and 2 after ALKBH5 knockdown. Supplemental Table S1 provides detailed gene quantitation information from the three replicate experiments. We next selected 33 genes based on transcriptome analysis for validation. Although there were differences in the fold changes of some genes between the RNA sequencing data and the qRT-qPCR validation data, 20 phenotypically relevant genes all exhibited changes in the same direction (Figures 3B-C), suggesting a significant impact of ALKBH5 on these mRNAs expression. Furthermore, HTR-8 cells were transfected with ALKBH5-expressing plasmid for 48 h, and qRT-PCR clearly showed that the MET, FGF1, WDFY1, and HEG1 transcripts were increased in trophoblasts from the ALKBH5 overexpression group. In contrast, FN1, CYR61, CTGF, and TPM4 transcripts were obviously decreased in trophoblast from the ALKBH5 overexpression group (Figure 3D). Furthermore, we determined the m6A modification of MET, FGF1, WDFY1, HEG1 FN1, CYR61, CTGF, and TPM4 by performing methylated RNA immunoprecipitation (MeRIP) experiments. Interestingly, CYR61 was significantly enriched in the anti-m6A group compared with the IgG group (Figure 3E). To define the potential functions of m6A-modified CYR61, using SRAMP online tool (A sequence-based N6-methyladenosine (m6A) modification site predictor) 30, we analyzed the m6A sites located in CYR61 and found five RRACH m6A sequence motifs in the 3'-untranslated regions (3'-UTR) proximal to the stop codon, which was previously described as a location for m6A deposition (Figure S2). To further characterize the m6A methylation of CYR61, we performed siRNA-mediated downregulation of ALKBH5, an important component of the m6A demethylase complex, in trophoblast. The MeRIP results showed that ALKBH5 knockdown significantly increased the enrichment of CYR61 mRNA in the anti-m6A group compared with the IgG group (Figure 3F). ALKBH5 overexpression decreased the enrichment of CYR61 mRNA (Figure 3G). We further detected the m6A methylation of CYR61 mRNA in villous from HCs and RM patients, the MeRIP-PCR result showed that enrichment of CYR61 mRNA was higher in HCs group compared with RM patients group (Figure 3H). These data indicate that ALKBH5 regulates the expression of CYR61 mRNA in an m6A-dependent manner.
Figure 3.
CYR61 is a downstream target of m6A in trophoblast. (A) Heatmap of normalized gene expression levels of HTR-8 cells transfected with siCtrl or siALKBH5. Blue indicates down-regulation of genes expression; Red indicates up-regulation of genes expression. (B and C) QRT-PCR was performed to determine the mRNA levels of the selected genes in HTR-8 cells transfected with siCtrl or siALKBH5. (D) QRT-PCR was performed to determine the mRNA levels of selected genes in HTR-8 cells transfected with vector or ALKBH5-expressing plasmid. (E) Enrichment of m6A-modified CYR61, CTGF, MET and WDFY1 in HTR-8 cells. The percentage of the input is shown. *P < 0.05 versus anti-IgG. (F) Changes in m6A-modified CYR61 level upon ALKBH5 knockdown. The percentage of the input is shown. *P < 0.05 versus siCtrl. (G) Changes in m6A-modified CYR61 level upon ALKBH5 overexpression. The percentage of the input is shown. *P < 0.05 versus vector. (H) Changes in m6A-modified CYR61 level upon HCs and RM. The percentage of the input is shown. *P < 0.05 versus HCs.
M6A methylation specifically controls CYR61 expression and stability in trophoblasts
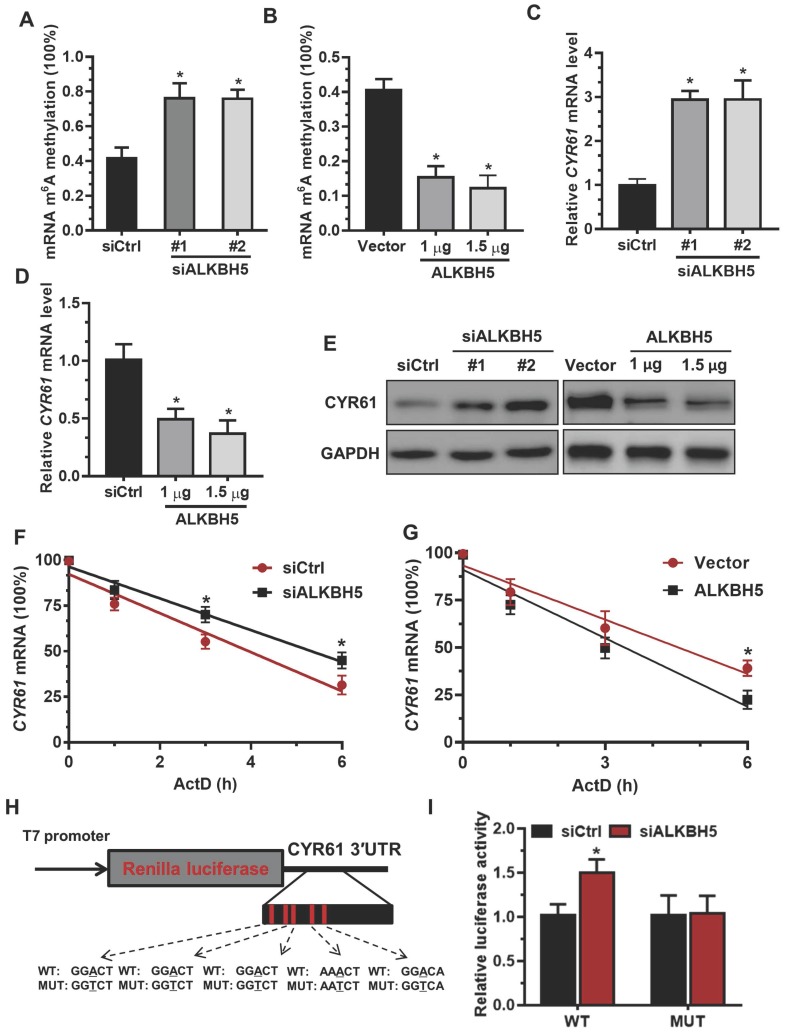
To determine the underlying mechanisms of decreased CYR61 expression upon increased m6A methylation, we measured the m6A levels in primary trophoblast transfected with ALKBH5-expressing plasmid or ALKBH5 siRNA. The results showed that ALKBH5 overexpression resulted in a significant reduction in the RNA m6A methylation of primary trophoblast, as indicated by m6A RNA methylation quantification (colorimetric) analysis (Figure 4A), whereas ALKBH5 knockdown increased the RNA m6A methylation level (Figure 4B). QRT-PCR results showed that ALKBH5 knockdown significantly promoted CYR61 mRNA expression in HTR-8 cells, while ALKBH5 overexpression decreased its expression (Figures 4C-D). Further, western blotting results showed that ALKBH5 knockdown significantly promoted expression of CYR61 protein in HTR-8 cells and JEG-3 cells, while overexpression of ALKBH5 decreased its expression (Figures 4E and Figure S3). We further explored the mechanism by which m6A methylation regulates mRNA stability. The mRNA half-life of CYR61, FGF1, CTGF and MET was assessed in the ALKBH5 siRNA-transfected trophoblast using an actinomycin D chase experiment and a one-phase exponential decay model with GAPDH mRNA normalization. This experiment revealed the strong impact of ALKBH5 on CYR61 mRNA stability, but did not affect FGF1, CTGF and MET mRNA stability. The half-life of CYR61 mRNA was approximately 4.3 h after siRNA control transfection; the half-life was increased when cells were transfected with siALKBH5 oligos (5.4 h) (Figure 4F and Figure S4). The half-life of CYR61 mRNA was further assessed in the ALKBH5-expressing trophoblast. The half-life of CYR61 was 4.5 h after vector control transfection, and the half-life was decreased by almost 1.5 times when cells were transfected with the ALKBH5 vector (3.2 h) (Figure 4G). To further elucidate the molecular mechanism of m6A regulation of CYR61 mRNA stability, we performed the CYR61 3' UTR-reporter luciferase assay and found that ALKBH5 knockdown increased the activity of the luciferase construct containing the CYR61 3'UTR. Mutation at all of these sites (A to T) almost completely rendered resistance to the luciferase activity of ALKBH5 knockdown (Figures 4H-I). These results suggest that ALKBH5 regulates CYR61 post-transcriptionally via an m6A methylation-dependent mechanism.
Figure 4.
The m6A mark is critical for CYR61 expression and stability in trophoblasts. (A and B) RNA m6A RNA methylation of primary trophoblast transfected with siALKBH5 or ALKBH5-expressing plasmid was assessed using the m6A RNA Methylation Quantification ELISA kit. (C-E) Western blotting and qRT-PCR analysis of CYR61 expression in HTR-8 cells transfected with siCtrl, siALKBH5 #1, siALKBH5 #2, control vector, or ALKBH5-expressing plasmid after 48 h. (F and G) HTR-8 cells were transfected with siCtrl, siALKBH5, vector or ALKBH5-expressing plasmid for 36 h; a time course for mRNA stability was initiated by adding an RNA-Polymerase II inhibitor [actinomycin D (5 µg/mL)]. Cells were harvested at the indicated time points. Expression levels were normalized to that at “0 hour”, and GAPDH mRNA was used as the reference gene. The results are shown as the mean of at least three independent experiments. *P < 0.05 versus the vector or siCtrl. (H and I) A schema for the constructs and co-transfection experiments, a fragment of 3'-UTR of CYR61 (wild-type and mutant) was cloned into a psiCheck2 vector, downstream of the renilla luciferase gene. These construct where was co-transfected with the siALKBH5 or siCtrl into HTR-8 cells. Cells were harvested after 24 h. Renilla luciferase activity was measured and normalized to that of firefly luciferase. The results are represented as the mean of three independent experiments. *P < 0.05 versus the siCtrl.
CYR61 is a functional target of ALKBH5 in trophoblast
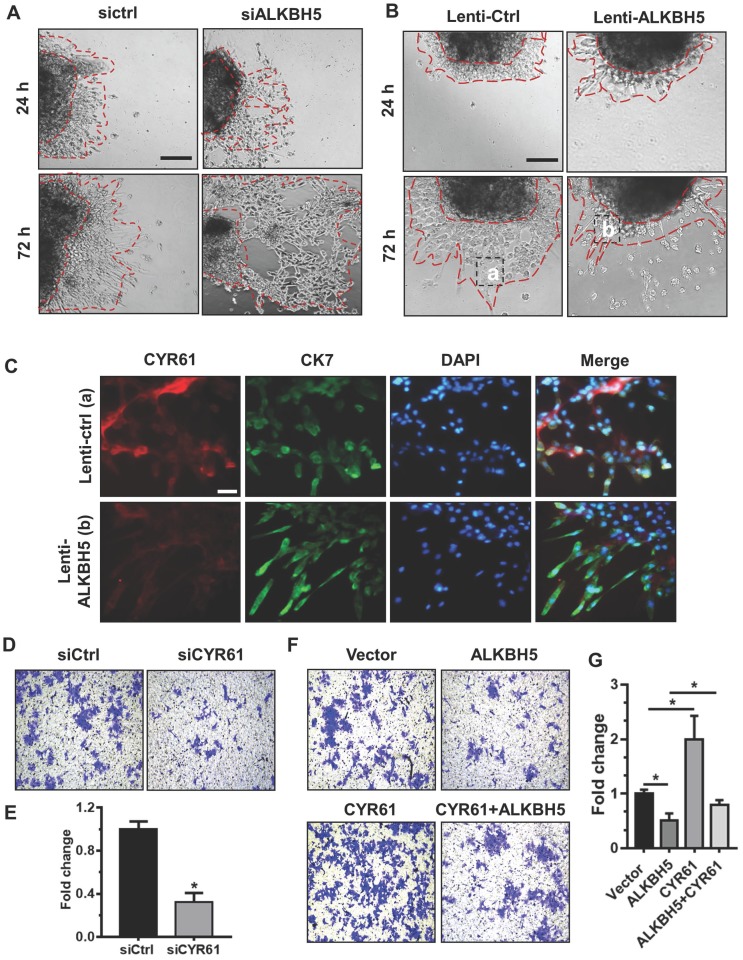
To further assess the relationship between ALKBH5 and the regulatory functions of CYR61 in trophoblast, we cultured explants freshly obtained from the HC villous tissue samples in 24-well dishes for 24 h, and then treated them with siCtrl, siALKBH5, lenti-ctrl or lenti-ALKBH5 lentivirus. Explants treated with lenti-ALKBH5 migrated significantly slower than the lenti-ctrl-treated explants group. By contrast, explants treated with siALKBH5 migrated significantly farther than the siCtrl-treated explants group (Figures 5A-B and Figure S5). Furthermore, whole-mount immunofluorescence staining of villous samples from both groups was performed to detect CYR61 expression and CK-7 was used as a marker of EVT invasion and migration. The CYR61 levels were significantly decreased in the EVT of villous tissues from the lenti-ALKBH5-treated group compared with the lenti-ctrl-treated group (Figure 5C).
Figure 5.
ALKBH5 promotes trophoblast invasion by upregulating CYR61 expression. (A and B) Extravillous explants from HCs were maintained in culture on Matrigel. Serial pictures of the explants incubated with siCtrl, siALKBH5, lenti-ctrl or lenti-ALKBH5 lentivirus were taken under a light microscope after 24 h and 72 h of culture in vitro. (C) Immunofluorescence staining using anti-CYR61 antibodies showed an obvious decrease in the CYR61 level in the ALKBH5 overexpression group (b) compared with the vector group (a). Green fluorescence signals indicate bound anti-CK-7 antibodies; CYR61 staining is visualized as red; and the DAPI-stained nuclei are blue. (D and E) HTR-8 cells were transfected with siCtrl or siCYR61 for 24 h. The invasive ability of the cells was assessed by Image-Pro Plus 6.0 software. *P < 0.05 compared with the siCtrl. (F and G) HTR-8 cells were transfected with vector or ALKBH5 and CYR61 expression vectors or with the ALKBH5 + CYR61 expression vector for 24 h. The invasive ability of the cells was assessed by Image-Pro Plus 6.0 software. *P < 0.05 versus the vector.
To further confirm the role of CYR61 in trophoblast invasion, western blotting analysis was used to analyze the level of CYR61 in HTR-8 cells transfected with siCYR61 or CYR61 overexpression vectors. CYR61 knockdown or ALKBH5 overexpression decreased expression of CYR61, while CYR61 overexpression significantly enhanced expression of CYR61. Overexpression of CYR61 could rescue the level of CYR61 protein decreased by ALKBH5 overexpression (Figure S6). Furthermore, the Matrigel cell invasion assays results revealed that CYR61 knockdown significantly decreased the invasive ability of trophoblast (Figures 5D-E). Additionally, CYR61 overexpression effectively reversed the reduction in trophoblast invasion caused by ALKBH5 overexpression (Figures 5F-G). These results indicate that ALKBH5 regulates trophoblast invasion via downregulation of CYR61 expression.
CYR61 expression correlates negatively with ALKBH5 expression in RM patients
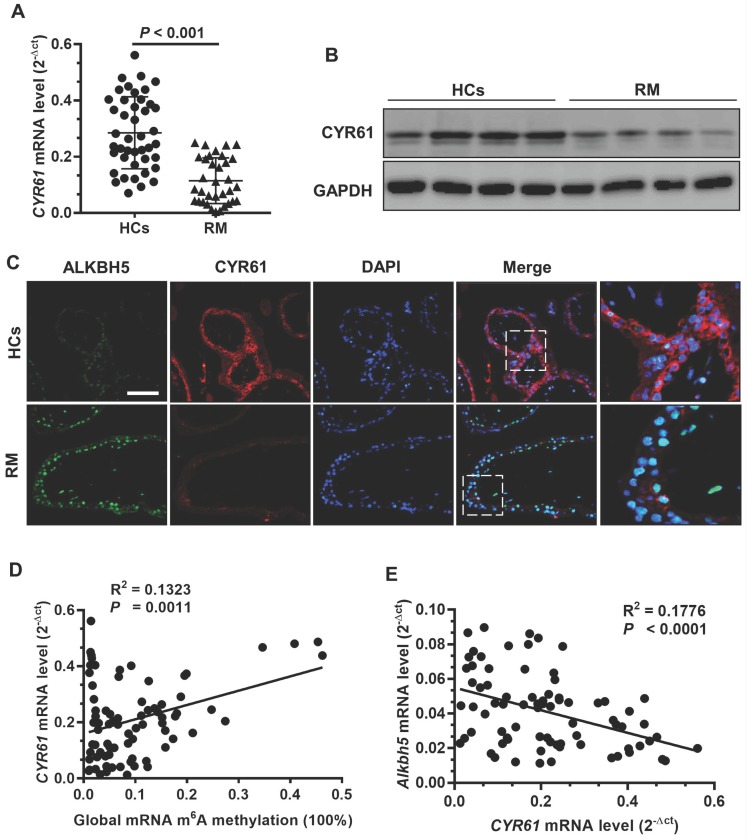
To further clarify the clinical significance of CYR61 in RM patients, we evaluated CYR61 expression using qRT -PCR and western blot analyses of first-trimester chorionic villi tissues to determine whether CYR61 is involved in the pathogenesis of RM. CYR61 expression was significantly downregulated in villous tissue of RM patients (Figures 6A-B). Double immunofluorescence staining for ALKBH5 and CYR61 revealed increased staining for ALKBH5 and decreased staining for CYR61 in CTBs from villous tissues of the RM group compared with that of the HC group (Figure 6C). Linear correlation analysis showed that the CYR61 mRNA level was positively correlated with global RNA m6A methylation in villous tissue (Figure 6D), while the CYR61 mRNA level was negatively correlated with the ALKBH5 mRNA level in villous tissue. These results suggest that ALKBH5 and CYR61 expression levels are negatively associated in the villous tissue of the RM group, which correlated with the pathogenesis of RM.
Figure 6.
ALKBH5 expression is negatively correlated with CYR61 expression in RM disease. (A) CYR61 mRNA levels in first-trimester human villous tissues from RM patients (n = 65) or in HCs (n = 57) were determined by qRT-PCR. (B) Levels of CYR61 expression in first-trimester human villous tissues from RM patients or in HCs were determined by western blotting. (C) Representative immunofluorescence images of ALKBH5 and CYR61 in frozen first-trimester villous sections obtained from RM patients and HCs. Fluorescence signals specific to anti-ALKBH5 antibodies appear green; anti-CYR61 antibodies appear red; and the DAPI-stained nuclei appear blue. (D) Global RNA m6A methylation level correlated positively with CYR61 mRNA levels in the villous tissue of RM patients and HCs. (E) ALKBH5 mRNA level correlated negatively with the CYR61 mRNA level in the villous tissues of RM patients and HCs.
Discussion
RM affects approximately 1-3% of women during their reproductive years and is usually defined as three or more consecutive spontaneous abortions before 20 weeks of gestation 31. RM is associated with substantial adverse clinical and psychological consequences for women and their families. Various therapeutic strategies to increase the rate of live births among these women have been evaluated, but no effective treatment has been identified 32. Moreover, RM is also associated with a high level of psychological distress. Thus, comprehensive molecular studies are needed to elucidate the causes of RM and to develop potential targets. However, the underlying molecular mechanism of RM remains poorly understood. Currently, many studies have found that insufficient proliferation and invasion of trophoblasts are associated with early and late RM 29, 33. In this study, we explored the function of ALKBH5 in human villi and trophoblast during the early stage of pregnancy.
ALKBH5 RNA demethylase belongs to the AlkB subfamily of the Fe (II)/2-oxoglutarate (2OG) dioxygenase superfamily and is primarily localized in the nucleus. This enzyme is upregulated under hypoxic conditions by the hypoxia-inducible factor (HIF) transcription factor pathway 17. ALKBH5 was observed to be highly expressed in the lung, followed by the testis, pancreas, spleen and ovary, but has low expression in the heart and brain 16, 34. Interestingly, ALKBH5 may function differently in distinct types of diseases. ALKBH5 functions as an oncoprotein in GBM and breast cancer 17, 18; it may exert a tumor-suppressor role in AML as implied by its frequent copy number loss and low expression level in AML 35. In this study, we first found that the ALKBH5 level was higher in the placental tissue from RM patients than that of age- and gestational-matched HCs, suggesting that ALKBH5 is disrupted under pathological conditions. Moreover, multiple lines of evidence, including in vitro cell migration and invasion assays, in vivo extravillous explant culture experiments, and analyses of human RM specimens, support the role of ALKBH5 in mediating trophoblast via regulating CYR61 expression. Therefore, these data demonstrate that ALKBH5 could play an important role in trophoblast invasion. This is in contrast to previous result, which reports ALKBH5 functions as an oncogene in GBM and breast cancer. It is possible that ALKBH5 could play different functions in the different cell context, likely through regulating distinct sets of targets. Thus, more detailed studies are warranted to explore the biological function of each individual m6A regulatory genes in different types of cell, and to identify their core target genes to elucidate the underlying molecule mechanisms.
During placental development, continuous invasion of trophoblasts into the maternal compartment depends on the support of proliferating EVT. Impairment of EVT invasion predisposes a pregnancy to uteroplacental insufficiency and results in a significantly increased risk of preeclampsia, fetal growth restriction (FGR), and early RM 36-38. Recently, CYR61 was shown to facilitate the migration and invasion of trophoblastic cells. CYR61 enhanced the migratory capability of trophoblast cells via the activation of FAK and Akt kinase 24. Here, we found that elevated ALKBH5 impaired trophoblast invasion by inhibiting the RNA m6A methylation level, which reduced the expression of CYR61. We further used gain- and loss-of-function analyses, MeRIP, and RNA decay analyses to demonstrate that ALKBH5 regulates CYR61 stability and expression in trophoblast via an m6A methylation-dependent mechanism. Together, these findings indicate that ALKBH5 might be involved in the progression of RM by modulating CYR61 expression.
In summary, this study provides new insight into the pathogenesis of RM by elucidating the role of ALKBH5, which was found to regulate trophoblast invasion in early pregnancy through its effects on mRNA m6A methylation. We also showed that ALKBH5 controlled CYR61 mRNA stability by regulating m6A methylation and that the downstream effects of ALKBH5 and CYR61 might be related. Thus, ALKBH5 and CYR61 may be involved in the pathogenesis of RM, and therefore, therapeutic methods that target these two factors may be effective for the treatment of RM.
Supplementary Material
Supplementary materials and methods, figures, and primers.
Supplementary table 1.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81401218 to F.-J.T., 81501250 to X.-C.L.), the key projects of the Shanghai Municipal Health and Family Planning Commission (201640012 to F.-J.T.), the Family Planning Project of the Shanghai Health and Family Planning Commission (20164037 to X.-C.L.), Shanghai Outstanding Young Medical Talents Training Program (X.-C.L.) and the Interdisciplinary Program of Shanghai Jiao Tong University (grant number YG2017MS38 to F.-J.T.).
Author Contributions
F.-J.T., X.-C.L., and F.J. performed most of the experiments. The molecular experiments were carried out by B.-Y.W., X.-J.Y., and W. H. The manuscript was written by F.-J.T., and X.-C.L. All the work was planned and supervised by F.-J.T. All authors have read and approved the final manuscript.
Abbreviations
- RM
recurrent miscarriage
- HCs
healthy controls
- m6A
N6-methyladenosine
- ALKBH5
alkylation repair homolog 5
- CYR61
cysteine-rich 61
- MeRIP
methylated RNA immunoprecipitation
References
- 1.Wei CM, Gershowitz A. Moss B. Methylated nucleotides block 5' terminus of HeLa-cell messenger-RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 2.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3´ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D. et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–105. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 6.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 7.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N 6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m6A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–5. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 11.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Bio. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer KD, Jaffrey SR. Rethinking m6A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer J1, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC. et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–8. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A- demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z. et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115:E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsube K, Sakamoto K, Tamamura Y, Yamaguchi A. Role of CCN, a vertebrate specific gene family, in development. Dev Growth Differ. 2009;51:55–67. doi: 10.1111/j.1440-169X.2009.01077.x. [DOI] [PubMed] [Google Scholar]
- 21.Brigstock DR. The CCN family: a new stimulus package. J Endocrinol. 2003;178:169–75. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Du XY. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007;100:1337–45. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- 23.Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kipkeew F, Kirsch M, Klein D, Wuelling M, Winterhager E, Gellhaus A. CCN1 (CYR61) and CCN3 (NOV) signaling drives human trophoblast cells into senescence and stimulates migration properties. Cell Adhes Migr. 2016;10:163–178. doi: 10.1080/19336918.2016.1139265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gellhaus A, Schmidt M, Dunk C, Lye SJ, Winterhager E. The circulating proangiogenic factors CYR61 (CCN1) and NOV (CCN3) are significantly decreased in placentae and sera of preeclamptic patients. Reprod Sci. 2007;14:46–52. doi: 10.1177/1933719107309816. [DOI] [PubMed] [Google Scholar]
- 26.Tian FJ, Cheng YX, Li XC, Wang F, Qin CM, Ma XL. et al. The YY1/MMP2 axis promotes trophoblast invasion at the maternal-fetal interface. J Pathol. 2016;239:36–47. doi: 10.1002/path.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham CH, Hawley TS, Hawley RG, MacDougall JR, Kerbel RS, Khoo N. et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206:204–211. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Jin F, Li XC, Shen FJ, Ma XL, Wu F. et al. The YY1-HOTAIR-MMP2 Signaling Axis Controls Trophoblast Invasion at the Maternal-Fetal Interface. Mol Ther. 2017;25:2394–2403. doi: 10.1016/j.ymthe.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Cakmak H, Taylor HS. Implantation failure: molecular mechanisms and clinical treatment. Hum Reprod Update. 2011;17:242–253. doi: 10.1093/humupd/dmq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj Rai, Lesley Regan. Recurrent miscarriage. The Lancet. 2006;368:601–611. doi: 10.1016/S0140-6736(06)69204-0. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer S, Fritz M, Goldberg J, McClure R, Thomas M, Widra E. et al. Practice Committee of the American Society for Reproductive Medicine Evaluation and treatment for recurrent pregnancy loss: a committee opinion. Fertil Steril. 2012;98:1103–1111. doi: 10.1016/j.fertnstert.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 33.Malassiné A, Frendo JL, Evain-Brion D. A comparison of placental development and endocrine functions between the human and mouse model. Hum Reprod Update. 2003;9:531–539. doi: 10.1093/humupd/dmg043. [DOI] [PubMed] [Google Scholar]
- 34.Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T. et al. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39–46. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M. et al. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114:744–754. doi: 10.1172/JCI22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staun-Ram E, Shalev E. Human trophoblast function during the implantation process. Reprod Biol Endocrinol. 2005;3:56–62. doi: 10.1186/1477-7827-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strickland S, Richards WG. Invasion of the trophoblasts. Cell. 1992;71:355–357. doi: 10.1016/0092-8674(92)90503-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods, figures, and primers.
Supplementary table 1.