Abstract
Polymorphic duplications in humans have been shown to contribute to phenotypic diversity. However, the evolutionary forces that maintain variable duplications across the human genome are largely unexplored. We developed a linkage-disequilibrium based method to detect insertion sites of polymorphic duplications not represented in reference genomes. This method also allows resolution of haplotypes harboring the duplications. Using this approach, we conducted genome-wide analyses and identified the insertion sites of 22 common polymorphic duplications. We found that the majority of these duplications is intrachromosomal and only one of them is an interchromosomal insertion. Further characterization of these duplications revealed significant associations to blood and skin phenotypes. On the basis of population genetics analyses, we found that the duplication of a well-characterized pigmentation-related region, including the HERC2 gene, may be selected against in European populations. We further demonstrated that the haplotype harboring this duplication significantly affects the expression of the HERC2P9 gene in multiple tissues. Our study sheds light onto the evolutionary impact of understudied polymorphic duplications in human populations and presents methodological insights for future studies.
Keywords: structural variants, copy number variation, KRT, natural selection
Introduction
Genomic structural variation (duplications, deletions, translocations, and inversions of genomic segments) has increasingly been appreciated as a driver of human phenotypic variation, accounting for several key adaptive phenotypes, as well as disease susceptibility (Zhang et al. 2009; Weischenfeldt et al. 2013). One of the best-known examples of the evolutionary impact of structural variation is the CCR5-Δ32 deletion polymorphism which is associated with HIV resistance (Dean et al. 1996; Sabeti et al. 2005). Another recent example that also invokes the adaptive role of structural variations to resistance to pathogens is the reassessment of the haplotypic architecture of the structural variants involving haptoglobin Glycophorin A and Glycophorin B genes. This study revealed multiple instances of recurrent evolution of structural variants in this locus that are associated with malaria resistance in African populations (Leffler et al. 2017). Even though its timing and nature is under scrutiny (Inchley et al. 2016; Fernández and Wiley 2017), another example of likely adaptation involving structural variations is the expansion of salivary amylase gene copy number among humans, likely driven by high starch consumption (Meisler and Ting 1993; Perry et al. 2007).
Despite their genomic and phenotypic impacts exemplified by these interesting examples, few studies have addressed the evolutionary forces that shape the evolutionary trajectories of polymorphic duplications. We argue that the main reason for the paucity of evolutionary studies on polymorphic duplications is that current, short-read based discovery and genotyping approaches are unable to resolve the genomic locations of inserted duplicated gene copies; consequently, the haplotypic variation associated with a given duplication cannot be fully studied. The two commonly used approaches to detect polymorphic duplications based on short-read sequences depend on paired-end mapping and read-depth (Mills et al. 2011; Zhao et al. 2013). Paired-end mapping approach depends on discordantly mapped paired-reads sequences where the distances between these two sequences are different from the expected. This method can detect some of the tandem duplications (Sudmant et al. 2015) and can be modified to detect mobile element insertions (Lee et al. 2012). However, this method is highly prone to false negatives as the short reads often fail to map to repetitive sequences (Narzisi and Schatz 2015). This problem is further aggravated by the complexity of a considerable portion of the loci harboring duplications, that is, they involve highly repetitive sequences (Sudmant et al. 2015). The more sensitive approaches to detect polymorphic duplications depend on read-depth, where deviations in the depth of coverage in a genomic region as compared with genome-wide expectations can signal copy number gain and loss of that particular sequence (Alkan et al. 2011). This method is relatively robust especially if the duplication is large. However, read-depth methods alone cannot detect the insertion site of the duplicated sequence. In summary, currently available methods using short-reads to detect polymorphic duplications are limited in their ability to detect the insertion sites of the duplicated sequences. Thus, the haplotypes harboring polymorphic duplications, which are crucial to conduct neutrality tests and functional analyses, often remain elusive. Here, by applying a novel linkage disequilibrium based method to the 1000 Genome Project phase 3 data set (Sudmant et al. 2015), we detected the insertion sites of 22 common human polymorphic duplications. This data set allowed us to more thoroughly investigate the potential adaptive contributions of some of these duplications on human phenotypic diversity.
Results
Detecting Putatively Adaptive Duplications and Their Insertion Sites
To detect insertion sites of polymorphic duplications, we utilized genome-wide linkage disequilibrium between the genotyped duplications (for which the insertion site is unknown) and single nucleotide variants (SNVs) across the human genome. Specifically, we assumed that when a duplicated sequence was inserted in a certain genomic region and subsequently increased in allele frequency, the flanking SNVs would show linkage disequilibrium with the duplication (fig. 1A). This method can only detect the insertion sites of gene duplications with relatively high allele frequency. The signal weakens considerably if the haplotype harboring the duplicated sequence undergoes recombination or gene conversion as expected from the previous studies (Saitou et al. 2018). It is also important to note here that our method’s power depends on the accuracy of variation calls and phasing of the database that we are using.
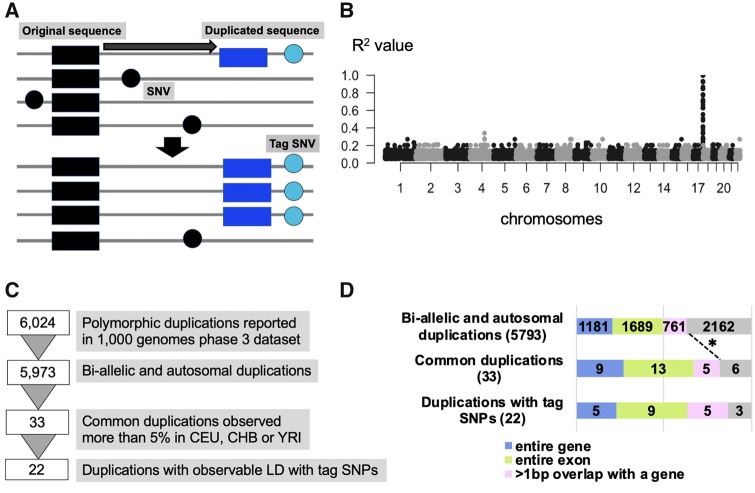
Fig. 1.
—The strategy of the linkage disequilibrium-based method to detect the insertion region of the polymorphic duplication. (A) The schematic representation of our approach to detect insertion sites and haplotypes harboring polymorphic duplications. (B) One example of the linkage disequilibrium-peak between and duplication and SNVs. Each dot indicates a SNV. The X-axis shows chromosomal locations and Y-axis shows the linkage disequilibrium between each SNV and a specific polymorphic duplication, in this case, esv3641421, in the CEU population. (C) The filtering process of the duplications. (D) The breakdown of the number of duplications based on their exonic content and allele frequency. The legend below indicates the color-coded functional categories. We observed an enrichment of genic content among the very common (>5% allele frequency) duplications when compared with the genic content of all polymorphic bi-allelic duplications (P-value = 0.03684, one-tail Pearson's Chi-squared test with Yates' continuity correction).
We chose to apply this method to the data provided by the 1000 Genome Project phase 3 data set (Sudmant et al. 2015), which reports 6,024 polymorphic duplications. We chose this data set as it remains the most accurate population-level compilation of human variable duplications and phased SNVs essential for our analysis. Specifically, the 1000 Genomes consortium applied multiple algorithms to detect polymorphic duplications including Delly (Rausch et al. 2012) and Genome STRiP (Handsaker et al. 2015). More importantly, comprehensive external validation of the discovered structural variants was used to minimize the false positive rate. Last but not least, the whole genome sequencing from thousands of individuals allows integrative phasing of all variants, which provided haplotype context of the structural variations (The 1000 Genomes Project Consortium et al. 2015). Therefore, we argue that the 1000 Genome Project phase 3 data set provides one of the most accurate short-read sequence based population-level structural variants callsets available along with the SNV information from the same individuals.
To further minimize false-positive structural variant calls and to avoid complicating our data set, we conducted some preliminary filtering (fig. 1B). First, we eliminated multiallelic copy number variations and focused only on bi-allelic duplications reported as 2, 3, or 4 diploid copies in humans. To increase our power for detecting linkage disequilibrium, we focused on common duplications observed in >5% in any of Central Europeans from Utah (CEU), Yoruba from Ibadan (YRI), or Han Chinese from Beijing (CHB). After this filtering, we were left with 33 common, bi-allelic duplications for this study. We identified observable peak(s) of linkage disequilibrium for 22 out of 33 common duplications (fig. 1C) with SNVs across the genome (table 1, supplementary table S1, Supplementary Material online). For the other 11 common duplications, we were not able to identify a linkage disequilibrium peak. Previous studies have shown that gene conversion and recurrence can explain this pattern. For example, Boettger (2016) reported the recurrent exonic deletions of the haptoglobin locus, for which the haplotype architecture was complex. Similarly, our own work showed the joint effect of recurrence and gene conversion in complicating the haplotypic background of structural variation in the GSTM1 locus (Saitou et al. 2018). Thus, similar characterization efforts to resolve the haplotypes harboring these 11 duplication polymorphisms would provide important venues for future research. Nevertheless, in this study, we focused our analysis on the 22 duplications for which we were able to detect linked haplotypic variation.
Table 1.
All the 22 Duplications and One of Their Tag SNVs, R2 Value, and the Phenotypic Information by Gene ATLAS
| ID | chr | gene | freq_CEU | freq_CHB | freq_YRI | overlap | tag SNP | TagSNP_hg19 | R 2 | GENEATLAS |
|---|---|---|---|---|---|---|---|---|---|---|
| esv3584976 | chr1 | FAM41C, FAM87B | — | 0.058 | — | Whole-gene | rs528265132 | chr1_844674 | 0.406 | NA |
| esv3585141 | chr1 | nongenic | 0.056 | — | — | Nongenic | rs74865018 | chr1_8208573 | 0.496 | Skin color, P-value = 1.4E–09 |
| esv3589561 | chr1 | OR2T27 | — | — | 0.343 | Partial-exon | rs28502564 | chr1_248831191 | 0.514 | NA |
| esv3590421 | chr2 | GALM, SRSF7 | 0.076 | — | — | Whole-exon | rs112011213 | chr2_38967847 | 0.857 | NS |
| esv3590859 | chr2 | PNPT1 | — | — | 0.069 | Whole-exon | rs115094228 | chr2_55731753 | 0.626 | NA |
| esv3592511 | chr2 | GPR39 | — | — | 0.069 | Intronic | rs77354775 | chr2_133345124 | 0.751 | NA |
| esv3594536 | chr2 | TM4SF20 | 0.076 | — | — | Whole-exon | rs80058427 | chr2_228399781 | 0.425 | NS |
| esv3599142 | chr3 | FGF12, FGF12-AS1 | — | 0.058 | 0.005 | Intronic | rs6788805 | chr3_192012511 | 0.569 | NA |
| esv3599420 | chr4 | HTT-AS | — | — | 0.051 | Whole-exon | rs1557213 | chr4_3038415 | 0.912 | NA |
| esv3601317 | chr4 | nongenic | 0.136 | 0.049 | — | Nongenic | rs74797043 | chr4_90102254 | 1.000 | NS |
| esv3603011 | chr4 | TRIM61 | — | — | 0.116 | Whole-gene | rs78990101 | chr4_161987911 | 0.880 | NA |
| esv3620370 | chr9 | UNC13B | — | — | 0.083 | Intronic | rs111637861 | chr9_35230046 | 0.942 | NA |
| esv3620559 | chr9 | APBA1 | — | — | 0.111 | Whole-exon | rs186797639 | chr9_72022475 | 0.511 | NA |
| esv3631000 | chr12 | ZNF664, ZNF664-FAM101A | 0.096 | — | 0.037 | Whole-exon | rs73131333 | chr2_3953369 | 1.000 | NS |
| esv3631499 | chr13 | nongenic | — | — | 0.065 | Nongenic | rs115022408 | chr13_23424799 | 1.000 | NA |
| esv3632749 | chr13 | COMMD6 | — | — | 0.134 | Whole-exon | rs61645976 | chr13_76107661 | 0.718 | NA |
| esv3635993 | chr15 | HERC2 | 0.025 | 0.66 | 0.282 | Intronic | rs376191081 | chr15_28549862 | 0.751 | NA |
| esv3640164 | chr17 | TRIM16L | 0.056 | 0.083 | 0.079 | Whole-exon | rs199526489 | chr17_15546785 | 0.950 | NS |
| esv3640585 | chr17 | KRT34 | 0.025 | 0.029 | 0.13 | Whole-gene | rs9914283 | chr17_39541260 | 0.959 | NS |
| esv3641421 | chr17 | TEX19 | 0.071 | 0.005 | — | Whole-gene | rs74001624 | chr17_80314483 | 1.000 | Monocyte percentage, P-value = 2.8E–16 |
| esv3643776 | chr19 | CYP4F12 | — | — | 0.069 | Whole-gene | rs112344570 | chr19_15831904 | 1.000 | NS |
| esv3645658 | chr20 | TTLL9 | — | — | 0.056 | Whole-exon | rs73903650 | chr20_30391721 | 0.928 | NS |
Note.—We described one tag SNV for each polymorphic duplication, with the highest linkage disequilibrium in table 1. We provide the highest R2 values observed in CEU, CHB, or YRI populations (the frequency column is bolded). The tag SNVs, thus, can be population specific. We bolded the allele frequency column to designate the populations where we identified the tag SNPs for the particular duplications. When we found multiple SNVs with the same R2 value, we chose one SNV which reported the SNV that is physically located in the middle of the most upstream and downstream SNV with equally high R2 values. All the tag SNVs are reported in table S1, Supplementary Material online.
We found that 21 out of 22 (∼96%) of duplication insertion sites are found on the same chromosome as where the duplicated sequence is found. Further scrutinization of the haplotypes harboring intrachromosomal duplications revealed that five of them overlap with the original duplicated region, six of them were located (>1 kb) upstream of the region and eight of them located (>1 kb) downstream of the region (table 1 and supplementary fig. S1, Supplementary Material online). Additionally, we found that one duplication (esv3631000), which contains the gene ZNF664 located on chromosome 12, is inserted into chromosome 2. This observation was supported by 17 SNVs on chromosome 2 in strong linkage disequilibrium (R2 > 0.8) with the duplication (supplementary table S1, Supplementary Material online). ZNF664 is classified as retro-duplication (Abyzov et al. 2013). Thus, the retroposon machinery may facilitate a copy and paste mechanism of the reverse transcribed mRNA of the original gene to a random insertion point, in this case, chromosome 2.
We then scrutinized the genic content of the filtered duplications. Of the 22 duplicated sequences, 5 contain whole genes, 9 contain coding exonic sequences, 5 contain intronic sequences, and 3 contain only intergenic sequences (table 1). We then asked whether the high proportion of duplications containing coding sequences is more than expected, especially given that a previous study reported that only ∼20% of common duplications overlap with coding sequences (Conrad et al. 2010). This contrasts with the >50% of duplications we outlined that overlap with an entire gene or coding exon (fig. 1D). We observed the enrichment of genic duplication in the common duplications compared with the initial duplication set (P-value = 0.03684, one-tail Pearson's chi-squared test) (fig. 1D). We found that duplications associated with strong linkage disequilibrium with SNVs do not significantly differ in their coding sequence content from duplications that do not (P-value = 0.8845, one-tail Pearson's chi-squared test) (fig. 1D). We further confirmed the general consensus that the allele frequency is negatively correlated with genic content among the 1000 Genome Project phase 3 data set duplications. However, we found an increase of genic duplications among very common (>5% allele frequency) duplications in general (supplementary fig. S2, Supplementary Material online). Thus, the highly genic nature of the 22 duplications that we focus on this study is a property of their high allele frequency and the underlying evolutionary reasons for this overall increase remains an open question.
Partial HERC2 Duplication May Be Selected against in European Populations
Our main goal in this paper is to leverage the haplotypes of the polymorphic duplications to identify potential selective forces acting on specific polymorphic duplications. To achieve this, we first calculated the allele frequency differences between populations for the 22 polymorphic duplications that we focus in this study. Then we compared these differences to those calculated for randomly selected 3,102 very common (>5% alternative allele frequency in CEU, YRI, or CHB to match our initial filtering) SNVs extracted from 1000 Genomes phase 3 data set (The 1000 Genomes Project Consortium et al. 2015) (fig. 2A). We found that a partial duplication of a well-characterized gene, the HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 2 gene (HERC2) (esv3635993) showed apparently higher allele frequency differentiation from the other gene duplications as well as the majority of random SNVs analyzed as a null background (table 1, fig. 2A and B, supplementary fig. S3, Supplementary Material online). The HERC2 partial duplication was also reported as the top population-stratified structural variants among 5,887 polymorphic duplications analyzed in a previous study (Sudmant et al. 2015) based on VST statistics (Redon et al. 2006).
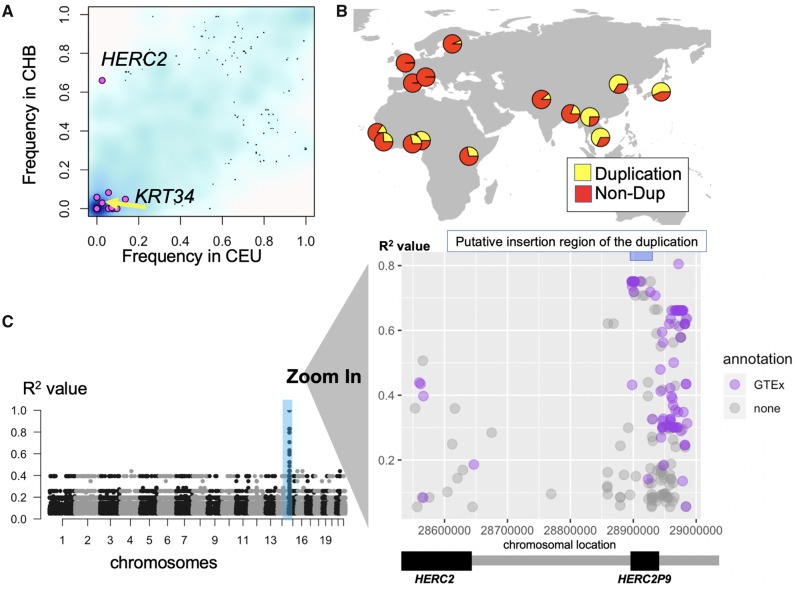
Fig. 2.
—The population differentiation of the partial HERC2 duplication. (A) The frequency of the target duplications which was observed either CHB or CEU (pink dots) and randomly selected 3,000 SNVs (>5% in CEU, YRI, or CHB) (blue background cloud). The density of the blue color reflects the density observations. The x-axis shows the frequency of the variation in CEU and the y-axis shows the frequency of the variation in CHB. (B) The geographical distribution of the HERC2 gene duplication allele. Yellow refers to the frequency of duplication allele and red refers to the frequency of the nonduplication allele. (C) Left: the putative location of the HERC2 duplication based on the linkage disequilibrium in the European populations. Right: the magnified version of the chromosomal location of HERC2 on chromosome 15. Dots are SNVs with R2 > 0.05 with the duplication in this location. The X-axis shows the chromosomal location and Y-axis shows the R2 between the SNV and the HERC2 duplication. The pale blue bar at upper-right indicates the haplotype block, which contains the SNVs with high linkage disequilibrium (R2 > 0.7) with the HERC2 duplication (hg19 chr15: 28894038-28927368). We assume that the insertion site of the duplication resides in this haplotype block and used this region for the subsequent analysis. The purple colored dots indicate SNVs that show significant association (P-value < 0.0001) with expression levels of neighboring genes based on GTEx portal (Lonsdale et al. 2013).
To further characterize this polymorphic duplication, we first manually confirmed this duplication by investigating the read-depth profiles of multiple samples from the 1000 Genomes phase 3 data set (supplementary fig. S3, Supplementary Material online). Then, we extended our linkage disequilibrium analysis to include the additional 1000 Genomes populations categorized across continental meta-populations (see Materials and Methods). On the basis of this analysis, we narrowed down the insertion site of the HERC2 partial gene duplication to hg19 chr15: 28894038-28927368 (R2 > 0.75), and observed a detectable increase in linkage disequilibrium between the duplication and flanking SNVs in all three continental populations (fig. 2C, supplementary fig. S4, Supplementary Material online). In addition, we attempted to resolve the breakpoints of the insertion site. To do this, we searched the recently available long-read sequence data sets including fosmid sequence data (Kidd et al. 2010) and long-read sequence data sets (Seo et al. 2016; Audano et al. 2019; Levy-Sakin et al. 2019; Nagasaki et al. forthcoming). However, none of these studies has reported this particular duplication. In addition, we were not able to locate this duplication among recently available segmental duplications in de novo genome assemblies (Vollger et al. 2019, also Volger M, personal communication). Two issues should be noted here. First, these long-read based sequence data sets focus on a small number of samples and thus it is plausible that genomes that are carrying this particular duplication are not represented in the data sets that we investigated. A second issue is that long-read sequences, even though substantially lnger than Illumina-based sequences, may have failed to cover the large ∼14 kb HERC2 duplication that we are focusing on. Last but not least, it is also possible that this duplication is a false-positive. However, the fact that we detected clear read-depth difference among genomes (supplementary fig. S5, Supplementary Material online) and the haplotype-level linkage disequilibrium between the duplication and SNVs strongly support the presence of a polymorphic duplication.
We found that linkage disequilibrium was strongest in European populations and weaker in East Asian and African populations. To investigate if the duplication is ancestral or derived, we compared the ∼100 kb region around the putative insertion site (hg19 chr15: 28894038-28927368) to the orthologous section in the chimpanzee reference assembly (determined by lift-over [Hinrichs et al. 2006], Pantro6, chr15: 1879453-1910950). We deduced that if the duplication is ancestral, we would identify an additional ∼14 kb sequence in the chimpanzee reference assembly that does not exist in the human reference genome. We failed to identify such a sequence in the chimpanzee assembly, strongly suggesting that duplication is the derived allele in the human lineage (supplementary fig. S6, Supplementary Material online). Further, we found that chimpanzees and Denisovan genomes do not harbor the 17 alleles that are linked with the duplication (R2 > 0.75 in European populations) (supplementary table S2, Supplementary Material online). This analysis supports our initial conclusion that the haplotype harboring the duplication is likely derived in the human lineage as compared with chimpanzees. Intriguingly we found that the Neanderthal genome is heterozygous at this locus, carrying both the haplotype associated with the duplication and those do not (supplementary table S2, Supplementary Material online). Given that we do not observe any deviation from the expected read-depth in Neanderthals, it is possible that the haplotype that harbors the duplication in humans have evolved before Humans and Neanderthals diverge and the duplication has evolved after their split. However, given the repetitive nature of this locus, future work is needed to definitively resolve the ancestral haplotype.
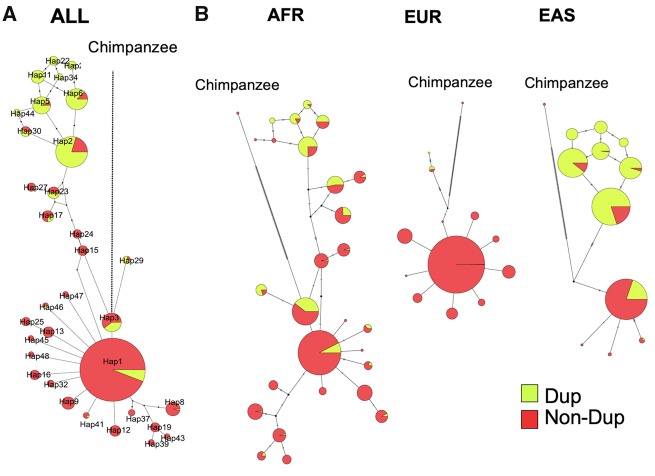
Next, we used VCFtoTree (Xu et al. 2017) to obtain an alignment file for the HERC2 duplication haplotype (hg19, chr15: 28898098-28902929), containing 2,504 samples available in the 1000 Genome phase 3 data set (Sudmant et al. 2015), as well as the reference chimpanzee genome (The Chimpanzee Sequencing Consortium 2005). We then constructed haplotype networks using PopART (version 1.7) (Leigh and Bryant 2015) using the Median Joining method (Bandelt et al. 1999) (fig. 3A). This network reveals an apparent reduction of haplotypic diversity in European populations as compared with East Asian and African populations (fig. 3B). This observation is consistent with the dramatically lower allele frequency of the duplication in European populations, which initially led us to focus on this locus (fig. 2A and B).
Fig. 3.
—Haplotype networks of the HERC2 duplication insertion region. (A) Merged haplotype network of the three meta-populations (AFR, EUR, EAS) constructed from 3,336 haplotypes from hg19 chr15: 28898098-28902929 (represented in fig. 2C). (B) Breakdown of individual networks to help visualization of the distribution of alleles in each meta-population. Yellow refers to the frequency of duplication allele and red refers to the frequency of the nonduplication allele.
We then asked whether population-specific selective forces can explain the reduction in haplotypic diversity at this locus in European populations. We calculated several neutrality measures at the locus harboring the HERC2 duplication and compared these to empirical distributions constructed from 26,283 3 kb regions across chromosome 15 from the 1000 Genomes Selection Browser (Last accessed, March 21, 2019) (Pybus et al. 2014). We found Tajima’s D scores in this genomic region to be lower than 90% of the values of control regions on chromosome 15 for European populations (fig. 4A). However, in East Asian and African populations, we observed the opposite trend, where Tajima’s D scores fell within an expected range, if not slightly higher, based on the empirical distribution. Tajima’s D measures deviations in the allele frequency spectrum (Tajima 1993); negative values indicate an excess of rare alleles, which may be a consequence of negative or positive selection. In this case, based on network analysis (fig. 3), we argue that this signal is primarily driven by the reduction of the frequency of haplotypes harboring the duplication in the European populations. One model that is consistent with the observed Tajima’s D values is negative selection against the duplication allele acting specifically in European populations.
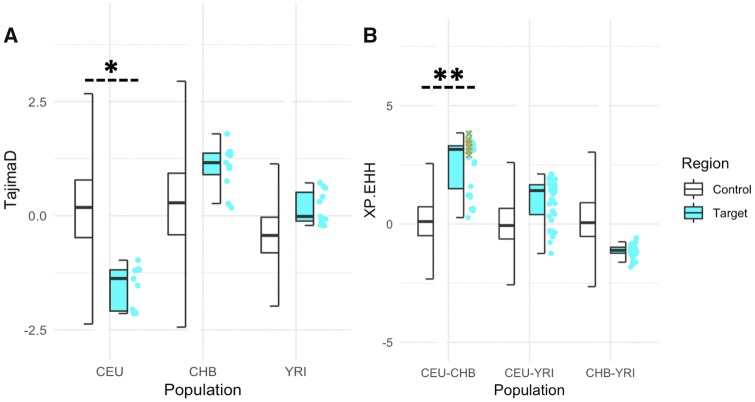
Fig. 4.
—Neutrality test on the putative insertion region of the partial HERC2 duplication. All values were obtained through the 1000 Genomes selection browser (Pybus et al. 2014). (A) Tajima’s D (11 bins of 3 kb window) and (B) XP-EHH values calculated for the HERC2 target region (hg19 chr15: 28894038-28927368, represented in fig. 2C), compared with the distributions calculated for all the accessible regions on the chromosome 15 on the 1000 Genomes selection browser (Pybus et al. 2014). * Represents that the mean value of the target region was within the lower 10% of the control region and there was a significant difference between control and target region (P-value = 6.18E–07, Wilcoxon rank sum test). ** Represents the mean value of the target region was within the upper 5% of the control region and there was a significant difference between control and target region (P-value < 2.2E–16, Wilcoxon rank sum test). Yellow cross represents SNVs with R2 > 0.75 with the HERC2 duplication in the European populations in the CEU–CHB comparison.
To test this hypothesis, we calculated XP-EHH scores between the three representative populations in a pairwise fashion for the SNVs from the same region that we calculate Tajima’s D values. Then, we compared these values to the empirical distribution of XP-EHH values constructed from the same 26,283 randomly chosen regions across chromosome 15 (fig. 4B). XP-EHH calculates the probability of runs of homozygosity around a given locus assuming that there is the same allele between two populations. A positive XP-EHH score is indicative of positive selection in the first population, whereas a negative score indicates positive selection in the second population (Sabeti et al. 2007). On the basis of this calculation, we found the average XP-EHH scores in this genomic region to be higher than 5% of the control regions on chromosome 15 in CEU versus CHB comparison (fig. 4B). In contrast, we found no clear population differentiation between other comparisons (fig. 4B). It should be noted here that if the selective pressure is on the duplication, it is plausible that the lack of XP-EHH signal in YRI and CHB populations may be due to lack of linkage disequilibrium between SNVs and the duplication in these two populations. In fact, a more focused analysis revealed that the high XP-EHH values are driven by SNVs that are linked with the duplication allele in the European population (fig. 4B). This means that there are relatively long runs of homozygosity in this region in CEU population as compared with CHB and YRI, concordant with the excess of rare variants suggested by Tajima’s D comparisons. In sum, these results are in line with a scenario that a recent selection event in Europe favors nonduplicated haplotypes over duplicated-haplotypes.
Next, we investigated the potential functional impact of the duplication haplotype in Europeans. We noted that HERC2 duplication is likely inserted within the neighboring HERC2P9 gene (fig. 2C). It is intriguing that a much more recent duplication of the HERC2 gene is inserted into an older paralog of the HERC2, which is expressed in multiple tissues. It is possible that recombination-based mechanisms facilitated by sequence homology between these genes led to the insertion of the duplication into HERC2P9. Eight HERC2 pseudogenes are reported in Ensembl (Zerbino et al. 2018) distributed across chromosomes 15 and 16, suggesting frequent duplication of this gene. On the basis of the GTEx portal (https://www.gtexportal.org/home/ Last accessed, March 21, 2019, Lonsdale et al. 2013), HERC2P2, HERC2P3, and HERC2P9 are expressed, as well as the intact HERC2 (supplementary fig. S7, Supplementary Material online).
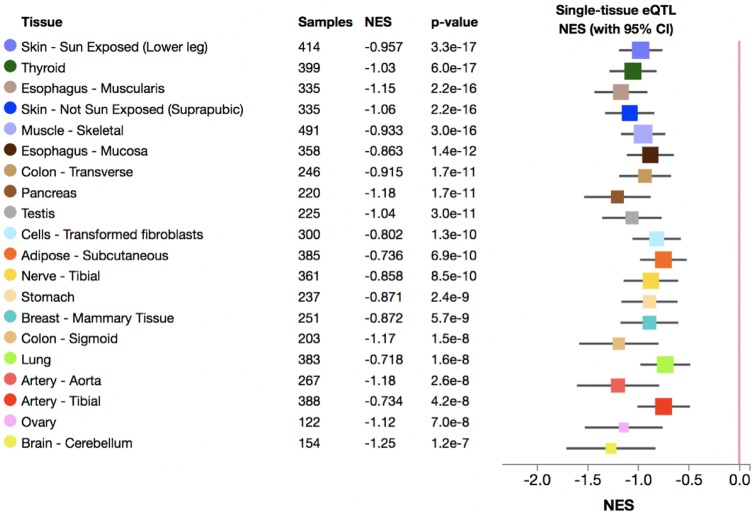
Furthermore, we found that the duplication haplotype (imputed by rs77868920, R2 = 0.75 in European populations) downregulates the expression of HERC2P9 in various tissues (fig. 5). The most significant effect observed for downregulation was in the sun-exposed skin (P-value = 3.3E–17, Normalized effect size = –0.96). It is possible that the polymorphic duplication may affect not only the expression levels but also alter the sequence in this region and change the transcribed RNA sequence of the HERC2P9. This remains an interesting area for further study. While there are multiple SNVs associated with skin color (Crawford et al. 2017) and iris color (Eiberg et al. 2008; Kayser et al. 2008; Sturm et al. 2008) in this region of the genome, the HERC2 duplication haplotype does not harbor any of them (MacArthur et al. 2017) (fig. 2C). It is important to note here that the duplication polymorphism is more common outside of Western Eurasia and thus association studies in nonEuropean populations will be key to resolve the putative functional impact of this duplication and associated haplotypes.
Fig. 5.
—The expression change of the HERC2P9 gene in various tissues associated with the HERC2 duplication tag SNV (rs77868920). The top 20 tissues on the GTEx (Lonsdale et al. 2013) with the lowest P-value are shown. Normalized effect size is defined as the slope of the linear regression of the expression of the HERC2P9 gene for three genotypes of the tag SNV. Normalized effect size is computed as the effect of the alternative allele relative (tagged to duplication) to the reference allele (tagged to nonduplication) in the human genome reference GRCh37/hg19. The whiskers on the plot represent the 95% confidence intervals.
The Functional Impact of Haplotypes Harboring Common Duplications
Our approach resolved the tag SNVs that are in strong linkage disequilibrium with common polymorphic gene duplications that may have important functional consequences (table 1). The ascertainment bias in most functional databases limits further scrutinization of the functional impact of polymorphisms to some extent. Specifically, most comprehensive data sets for expression quantitative trait loci analysis and most genome-wide association studies (e.g., GeneATLAS) were constructed mostly by data gathered from western European individuals. Majority of gene duplications for which we were able to resolve the haplotypes were found in African populations only (table 1). Still, we were able to search for specific associations of eight gene duplication haplotypes with >5% allele frequency in European populations with gene expression levels documented in GTEx (Last accessed, March 21, 2019) (Lonsdale et al. 2013), as well as with 778 traits documented in GeneATLAS (http://geneatlas.roslin.ed.ac.uk/, Last accessed March 21, 2019; Canela-Xandri et al. 2018). We found two significant associations. First, we found the exonic duplication involving TEX19 gene (esv3641421, tag-variant: rs74001624 [R2 = 1, G-allele is associated with the duplication]) is significantly associated with lower levels of expression of the adjacent gene SECTM1 on GTEx (P-value = 4.5E–10). Further, we found that the duplication haplotype is significantly (P-value = 2.8E–16) associated with Monocyte percentage (table 1). This finding is concordant with the previous findings that SECTM1 is involved in hematopoietic processes (Slentz-Kesler et al. 1998).
Second, we found that the haplotype harboring esv3585141 duplication, which involves nongenic sequences only (tag-variant: rs74865018 [R2 = 0.5, A-allele is associated with the duplication]), is associated with skin color in GeneATLAS Phenome-Wide Association Study database (P-value = 1.3E–09). However, we have not found a significant association with the expression levels of any neighboring genes based on our search in the GTEx database.
Our previous research has shown that copy number variants, including gene duplications, may be important factors in shaping skin/hair phenotypes (Eaaswarkhanth et al. 2014, 2016; Pajic et al. 2016). Indeed, we found that one of the haplotypes in our study harbors the whole gene duplication of the KRT34 (RefSeq: NM_021013), a member of the keratin gene family, which is important for hair phenotypes and is shown to be affected by copy number variation. This whole gene duplication is common in African populations, but not observed in Eurasian populations (table 1). Gene expression of KRT34 in human hair follicles is higher in young individuals than that in old individuals (Giesen et al. 2011). On the basis of GTEx data (Lonsdale et al. 2013), we demonstrate that the haplotype harboring the duplication led to an increase in the dosage of the KRT34 expression (supplementary fig. S10, Supplementary Material online). Interestingly, this duplication shows high linkage disequilibrium (R2 = 0.80) with the adjacent deletion of the KRT33B (esv3640584, chr17: 39506753-39525903), which may suggest that KRT34 replaced KRT33B through gene conversion. Overall, resolving the haplotypes harboring gene duplications provide a powerful framework to further scrutinize the functional impact, if any, of these variants. Our observations involving the highlighted genes provide candidates for future evolutionary and biomedical studies.
Discussion
One of the major questions in the population genetics is the impact of polymorphic duplications on phenotypic variation and the evolutionary consequences of this impact. Only a few studies have investigated associations between polymorphic duplications to phenotypic variation (Stranger et al. 2007; Wellcome Trust Case Control Consortium et al. 2010; Yang et al. 2015). Similarly, standard population genetics tools are often designed for analysis of SNV and thus cannot directly be applied to scrutinize the evolution of duplications (Iskow et al. 2012). To resolve these problems, we first identified 22 common polymorphic duplications that show linkage disequilibrium with SNVs across the human genome (table 1). By investigating the SNVs, we were able to investigate the evolutionary trajectories of the haplotypes harboring the duplications. On the basis of such analysis, we here present multiple lines of evidence that the haplotype harboring the partial HERC2 gene duplication is selected against in European populations. We found that this haplotype affects the expression level of HERC2P9 significantly, even though the exact phenotype that is under selection is not clear. This methodology enabled us to resolve the haplotypes that harbor these duplications. Similarly, using SNV information, we were able to associate two of these haplotypes to skin color and monocyte percentage. Given that these duplications are major mutation events potentially affecting thousands of base pairs, it is likely that they are the causal variants that affect these phenotypes.
We argue that as more complete variation data sets and accompanying databases with expression and phenotype data become available, the haplotype level analysis of gene duplications, in particular, and structural variation, in general, will become more commonplace. Our study represents a first step in integrating multiple data types to understand the evolutionary impact of gene duplications. It is important to note here that we did not find high linkage disequilibrium between some polymorphic duplications and tag SNVs (table 1), reducing the statistical power of imputation of these duplications in both genome-wide association studies and evolutionary inquiries. As Hong and Park (2012) demonstrated that the statistical power to detect an association largely depends on the strength of the linkage disequilibrium between the casual and tag variants. We believe that this is a major issue that leads to a general underappreciation of the biomedical and evolutionary impact of structural variation. Eventually direct genotyping of duplications will be a more straightforward and statistically powerful way of conducting such associations and understanding the evolutionary trends that underlie these variations.
Materials and Methods
The Linkage Disequilibrium Based Detection of Duplications
We modified VCFtools (0.1.16) (Danecek et al. 2011) to calculate the R2 between a target duplication and other variants in a genome-wide manner. We first made a custom genome-wide VCF file from 1000 Genomes phase 3 data set for CEU, YRI, and CHB population. We conducted population-specific analyses to increase the sensitivity of linkage disequilibrium. To reduce file size, we omitted variants which were not observed in the population of interest. Then we calculated the R2 between a target duplication and other variants in a genome-wide manner with VCFtools (0.1.16). We visualized linkage disequilibrium by using R qqman package (fig. 1B). To ensure the accuracy of these haplotypes, we manually verified the informative variants in the Integrated Genome Browser (Thorvaldsdóttir et al. 2013). For example, we verified one insertion–deletion polymorphism that is in strong linkage disequilibrium (R2 = 0.75) with a duplication (esv3635993), which provides a clear example of a likely true-positive variant calling in this region tagging the duplication polymorphism (supplementary fig. S8, Supplementary Material online). To identify the haplotype block that likely harbor the duplicated HERC2 sequence, we set the threshold R2 value as 0.75 in the Europeans and defined the putative insertion region “hg19 chr15: 28894038-28927368” accordingly. We conducted all the downstream analysis with these coordinates. To avoid analysis complications, we did not consider two SNVs (hg19 chr15: 28553017 and hg19 chr15: 28971921) as they are relatively distant outliers from the observed haplotype block (supplementary fig. S4, Supplementary Material online).
The Detection of Genic/Exonic Duplications
We used NCBI RefSeq track on UCSC Genome Table Browser (Last accessed March 21, 2019) to get the gene and exon information. By using Bedtools (v2.27.1) intersect (Quinlan and Hall 2010), we counted the number of duplications that overlap with 1) entire genes, 2) entire exons, 3) more than one base pair of a gene (including introns). Note that none of the 22 duplications that we scrutinized here partially overlap with a coding exon, that is, if a duplication overlap with a coding sequence, it contains at least one entire exon (table 1). The gene functions listed in supplementary table S1, Supplementary Material online are based on the genetic associations in GeneATLAS (Canela-Xandri et al. 2018).
We found one transchromosomal duplication, esv3631000, which contains ZNF664. To verify this, we checked all the 19 SNVs that have strong linkage disequilibrium (>0.8) with this duplication. We found that 17 of those clusters in the 50 kb region on chr2 (hg19, chr2: 3918719-3970271). The other two SNVs actually overlap with the original duplicated copy on chromosome 12. We thought that these may be false positive calls due to misalignment of the reads originating from the duplicated copy onto the original gene. If this is the case, we expect that the mapped reads to have a ⅓ ratio in a sample where there is a heterozygous duplication. We also expect that these SNVs are called as heterozygous in all cases. Indeed, we found that 235 out of 2,504 individuals are heterozygous for both of these two SNVs (rs80197353, rs78005948) and no homozygous variants were documented. Furthermore, we manually inspected these SNVs using exome data from 1,000 Genomes data set and found that reads carrying the nonreference alleles were found in ∼⅓ of the reads for both SNVs. This is not consistent with the expected 50–50 ratio for heterozygous variant calls (supplementary fig. S9, Supplementary Material online). Collectively, our analysis suggests that the SNVs on chromosome 2 are likely false positive variant calls due to erroneous read mapping and that the duplication insertion site is indeed on chromosome 12.
Getting Random Control Regions
To obtain random SNVs which match our initial filtering process for polymorphic duplications (>5% in CEU, YRI, or CHB), we first used bedtools (v2.27.1) (Quinlan and Hall 2010) for constructing random chromosomal coordinates. We then applied the random chromosomal coordinates to the 1000 Genome Project phase 3 data set variants (Sudmant et al. 2015) and used Vcftools (0.1.16) (Danecek et al. 2011) to retrieve the allele frequency information. We finally used 3,000 SNVs for the comparison between duplicated regions and random SNVs (fig. 2A). In a similar way, we used all the available coordinates on chromosome 15, on which the HERC2 is located, for the neutrality test on the selection browser (Pybus et al. 2014).
Population Genetics Analyses on the HERC2 Duplication
To increase the sensitivity and confirm the initial linkage disequilibrium calculation, we extended the linkage disequilibrium analysis on the original HERC2 gene—putative HERC2 duplication region (hg19, chr15: 28894038-28927368) from CEU to all the available European populations (Utah residents with Northern and Western European ancestry [CEU], Toscani in Italy [TSI], Finnish in Finland [FIN], British in England and Scotland [GBR], Iberian populations in Spain [IBS]), YRI to all the available African populations (Gambian in Western Division, The Gambia [GWD], Mende in Sierra Leone [MSL], Esan in Nigeria [ESN], Yoruba in Ibadan, Nigeria [YRI], Luhya in Webuye, Kenya [LWK]), CHB to all the available East Asian populations (Han Chinese in Beijing, China [CHB], Japanese in Tokyo, Japan [JPT], Southern Han Chinese, China [CHS], Chinese Dai in Xishuangbanna, China [CDX], Kinh in Ho Chi Minh City, Vietnam [KHV]). We observed similar peaks in all populations in hg19 chr15: 28894038-28927368 (fig. 2C, supplementary fig. S4, Supplementary Material online).
To visualize the geographic distribution of the HERC2 duplication allele before recent human migrations, we used data from 15 populations in the 1000 Genome Project: BEB, CDX, CHB, ESN, FIN, GBR, GWD, IBS, JPT, KHV, LWK, MSL, PJL, TSI, and YRI, which have not experienced recent population admixture or migration (fig. 2B). We used the “rworldmap” package (South 2011).
Neutrality Tests
Tajima’s D (Tajima 1993) and XP-EHH (Sabeti et al. 2007) values were downloaded from the 1000 Genomes selection browser (Pybus et al. 2014) for the bins containing the target region (hg19 chr15: 28894038-28927368) and control region (all the available 26,283 3 kb regions across the chromosome 15).
Haplotype Network Analysis
To draw the haplotype networks, we first converted the target region vcf file (chr15: 28898098-28902929) from the 1000 Genome Project phase 3 data set and hg19 reference genome to a fasta file by VCTtoTree (V3.0.0) (Xu et al. 2017). We also included the chimpanzee genome sequence (The Chimpanzee Sequencing Consortium 2005). We manually checked informative alleles in Neanderthal and Denisovan genomes (Reich et al. 2010; Prüfer et al. 2014). We used PopART (Version 1.7) (Leigh and Bryant 2015) for the visualization.
Association Analysis
To assess the phenotypic effect of the polymorphic duplications, we searched the tag SNV of the polymorphic duplications (supplementary table S1, Supplementary Material online). Then we searched the tag SNVs on GeneATLAS phewas (http://geneatlas.roslin.ed.ac.uk/phewas/). However, since GeneATLAS is based on the UK population specifically, neither the East-Asian specific variants nor African-specific variants are included in the data set. We used the nominal P-value 10−8 as a threshold of significant association using the GeneATLAS phewas. Given that we are investigating the association between SNVs and 778 traits, this significance threshold can be considered conservative. If the tag variants are not reported in the GeneATLAS database, we reported them as NA and if no significant phenotype association was found, we described these as NS.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study is supported by OG’s funds from National Science Foundation Grant # 1714867. M.S. is funded by Astellas Foundation for Research on Metabolic Disorders. We would like to thank Izzy Starr, Recep Ozgur Taskent, Dr Rebecca Torene Iskow, and Dr Yoko Satta for careful reading of this manuscript.
Literature Cited
- Abyzov et al. 2013Analysis of variable retroduplications in human populations suggests coupling of retrotransposition to cell division. Genome Res. 23:2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE.. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet. 12(5):363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, et al. 2019. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 176(3):663–675.e19. doi: 10.1016/j.cell.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Boettger LM, et al. 2016. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet. 48(4):359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela-Xandri O, Rawlik K, Tenesa A.. 2018. An atlas of genetic associations in UK Biobank. Nat Genet. 50(11):1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464(7289):704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, et al. 2017. Loci associated with skin pigmentation identified in African populations. Science 358. doi:10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273(5283):1856–1862. [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M, et al. 2016. Atopic dermatitis susceptibility variants in Filaggrin Hitchhike Hornerin Selective Sweep. Genome Biol Evol. 8(10):3240–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M, Pavlidis P, Gokcumen O.. 2014. Geographic distribution and adaptive significance of genomic structural variants: an anthropological genetics perspective. Hum Biol. 86(4):260–275. [DOI] [PubMed] [Google Scholar]
- Eiberg H, et al. 2008. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 123(2):177–187. [DOI] [PubMed] [Google Scholar]
- Fernández CI, Wiley AS.. 2017. Rethinking the starch digestion hypothesis for AMY1 copy number variation in humans. Am J Phys Anthropol. 163(4):645–657. [DOI] [PubMed] [Google Scholar]
- Giesen M, et al. 2011. Ageing processes influence keratin and KAP expression in human hair follicles. Exp Dermatol. 20(9):759–761. [DOI] [PubMed] [Google Scholar]
- Handsaker RE, et al. 2015. Large multiallelic copy number variations in humans. Nat Genet. 47(3):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34(Database issue):D590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EP, Park JW.. 2012. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 10(2):117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inchley CE, et al. 2016. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci Rep. 6:37198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, Gokcumen O, Lee C.. 2012. Exploring the role of copy number variants in human adaptation. Trends Genet. 28(6):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, et al. 2008. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet. 82(2):411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, et al. 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 143(5):837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337(6097):967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, et al. 2017. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356(6343): eaam6393. doi:10.1126/science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D.. 2015. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116. [Google Scholar]
- Levy-Sakin M, et al. 2019. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun. 10(1):1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, et al. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, Ting CN.. 1993. The remarkable evolutionary history of the human amylase genes. Crit Rev Oral Biol Med. 4(3–4):503–509. [DOI] [PubMed] [Google Scholar]
- Mills RE, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470(7332):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narzisi G, Schatz MC.. 2015. The challenge of small-scale repeats for indel discovery. Front Bioeng Biotechnol. 3:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, et al. 2018. Amylase copy number analysis in several mammalian lineages reveals convergent adaptive bursts shaped by diet. bioRxiv 339457. doi:10.1101/339457. [Google Scholar]
- Pajic P, Lin Y-L, Xu D, Gokcumen O.. 2016. The psoriasis-associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence. BMC Evol Biol. 16(1):265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, et al. 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42(Database issue):D903–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, et al. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, et al. 2006. Global variation in copy number in the human genome. Nature 444(7118):444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468(7327):1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2005. The case for selection at CCR5-Delta32. PLoS Biol. 3:1963–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Satta Y, Gokcumen O, Ishida T.. 2018. Complex evolution of the GSTM gene family involves sharing of GSTM1 deletion polymorphism in humans and chimpanzees. BMC Genomics 19:293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, et al. 2016. Comprehensive identification of sexually dimorphic genes in diverse cattle tissues using RNA-seq. BMC Genomics 17:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slentz-Kesler KA, Hale LP, Kaufman RE.. 1998. Identification and characterization of K12 (SECTM1), a novel human gene that encodes a Golgi-associated protein with transmembrane and secreted isoforms. Genomics 47(3):327–340. [DOI] [PubMed] [Google Scholar]
- South A. 2011. rworldmap: a new R package for mapping global data. R J. 3:35–43. [Google Scholar]
- Stranger BE, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315(5813):848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RA, et al. 2008. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 82(2):424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135(2):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium et al. 2015. A global reference for human genetic variation. Nature 526:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollger MR, et al. 2019. Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. doi: https://doi.org/10.1101/635037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO.. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 14(2):125–138. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium et al. 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jaber Y, Pavlidis P, Gokcumen O.. 2017. VCFtoTree: a user-friendly tool to construct locus-specific alignments and phylogenies from thousands of anthropologically relevant genome sequences. BMC Bioinformatics 18(1):426.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z-M, et al. 2015. The roles of AMY1 copies and protein expression in human salivary α-amylase activity. Physiol Behav. 138:173–178. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR.. 2009. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 10:451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wang Q, Wang Q, Jia P, Zhao Z.. 2013. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 14(Suppl 11):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.