Abstract
Background
Lymph‐vascular space invasion (LVSI) is an unfavorable prognostic factor in cervical cancer. Unfortunately, there are no current clinical tools for the preoperative prediction of LVSI.
Purpose
To develop and validate an axial T1 contrast‐enhanced (CE) MR‐based radiomics nomogram that incorporated a radiomics signature and some clinical parameters for predicting LVSI of cervical cancer preoperatively.
Study Type
Retrospective.
Population
In all, 105 patients were randomly divided into two cohorts at a 2:1 ratio.
Field Strength/Sequence
T1 CE MRI sequences at 1.5T.
Assessment
Univariate analysis was performed on the radiomics features and clinical parameters. Multivariate analysis was performed to determine the optimal feature subset. The receiver operating characteristic (ROC) analysis was performed to evaluate the performance of prediction model and radiomics nomogram.
Statistical Tests
The Mann–Whitney U‐test and the chi‐square test were used to evaluate the performance of clinical characteristics and LVSI status by pathology. The minimum‐redundancy/maximum‐relevance and recursive feature elimination methods were applied to select the features. The radiomics model was constructed using logistic regression.
Results
Three radiomics features and one clinical characteristic were selected. The radiomics nomogram showed favorable discrimination between LVSI and non‐LVSI groups. The AUC was 0.754 (95% confidence interval [CI], 0.6326–0.8745) in the training cohort and 0.727 (95% CI, 0.5449–0.9097) in the validation cohort. The specificity and sensitivity were 0.756 and 0.828 in the training cohort and 0.773 and 0.692 in the validation cohort.
Data Conclusion
T1 CE MR‐based radiomics nomogram serves as a noninvasive biomarker in the prediction of LVSI in patients with cervical cancer preoperatively.
Level of Evidence: 4
Technical Efficacy: Stage 2
J. Magn. Reson. Imaging 2019;49:1420–1426.
Keywords: radiomics nomogram, cervical cancer, lymph‐vascular space invasion, MRI, prediction model
CANCER OF THE CERVIX, accounting for almost 12% of all female cancers, is the fourth most common cancer. There were an estimated 528,000 new cases and 266,000 deaths each year, making it the second leading cause of cancer death among women in developing countries, including China.1
Lymph‐vascular space invasion (LVSI) is defined as the presence of carcinoma cells within the lymphatic and/or blood vessels,2 and is a crucial step in the dissemination of tumor cells.3 LVSI is now widely accepted as the major high‐risk factor.4, 5 More fertility‐sparing operations could be applied to treat nulliparas with early‐stage cervical cancer6, 7, 8 if the presence of LVSI could be determined before surgery.
Magnetic resonance imaging (MRI) has been an essential part in diagnosing and staging of cervical carcinoma and assists in determining the tumor size, location, degree of invasion into adjacent organs, and lymph node metastasis.9 “Radiomics” refers to the process of converting medical images into high‐dimensional data from the high‐throughput extraction of quantitative imaging features, and subsequently analyzes the data for decision‐making.10, 11, 12 Objective and quantitative imaging descriptors can improve the accuracy in diagnosis, evaluation of prognosis, and prediction of therapeutic response.10 According to previous studies, radiomic mineable quantitative data could serve as predictive or prognostic biomarkers.13, 14, 15, 16 In the field of oncology, especially in lung cancer, radiomics is mostly used to facilitate the improvement diagnostic accuracy as well as making treatment decisions.16, 17, 18, 19 Therefore, the objective of our study was to build and evaluate an MR‐based radiomics nomogram that incorporates a radiomics signature and clinical parameters for preoperatively predicting the LVSI of cervical cancer.
Materials and Methods
Patients
This retrospective analysis of imaging data was approved by the Institutional Ethics Committee. Informed consent was not required for this study. A total of 105 patients with cervical cancer treated between December 2013 and September 2017 were enrolled. All patients met the following inclusion criteria: 1) pathologically confirmed cervical cancer; 2) pretreatment evaluation of 1.5T MRI scan (Signa HDxt; GE Medical Systems, Milwaukee, WI); 3) preoperative contrast‐enhanced T1‐weighted MRI 1 month before cervical biopsy; and 4) availability of clinical characteristics, such as age, red blood cell (RBC) count, white blood cell (WBC) count, platelet (PLT) count, alkaline phosphatase (ALP), squamous cell carcinoma antigen (SCC), cancer antigen 125 (CA‐125), cancer antigen 199 (CA‐199) and carcinoembryonic antigen (CEA). Patients were excluded from this study for the following reasons: i) history of preoperative therapy (neoadjuvant chemotherapy or radiotherapy); ii) absence of preoperative contrast‐enhanced MR in this hospital; iii) diagnosed with other tumor diseases at the same time; and iv) incomplete clinical data. Finally, 105 patients were included in our study. We divided these patients into two independent cohorts: The training cohort constituted of 70 patients treated between December 2013 and November 2016, whereas the validation cohort consisted of 35 patients treated between November 2016 and September 2017, and the ratio of training cohort to validation cohort was 2:1.
MRI Acquisition and Segmentation
For image segmentation and feature selection, axial contrast‐enhanced T1‐weighted Digital Imaging and Communications in Medicine (DICOM) images that had been archived in the Picture Archiving and Communication System (PACS) (Winning Soft 2.0, Shanghai, China) before any preprocessing or standardization were used. MRI of the abdomen and pelvis was acquired during the routine clinical workup using a 1.5T MR system (Signa HDxt; GE Medical Systems) with a pelvic array coil. Axial contrast‐enhanced T1‐weighted images were acquired with repetition time / echo time (TR/TE) = 4.10/1.95 msec, field‐of‐view (FOV) = 400 × 320 mm, number of excitations/averages (NEX) = 0.75, slice thickness = 4 mm, and spacing = 2 mm.
We used ITK‐SNAP (v. 3.6.0; www.itksnap.org;open source software) for segmentation of manual MR images. Regions of interest (ROIs) of tumors were manually segmented by a radiologist (H.W. with 20 years of experience), and a senior radiologist (G.Z. with 27 years of experience in gynecology MRI interpretation) to validate each processed segmentation.
Radiomics Feature Extraction
All feature extraction methods20 were implemented in Python (v. 3.6.5; https://www.python.org/). Since contrast‐enhanced images have some impact on imaging features, in Pyradiomics the image was normalized by centering it at the mean with standard deviation to eliminate the influence of the different ranges of gray values. Radiomic features were extracted from ROIs, including first‐order features, shape‐based features, and texture features. The detailed information about radiomic features is shown in the Supplementary Material. Combined with preoperative clinical parameters, the number of these features could be further reduced during the feature selection process.
Data Analysis
The statistical analysis program was written in the R language (v. 3.5.0; https://www.r-project.org/), and all statistical hypothesis tests were two‐sided.
Feature selection and model construction were only performed on the training cohort, and the validation cohort only for evaluating the model performance. All the cutoff values of receiver operating characteristics (ROCs) were determined by the principle of maximum Youden index.
Feature Selection
Since some of the features were relevant but redundant, we used the minimum redundancy maximum relevance (mRMR) method to select the features. The mRMR method trades off between relevancy and redundancy, and it not only calculates the mutual information between the two features, but also the mutual information between the feature and the label.21 Univariate analysis was used for the retained features; features significantly associated with LVSI status were selected. A Mann–Whitney U‐test was performed on the continuous features, and a chi‐square test was performed on the discrete features. Finally, the recursive feature elimination (RFE) was performed as a multivariate analysis method to select the LVSI‐related features. RFE constantly eliminates unimportant features to obtain the optimal feature set by calculating the importance of features.22
MRI Model and Radiomics Nomogram Construction
To investigate the LVSI status information contained in the MRI, an MRI model was constructed by using only the radiomics features extracted from the MRI. A logistic regression model was constructed to predict the LVSI status by fitting the selected radiomics features.
The radiomics model was constructed by combining the MR‐based radiomic features and clinical parameters. To facilitate the study, a radiomics signature was constructed using a linear combination of selected radiomics features according to the respective coefficient generated by the radiomics model formula.
For convenience of clinical application, a radiomics nomogram was constructed from the logistic regression model to predict the risk of LVSI. The radiomics nomogram is a visual representation of the radiomics model, where both have equal levels of prediction performance. The predictors of LVSI in the radiomics nomogram included the radiomics signature and selected clinical parameters.
Validation of Prediction Model and Radiomics Nomogram
ROC curve analysis was performed to illustrate the model performance in the training cohort and validation cohort. Meanwhile, we calculated the area under the curve (AUC), specificity, and sensitivity to further evaluate the performance of the MRI model and radiomics nomogram. To determine the clinical value of the radiomics nomogram, decision curve analysis (DCA) was performed by quantifying the net benefits for a range of threshold probabilities in the whole cohort.
Results
Patient Characteristics
Clinical characteristics of patients are given in Table 1. There was no significant difference between the training and validation cohorts (P = 0.2908, χ2 test), with LVSI positivity 38.7% and 43.3%., respectively.
Table 1.
Characteristics of Patients in the Training and Validation Cohorts
| Training cohort (n = 70) | Validation cohort (n = 35) | |||||
|---|---|---|---|---|---|---|
| Characteristic | LVSI(+) (n = 29) | LVSI(‐) (n = 41) | P | LVSI(+) (n = 13) | LVSI(–) (n = 22) | P |
| Age, mean ± SD, years | 49.34 ± 8.55 | 46.54 ± 9.66 | 0.387 | 54.54 ± 10.42 | 48.59 ± 10.69 | 0.194 |
| WBC, mean ± SD, × 109/L | 6.03 ± 0.96 | 6.64 ± 1.86 | 0.211 | 6.09 ± 2.83 | 6.57 ± 3.19 | 0.408 |
| RBC, mean ± SD, × 1012/L | 4.02 ± 0.47 | 4.30 ± 0.59 | 0.002* | 3.96 ± 0.38 | 4.16 ± 0.66 | 0.028* |
| PLT, mean ± SD, × 109/L | 253.62 ± 93.16 | 253.84 ± 75.15 | 0.587 | 216.00 ± 62.77 | 236.77 ± 80.88 | 0.413 |
| ALP, mean ± SD, U/L | 78.57 ± 27.33 | 78.69 ± 21.50 | 0.971 | 75.38 ± 23.80 | 78.05 ± 26.61 | 0.973 |
| SCC, No (%) | 0.800 | 0.204 | ||||
| Normal | 11(37.9) | 18(43.9) | 4(30.8) | 13(59.0) | ||
| Abnormal | 18(62.1) | 23(56.1) | 9(69.2) | 9(41.0) | ||
| CA‐125, No (%) | 0.081 | 0.987 | ||||
| Normal | 24(82.8) | 40(97.6) | 11(84.7) | 20(90.9) | 0.987 | |
| Abnormal | 5(17.2) | 1(2.4) | 2(15.3) | 2(9.1) | ||
| CA‐199, No (%) | 0.684 | 1.000 | ||||
| Normal | 26(89.7) | 39(95.1) | 12(92.3) | 19(86.4) | ||
| Abnormal | 3(10.3) | 2(4.9) | 1(7.7) | 3(13.6) | ||
| CEA, No (%) | 0.366 | 1.000 | ||||
| Normal | 24(82.8) | 38(92.7) | 12(92.3) | 19(86.4) | ||
| Abnormal | 5(17.2) | 3(7.3) | 1(7.7) | 3(13.6) | ||
P value was derived from the univariate association analyses between each clinical parameter and LVSI status.
LVSI, lymph‐vascular space invasion; WBC, white blood cell; RBC, red blood cell; PLT, platelet ;ALP, alkaline phosphatase; SCC, squamous cell carcinoma antigen; CEA, carcinoembryonic antigen; SD, standard deviation.
P < 0.05.
Radiomics Feature Extraction and Selection
The original feature set for the radiomics model included nine clinical parameters and 1392 radiomics features extracted from T1 CE MR images, and was constructed further by selecting the upcoming installment. The original feature set for the MRI model contained only 1392 radiomics features. First, the mRMR method was performed to remove the relevant and redundant features, and the top 1% features remained. In univariate analysis, P < 0.05 was considered to indicate a statistically significant difference. After performing the RFE method, three radiomics features and one clinical parameter were finally selected to fit the logistic regression model (Fig. 1). Note that the output of the model represents the LVSI risk score. If the risk score is greater than zero, the model will consider the corresponding case to be LVSI‐positive.
Figure 1.
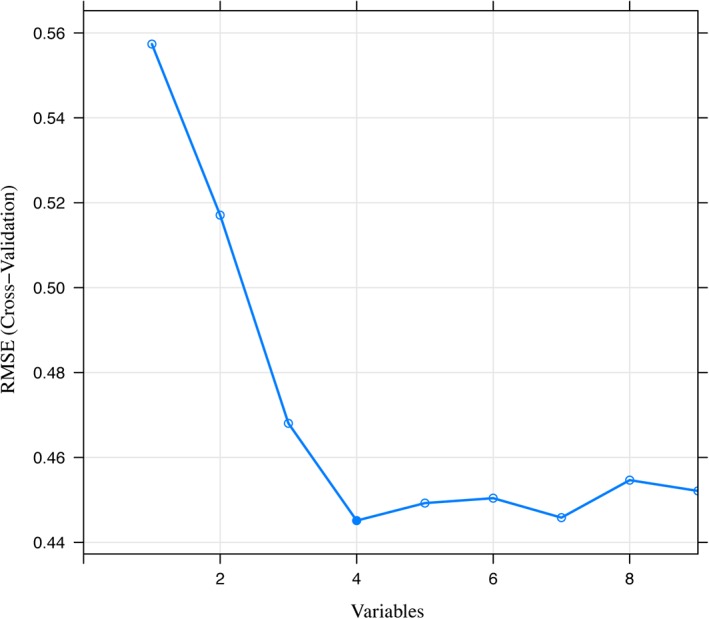
The feature selection process of the RFE method. Each iteration removes a feature that is considered least important and corresponds to a 10‐fold cross‐validation. After 10‐fold cross‐validation, the RMSE of the model in the training cohort was used to select the optimal feature set. Finally, four features were selected by the RFE method. RFE, recursive feature elimination; RMSE, root mean square error.
Development of Radiomics Signature and Radiomics Nomogram
The radiomics nomogram was plotted for clinical use based on the formula (Fig. 2) generated from the radiomics model. The formula was as follows:
Figure 2.
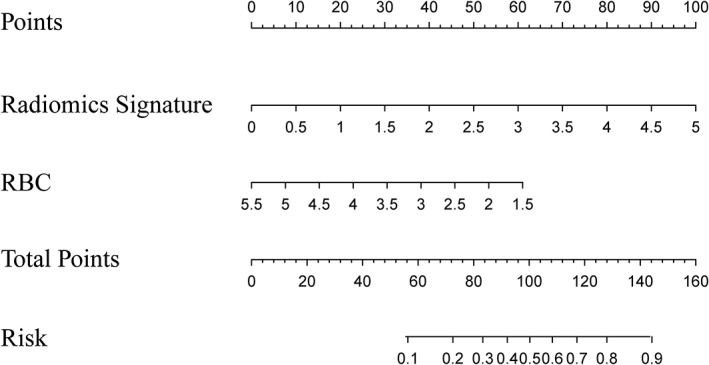
A radiomics nomogram integrated the radiomics signature from axial T1 contrast‐enhanced images with the RBC from complete blood count in the training cohort. The value of each predictor can be converted into a risk score according to the “Points” at the top of the nomogram. After adding up the individual risk score of these predictors in “Total Points,” the corresponding prediction probability at the bottom of the nomogram is the LVSI. The cutoff value in this nomogram is 0.606. The case would be diagnosed as LVSI when the total prediction probability is beyond the cutoff value. RBC, red blood cell.
As mentioned above, the radiomics signature consists of the above three radiomics features, and the formula after combining the radiomics signature was as follows:
where:
Diagnostic Validation of the MRI Model and Radiomics Nomogram
The MRI model showed a degree of prediction performance of LVSI status (Fig. 3A,B), which reached an AUC of 0.710 (95% confidence interval [CI], 0.5864–0.8333) in the training cohort, with a sensitivity of 0.854 and a specificity of 0.552, and the AUC of 0.633 (95% CI, 0.4401–0.8256) in the validation cohort, with a sensitivity of 0.818 and a specificity of 0.231.
Figure 3.
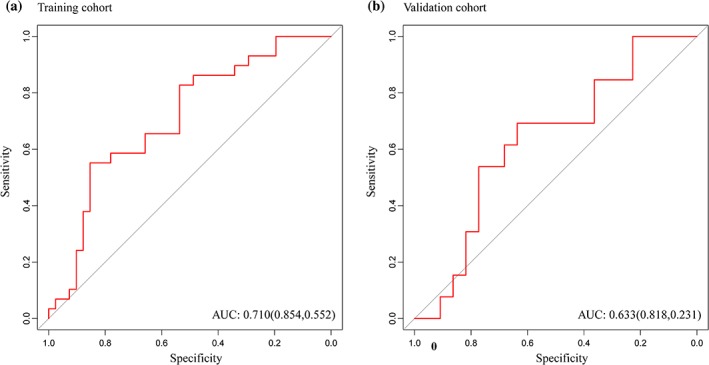
(A) MRI model reached AUC of 0.710 in the training cohort, with a sensitivity of 0.854 and a specificity of 0.552, and (B) the AUC of 0.633 in the validation cohort, with a sensitivity of 0.818 and a specificity of 0.231. AUC: area under the receiver operating characteristic curve.
The radiomics nomogram showed good forecasting ability, with an AUC of 0.754 (95% CI, 0.6326–0.8745] in the training cohort and 0.727 (95% CI, 0.5449–0.9097) in the validation cohort (Fig. 4A,B). The specificity and sensitivity were 0.756 and 0.828 in the training cohort and 0.773 and 0.692 in the validation cohort.
Figure 4.
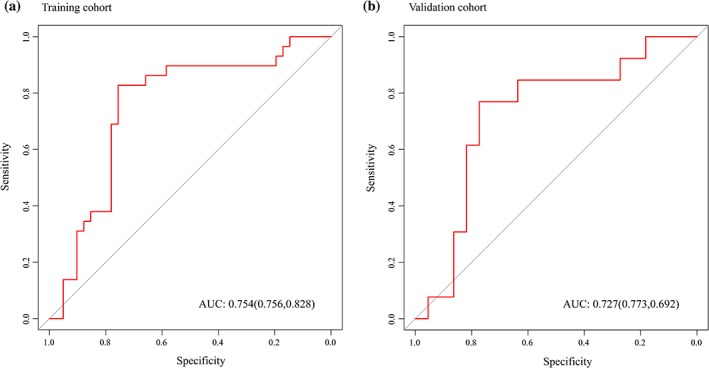
(A) Radiomics nomogram reached the highest AUC of 0.754 in the training cohort, with a sensitivity of 0.756 and a specificity of 0.828, and (B) the highest AUC of 0.727 in the validation cohort, with a sensitivity of 0.773 and a specificity of 0.692. AUC: area under the receiver operating characteristic curve.
The decision curve23 also showed favorable performance of the radiomics nomogram (Fig. 5). This reflected greater benefit for the cervical cancer patient cohort by radiomics nomogram in the prediction of LVSI if the threshold probability of patients or doctors was greater than 0.23.
Figure 5.
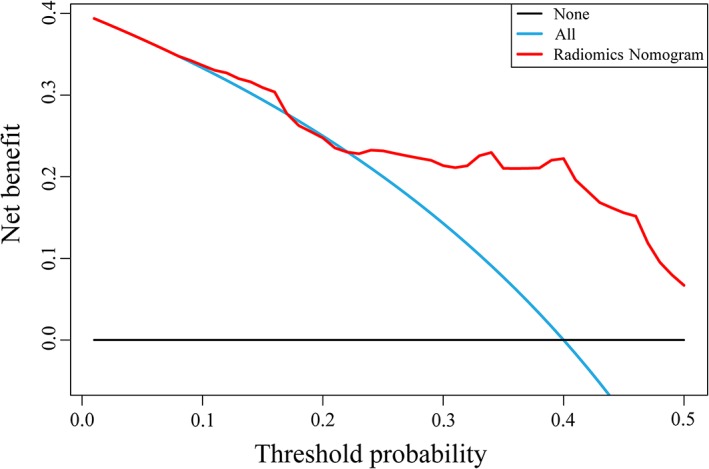
Decision curve analysis for the radiomics nomogram. On the horizontal axis is the net benefit. The threshold probability is on the vertical axis. The blue line represents the assumption that all patients have LVSI. The black line represents the assumption that no patients have LVSI. The red line represents the radiomics nomogram. LVSI, lymph‐vascular space invasion.
Discussion
LVSI is an unfavorable prognostic factor in cervical cancer4, 5 and remains a crucial element in the application of fertility‐sparing operations.24, 25, 26 According to the National Comprehensive Cancer Network Guidelines V. 1.2019 Cervical Cancer, primary treatment is different for clinical stage IA1 patients with or without LVSI.27 Moreover, following primary hysterectomy the presence of LVSI may warrant the use of adjuvant radiotherapy. Accordingly, the development of noninvasive biomarkers with the potential to provide prediction of LVSI preoperatively in patients with cervical cancer is urgent. There has been no previous report about the preoperative prediction of LVSI in cervical cancer.
Therefore, in the current study we considered the clinical parameters and attempted to develop an MR‐based radiomics nomogram and assessed its ability for predicting LVSI preoperatively in patients with cervical cancer. Our radiomics nomogram showed favorable discrimination, with high AUCs in the training cohort validation cohort. In addition, with the addition of clinical parameters in the combined model, the predictive performance was improved, especially in terms of specificity, indicating that the radiomics features have the potential to predict performance with MRI. Also, the combination of a clinical parameter (RBC) and MR‐based radiomics features contributed to improve the predictive performance. Moreover, negative correlations were found between RBC and LVSI, suggesting that LVSI may lead to chronic blood loss, and requires further consideration and exploration. We also provided an easy‐to‐use tool for clinicians as well as patients by establishing a radiomics nomogram based on the multivariate logistic regression model that showed a favorable standardization and discrimination in both the training and validation datasets. In the validation cohort, the prediction accuracy of the combined radiomics nomogram is 0.74, while the accuracy of the MRI model and RBC is only 0.60 and 0.54, respectively, which demonstrated that the nomogram achieved better predictive efficacy than either the MR‐based radiomics features or RBC alone. The decision curve analysis of the radiomics nomogram in a substantial range of threshold probability demonstrated greater benefit for the cervical cancer patient cohort, compared with the unpredictable situation of LVSI at present.
The selected features in the radiomics nomogram were the first‐order statistics and shape‐based features, which are explained in detail in the Supplementary Materials. This reflected that the gray level changes and the shape of the tumor boundaries, as shown in the MRI, are very important for predicting LVSI status. But they are too difficult to quantify for a clinical user, and hence it was almost impossible to discriminate the LVSI status before our study. Therefore, the objectivity and the accuracy of the combination of MR‐based radiomics and clinical parameters are reflected in this study, and remain promising for more MRI studies.
Currently, neoadjuvant chemotherapy (NACT) can reduce the tumor volume preoperatively, increase the resection rate,28 and make the LVSI shrink or even disappear.29 Several research studies have proved the ability of NACT in improving the prognosis of cervical cancer.30, 31 However, controversy regarding the NACT should be further investigated. A systematic review of 21 studies failed to confirm the conclusion that NACT before radical surgery (RS) produced a survival benefit.32 Recently, some investigations comparing NACT + RS with RS alone have been reported, but the results are inconsistent.33, 34, 35, 36 Therefore, the lack of techniques before NACT to determine the existence of LVSI led us to underestimate the role of NACT in the improvement of prognosis. Next, ongoing multicenter prospective randomized clinical trials are required to more objectively evaluate the effect of NACT in patients with cervical cancer by applying the MRI‐based radiomics nomogram for the prediction of LVSI before NACT.
There are a few limitations to our study, including the fact that genomic characteristics have not been incorporated in our nomogram. In recent years, genetic polymorphism of C‐reactive protein (CRP) 1846C>T was associated with severe LVSI in endometrial cancer37 and a high DLL4 protein level correlated with LVSI in early‐stage cervical cancer38 has been reported. In addition, the lack of external validation for the model, the relatively small sample size, and inherent biases in retrospective studies should also be considered. It is necessary to further conduct a multicenter validation with a larger sample size to provide favorable evidence for clinical application.
In conclusion, this study presented a radiomics nomogram, incorporating both the radiomics signature as well as clinical parameters, as a noninvasive biomarker that can predict LVSI in patients with cervical cancer preoperatively. However, further retrospective and even prospective validation analysis should be conducted to confirm its predictive properties in subsequent studies.
Conflict of Interest
The authors declare no potential conflicts of interest.
Supporting information
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3
Contributor Information
Siwen Wang, Email: wsf_sfph@sjtu.edu.cn.
Jie Tian, Email: jie.tian@ia.ac.cn.
Sufang Wu, Email: wsf_sfph@sjtu.edu.cn.
Han Wang, Email: han.wang@shsmu.edu.cn.
References
- 1. Koh WJ, Greer BE, Abu‐Rustum NR, et al. Cervical Cancer, Version 2.2015. J Natl Compreh Cancer Netw 2015;13:395–404; quiz 404. [DOI] [PubMed] [Google Scholar]
- 2. Padera TP, Kadambi A, di Tomaso E, et al. Lymphatic metastasis in the absence of functional intratumor lymphatics. Science 2002;296:1883–1886. [DOI] [PubMed] [Google Scholar]
- 3. Kikuchi E, Margulis V, Karakiewicz PI, et al. Lymphovascular invasion predicts clinical outcomes in patients with node‐negative upper tract urothelial carcinoma. J Clin Oncol 2009;27:612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biewenga P, van der Velden J, Mol BW, et al. Prognostic model for survival in patients with early stage cervical cancer. Cancer 2011;117:768–776. [DOI] [PubMed] [Google Scholar]
- 5. Yu Q, Lou XM, He Y. Prediction of local recurrence in cervical cancer by a Cox model comprised of lymph node status, lymph‐vascular space invasion, and intratumoral Th17 cell‐infiltration. Med Oncol 2014;31:795. [DOI] [PubMed] [Google Scholar]
- 6. Vranes B, Milenkovic S, Radojevic M, Soldatovic I, Kesic V. Risk of parametrial spread in small stage I cervical carcinoma: pathology review of 223 cases with a tumor diameter of 20 mm or less. Int J Gynecol Cancer 2016;26:416–421. [DOI] [PubMed] [Google Scholar]
- 7. Robova H, Rob L, Halaska MJ, Pluta M, Skapa P. Review of neoadjuvant chemotherapy and trachelectomy: which cervical cancer patients would be suitable for neoadjuvant chemotherapy followed by fertility‐sparing surgery? Curr Oncol Rep 2015;17:446. [DOI] [PubMed] [Google Scholar]
- 8. Nick AM, Frumovitz MM, Soliman PT, Schmeler KM, Ramirez PT. Fertility sparing surgery for treatment of early‐stage cervical cancer: open vs. robotic radical trachelectomy. Gynecol Oncol 2012;124:276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kusmirek J, Robbins J, Allen H, Barroilhet L, Anderson B, Sadowski EA. PET/CT and MRI in the imaging assessment of cervical cancer. Abdom Imaging 2015;40:2486–2511. [DOI] [PubMed] [Google Scholar]
- 10. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016;34:2157–2164. [DOI] [PubMed] [Google Scholar]
- 13. Zhou H, Vallieres M, Bai HX, et al. MRI features predict survival and molecular markers in diffuse lower‐grade gliomas. Neuro Oncol 2017;19:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Y, He L, Huang Y, et al. CT‐based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol 2017;42:1695–1704. [DOI] [PubMed] [Google Scholar]
- 15. Zhang B, Tian J, Dong D, et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res 2017;23:4259–4269. [DOI] [PubMed] [Google Scholar]
- 16. Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease‐free survival in early‐stage (I or II) non‐small cell lung cancer. Radiology 2016;281:947–957. [DOI] [PubMed] [Google Scholar]
- 17. Yip SS, Kim J, Coroller TP, et al. Associations between somatic mutations and metabolic imaging phenotypes in non‐small cell lung cancer. J Nucl Med 2017;58:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res 2017. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiangdian S, Di D, Yanqi H, Yali Z, Zaiyi L, Jie T. Association between tumor heterogeneity and progression‐free survival in non‐small cell lung cancer patients with EGFR mutations undergoing tyrosine kinase inhibitors therapy. Conf Proc IEEE Eng Med Biol Soc 2016;2016:1268–1271. [DOI] [PubMed] [Google Scholar]
- 20. Alcazar JL, Gaston B, Navarro B, Salas R, Aranda J, Guerriero S. Transvaginal ultrasound versus magnetic resonance imaging for preoperative assessment of myometrial infiltration in patients with endometrial cancer: a systematic review and meta‐analysis. J Gynecol Oncol 2017;28:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005;3:185–205. [DOI] [PubMed] [Google Scholar]
- 22. Granitto PM, Furlanello C, Biasioli F, Gasperi F. Recursive feature elimination with random forest for PTR‐MS analysis of agroindustrial products. Chemometr Intell Lab 2006;83:83–90. [Google Scholar]
- 23. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang C, Havrilesky LJ, Broadwater G, et al. Relationship between minimally invasive hysterectomy, pelvic cytology, and lymph vascular space invasion: a single institution study of 458 patients. Gynecol Oncol 2014;133:211–215. [DOI] [PubMed] [Google Scholar]
- 25. Wang W, Shang C, Huang J, Chen S, Shen H, Yao S. [Long‐term oncological outcomes after laparoscopic versus abdominal radical hysterectomy in stage I a2‐ II a2 cervical cancer: a matched cohort study]. Zhonghua fu chan ke za zhi 2015;50:894–901. [PubMed] [Google Scholar]
- 26. Togami S, Kamio M, Kobayashi H, Douchi T. Is it possible to perform less radical surgery for invasive uterine cervical cancer? Gynecol Obstet Invest 2016;81:251–255. [DOI] [PubMed] [Google Scholar]
- 27. NCCN Guidelines Version 1.2019 Cervical Cancer. https://www.nccn.org/professionals/physician_gls/pdf/cervical.pdf
- 28. Ki KD, Song DH, Tong SY, Lim MC, Lee JM, Lee SK. Neoadjuvant chemotherapy in bulky stage IB‐IIA cervical cancer: results of a quick course with vincristine, bleomycin, and cisplatin. Int J Gynecol Cancer 2009;19:50–53. [DOI] [PubMed] [Google Scholar]
- 29. Wang Y, Wang G, Wei LH, et al. Neoadjuvant chemotherapy for locally advanced cervical cancer reduces surgical risks and lymph‐vascular space involvement. Chin J Cancer 2011;30:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghezzi F, Cromi A, Ditto A, et al. Laparoscopic versus open radical hysterectomy for stage IB2‐IIB cervical cancer in the setting of neoadjuvant chemotherapy: a multi‐institutional cohort study. Ann Surg Oncol 2013;20:2007–2015. [DOI] [PubMed] [Google Scholar]
- 31. Vizza E, Corrado G, Zanagnolo V, et al. Neoadjuvant chemotherapy followed by robotic radical hysterectomy in locally advanced cervical cancer: a multi‐institution study. Gynecol Oncol 2014;133:180–185. [DOI] [PubMed] [Google Scholar]
- 32. Neoadjuvant chemotherapy for locally advanced cervical cancer: a systematic review and meta‐analysis of individual patient data from 21 randomised trials. Eur J Cancer 2003;39:2470–2486. [DOI] [PubMed] [Google Scholar]
- 33. Bogani G, Cromi A, Serati M, et al. A prospective case‐control study on the impact of neoadjuvant chemotherapy on surgery‐related outcomes of laparoscopic radical hysterectomy. Anticancer Res 2014;34:5703–5708. [PubMed] [Google Scholar]
- 34. Katsumata N, Yoshikawa H, Kobayashi H, et al. Phase III randomised controlled trial of neoadjuvant chemotherapy plus radical surgery vs radical surgery alone for stages IB2, IIA2, and IIB cervical cancer: a Japan Clinical Oncology Group trial (JCOG 0102). Br J Cancer 2013;108:1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai HB, Chen HZ, Yin HH. Randomized study of preoperative chemotherapy versus primary surgery for stage IB cervical cancer. J Obstet Gynaecol Res 2006;32:315–323. [DOI] [PubMed] [Google Scholar]
- 36. Chen H, Liang C, Zhang L, Huang S, Wu X. Clinical efficacy of modified preoperative neoadjuvant chemotherapy in the treatment of locally advanced (stage IB2 to IIB) cervical cancer: randomized study. Gynecol Oncol 2008;110:308–315. [DOI] [PubMed] [Google Scholar]
- 37. Kito M, Motoyama S, Fujita K, et al. CRP 1846C>T genetic polymorphism is associated with lymph node metastasis and/or severe lymphatic invasion in endometrial cancer. Tohoku J Exp Med 2015;237:25–30. [DOI] [PubMed] [Google Scholar]
- 38. Yang S, Liu Y, Xia B, et al. DLL4 as a predictor of pelvic lymph node metastasis and a novel prognostic biomarker in patients with early‐stage cervical cancer. Tumour Biol 2016;37:5063–5074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3


