Summary
The inflammatory response to transformed cells forms the cornerstone of natural or therapeutically induced protective immunity to cancer. Regulatory T (Treg) cells are known for their critical role in suppressing inflammation, and therefore can antagonize effective anti‐cancer immune responses. As such, Treg cells can play detrimental roles in tumour progression and in the response to both conventional and immune‐based cancer therapies. Recent advances in our understanding of Treg cells reveal complex niche‐specific regulatory programmes and functions, which are likely to extrapolate to cancer. The regulation of Treg cells is reliant on upstream cues from haematopoietic and non‐immune cells, which dictates their genetic, epigenetic and downstream functional programmes. In this review we will discuss how Treg cells are themselves regulated in normal and transformed tissues, and the implications of this cross talk on tumour growth.
Keywords: cancer, chemokine/chemokine receptors, cytokines/cytokine receptors, regulatory T cells, tumour immunology
Introduction
The interplay between cancer and inflammation is noteworthy in both its importance and its complexity. It is now appreciated that the immune system has a critical importance in both carcinogenesis and tumour rejection. Whereas inflammation can contribute to carcinogenesis and thereby serve a deleterious role, the effect of immune activation during tumour progression is often beneficial and can promote tumour rejection. Although superficially paradoxical, this disparity can be explained by differences in disease stage, location and host, among many other factors.1 Nevertheless, it is generally accepted that certain inflammatory signatures correspond with either pro‐ or anti‐tumorigenic potential. Cytotoxic lymphocytes, such as natural killer or CD8+ T cells, or type 1 helper CD4+ T (Th1) cells, have potent direct and indirect anti‐cancer tumour activity and their presence in human tumours is frequently associated with favourable outcomes (reviewed in ref. 2). In contrast, regulatory T (Treg) cells suppress the function of conventional T (Tconv) cells including CD4+ and CD8+ T cells and although this function is required to prevent unwanted autoimmune and allergic inflammation, it is now known that Treg cells play a critical role in suppressing immune responses in cancer.3, 4 Treg cells also have a number of non‐classical functions, some of which may directly influence tumour cell biology, or act in a tissue‐specific manner. Hence, it is imperative to resolve the role of Treg cells at each distinct stage of cancer, from carcinogenesis to tumour progression and metastasis. In this review we will focus on the known and proposed mechanisms that regulate the recruitment, local homeostasis, and function of Treg cells during cancer progression.
Identification of Treg cells
Treg cells are a suppressive subset of CD4+ T cells required to prevent lethal inflammation.5, 6 In mice, Treg cells arising early in neonatal life are required for tolerance to self‐antigens, a conclusion made from the paradoxical observation that neonatal thymectomy results in a lethal inflammatory disorder, that is reversible by the transfer of CD4+ CD25+ Treg cells.7 Treg cells are dependent upon the lineage‐specifying transcription factor Foxp3 and mice and humans carrying inactivating mutations of Foxp3 succumb to lethal inflammatory disease within approximately 3 weeks and in early infancy, respectively.8, 9, 10, 11, 12, 13, 14 Whereas Foxp3 expression is predominantly restricted to Treg cells in mouse, FOXP3 is expressed in both Treg and Tconv cells upon acute activation in humans.15, 16, 17 Hence, although Foxp3 expression usefully distinguishes murine Treg cells from CD4+ Tconv cells, human FOXP3+ T cells may comprise both Treg and Tconv cells, especially during ongoing immune responses. Instead, demethylation of the intronic Foxp3 cis‐regulatory element CNS2 (conserved non‐coding sequence 2) is a cardinal feature of Treg cells, both in human and mouse.18, 19
Dual ontogeny of Treg cells
Thymus‐derived (tTreg) cells develop in response to high‐affinity interactions between the T‐cell receptor (TCR) of double‐positive and CD4 single‐positive thymocytes and self‐peptide–major histocompatibility complex (MHC) complexes in the thymus, and, among other functions, suppress autoimmune reactions directed against self‐antigens. As a product of this selection process in the thymus, tTreg cells have a largely distinct TCR repertoire to conventional CD4+ T cells.20, 21 Thymic selection results in differentiation of Treg cells with specificity for self‐antigens, but tolerance to innocuous foreign antigens unanticipated in the thymus is mediated by peripheral Treg (pTreg) cells, induced in peripheral tissues.22 This occurs when antigens are encountered by naive CD4+ T cells in the absence of optimal co‐stimulation or in the presence of transforming growth factor‐β (TGF‐β) abundant at mucosal sites but also within tumours. As a result, pTreg cells prevent inflammation directed against innocuous antigens often found at mucosal sites, including antigens expressed by commensal microflora or dietary components. Whereas tTreg and pTreg cell development have important similarities, such as their dependency upon the activity of the transcription factors FOXP3 and BACH2,23, 24, 25, 26 their distinct ontogeny is reflected in differences in gene regulatory mechanisms underlying their development. This is best illustrated by the finding that an intronic Foxp3 cis‐regulatory element, CNS1, which contains SMAD3 binding sites, is necessary for pTreg cell differentiation but dispensable for tTreg cell differentiation.27 Although deletion of Foxp3 leads to loss of both tTreg and pTreg cells and to lethal multi‐organ auto‐inflammation, selective ablation of the pTreg cell pool resulting from deletion of the CNS1 locus only leads to a mild late‐onset mucosal type 2 inflammation in the gut and the lungs.28 Additionally, the TCR specificity of tTreg cells and pTreg cells is largely distinct.22, 29
Functional specialization of Treg cells
Treg cells suppress inflammation through a number of mechanisms and it is now apparent that Treg cells undergo functional specialization to share some of the molecular characteristics of the cell types that they control.30, 31 For example, expression of the Th1 cell‐associated transcription factor, T‐bet, promotes Treg‐mediated restraint of type 1 inflammation.32, 33 Similarly, Treg cells expressing other CD4+ helper T lineage‐specific transcription factors such as ROR‐γt, GATA3 and IRF4 exert specialized functions suited to suppression of inflammation driven by their cognate Th cell counterparts.34, 35, 36, 37 It is notable that despite expressing lineage‐specifying transcription factors in some cases required for production of helper cytokines by Th cells, Treg cell subsets are configured to suppress these cell types. Mechanisms by which specialized Treg cell subsets suppress their cognate T helper counterparts are unclear, expression of lineage‐specifying transcription factors such as T‐bet or GATA3 may drive Treg cells to express a similar spectrum of chemokine receptors permitting more effective co‐localization with cognate Th cells. It may also promote a similar responsiveness to environmental cues as their cognate Th counterparts.
Non‐classical functions of Treg cells
Recent research has uncovered a pleiotropy of non‐classical mechanisms by which Treg cells contribute to homeostasis with diverse roles in physiological processes as control of tissue metabolism, stem cell maintenance and wound healing. It is significant that these processes also play fundamental roles in cancer pathophysiology. For instance, pioneering work on adipose tissue Treg cells, which express the transcription factor PAPR γ, demonstrate that these resident cells play critical roles in controlling tissue metabolism and insulin sensitivity.38, 39 Importantly, adipose tissue Treg cells exert their function in concert with a number of other tissue‐resident immune cells, such as macrophages, and group 2 innate lymphoid cells (reviewed in ref. 40). Other tissues, such as the muscle, harbour similar tissue‐resident Treg cell populations, which, upon detection of tissue‐damage, activate tissue‐regenerative programmes.41 Skin Treg cells have also been observed in close association with the stem‐cell‐containing dermal follicular regions.42 Surprisingly, Treg cells were in dynamic equilibrium with hair‐regrowth phases, and influenced follicular stem cell quiescence by expression of the Notch1 ligand Jagged 1. Similarly, Treg cells play an active role in protecting the intestinal epithelial, and bone marrow stem cell niche.43, 44 Finally, tissue‐resident Treg cells also express certain genes involved in epithelial cell repair, such as the growth factor amphiregulin (Areg),41, 45 and thereby contribute to wound healing. Hence, our appreciation of Treg cell biology is rapidly evolving, and it is likely that ‘non‐classical’ functions of Treg cells are co‐opted by cancer.
Regulation of Treg cells in cancer
Origin of Treg cells within tumours
In human tumours, the frequency of FOXP3+ cells relative to total CD3+ T cells or CD8+ T cells is negatively correlated with survival in multiple cancer types, including renal cell carcinoma,46 non‐small‐cell lung carcinoma,47 hepatocellular carcinoma,48 pancreatic cancer,49 gastric cancer,50 cervical cancer,51 ovarian cancer,52, 53 breast cancer54 and colorectal cancer.55 The frequency of Treg cells as a ratio of total CD4+ T cells can be extremely high – as high as 60–80% of total CD4+ T cells in murine orthotopic B16 tumours where Treg cells can be unambiguously defined using intranuclear Foxp3 staining.56 The size of Treg cell populations within tumours can be affected by a number of processes: the conversion of conventional CD4+ Foxp3− (Tconv cells) into pTreg cells under the influence of tumour‐derived factors including TGF‐β, recruitment of Treg cells from the periphery into tumours, the rate of proliferation and survival of recruited, peripherally induced or tissue‐resident Treg cells (Fig. 1). We will hereafter review the evidence in favour or against each of these hypotheses.
Figure 1.
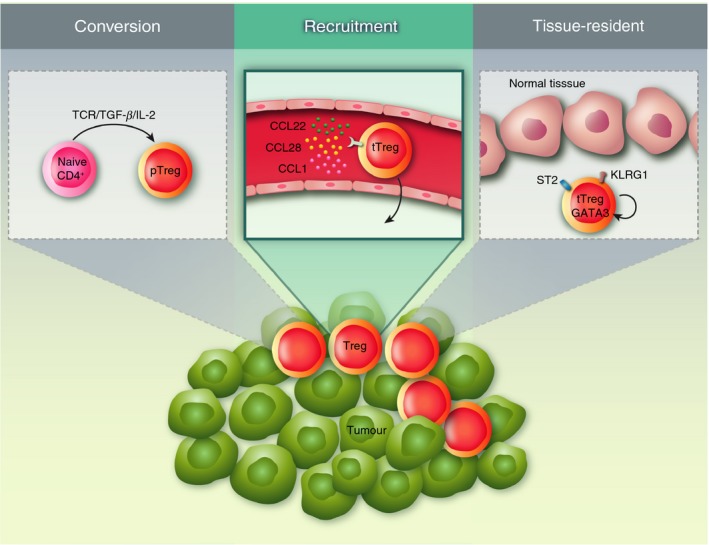
Origin of regulatory T (Treg) cells in tumours. Depicted are the three scenarios – not mutually exclusive – that could account for the presence of Treg cells within tumours. Left, conversion of naive CD4+ T cells into peripheral Treg (pTreg) cells; centre, recruitment of thymus‐derived Treg (tTreg) cells from the circulation; right, expansion of tissue‐resident Treg cells.
Conversion of Tconv into pTreg cells
An initial study by Valzasina et al.57 revealed that CD4+ CD25− transferred into tumour‐bearing hosts could convert into Foxp3+ Treg cells within tumours. In addition, conditioned media from murine tumour cells could convert CD4+ CD25− in CD4+ CD25+ Foxp3+ T cells.58 Because a blocking antibody to TGF‐β reversed this effect, the authors concluded that the secretion of TGF‐β by tumour cells promotes an environment favourable to the peripheral conversion of Tconv cells into tolerogenic pTreg cells. Along the same lines, the group of Zitvogel showed that rodent tumour cells induced the production of TGF‐β by immature myeloid dendritic cells that in turn sustain the proliferation of Treg cells.59 Similarly, myeloid‐derived suppressor cell‐derived interleukin‐10 (IL‐10) and TGF‐β supports Treg cell development both in vitro and in experiments where myeloid‐derived suppressor cells and T cells are adoptively transferred into irradiated tumour‐bearing mice.60
Recently, expression of the surface protein Neuropilin‐1 and the transcription factor Helios have been proposed to distinguish tTreg cells from pTreg cells,61, 62 although whether expression of these markers faithfully reports tTreg cells has been called into question.63, 64 This may explain contradictory reports regarding the relative frequency of pTreg and tTreg cells assessed using these markers in transplantable tumour models in mice.65, 66, 67 TCR repertoire analyses of mouse and human tumour‐infiltrating lymphocytes have been proposed to provide evidence of a low frequency of Tconv to pTreg conversion within tumours, as the TCR repertoire of tumour‐infiltrating Treg cells was found to be largely distinct from that of tumour‐infiltrating Tconv cells.68, 69, 70, 71, 72 However, the efficiency of pTreg induction is affected by antigen dose in addition to cytokine signalling73 and it is not inconceivable that pTreg cells arising from Tconv cells could have a substantially skewed repertoire to Tconv cells, having expanded from a small fraction of naive CD4+ T cells among the total Tconv pool.
Hence, it has been difficult to precisely discern the role of pTreg cells in Treg‐mediated tumour immunosuppression, although comparison of tumour growth and immune infiltrates in wild‐type and Foxp3 CNS1 knockout mice should allow the functional contribution of pTreg cells to be defined. However, it has been proposed that decreased susceptibility of mice lacking T‐cell intrinsic expression of all three isoforms of the oxygen‐sensing prolyl hydroxylase family is attributable to defective pulmonary pTreg cell differentiation.74
Evidence that tTreg cells can contribute to tumour immunosuppression is much clearer. Malchow et al.72 used an autochthonous mouse model of prostate cancer driven by the restricted expression of the SV40 oncogene in the prostate to study the TCR reactivity of tumour‐associated Treg cells. On a fixed TCR‐β background, TCR‐α sequencing revealed that one TCR‐α sequence was recurrently expressed by Treg cells isolated from tumours of distinct animals, suggesting that Treg cells of a single specificity are recurrently enriched within these prostate tumours. They further showed that this sequence was only overrepresented in the prostate tumour and draining lymph node, but not in the spleen or the thymus, in line with the idea of a tumour‐specific enrichment. Repertoire analysis also confirmed the absence of overlap between Tconv and Treg cells isolated from these tumours. Interestingly, transgenic expression of this TCR on a Rag1 −/− background led to the spontaneous accumulation of activated transgenic T cells in the prostate and draining lymph nodes of male tumour‐free mice, whereas in female mice transgenic T cells showed no signs of activation. Further analysis showed that thymic development of Treg cells expressing the transgenic TCR was dependent on the autoimmune regulator Aire. Altogether, these results clearly demonstrate that pre‐existing tTreg cells reactive to self‐antigens are expanded upon tumorigenesis.
Recruitment and retention of Treg cells to tumours
Identification of the chemokines and their cognate receptors involved in specific Treg cell recruitment and/or retention into tumour lesions is an active area of research, given their potential as druggable targets. In human ovarian cancer, Curiel et al.53 showed that, similar to blood CD4+ CD25+ Treg cells, CD4+ CD25+ T cells isolated from malignant ascites express the chemokine receptor CCR4. Furthermore, the authors found that CCL22, the ligand of CCR4, is highly expressed in the ascites and tumour tissue of patients with ovarian cancer in comparison with control groups. They demonstrated with in vitro migration assays that CD4+ CD25+ Treg cells migrate to malignant ascites, and that this effect is abolished upon addition of an anti‐CCL22 antibody. Other authors later reported the same observation in prostate, breast and gastric tumours.75, 76, 77 In the context of cancer, CCL22 is probably derived from activated myeloid cells, although lymphocytes and tumour cells are potential alternative sources.78, 79, 80 Upstream, CCL22 is strongly induced by IL‐4 and IL‐13, which can be produced by adaptive and innate lymphoid cells, as well as some myeloid cell types.81 CCL17, another ligand for CCR4, is also implicated as a chemoattractant in cancer,76 although it is suggested that, due to different affinities for conformational isoforms of CCR4 leading to distinct signalling characteristics, CCL17 is more important in recruiting CCR4+ effector T cells.82, 83 Additionally, the Sakaguchi group further ascribed high CCR4 expression in human T cells to a subset of effector Treg cells, defined as FOXP3hi CD45RA−, in both melanoma tissues and peripheral blood.84 Ex vivo depletion of CCR4+ cells using an anti‐CCR4 monoclonal antibody selectively ablated effector Treg cells in healthy donors and individuals with melanoma, and was associated with an increase in CD4+ and CD8+ T‐cell responses to the cancer‐germline antigen NY‐ESO‐1 upon in vitro re‐stimulation. Furthermore, in vivo administration of Mogamulizumab, a CCR4‐depleting monoclonal antibody, in two adults with T‐cell leukaemia‐lymphoma diminished the percentage of blood effector Treg cells. In one patient whose leukaemic cells expressed NY‐ESO‐1, the reduction in effector Treg cells was further associated with an enhanced NY‐ESO‐1‐specific CD8+ T‐cell response.
Apart from CCL22, CCL28 produced by ovarian tumour cells under hypoxia has also been implicated in the preferential recruitment of Treg cells, both in in vitro migration assays using mouse and human Treg cells and in an in vivo model of ascitic ovarian tumours.85 CCR10, rather than CCR3, was shown to be the receptor involved in this effect.
In human breast cancer, RNA‐seqencing analyses of tumour‐infiltrating CD4+ CD25+ Treg cells revealed high expression of the genes encoding the chemokine receptors CCR5, CCR8, CCR10, CX3CR1, CXCR3 and CXCR6 compared with blood CD4+ CD25+ Treg cells.70 Among these, some were also shared with tumour‐infiltrating Tconv cells (CCR5, CXCR3 and CXCR6), suggesting that Treg cells may employ both unique and shared pathways to migrate to breast tumour lesions. The authors further focused on CCR8, and showed that its surface expression was restricted to Treg cells and to a subset of natural killer T cells among CD45+ and non‐CD45 cells within tumours. CCR8 expression was shared by Treg cells infiltrating breast, lung and colorectal cancers, as well as in melanoma and in angiosarcoma.70, 86 Functionally, sorted tumour‐infiltrating CD4+ CD25+ Treg cells were able to migrate more robustly than Tconv cells towards the CCR8 ligand CCL1.70 Consistent with its tumour‐specific pattern of expression, robust CCR8 expression by Treg cells was shown to require TCR engagement as well as soluble tumour‐derived factors using tumour explant co‐cultures. Finally, analysis of the breast samples from the The Cancer Genome Atlas data sets revealed a strong association of CCR8 mRNA amounts normalized to FOXP3, but not FOXP3 mRNA amounts alone, with poor prognosis, suggesting a detrimental role for CCR8‐expressing Treg cells in breast cancer progression.
High endothelial venules (HEV) which can be associated with tertiary lymphoid structures are present within tumours. The presence of HEV is promoted by activation of Tconv cells and dependent upon tumour necrosis factor receptor signalling.87 As HEV themselves drive further T‐cell recruitment, it has been proposed that this supportive relationship between Tconv cells and HEV forms the potential for a self‐amplifying loop that can drive tumour destruction. HEV neogenesis is indirectly inhibited by Treg cells through their suppression of Tconv cell activation.87 However, tertiary lymphoid structures contain Treg cells88 and whether and how HEV play an active role in recruitment of Treg cells is unclear. L‐selectin expression is required for appropriate trafficking and tissue distribution of Treg cells under physiological conditions.89 Whereas a majority of Treg cells in tumours are activated and express low levels of L‐selectin, it is possible that L‐selectin+ Treg cells are recruited by tumour HEV, which subsequently down‐regulate L‐selectin expression in response to the activating environment of the tumour. This may represent a physiological regulatory feedback mechanism in inflamed tissues that is co‐opted by tumours to counteract an otherwise beneficial self‐reinforcing feedback loop of Tconv‐driven HEV neogenesis in cancer, which could be subject to therapeutic intervention.
It is worth noting that the pathways described above may not only account for Treg cell recruitment but also for their retention within the tumour, thanks to the continuous secretion of chemo‐attractants by either tumour cells or their associated stroma. In addition, recent evidence suggests that the early activation marker CD69 could play a role in Treg cell retention within tumours, as a large proportion of tumour‐infiltrating Treg cells express high levels of this protein.90 Indeed, CD69 has been shown to be crucial for T‐cell trafficking by interfering with the expression of S1P1, so preventing lymphocyte egress from peripheral tissues.91, 92 Interestingly, CD69 is also linked to Treg cell function as CD69‐deficient Treg cells display an altered suppressive function in vitro and in vivo.90, 93 How CD69 affects Treg cell retention and function at the molecular level remains to be addressed.
There is now evidence to support the rationale of targeting chemokine receptors to alter Treg cell accumulation and/or retention in several types of tumours. However, it is worth noting that these pathways may also be shared with Tconv,94 and may not be unique to cancer, providing a potential for on‐target side effects. Moreover, the mechanisms driving chemokine receptor expression among Treg cells are unclear but there may be considerable overlap with the mechanisms that drive such receptor expression among cognate T helper counterparts, including the involvement of lineage‐specifying transcription factors of the T helper lineages that are also expressed in Treg cells.
Local expansion of tissue‐resident Treg cells
Recent developments in the field have led to the emerging idea that tissue‐resident Treg cells may contribute to the accumulation of Treg cells seen within tumour lesions. Although some markers of tissue‐resident Treg cells appear to be present in Treg cells in multiple tissues, large‐scale transcriptional analyses also suggest that tissue‐resident Treg cells within distinct tissues have unique phenotypes.95, 96 Pan‐tissue tissue‐resident Treg markers include the IL‐33 receptor ST2 (encoded by gene Il1rl1), the activation marker KLRG1, the transcription factor GATA‐3 and the growth factor Amphiregulin.
In searching for a potential role of tissue‐resident Treg cells in tumour progression, Green et al.97 analysed transplantable lung tumours in mice and found that tumour‐infiltrating Treg cells expressed higher levels of Amphiregulin compared with normal lung Treg cells. In one of the two lung tumour models tested, conditional deletion of Areg in Treg cells resulted in a decreased tumour volume. Although the authors excluded both immune and tumour cells, the precise identity of the cells targeted by the Treg‐derived Amphiregulin remains to be addressed. In line with these data, an increased proportion of Treg cells were shown to be ST2+ in both primary orthotopic mouse mammary carcinoma and lung metastases, and ST2+ lung tumour Treg cells, but not ST2−, were shown to produce Amphiregulin.98
In human breast tumour and healthy adjacent tissue, RNA‐sequencing analyses from the Plitas et al.70 study described above revealed that the gene expression profile of tumour‐infiltrating Treg cells was similar to those of the tissue‐resident Treg cells of the normal breast parenchyma, but distinct from the profile of the CD45RO+ activated Treg cells isolated from peripheral blood, used here as a reference of activated Treg cells. Surprisingly though, extraction of the TCR repertoire of these cells from RNA‐sequencing data did not support the hypothesis of local tissue‐resident Treg cell expansion: only low clonal overlap was found between the tumour‐infiltrating Treg cells and those from the normal adjacent tissue.
Altogether, there is not much evidence supporting the hypothesis of the local amplification of tissue‐resident Treg cells within tumour lesions. However, this does not imply that they do not make a significant contribution to tumour progression. Alternatively, their pre‐existence in the tissue‐of‐origin of cancer may position them to contribute to early events in carcinogenesis and metastasis through classical or non‐classical functions. Further studies will be needed to address these hypotheses.
Signals regulating Treg cells within tumours
Co‐stimulatory and co‐inhibitory receptors
Tumour‐associated Treg cells are known to express numerous co‐stimulatory (i.e. ICOS, OX40, GITR) and co‐inhibitory (i.e. Lag‐3, Klrg1, Tim‐3, TIGIT, PD‐1) receptors that modulate their function (Fig 2). Resolving the functional role of such receptors is important, but is complicated by their frequent expression on conventional T and other immune cell‐types. Hence, lineage‐specific deletion experiments are often required to fully understand their role in cancer. Many potential therapeutic strategies targeting these co‐receptors are primarily aimed at promoting effector T cells in cancer but could induce changes that either help or hinder the therapeutic response, highlighting the importance of considering their role on tumour‐associated Treg cells.
Figure 2.
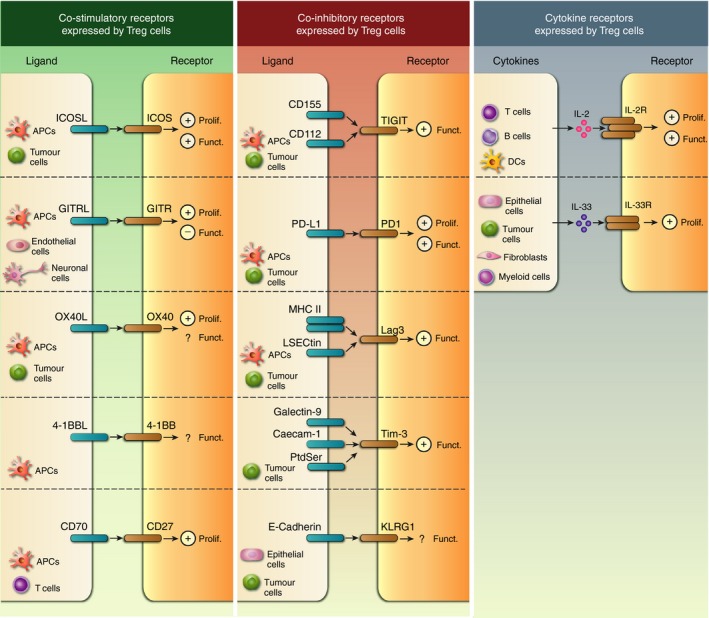
Co‐stimulatory, co‐inhibitory and cytokine receptors expressed by regulatory T (Treg) cells in tumours. Co‐stimulatory (left) and co‐inhibitory (middle) molecules, as well as cytokine receptors (right), deliver signals to Treg cells following engagement by their ligands expressed by immune and/or non‐immune cell types within the tumour. Positive and negative signs in the Treg cell indicate either a positive effect on proliferation (Prolif.) or function (Funct.) or an inhibitory effect (i.e. reduced function), respectively, after receptor engagement. Question marks (?) indicate that the outcome of the receptor engagement in the Treg cell warrants further investigation.
Co‐stimulatory receptors
Inducible T‐cell co‐stimulator (ICOS) is important in Treg cell homeostasis and function,99, 100, 101 and is highly expressed by activated Treg cells in prostate cancer77 and melanoma.102 In the context of cancer, ICOS ligand (ICOSL) can be expressed by tumour cells103 as well as myeloid infiltrates, primarily antigen‐presenting cells, including transformed follicular lymphoma B cells104 and dendritic cells.105 These findings, in conjunction with studies on the function of ICOS/ICOSL signalling in non‐tumour settings, suggest that this pathway is instructive in supporting local Treg cell expansion and function.
The tumour necrosis factor receptor superfamily (TNFRSF) members, including GITR, OX40, CD27 and 4‐1BB are important co‐stimulatory molecules on T cells. In addition to influencing Treg cell development in the thymus, TNFRSF members are critical for the maintenance of Treg cells in peripheral tissues, in part via nuclear factor‐κB/RelA‐mediated signalling.106
GITR is highly expressed by Treg cells, and plays an important role in Treg cell expansion.107 However, GITR signalling has been proposed to impair Treg cell suppressive capacity, although interrogation of this receptor is complex because of concurrent effects on conventional CD4+ and CD8+ T cells.108 As for ICOSL, GITR ligand (GITRL) is mainly expressed by antigen‐presenting cells, but also endothelial and neuronal cells.109, 110 In the context of cancer, in vitro studies show that tumour‐derived TGF‐β can induce GITRL on dendritic cells, which subsequently support expansion of Treg cells.111 Interestingly, an association was observed between single nucleotide polymorphisms in GITR (and OX40) and poor survival in ovarian cancer.112
OX40 is expressed constitutively by a subset of Treg cells, but also on activated non‐Treg cells.113 In cancer, Treg cells can comprise a significant proportion of tumour‐resident OX40+ cell types.114, 115, 116 Although OX40‐agonistic reagents are used to stimulate anti‐tumour T‐cell responses, the effect on Treg cells in cancer is not well understood. In wild‐type mice, OX40 is important for Treg cell development in the thymus, as well as expansion in the periphery, although its effect on Treg cell function is less clear. Administration of agonistic reagents demonstrates that OX40‐signalling reduces Treg cell suppressive function in vitro and in vivo;117 however, other studies suggest that Treg cells are not impaired.118 One potential explanation is that the context of OX40 ligand (OX40L) expression can have different effects on Treg cells.113 In the context of cancer, OX40L is expressed by glioblastoma, especially under hypoxic conditions. In vitro experiments indicated that OX40L expression promoted OX40‐driven activation of Treg cells.119 In another study of patients with hepatocellular carcinoma, liver‐resident monocytes and macrophages expressed OX40L (in response to concurrent hepatitis C virus infection), which in turn promoted liver Treg cell expansion.120 Moreover, two studies identified an association between single nucleotide polymorphisms in OX40L and increased occurrence of breast cancer.121, 122
4‐1BB is expressed by both lymphoid and myeloid cell types and among T cells, it is expressed by Treg cells and activated CD4+ and CD8+ T cells.123 4‐1BB activation provides a potent stimulus for anti‐tumour natural killer, CD4+ and CD8+ T cells and 4‐1BB agonistic antibodies can provoke rejection of tumours in multiple mouse models but non‐specific agonism results in generalized T‐cell activation, cytokine release and systemic inflammation.123 Despite initial signs of efficacy, clinical development of agonistic clinical 4‐1BB agonists urelumab and utomilumab has been hampered by inflammatory liver toxicity at moderate systemic doses.124 Similar to OX40, 4‐1BB is expressed both on Treg cells and activated CD4+ and CD8+ Tconv cells. Consequently, it has been difficult to distinguish the effect of therapy on either subset alone and there is contradictory evidence for the effect of 4‐1BB ligation on Treg cells, with some studies demonstrating an inhibitory effect on their immunoregulatory function125, 126 and others suggesting a stimulatory effect on proliferation.127, 128 Hence, there is a need to dissect the effect of 4‐1BB agonists on Treg and Tconv cells both in the context of anti‐tumour immunity and the on‐target toxicity that they provoke.
CD27 is expressed mainly by naive and subsets of memory CD4+ and CD8+ T cells, as well as Treg cells.113 CD70 can be expressed by antigen‐presenting cells, but also by tumour‐infiltrating Treg and effector T cells.129 Engagement of CD27 with CD70 within tumours is important for expansion of Treg cells within tumours.129 Intriguingly, it was proposed that this activity is indirect, and acts through the ability of CD27‐CD70 signalling to drive IL‐2 production by Tconv cells, although evidence supporting this hypothesis was derived from in vitro experiments.
In summary, it is clear that co‐stimulatory receptor signalling is critical not only for anti‐tumour immunity, but also influences local Treg cell biology. It is likely that contact‐mediated regulation of Treg cells varies greatly depending on tissue‐ and tumour‐type, and the presence of a shared niche containing cell‐types that express co‐stimulatory ligands.
Co‐inhibitory receptors
The co‐inhibitory receptor T‐cell immunoglobulin and ITIM domain (TIGIT) marks a population of Treg cells with enhanced suppressive capacity in tumours.130, 131 Interestingly, TIGIT+ Treg cells preferentially suppress Th1 and Th17 (but not Th2) cells, the former being important in anti‐tumour immunity. Moreover, TIGIT competes for its ligands CD112 and CD155 with the co‐stimulatory receptor CD226 which, unlike conventional T cells, is associated with functionally suppressed Treg cells.132 This study suggests that the CD226/TIGIT ratio correlates with Treg cell stability, and clinical outcome in individuals with melanoma. Moreover, both CD112 and CD155 over‐expression is reported in cancer,133 although further work is required to delineate the interaction dynamics with the multiple TIGIT‐expressing cell‐types.
Although the role of the inhibitory receptor programmed‐death 1 (PD‐1) on conventional T cells is well established, its function in Treg cells is less clear. Work by the Sharpe laboratory demonstrated that PD‐1 and its ligand PD‐L1 are important for pTreg cell development and function.134 Subsequently, PD‐1 was demonstrated to contribute to tTreg cell stability in Foxp3low conditions.135 Moreover, PD‐L1 expression on antigen‐presenting cells can expand Treg cells in patients after allogeneic bone marrow transplants.136 However, in the context of cancer, PD‐1 expression by Treg cells has been associated with their dysfunction.137
The inhibitory receptor lymphocyte activation gene 3 (Lag‐3) is expressed on Treg cells, in addition to other lymphocytes, and binds to two known ligands (MHC‐II and LSECtin). Lag‐3 also promotes suppressive function of Treg cells in homeostasis and cancer.138, 139 In addition to Treg cell modulation, Lag‐3/MHC‐II interactions are proposed to suppress the maturation of MHC‐II+ dendritic cells.140 Moreover, LSECtin expression has been reported on tumour cells, suggesting potential Lag‐3‐dependent mechanisms of Treg cell regulation.141 T‐cell immunoglobin and mucin domain 3 (Tim‐3) is a co‐inhibitory receptor expressed by different myeloid and lymphoid cells, including Treg cells, and binds to several identified ligands (i.e. galectin‐9, HMGB1, caecam‐1, phosphatidyl serine, reviewed in ref. 142). Tim‐3+ Treg cells are detected in many murine and human tumour samples, and have increased suppressive capacity. Conversely, Tim‐3 ligands are widely expressed on both immune and non‐immune cell types in cancer and homeostasis. Another co‐inhibitory receptor, killer cell lectin‐like receptor G1 (Klrg1) is expressed by many immune cells including Treg cells, and binds to its ligands (E‐ and N‐cadherin). Although Klrg1 is used to identify Treg cell subsets, little is known about its functional role. In the gut, Klrg1‐signalling on Treg cells impairs their suppressive function.143 Interestingly, loss of E‐cadherin has been shown in cancer progression, suggesting a potential mechanism by which tumour cells promote Treg cell‐mediated immunosuppression.144
In summary, cell‐to‐cell interactions via co‐stimulatory or co‐inhibitory receptors are important for local regulation of Treg cell function in cancer. Importantly, the function of these receptors also frequently differs in Treg cells compared with Tconv cells. Moreover, their expression often overlaps with other immune cell‐types, highlighting the need for careful dissection of functional effects on Treg cells.
Cytokines
Interleukin‐2
Treg cells are mainly dependent upon IL‐2 signalling for both their thymic and peripheral differentiation and for their survival, although IL‐15 and IL‐7 can partially substitute for IL‐2 in maintaining Treg cell survival in the genetic absence of IL‐2145or following Treg‐specific disruption of CD25 expression,146 respectively. This leads to the question of whether IL‐2 production within tumours is required for maintenance of intratumoural Treg cell populations and what the dominant cellular source of IL‐2 is within tumours (reviewed in ref. 147). There is contradictory evidence regarding the role of IL‐2 in intratumoural Treg homeostasis. Consistent with a role for IL‐2 in tumour immunosuppression, IL‐2 neutralization has been shown to retard the growth of implanted renal cell carcinoma tumours.148 Administration of IL‐2 to mice bearing syngeneic B16 melanoma tumours results in an increase in the frequency of Foxp3+ cells within the intratumoural CD4+ T‐cell pool.149 However, the frequency of Treg cells in tumours of mice bearing methylcholanthrene‐induced fibrosarcomas was not affected by administration of IL‐2/anti‐IL‐2 complexes though this does not exclude the possibility that endogenous intratumoural IL‐2 levels are already at functionally saturating levels in such tumours or that other limits to the size of the Treg pool prevent further Treg cell expansion.90 In humans, whereas high‐dose IL‐2 therapy is an established treatment for metastatic melanoma and can drive striking clinical responses in a small subset of patients, IL‐2 therapy can also drive expansion of ICOS+ Treg cells, the extent of which correlates with worse clinical outcomes following therapy.150
In general, CD4+ and CD8+ Tconv cells are the major cellular source of IL‐2 in vivo although the cytokine is also expressed by B cells and dendritic cells.151 It is important to note that not all Tconv cells produce IL‐2. Cells in the early stages of differentiation, such as naive and memory CD8+ T cells, produce IL‐2 upon stimulation whereas cells that have undergone full effector differentiation do not.147 Indeed, loss of the ability of Tconv populations to produce IL‐2 upon full effector differentiation is associated with a catastrophic decline in Treg cell numbers during infection of wild‐type mice with Toxoplasma gondii and of perforin‐deficient mice with lymphocytic choriomeningitis virus.152, 153 Given this exclusively supportive role of Tconv cells in the early stages of differentiation to Treg cell survival, it is important to test whether such cells are present and functionally relevant within tumours. Indeed, a proportion of CD8+ T cells within tumours have an early memory phenotype154, 155 and it will be interesting to determine whether these cells produce IL‐2 and whether similar early memory cells contribute to intratumoural Treg cell maintenance using mouse models. Given the ability of IL‐15 and IL‐7 to act as surrogates for the absence of IL‐2 signalling, it would also be interesting to determine whether these cytokines play a role in Treg cell maintenance in tumours. Finally, Treg cells differ in their requirement for IL‐2 signalling, with CD25lo Treg cells less dependent and intrinsically short‐lived compared with CD25hi cells whose longevity requires CD25 signalling. Hence, the role of IL‐2 will need to be considered in the context of heterogeneity of Treg cell populations and differential requirements for IL‐2.
Interleukin‐33
Accumulating evidence indicates that IL‐33 is an important homeostatic factor for Treg cells in multiple tissue sites, in line with their expression of the IL‐33 receptor ST2. Direct action of IL‐33 on Treg cells has been shown to enhance their expansion in the colonic lamina propria, or in the visceral adipose compartment for example.156, 157 Interleukin‐33 is mostly produced by non‐haematopoietic cells, including fibroblasts and epithelial cells, but also by some activated myeloid cells. Although the precise mechanism remains unclear, it is believed that IL‐33 is released from cells upon certain types of cell death. IL‐33 further functions as an alarmin through the binding to its receptor complex, composed of ST2 and IL1RAcP, expressed on a variety of immune cells, so including Treg cells. In cancer, the role of IL‐33 remains controversial, with both pro‐tumorigenic and anti‐tumorigenic effects reported across different cancer types and cellular sources. Whether the IL‐33/ST2 axis on Treg cells plays a role in tumour progression has yet to be established. Interestingly, administration of IL‐33 to tumour‐bearing mice was shown to expand tumour‐infiltrating Treg cells,98, 158 in line with the aforementioned IL‐33‐driven expansion of Treg cells in healthy tissues. Recent generation of Foxp3‐Cre × Il1rl1 fl/fl mice159 should shed light on the relevance of this pathway in vivo.
Metabolic fitness
Co‐stimulatory ligands, cytokines and chemokines are well‐established contributors to tTreg cell preferential expansion/maintenance in the tumour microenvironment, but recent evidence points also towards a peculiar cell‐intrinsic metabolism as a means by which tTreg cells survive, expand and exert their function within tumours.160 Importantly, tumour‐infiltrating tTreg cells display high expression of the glucose transporter Glut1 compared with splenic Treg cells, and are capable of increased glucose uptake in mouse tumour models.161, 162 Such an improved glucose usage may in turn fuel fatty acid biosynthesis, in line with observations of a tTreg high neutral lipid content.162 Given the important role of both the glycolytic and lipid pathways in tTreg proliferation, suppressive function and trafficking,160, 163, 164 it is tempting to speculate that the adaptation of Treg cell metabolism to ensure exquisite function at the expense of other T cells within the tumour bed is a feature that may be promoted under the influence of tumour and/or stromally derived factors. Identification of such specific cues and pathways may reveal promising therapeutic targets in the near future.
Future direction and conclusion
Our understanding of Treg cells is rapidly evolving, addressing both long‐standing questions in Treg cell ontogeny and antigen‐specificity while simultaneously exploring new frontiers in the realm of tissue‐residency and interactions with non‐haematopoietic cell types. These new findings may have significant impact in our understanding of tumour immunology, given the clear evidence of Treg cell enrichment, association with poor prognosis, and immune‐regulatory functions in cancer. Importantly, it is clear that Treg cell identity and function are influenced by their niche. In conclusion, our understanding of how Treg cells are themselves regulated will be essential to design novel immunotherapies and leverage existing cancer treatments with greater effect.
Disclosures
None.
Acknowledgements
This study is supported by the Wellcome Trust (TH – 204622/Z/16/Z) and Cancer Research UK (TH/JS – A24995).
References
- 1. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140:883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fridman WH, Pages F, Sautes‐Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12:298–306. [DOI] [PubMed] [Google Scholar]
- 3. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol 2008; 8:523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shevach EM. Mechanisms of foxp3+ T regulatory cell‐mediated suppression. Immunity 2009; 30:636–45. [DOI] [PubMed] [Google Scholar]
- 5. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 2012; 30:531–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell 2008; 133:775–87. [DOI] [PubMed] [Google Scholar]
- 7. Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self‐tolerance maintained by activated T cells expressing IL‐2 receptor α‐chains (CD25). Breakdown of a single mechanism of self‐tolerance causes various autoimmune diseases. J Immunol 1995; 155:1151–64. [PubMed] [Google Scholar]
- 8. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003; 299:1057–61. [DOI] [PubMed] [Google Scholar]
- 9. Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 2003; 4:337–42. [DOI] [PubMed] [Google Scholar]
- 10. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003; 4:330–6. [DOI] [PubMed] [Google Scholar]
- 11. Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L et al The immune dysregulation, polyendocrinopathy, enteropathy, X‐linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 2001; 27:20–1. [DOI] [PubMed] [Google Scholar]
- 12. Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA et al Disruption of a new forkhead/winged‐helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 2001; 27:68–73. [DOI] [PubMed] [Google Scholar]
- 13. Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C et al JM2, encoding a fork head‐related protein, is mutated in X‐linked autoimmunity‐allergic disregulation syndrome. J Clin Invest 2000; 106:R75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N et al X‐linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 2001; 27:18–20. [DOI] [PubMed] [Google Scholar]
- 15. Walker MR, Kasprowicz DJ, Gersuk VH, Benard A, Van Landeghen M, Buckner JH et al Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25− T cells. J Clin Invest 2003; 112:1437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roncador G, Brown PJ, Maestre L, Hue S, Martinez‐Torrecuadrada JL, Ling KL et al Analysis of FOXP3 protein expression in human CD4+CD25+ regulatory T cells at the single‐cell level. Eur J Immunol 2005; 35:1681–91. [DOI] [PubMed] [Google Scholar]
- 17. Morgan ME, van Bilsen JH, Bakker AM, Heemskerk B, Schilham MW, Hartgers FC et al Expression of FOXP3 mRNA is not confined to CD4+CD25+ T regulatory cells in humans. Hum Immunol 2005; 66:13–20. [DOI] [PubMed] [Google Scholar]
- 18. Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J et al DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3+ conventional T cells. Eur J Immunol 2007; 37:2378–89. [DOI] [PubMed] [Google Scholar]
- 19. Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J et al Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 2007; 5:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self‐reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol 2006; 7:401–10. [DOI] [PubMed] [Google Scholar]
- 21. Wong J, Obst R, Correia‐Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self‐peptides in regulatory and nonregulatory CD4+ T cells. J Immunol 2007; 178:7032–41. [DOI] [PubMed] [Google Scholar]
- 22. Yadav M, Stephan S, Bluestone JA. Peripherally induced Tregs – role in immune homeostasis and autoimmunity. Front Immunol 2013; 4:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol 2006; 24:209–26. [DOI] [PubMed] [Google Scholar]
- 24. Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M et al BACH2 represses effector programs to stabilize Treg‐mediated immune homeostasis. Nature 2013; 498:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Igarashi K, Kurosaki T, Roychoudhuri R. BACH transcription factors in innate and adaptive immunity. Nat Rev Immunol 2017; 17:437–50. [DOI] [PubMed] [Google Scholar]
- 26. Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol 2014; 192:985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non‐coding DNA elements in the Foxp3 gene in regulatory T‐cell fate. Nature 2010; 463:808–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y et al Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 2012; 482:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen‐specific peripheral shaping of the natural regulatory T cell population. J Exp Med 2008; 205:3105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaudhry A, Rudensky AY. Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest 2013; 123:939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shevach EM. Foxp3+ T regulatory cells: still many unanswered questions‐A perspective after 20 years of study. Front Immunol 2018; 9:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koch MA, Tucker‐Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T‐bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 2009; 10:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levine AG, Mendoza A, Hemmers S, Moltedo B, Niec RE, Schizas M et al Stability and function of regulatory T cells expressing the transcription factor T‐bet. Nature 2017; 546:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sefik E, Geva‐Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM et al Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ + regulatory T cells. Science 2015; 349:993–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y et al Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 2015; 349:989–93. [DOI] [PubMed] [Google Scholar]
- 36. Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT et al Regulatory T‐cell suppressor program co‐opts transcription factor IRF4 to control TH2 responses. Nature 2009; 458:351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA‐3 for the function of regulatory T cells. Immunity 2011; 35:337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A et al Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 2009; 15:930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE et al PPAR‐γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012; 486:549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cautivo KM, Molofsky AB. Regulation of metabolic health and adipose tissue function by group 2 innate lymphoid cells. Eur J Immunol 2016; 46:1315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y et al A special population of regulatory T cells potentiates muscle repair. Cell 2013; 155:1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K et al Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell 2017; 169:1119–29 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Agudo J, Park ES, Rose SA, Alibo E, Sweeney R, Dhainaut M et al Quiescent tissue stem cells evade immune surveillance. Immunity 2018; 48:271–85 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirata Y, Furuhashi K, Ishii H, Li HW, Pinho S, Ding L et al CD150high bone marrow Tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell 2018; 22:445–53 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S et al A distinct function of regulatory T cells in tissue protection. Cell 2015; 162:1078–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Griffiths RW, Elkord E, Gilham DE, Ramani V, Clarke N, Stern PL et al Frequency of regulatory T cells in renal cell carcinoma patients and investigation of correlation with survival. Cancer Immunol Immunother 2007; 56:1743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petersen RP, Campa MJ, Sperlazza J, Conlon D, Joshi MB, Harpole DH Jr et al Tumor infiltrating Foxp3+ regulatory T‐cells are associated with recurrence in pathologic stage I NSCLC patients. Cancer 2006; 107:2866–72. [DOI] [PubMed] [Google Scholar]
- 48. Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS et al Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007; 25:2586–93. [DOI] [PubMed] [Google Scholar]
- 49. Hiraoka N, Onozato K, Kosuge T, Hirohashi S. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res 2006; 12:5423–34. [DOI] [PubMed] [Google Scholar]
- 50. Perrone G, Ruffini PA, Catalano V, Spino C, Santini D, Muretto P et al Intratumoural FOXP3‐positive regulatory T cells are associated with adverse prognosis in radically resected gastric cancer. Eur J Cancer 2008; 44:1875–82. [DOI] [PubMed] [Google Scholar]
- 51. Jordanova ES, Gorter A, Ayachi O, Prins F, Durrant LG, Kenter GG et al Human leukocyte antigen class I, MHC class I chain‐related molecule A, and CD8+/regulatory T‐cell ratio: which variable determines survival of cervical cancer patients? Clin Cancer Res 2008; 14:2028–35. [DOI] [PubMed] [Google Scholar]
- 52. Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F et al Intraepithelial CD8+ tumor‐infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA 2005; 102:18538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P et al Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004; 10:942–9. [DOI] [PubMed] [Google Scholar]
- 54. Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL et al Quantification of regulatory T cells enables the identification of high‐risk breast cancer patients and those at risk of late relapse. J Clin Oncol 2006; 24:5373–80. [DOI] [PubMed] [Google Scholar]
- 55. Houart C, Szpirer J, Szpirer C. The α‐foetoprotein proximal enhancer: localization, cell specificity and modulation by dexamethasone. Nucleic Acids Res 1990; 18:6277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roychoudhuri R, Eil RL, Clever D, Klebanoff CA, Sukumar M, Grant FM et al The transcription factor BACH2 promotes tumor immunosuppression. J Clin Invest 2016; 126:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Valzasina B, Piconese S, Guiducci C, Colombo MP. Tumor‐induced expansion of regulatory T cells by conversion of CD4+CD25− lymphocytes is thymus and proliferation independent. Cancer Res 2006; 66:4488–95. [DOI] [PubMed] [Google Scholar]
- 58. Liu VC, Wong LY, Jang T, Shah AH, Park I, Yang X et al Tumor evasion of the immune system by converting CD4+CD25− T cells into CD4+CD25+ T regulatory cells: role of tumor‐derived TGF‐β . J Immunol 2007; 178:2883–92. [DOI] [PubMed] [Google Scholar]
- 59. Ghiringhelli F, Puig PE, Roux S, Parcellier A, Schmitt E, Solary E et al Tumor cells convert immature myeloid dendritic cells into TGF‐β‐secreting cells inducing CD4+CD25+ regulatory T cell proliferation. J Exp Med 2005; 202:919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J et al Gr‐1+CD115+ immature myeloid suppressor cells mediate the development of tumor‐induced T regulatory cells and T‐cell anergy in tumor‐bearing host. Cancer Res 2006; 66:1123–31. [DOI] [PubMed] [Google Scholar]
- 61. Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y et al Expression of Helios, an Ikaros transcription factor family member, differentiates thymic‐derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 2010; 184:3433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yadav M, Louvet C, Davini D, Gardner JM, Martinez‐Llordella M, Bailey‐Bucktrout S et al Neuropilin‐1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo . J Exp Med 2012; 209:1713–22, s1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gottschalk RA, Corse E, Allison JP. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol 2012; 188:976–80. [DOI] [PubMed] [Google Scholar]
- 64. Szurek E, Cebula A, Wojciech L, Pietrzak M, Rempala G, Kisielow P et al Differences in expression level of Helios and neuropilin‐1 do not distinguish thymus‐derived from extrathymically‐induced CD4+Foxp3+ regulatory T cells. PLoS One 2015; 10:e0141161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN et al Neuropilin 1, is expressed on thymus‐derived natural regulatory T cells, but not mucosa‐generated induced Foxp3+ Treg cells. J Exp Med 2012; 209:1723–42, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hansen W, Hutzler M, Abel S, Alter C, Stockmann C, Kliche S et al Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. J Exp Med 2012; 209:2001–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE et al Stability and function of regulatory T cells is maintained by a neuropilin‐1‐semaphorin‐4a axis. Nature 2013; 501:252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hindley JP, Ferreira C, Jones E, Lauder SN, Ladell K, Wynn KK et al Analysis of the T‐cell receptor repertoires of tumor‐infiltrating conventional and regulatory T cells reveals no evidence for conversion in carcinogen‐induced tumors. Cancer Res 2011; 71:736–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sainz‐Perez A, Lim A, Lemercier B, Leclerc C. The T‐cell receptor repertoire of tumor‐infiltrating regulatory T lymphocytes is skewed toward public sequences. Cancer Res 2012; 72:3557–69. [DOI] [PubMed] [Google Scholar]
- 70. Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV et al Regulatory T cells exhibit distinct features in human breast cancer. Immunity 2016; 45:1122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q et al Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018; 564:268–72. [DOI] [PubMed] [Google Scholar]
- 72. Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP et al Aire‐dependent thymic development of tumor‐associated regulatory T cells. Science 2013; 339:1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M et al T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA 2008; 105:7797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clever D, Roychoudhuri R, Constantinides MG, Askenase MH, Sukumar M, Klebanoff CA et al Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell 2016; 166:1117–31.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gobert M, Treilleux I, Bendriss‐Vermare N, Bachelot T, Goddard‐Leon S, Arfi V et al Regulatory T cells recruited through CCL22/CCR75 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res 2009; 69:2000–9. [DOI] [PubMed] [Google Scholar]
- 76. Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H et al CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer 2008; 122:2286–93. [DOI] [PubMed] [Google Scholar]
- 77. Miller AM, Lundberg K, Ozenci V, Banham AH, Hellstrom M, Egevad L et al CD4+ CD25high T cells are enriched in the tumor and peripheral blood of prostate cancer patients. J Immunol 2006; 177:7398–405. [DOI] [PubMed] [Google Scholar]
- 78. Li YQ, Liu FF, Zhang XM, Guo XJ, Ren MJ, Fu L. Tumor secretion of CCL22 activates intratumoral Treg infiltration and is independent prognostic predictor of breast cancer. PLoS One 2013; 8:e76379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Godiska R, Chantry D, Raport CJ, Sozzani S, Allavena P, Leviten D et al Human macrophage‐derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte‐derived dendritic cells, and natural killer cells. J Exp Med 1997; 185:1595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Andrew DP, Chang MS, McNinch J, Wathen ST, Rihanek M, Tseng J et al STCP‐1 (MDC) CC chemokine acts specifically on chronically activated Th2 lymphocytes and is produced by monocytes on stimulation with Th2 cytokines IL‐4 and IL‐13. J Immunol 1998; 161:5027–38. [PubMed] [Google Scholar]
- 81. Bao K, Reinhardt RL. The differential expression of IL‐4 and IL‐13 and its impact on type‐2 immunity. Cytokine 2015; 75:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mariani M, Lang R, Binda E, Panina‐Bordignon P, D'Ambrosio D. Dominance of CCL22 over CCL17 in induction of chemokine receptor CCR82 desensitization and internalization on human Th2 cells. Eur J Immunol 2004; 34:231–40. [DOI] [PubMed] [Google Scholar]
- 83. Viney JM, Andrew DP, Phillips RM, Meiser A, Patel P, Lennartz‐Walker M et al Distinct conformations of the chemokine receptor CCR83 with implications for its targeting in allergy. J Immunol 2014; 192:3419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sugiyama D, Nishikawa H, Maeda Y, Nishioka M, Tanemura A, Katayama I et al Anti‐CCR84 mAb selectively depletes effector‐type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc Natl Acad Sci USA 2013; 110:17945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang LP et al Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature 2011; 475:226–30. [DOI] [PubMed] [Google Scholar]
- 86. De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C et al Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor‐infiltrating T regulatory cells. Immunity 2016; 45:1135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Colbeck EJ, Jones E, Hindley JP, Smart K, Schulz R, Browne M et al Treg depletion licenses T cell‐driven HEV neogenesis and promotes tumor destruction. Cancer Immunol Res 2017; 5:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Joshi NS, Akama‐Garren EH, Lu Y, Lee DY, Chang GP, Li A et al Regulatory T cells in tumor‐associated tertiary lymphoid structures suppress anti‐tumor T cell responses. Immunity 2015; 43:579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Venturi GM, Conway RM, Steeber DA, Tedder TF. CD25+CD4+ regulatory T cell migration requires L‐selectin expression: L‐selectin transcriptional regulation balances constitutive receptor turnover. J Immunol 2007; 178:291–300. [DOI] [PubMed] [Google Scholar]
- 90. Colbeck EJ, Hindley JP, Smart K, Jones E, Bloom A, Bridgeman H et al Eliminating roles for T‐bet and IL‐2 but revealing superior activation and proliferation as mechanisms underpinning dominance of regulatory T cells in tumors. Oncotarget 2015; 6:24649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL et al CD69 acts downstream of interferon‐α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 2006; 440:540–4. [DOI] [PubMed] [Google Scholar]
- 92. Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S et al Cutting edge: CD69 interference with sphingosine‐1‐phosphate receptor function regulates peripheral T cell retention. J Immunol 2015; 194:2059–63. [DOI] [PubMed] [Google Scholar]
- 93. Cortes JR, Sanchez‐Diaz R, Bovolenta ER, Barreiro O, Lasarte S, Matesanz‐Marin A et al Maintenance of immune tolerance by Foxp3+ regulatory T cells requires CD69 expression. J Autoimmun 2014; 55:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ondondo B, Colbeck E, Jones E, Smart K, Lauder SN, Hindley J et al A distinct chemokine axis does not account for enrichment of Foxp3+ CD4+ T cells in carcinogen‐induced fibrosarcomas. Immunology 2015; 145:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. DiSpirito JR, Zemmour D, Ramanan D, Cho J, Zilionis R, Klein AM et al Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol 2018; 3:eaat5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz‐Wagenblatt A, Trager U et al Genome‐wide DNA‐methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol 2017; 18:1160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Green JA, Arpaia N, Schizas M, Dobrin A, Rudensky AY. A nonimmune function of T cells in promoting lung tumor progression. J Exp Med 2017; 214:3565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Halvorsen EC, Franks SE, Wadsworth BJ, Harbourne BT, Cederberg RA, Steer CA et al IL‐33 increases ST2+ Tregs and promotes metastatic tumour growth in the lungs in an amphiregulin‐dependent manner. Oncoimmunology 2018; 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ito T, Hanabuchi S, Wang YH, Park WR, Arima K, Bover L et al Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity 2008; 28:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Herman AE, Freeman GJ, Mathis D, Benoist C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J Exp Med 2004; 199:1479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Burmeister Y, Lischke T, Dahler AC, Mages HW, Lam KP, Coyle AJ et al ICOS controls the pool size of effector‐memory and regulatory T cells. J Immunol 2008; 180:774–82. [DOI] [PubMed] [Google Scholar]
- 102. Strauss L, Bergmann C, Szczepanski MJ, Lang S, Kirkwood JM, Whiteside TL. Expression of ICOS on human melanoma‐infiltrating CD4+CD25highFoxp3+ T regulatory cells: implications and impact on tumor‐mediated immune suppression. J Immunol 2008; 180:2967–80. [DOI] [PubMed] [Google Scholar]
- 103. Martin‐Orozco N, Li Y, Wang Y, Liu S, Hwu P, Liu YJ et al Melanoma cells express ICOS ligand to promote the activation and expansion of T‐regulatory cells. Cancer Res 2010; 70:9581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Le KS, Thibult ML, Just‐Landi S, Pastor S, Gondois‐Rey F, Granjeaud S et al Follicular B lymphomas generate regulatory T cells via the ICOS/ICOSL pathway and are susceptible to treatment by anti‐ICOS/ICOSL therapy. Cancer Res 2016; 76:4648–60. [DOI] [PubMed] [Google Scholar]
- 105. Faget J, Bendriss‐Vermare N, Gobert M, Durand I, Olive D, Biota C et al ICOS‐ligand expression on plasmacytoid dendritic cells supports breast cancer progression by promoting the accumulation of immunosuppressive CD4+ T cells. Cancer Res 2012; 72:6130–41. [DOI] [PubMed] [Google Scholar]
- 106. Vasanthakumar A, Liao Y, Teh P, Pascutti MF, Oja AE, Garnham AL et al The TNF receptor superfamily‐NF‐κB axis is critical to maintain effector regulatory T cells in lymphoid and non‐lymphoid tissues. Cell Rep 2017; 20:2906–20. [DOI] [PubMed] [Google Scholar]
- 107. van Olffen RW, Koning N, van Gisbergen KP, Wensveen FM, Hoek RM, Boon L et al GITR triggering induces expansion of both effector and regulatory CD4+ T cells in vivo . J Immunol 2009; 182:7490–500. [DOI] [PubMed] [Google Scholar]
- 108. Knee DA, Hewes B, Brogdon JL. Rationale for anti‐GITR cancer immunotherapy. Eur J Cancer 2016; 67:1–10. [DOI] [PubMed] [Google Scholar]
- 109. Kim JD, Choi BK, Bae JS, Lee UH, Han IS, Lee HW et al Cloning and characterization of GITR ligand. Genes Immun 2003; 4:564–9. [DOI] [PubMed] [Google Scholar]
- 110. O'Keeffe GW, Gutierrez H, Pandolfi PP, Riccardi C, Davies AM. NGF‐promoted axon growth and target innervation requires GITRL‐GITR signaling. Nat Neurosci 2008; 11:135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN, Gao FG. TGF‐β of lung cancer microenvironment upregulates B7H1 and GITRL expression in dendritic cells and is associated with regulatory T cell generation. Oncol Rep 2012; 28:615–21. [DOI] [PubMed] [Google Scholar]
- 112. Goode EL, DeRycke M, Kalli KR, Oberg AL, Cunningham JM, Maurer MJ et al Inherited variants in regulatory T cell genes and outcome of ovarian cancer. PLoS One 2013; 8:e53903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Buchan SL, Rogel A, Al‐Shamkhani A. The immunobiology of CD27 and OX40 and their potential as targets for cancer immunotherapy. Blood 2018; 131:39–48. [DOI] [PubMed] [Google Scholar]
- 114. Timperi E, Pacella I, Schinzari V, Focaccetti C, Sacco L, Farelli F et al Regulatory T cells with multiple suppressive and potentially pro‐tumor activities accumulate in human colorectal cancer. Oncoimmunology 2016; 5:e1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Montler R, Bell RB, Thalhofer C, Leidner R, Feng Z, Fox BA et al OX40, PD‐1 and CTLA‐4 are selectively expressed on tumor‐infiltrating T cells in head and neck cancer. Clin Transl Immunology 2016; 5:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lai C, August S, Albibas A, Behar R, Cho SY, Polak ME et al OX40+ regulatory T cells in cutaneous squamous cell carcinoma suppress effector T‐cell responses and associate with metastatic potential. Clin Cancer Res 2016; 22:4236–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Valzasina B, Guiducci C, Dislich H, Killeen N, Weinberg AD, Colombo MP. Triggering of OX40 (CD134) on CD4+CD25+ T cells blocks their inhibitory activity: a novel regulatory role for OX40 and its comparison with GITR. Blood 2005; 105:2845–51. [DOI] [PubMed] [Google Scholar]
- 118. Bhattacharya P, Gopisetty A, Ganesh BB, Sheng JR, Prabhakar BS. GM‐CSF‐induced, bone‐marrow‐derived dendritic cells can expand natural Tregs and induce adaptive Tregs by different mechanisms. J Leukoc Biol 2011; 89:235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Shibahara I, Saito R, Zhang R, Chonan M, Shoji T, Kanamori M et al OX40 ligand expressed in glioblastoma modulates adaptive immunity depending on the microenvironment: a clue for successful immunotherapy. Mol Cancer 2015; 14:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Piconese S, Timperi E, Pacella I, Schinzari V, Tripodo C, Rossi M et al Human OX40 tunes the function of regulatory T cells in tumor and nontumor areas of hepatitis C virus‐infected liver tissue. Hepatology 2014; 60:1494–507. [DOI] [PubMed] [Google Scholar]
- 121. Vakil Monfared R, Mashayekhi F. OX40L gene polymorphism and breast cancer in Iranian population. Exp Oncol 2018; 40:132–5. [PubMed] [Google Scholar]
- 122. Weiguang Y, Dalin L, Lidan X, Yonggang C, Shuang C, Yanhong L et al Association of OX40L polymorphisms with sporadic breast cancer in northeast Chinese Han population. PLoS One 2012; 7:e41277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bartkowiak T, Curran MA. 4‐1BB agonists: multi‐potent potentiators of tumor immunity. Front Oncol 2015; 5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chester C, Sanmamed MF, Wang J, Melero I. Immunotherapy targeting 4‐1BB: mechanistic rationale, clinical results, and future strategies. Blood 2018; 131:49–57. [DOI] [PubMed] [Google Scholar]
- 125. Smith SE, Hoelzinger DB, Dominguez AL, Van Snick J, Lustgarten J. Signals through 4‐1BB inhibit T regulatory cells by blocking IL‐9 production enhancing antitumor responses. Cancer Immunol Immunother 2011; 60:1775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Choi BK, Bae JS, Choi EM, Kang WJ, Sakaguchi S, Vinay DS et al 4‐1BB‐dependent inhibition of immunosuppression by activated CD4+CD25+ T cells. J Leukoc Biol 2004; 75:785–91. [DOI] [PubMed] [Google Scholar]
- 127. Zheng G, Wang B, Chen A. The 4‐1BB costimulation augments the proliferation of CD4+CD25+ regulatory T cells. J Immunol 2004; 173:2428–34. [DOI] [PubMed] [Google Scholar]
- 128. Elpek KG, Yolcu ES, Franke DD, Lacelle C, Schabowsky RH, Shirwan H. Ex vivo expansion of CD4+CD25+FoxP3+ T regulatory cells based on synergy between IL‐2 and 4‐1BB signaling. J Immunol 2007; 179:7295–304. [DOI] [PubMed] [Google Scholar]
- 129. Claus C, Riether C, Schurch C, Matter MS, Hilmenyuk T, Ochsenbein AF. CD27 signaling increases the frequency of regulatory T cells and promotes tumor growth. Cancer Res 2012; 72:3664–76. [DOI] [PubMed] [Google Scholar]
- 130. Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C et al Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 2014; 40:569–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kurtulus S, Sakuishi K, Ngiow SF, Joller N, Tan DJ, Teng MW et al TIGIT predominantly regulates the immune response via regulatory T cells. J Clin Invest 2015; 125:4053–62. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132. Fourcade J, Sun Z, Chauvin JM, Ka M, Davar D, Pagliano O et al CD226 opposes TIGIT to disrupt Tregs in melanoma. JCI Insight 2018; 3:121157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kucan Brlic P, Lenac Rovis T, Cinamon G, Tsukerman P, Mandelboim O, Jonjic S. Targeting PVR (CD155) and its receptors in anti‐tumor therapy. Cell Mol Immunol 2019; 16:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK et al PD‐L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med 2009; 206:3015–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhang B, Chikuma S, Hori S, Fagarasan S, Honjo T. Nonoverlapping roles of PD‐1 and FoxP3 in maintaining immune tolerance in a novel autoimmune pancreatitis mouse model. Proc Natl Acad Sci USA 2016; 113:8490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yi T, Li X, Yao S, Wang L, Chen Y, Zhao D et al Host APCs augment in vivo expansion of donor natural regulatory T cells via B7H1/B7.1 in allogeneic recipients. J Immunol 2011; 186:2739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lowther DE, Goods BA, Lucca LE, Lerner BA, Raddassi K, van Dijk D et al PD‐1 marks dysfunctional regulatory T cells in malignant gliomas. JCI Insight 2016; 1:e85935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G et al Role of LAG‐3 in regulatory T cells. Immunity 2004; 21:503–13. [DOI] [PubMed] [Google Scholar]
- 139. Camisaschi C, Casati C, Rini F, Perego M, De Filippo A, Triebel F et al LAG‐3 expression defines a subset of CD4+CD25highFoxp3+ regulatory T cells that are expanded at tumor sites. J Immunol 2010; 184:6545–51. [DOI] [PubMed] [Google Scholar]
- 140. Liang B, Workman C, Lee J, Chew C, Dale BM, Colonna L et al Regulatory T cells inhibit dendritic cells by lymphocyte activation gene‐3 engagement of MHC class II. J Immunol 2008; 180:5916–26. [DOI] [PubMed] [Google Scholar]
- 141. Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z et al LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T‐cell responses. Cancer Res 2014; 74:3418–28. [DOI] [PubMed] [Google Scholar]
- 142. Anderson AC, Joller N, Kuchroo VK. Lag‐3, Tim‐3, and TIGIT: co‐inhibitory receptors with specialized functions in immune regulation. Immunity 2016; 44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Meinicke H, Bremser A, Brack M, Schrenk K, Pircher H, Izcue A. KLRG1 impairs regulatory T‐cell competitive fitness in the gut. Immunology 2017; 152:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kourtidis A, Lu R, Pence LJ, Anastasiadis PZ. A central role for cadherin signaling in cancer. Exp Cell Res 2017; 358:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Vang KB, Yang J, Mahmud SA, Burchill MA, Vegoe AL, Farrar MA. IL‐2, ‐7, and ‐15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J Immunol 2008; 181:3285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fan MY, Low JS, Tanimine N, Finn KK, Priyadharshini B, Germana SK et al Differential roles of IL‐2 signaling in developing versus mature Tregs. Cell Rep 2018; 25:1204–13.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol 2015; 33:101–11. [DOI] [PubMed] [Google Scholar]
- 148. Fukuhara H, Matsumoto A, Kitamura T, Takeuchi T. Neutralization of interleukin‐2 retards the growth of mouse renal cancer. BJU Int 2006; 97:1314–21. [DOI] [PubMed] [Google Scholar]
- 149. Kryczek I, Wei S, Zou L, Altuwaijri S, Szeliga W, Kolls J et al Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL‐2 in the tumor microenvironment. J Immunol 2007; 178:6730–3. [DOI] [PubMed] [Google Scholar]
- 150. Sim GC, Martin‐Orozco N, Jin L, Yang Y, Wu S, Washington E et al IL‐2 therapy promotes suppressive ICOS+ Treg expansion in melanoma patients. J Clin Invest 2014; 124:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Owen DL, Mahmud SA, Vang KB, Kelly RM, Blazar BR, Smith KA et al Identification of cellular sources of IL‐2 needed for regulatory T cell development and homeostasis. J Immunol 2018; 200:3926–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L et al Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 2009; 31:772–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Humblet‐Baron S, Franckaert D, Dooley J, Bornschein S, Cauwe B, Schonefeldt S et al IL‐2 consumption by highly activated CD8 T cells induces regulatory T‐cell dysfunction in patients with hemophagocytic lymphohistiocytosis. J Allergy Clin Immunol 2016; 138:200–9.e8. [DOI] [PubMed] [Google Scholar]
- 154. Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J et al High‐dimensional single cell analysis identifies stem‐like cytotoxic CD8+ T cells infiltrating human tumors. J Exp Med 2018; 215:2520–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sheng SY, Gu Y, Lu CG, Tang YY, Zou JY, Zhang YQ et al The characteristics of naive‐like T cells in tumor‐infiltrating lymphocytes from human lung cancer. J Immunother 2017; 40:1–10. [DOI] [PubMed] [Google Scholar]
- 156. Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S et al The transcriptional regulators IRF4, BATF and IL‐33 orchestrate development and maintenance of adipose tissue‐resident regulatory T cells. Nat Immunol 2015; 16:276–85. [DOI] [PubMed] [Google Scholar]
- 157. Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA et al The alarmin IL‐33 promotes regulatory T‐cell function in the intestine. Nature 2014; 513:564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jovanovic IP, Pejnovic NN, Radosavljevic GD, Pantic JM, Milovanovic MZ, Arsenijevic NN et al Interleukin‐33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer 2014; 134:1669–82. [DOI] [PubMed] [Google Scholar]
- 159. Li C, DiSpirito JR, Zemmour D, Spallanzani RG, Kuswanto W, Benoist C et al TCR transgenic mice reveal stepwise, multi‐site acquisition of the distinctive fat‐Treg phenotype. Cell 2018; 174:285–99 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Newton R, Priyadharshini B, Turka LA. Immunometabolism of regulatory T cells. Nat Immunol 2016; 17:618–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R et al Phosphoenolpyruvate is a metabolic checkpoint of anti‐tumor T cell responses. Cell 2015; 162:1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Pacella I, Procaccini C, Focaccetti C, Miacci S, Timperi E, Faicchia D et al Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci USA 2018; 115:E6546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Procaccini C, Carbone F, Di Silvestre D, Brambilla F, De Rosa V, Galgani M et al The proteomic landscape of human ex vivo regulatory and conventional T cells reveals specific metabolic requirements. Immunity 2016; 44:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kishore M, Cheung KCP, Fu H, Bonacina F, Wang G, Coe D et al Regulatory T cell migration is dependent on glucokinase‐mediated glycolysis. Immunity 2017; 47:875–89 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]


