Abstract
Background
Overtreatment is a well‐known clinical challenge in local prostate cancer (PCa). Although risk assessment models have contributed to a better stratification of patients with local PCa, a tailored management is still in its infancy. Over the last few decades, microRNAs (miRNAs) have shown promising results as biomarkers in PCa. The aim of this study was to investigate circulating miRNAs after management of local PCa.
Methods
The relative expression of four miRNAs (miRNA‐21, ‐93, ‐125b, and miRNA‐221) was assessed in plasma from 149 newly diagnosed patients with local or locally advanced PCa. Real‐time polymerase chain reaction was used for analysis. A baseline sample at time of diagnosis and a follow‐up sample after 6 months were assessed. The patients were grouped in an interventional cohort (radical prostatectomy, curative intent radiotherapy, or androgen‐deprivation therapy alone) and an observational cohort (watchful waiting or active surveillance).
Results
In the interventional cohort, levels of both miRNA‐93 and miRNA‐221 were significantly lower in the follow‐up samples compared to baseline z = −2.738, P = 0.006, and z = −4.498, P < 0.001, respectively. The same observation was recorded for miRNA‐125b in the observational cohort (z = −2.656, P = 0.008). Both miRNA‐125b and miRNA‐221 were correlated with risk assessment r = 0.23, P = 0.015, and r = 0.203, P = 0.016 respectively, while miRNA‐93 showed tendency to significant correlation with the prostatectomy Gleason score (r = 0.276, P = 0.0576).
Conclusions
The current results indicate a possible role of miRNA‐93 and miRNA‐221 in disease monitoring in localized and locally advanced PCa. Larger studies are warranted to assess the clinical impact of these biomarkers.
Keywords: liquid biopsy, microRNAs, PCR, prostate cancer, risk profile
1. INTRODUCTION
Prostate cancer (PCa) accounts for almost one in every five new non‐melanoma cancer diagnoses in men, and although reduced by more than 50% within the last three decades, PCa is still the second most cancer‐related mortality in men in the Western countries.1, 2
While the incidence of primary metastatic PCa decreased about 20% by the introduction of prostate specific antigen (PSA) in the late 1980's,3 two other clinical challenges emerged, that is, overdiagnosis and overtreatment.4 Therefore, an accurate risk stratification tool at the time of diagnosis is the cornerstone of clinical decision making paving the way for personalized management of PCa.
Since the mid‐1990's there was a plethora of pre‐and post‐treatment predictive models to propose risk assessment of localized PCa of both clinical and biochemical outcome.5 These models depend either completely or partially on PSA, clinical tumor stage (cT), and Gleason score (GS).
Whereas GS is still the best prognostic indicator in PCa management,6 discrepancy between GS in the diagnostic needle biopsies and the prostatectomy specimens,7 inter‐ and intraobserver variability,8 and sampling error9 are among its most common pitfalls in daily practice.
These models are challenged by the nature of PCa as a heterogeneous disease. Patients with high risk PCa have an approximate 50% risk of progression at 5 years10 while 70% of the patients with PCa under the active surveillance strategy (AS) can avoid active treatment during 15 years of follow‐up.11 Moreover, throughout treatment it may also be difficult as well as doubtful to rely on a model based only on the PCa characteristics at the time of diagnosis.
More robust risk assessment tools are therefore needed in the management of PCa and with the advent of bio‐informatics, temporal genomic analysis may now be incorporated into the predictive tools to improve stratification accuracy. Since heterogeneity of PCa tissue is a well‐known clinical challenge in PCa management, liquid biomarkers may represent an alternative diagnostic and prognostic tool in PCa.12
MicroRNAs (miRNAs) are non‐coding single‐stranded evolutionarily conserved RNA molecules comprising 19–24 nucleotides in length that regulate gene expression both at the transcriptional and post transcriptional level.13, 14 Recently, more than 1900 human miRNAs have been identified (www.mirbasae.org, March 2018), their genes being located within protein‐coding genes or in intergenic regions; either alone or in clusters.15 MicroRNAs play a crucial rule in both physiological and pathological cellular processes such as cell cycle regulation,16 development,17 and apoptosis,18 and in cancer development and progression.19 They are quite stable in different biological materials and in various storage conditions.20 They are dysregulated in many type of cancers,21 including PCa22 and available for sampling through a variety of body fluids.23 Consequently, miRNAs have attracted attention as candidates for minimally invasive biomarkers in PCa.
The aim of this study was to illustrate the change of circulating miRNAs after management of newly diagnosed patients with localized/locally advanced PCa and to analyze the correlation between circulating miRNAs and common risk parameters (PSA, GS, cT, and the European Association of Urology (EAU) risk profile24).
2. MATERIAL AND METHODS
2.1. Patients
As described in our previous work (validation cohort),25 all patients were referred to the Department of Urology either at Vejle Hospital or Esbjerg Hospital, Denmark, between September 2015 and May 2017. All participants provided informed written consent.
In total, 149 patients were prospectively included in this study. Patients were referred for further diagnostic evaluation due to high PSA levels with or without lower urinary tract symptoms. All patients were diagnosed with localized/locally advanced adenocarcinoma of the prostate based on transrectal ultrasound biopsy (TRUS) with no clinical evidence of distant metastases on the standard staging radiological examination (bone scan +/− CT scan). The cohort was divided into two groups according to national guidelines26; an intervention group and an observation group. Three different treatment strategies were applied to the intervention group; robot assisted radical prostatectomy (RARP) curative intent radiotherapy (RT), or palliative treatment with androgen‐deprivation therapy (ADT) alone. In the observation group patients were managed either by AS or the watchful waiting (WW) strategy. Baseline plasma samples were collected at the time of diagnosis and approximately 6 months later.
2.2. Blood collection and storage
Two venous blood samples were collected from all patients; baseline (prior to TRUS) and follow‐up (after 6 months). Sampling was performed by skilled phlebotomists using a minimum of venous stasis to prevent hemolysis. Whole blood was collected into 9 mL ethylenediaminetetraacetic acid (EDTA) containing tubes (Becton‐Dickinson, Franklin Lakes, NJ, USA). Platelet‐poor plasma (PPP) was prepared by dual centrifugation within 2 h from blood sampling. The samples were centrifuged at 3000 g for 15 min. and carefully transferred to another tube, leaving approximately 1 mL of plasma on top of the buffy coat. The centrifugation step was repeated and again approximately 1 mL was left at the bottom of the tube when the PPP was transferred into cryo‐tubes and stored at −80°C.
To reduce the risk of hemolysis and diminish the risk of release of miRNAs from other intravascular cell compartments, all blood samples were handled within the first 2 h after sample collection. Also, all samples were evaluated for hemolysis by A414 measure and approx. 85% of all values were ≤ log (abs) = 0.25, which corresponds to less than 1% hemolysis when comparing to recommendations. There were 32 samples (11%) above 0.25 and below 0.5. Evaluation of these samples did not show any trends toward neither higher nor lower Ct values and therefore, they were not excluded. In only 11 samples (4%) the hemolysis index was relatively high (≥ 0.5) and these samples were excluded from further data analysis.
2.3. Selected miRNA targets
In the present study a panel of four microRNAs was selected; miRNA‐21, ‐93, ‐125b, and miRNA‐221. This choice was based on results from our previous work25, 27 and a comprehensive review of the literature (Supplementary Table SI).
2.4. RNA extraction
2.4.1. Plasma samples
Based on the validation cohort from our previous work,25 microRNA purification from PPP was performed using the Maxwell RSC miRNA Tissue Kit (AS1460) verified for plasma use (Scientific Style and Format, 7th edition, 2006). Two hundred microliter plasma was mixed with 200 μL chilled 1‐Thioglycerol/Homogenization Solution and 200 μL lysis solution. After incubation for 10 min, spike‐in Cel‐miRNA‐39 (5′ phosphorylated) was added (final concentration 1 pM) as a purification control and then loaded into the Maxwell RSC Instrument. Elution was done using 60 μL elution buffer.
2.4.2. Reverse transcription, pre‐amplification, and qPCR
All preparations took place according to the standard protocol from Life Technologies, Custom Taqman® Advanced miRNA Assays single tube assays (A25576) and the qPCR instrument Roche Lightcycler®96. In brief, reverse transcription (RT) was performed in three steps, first a poly‐A tailing, then adaptor ligation reaction followed by a universal RT. After RT, we performed targeted pre‐amplification according to protocol and the final cDNA was diluted 10 times before mixing the qPCR reaction. All qPCR reactions were performed in triplicate. Results were analyzed using Roche Lightcycler®96 Software.
We have previously25 established both miRNA‐17 and miRNA‐191 as suitable internal controls, that is, they displayed stable expression levels throughout all previous samples as judged by manual review and according to the Normfinder algorithm.28 Both miRNAs had previously proven suitable as reference RNA targets.29 In this study, however, the commercial miRNA‐191 single assay did not perform satisfactorily and was discarded from further use leaving miRNA‐17 as the internal control miRNA. Samples were distributed across several qPCR plates and in order to facilitate individual comparison between samples on different plates we used plate‐to‐plate variation controls.
2.5. Statistical analysis
Data are presented as means of three technical replicates per plasma sample from each patient. The fold change of plasma miRNA was calculated based on the threshold cycle (Ct) value using the following formula:
Results were linearized using 2−(ΔCT), which was subsequently applied in the statistical analysis.
Samples with missed Ct measurements in either baseline or follow‐up samples were dismissed. However, Ct values above 35 were dismissed only from baseline plasma samples, since a change in miRNAs in plasma was expected in follow‐up samples. A summary of all dismissed values is showed in supplementary Table SII.
Wilcoxon signed rank sum test was used to analyze the difference between plasma miRNA levels in follow‐up and baseline samples. For assessment of the difference between baseline plasma miRNAs in the different groups Wilcoxon rank sum test was applied.
Spearman's rank‐order correlation coefficient (univariate analysis) was used to investigate the correlation between baseline plasma miRNA levels and clinical characteristics (PSA, GS, and the updated EAU risk profile24).
Up/downregulation of plasma miRNA levels in baseline compared with follow‐up samples in both the observational and the interventional groups was illustrated by boxplots.
All analyses were performed in STATA version 15.1 (STATA Corp LLC, TX, USA), and correlations/differences were considered statistically significant when the P‐value was less than 0.05.
3. RESULTS
3.1. Patient characteristics
While mean PSA level was significantly lower, mean prostate volume was significantly higher in the observation cohort compared to the intervention cohort.
The clinicopathological characteristics of all PCa patients are presented in Table 1.
Table 1.
Clinicopathological characteristics of the observational and interventional group
| Interventional cohort n = 68 | Observational cohort n = 81 | P‐value | |
|---|---|---|---|
| Mean age, years (range) | 69 (48–86) | 70 (51–90) | 0.28 |
| Reason for management | |||
| High PSA | 50 (74) | 55 (68) | |
| LUTS + high PSA | 17 (25) | 23 (28) | |
| Accidental | 1 (1) | 3 (4) | |
| Mean prostate volume, cm3 (range) | 41 (20–65) | 53 (11–176) | 0.03 |
| Mean PSA level, ng/mL (range) | 32 (4.5–250) | 10 (2.5–53) | <0.01 |
| GS a | |||
| 6 (3+3) | 5 (7) | 52 (64) | |
| 7 (3+4) | 33 (49) | 20 (25) | |
| 7 (4+3) | 14 (21) | 4 (5) | |
| 8 | 3 (4) | 3 (4) | |
| ≥9 | 13 (19) | 2 (2) | |
| cT | |||
| ≤ cT2b | 47 (69) | 70 (86) | |
| cT2c‐cT3a | 15 (22) | 9 (11) | |
| ≥ cT3b | 6 (9) | 2 (3) | |
| cN stage | |||
| cNx | 43 (63) | 73 (90) | |
| cN0 | 23 (34) | 8 (10) | |
| cN1 | 2 (3) | 0 (0%) | |
| Management | |||
| RARP 36 (53) | AS 52 (64) | ||
| RT+/ − ADT 8 (12) | WW 29 (36) | ||
| ADT 24 (35) | |||
| Death of any cause | 3 (4) | 3 (4) | |
| Another cancer | 5 (7) | 9 (11) |
ADT, androgen deprivation therapy; cT, clinical tumour stage; cN, clinical lymph node stage; GS, Gleason score; LUTS, lower urinary tract symptoms; PSA, prostate specific antigen; RARP, robot‐assisted radical prostatectomy; RT, Radiotherapy.
The highest GS.
3.2. Changes in circulating miRNAs from baseline to follow‐up
The plasma levels of both miRNA‐93 and miRNA‐221 were significantly downregulated in the follow‐up samples compared to the baseline samples in the interventional group (z = −2.738, P = 0.006 and z = −4.498, P < 0.001, respectively) (results regarding miRNA‐93 has been published in our previous work.25 The downregulation of miRNA‐93 was more significant in the RT subgroup (z = −2.366, P = 0.018) than in the RARP subgroup (−2.169, P = 0.030). On the other hand, plasma levels of miRNA‐221 was more downregulated in the RARP subgroup (z = −3.802, P = < 0.001) than in the RT subgroup (z = −2.197, P = 0.028).
In the observation group, miRNA‐125b was downregulated whereas miRNA‐221 tended to be upregulated in follow‐up plasma samples compared to baseline samples (z = −2.656, P = 0.008 and z = 1.863, P = 0.063, respectively) (Table 2).
Table 2.
Change in plasma levels of miRNA in follow‐up samples compared to baseline in both the observation and the intervention PCa group
| miRNA‐93 | miRNA‐21 | miRNA‐221 | miRNA‐125b | |||||
|---|---|---|---|---|---|---|---|---|
| Management type | z score | P‐value | z score | P‐value | z score | P‐value | z score | P‐value |
| Intervention | −2.738 a | 0.006 | 0.923 | 0.356 | −4.498 | <0.001 | 0.536 | 0.592 |
| RARP | −2.169 | 0.030 | 0.601 | 0.548 | −3.802 | <0.001 | −1.345 | 0.179 |
| RT | −2.366 | 0.018 | ‐0.447 | 0.654 | −2.197 | 0.028 | 1.782 | 0.075 |
| ADT | −0.743 | 0.457 | 0.805 | 0.420 | −1.802 | 0.072 | 0.592 | 0.554 |
| Observation | −1.683 a | 0.092 | 0.431 | 0.666 | 1.863 | 0.063 | −2.656 | 0.008 |
ADT, androgen deprivation therapy; RARP, robot‐assisted radical prostatectomy; RT, radiotherapy.
P‐value and z score are based on Wilcoxon signed rank sum test. Values in bold are statistically significant.
Data regarding miRNA‐93 in both intervention and observation cohort was published in our previous work (ref. 25).
There was no difference in baseline miRNA levels between the observation and intervention group (Supplementary Table SIII).
The difference between baseline and follow‐up samples in both groups is illustrated by box plots in Figure 1. For graphical reasons values with linear ΔCt higher than 30 has been removed from miRNA‐221 box plots.
Figure 1.
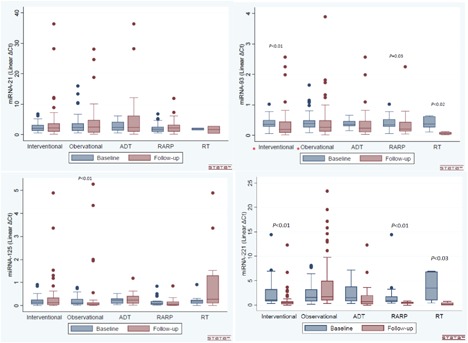
Box plots illustrating the change in plasma miRNAs between follow‐up samples and baseline in both the observation and intervention PCa group. Only significant fold change is supplied with a P‐value. *Box plots regarding miRNA‐93 in both intervention and observation cohort was published in our previous work (ref. 25). [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Correlation between miRNA plasma levels in baseline samples with diagnostic clinical characteristics
Baseline plasma levels of miRNA‐125b were significantly correlated with both prebiopsy PSA (rho = 0.198, P = 0.043) and EAU risk profile (rho = 0.230, P = 0.015). Moreover, there was a tendency toward a correlation with both needle GS and cT (rho = 0.171, P = 0.069, and rho = −0.170, P = 0.070, respectively). There was a significant correlation between plasma miRNA‐221 and the EAU risk profile (rho = 0.203, P = 0.016). The plasma level of miRNA‐21 showed an almost significant correlation with cT (rho = 0.155, P = 0.060).
Interestingly, the plasma levels of all miRNAs (except for miRNA‐93) were significantly correlated with patient age at the time of diagnosis (Table 3).
Table 3.
Correlation between baseline miRNA plasma levels and diagnostic characteristics in localized/locally advanced PCa patients
| PSA | Needle GS | cT | EAU risk profile | Prostate volume | Age | ||
|---|---|---|---|---|---|---|---|
| miRNA‐93 | rho: | −0.035 | 0.048 | 0.044 | 0.033 | 0.020 | 0.103 |
| P‐value | 0.674 | 0.570 | 0.601 | 0.693 | 0.814 | 0.220 | |
| miRNA‐21 | rho: | 0.050 | −0.008 | 0.155 | 0.112 | −0.080 | 0.212 |
| P‐value | 0.540 | 0.917 | 0.060 | 0.175 | 0.340 | 0.010 | |
| miRNA‐221 | rho: | 0.138 | 0.040 | 0.118 | 0.203 | 0.061 | 0.196 |
| P‐value | 0.103 | 0.642 | 0.165 | 0.016 | 0.475 | 0.020 | |
| miRNA‐125b | rho: | 0.198 | 0.171 | −0.170 | 0.230 | 0.087 | 0.329 |
| P‐value | 0.043 | 0.069 | 0.070 | 0.015 | 0.359 | 0.000 |
cT, Clinical tumour stage; EAU risk profile, European Association of Urology risk profile; GS, Gleason score; rho‐scores are from Spearman's rank correlation coefficient. Statistically significant values are marked in bold writing.
3.4. Correlation to post‐operative pathological characteristics in the RARP subgroup
In the subgroup analysis of the baseline plasma level of miRNAs in patients who underwent RARP, miRNA‐93 showed an almost significant correlation with prostatectomy GS (rho = 0.276, P = 0.058). Looking at pN, radicality of tumor resection (R), and pT, no correlation was observed with miRNAs levels in baseline plasma samples (Table 4).
Table 4.
Correlation of miRNA level in baseline plasma samples with pathological characteristics in localized/locally advanced PCa patients managed with curatively intended radical prostatectomy
| miRNA‐21 | miRNA‐93 | miRNA‐125b | miRNA‐221 | |
|---|---|---|---|---|
| pN | ||||
| z: | −1.032 | 0.049 | −1.611 | 0.133 |
| P‐value | 0.302 | 0.911 | 0.107 | 0.895 |
| R | ||||
| z: | 0.082 | 0.061 | −0.068 | 0.283 |
| P‐value | 0.935 | 0.951 | 0.946 | 0.777 |
| pT | ||||
| rho: | −0.130 | 0.049 | 0.076 | −0.093 |
| P‐value | 0.369 | 0.742 | 0.644 | 0.536 |
| pGS | ||||
| rho: | −0.023 | 0.276 | 0.155 | −0.117 |
| P‐value | 0.874 | 0.058 | 0.340 | 0.433 |
pN, pathological lymph nodes; R, free resection; pT, pathological tumour stage; pGS, prostatectomy Gleason score. rho‐scores are from Spearman's rank correlation coefficient.
4. DISCUSSION
In this study we analyzed circulating miRNAs in two cohorts of localized/locally advanced PCa patients; an intervention cohort (RARP, RT, or ADT) and an observation cohort (AS or WW). Both miRNA‐93 and miRNA‐221 demonstrated a significant decrease in plasma levels after both RARP and RT but not after ADT treatment. In the observation cohort, the plasma level of miRNA‐125b decreased significantly while there was a tendency toward an increase of miRNA‐221.
Accumulating evidence shows that RT in cancer treatment, including PCa, can significantly change miRNA expression levels.30, 31, 32 Leung et al found decrease levels of 16 miRNAs (including miRNA‐221) in PC‐3 PCa cells when exposed to different radiation doses compared to the parental cells.33 To the best of our knowledge, this is the first study to demonstrate a change of plasma miRNAs level in PCa patients having received curative intent RT.
Change of circulating miRNA levels after radical surgery has also been reported previously in breast cancer, colon cancer, and PCa.34, 35, 36 Mahn et al observed significant reduction of miRNA‐16, ‐26a, and miRNA‐195 serum levels after RARP in 37 localized PCa patients.36 Kelly et al found post‐RARP expression levels of miR‐141 to decline to levels similar to those of the controls.37 In prostatectomy specimens from 28 localized PCa patients, Lehmusvaara et al reported downregulation of 24 miRNAs and upregulation of another 28 miRNAs (including miRNA‐125b) after neoadjuvant bicalutamide and goserelin, respectively, before RARP.38 The mechanism of action of bicalutamide is quite different from goserelin.39 Therefore one of the reasons for non‐significant changes of miRNA plasma levels in our ADT cohort could be pooling of data from patients treated with either bicalutamide or goserelin in the same group. Another explanation could be different materials, as Lehmusvaara et al studied miRNA expression in formalin‐fixed and paraffin‐embedded PCa tissue while plasma samples were used in the present study.
Previous studies have shown that miRNA‐93 and miRNA‐221 levels are upregulated in blood samples from localized PCa patients compared to healthy men.25, 40, 41 We have observed a correlation between miRNA‐93 expression in cancerous tissue and paired plasma samples from mPCa.25 Therefore, the significant decline in both the miRNA‐93 and miRNA‐221 plasma level after intervention in this study underline the clinical importance of these miRNAs as onco‐miRNAs and their potential role as biomarkers for PCa management.
The upregulation of miRNA‐125b in PCa patients compared to healthy controls has been documented in many studies.25, 42, 43 Singh et al. observed an increased level of miRNA‐125b in PCa patients with biochemical relapse after RARP compared to those who were relapse free. As a result, it was hypothesized that the more aggressive the PCa, the higher the level of miRNA‐125b in the plasma.43 Therefore, the increase of miRNA‐125b in the observation cohort in this study may be associated with the risk of metastatic spread to regional LN. However, the exact role of miRNA‐125b in cancer is far from fully understood, yet.44
Both miRNA‐125b and miRNA‐221 were significantly correlated with the EAU risk assessment. The same observation was described in N0M0PCa patients where Shen et al documented a higher plasma level of miRNA‐221 in the intermediate and high‐risk groups compared to the low‐risk group according to the D'Amico risk profile.45 Moltzahn et al found a stepwise increase in serum miRNA‐93 from low to intermediate to high‐risk group according to the Cancer of the Prostate Risk Assessment (CAPRA) tool.
Currently, there are more than 100 statistical models available for risk stratification of localized/locally advanced PCa. However, many of them lack external validation,46 and even though the models are strictly used, misclassification rates are still around 30%, leaving a limitation to the accurate identification of low‐risk PCa patients.47 Therefore, the use of continuous risk variables in these models rather than defined risk categories increases the accuracy.48
Only miRNA‐125b correlated with prebiopsy PSA in our study. Guan et al did not observe any correlation between miRNA‐21 and the diagnostic characteristics in mPCa patients (initial PSA, cT, or GS). Investigating the serum level of 10 miRNAs (including miRNA‐93) in localized/locally advanced PCa, Moltzahn et al could not demonstrate any correlation with PSA, GS, or even age.41
However, we showed a significant correlation of miRNA‐21, ‐125b, and miRNA‐221 with patient age at time of PCa diagnosis. Both miRNA‐21 and miRNA‐125b have been shown to be involved in the aging process through an altered DNA damage response.49
Interestingly, the plasma level of miRNA‐93 was significantly correlated with prostatectomy GS but not with diagnostic GS, which emphasizes the discordance between needle biopsy GS and post‐operative GS.
The prospective collection of blood samples, the different treatment modalities, and a participation rate of 100% for both sampling time points are the strengths of the current study. However, the relatively small sample size of the RT and ADT subgroups is considered a limitation in our study and the results should be seen as hypothesis generating.
The clinical importance of any promising biomarker in cancer in general and specifically in PCa is derived from its ability to better distinguish indolent from aggressive cases in a more individualized pattern rather than the “one size fits all” principle. Large‐scale studies with long follow‐up periods are essential to get more reliable results. Prediction of treatment response is another challenge in PCa management where serial non‐invasive liquid biopsies could be of great help in monitoring and observing the patients, since early detection of relapse/progression of PCa is an important factor for better control of the disease.50
5. CONCLUSION
MiRNA‐93 and miRNA‐221 plasma levels decreased significantly after intervention compared to observation in localized/locally advanced PCa. While miRNA‐125b and miRNA‐221 were correlated with EAU risk assessment, there was a tendency toward a significant correlation between miRNA‐93 with GS in prostatectomy specimens. These findings emphasize the clinical significance of miRNAs as potential biomarkers in PCa. Further studies illustrating the prognostic and predictive value of miRNAs in different stages of PCa are required.
ETHICS APPROVAL
The study was approved by The Regional Committees on Health Research Ethics for Southern Denmark (S‐20170006) and The Danish Data Protection Agency according to Danish law. The Danish Registry of Tissue Utilization was screened prior to study initiation.
AUTHORS’ CONTRIBUTION
Urologist Søren Sørensen Madsen and pathologist Niels Korsgaard, Esbjerg Hospital, have contributed to this study by recruiting patients and preparing samples, respectively.
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Supporting Table S1.
Supporting Table S2.
Supporting Table S3.
ACKNOWLEDGMENTS
Many thanks to Lone Frischknecht for excellent technical assistance. Special thanks to Karin Larsen for linguistic editing of the manuscript. We would also like to thank OPEN, Odense Patient Data Explorative Network, Odense University Hospital, Odense, Denmark for data management. This study was funded by the Research Council of Lillebaelt Hospital (AHZ) and the Japanese‐Swedish Research Foundation.
Zedan AH, Hansen TF, Assenholt J, Madsen JS, Osther PJS. Circulating miRNAs in localized/locally advanced prostate cancer patients after radical prostatectomy and radiotherapy. The Prostate. 2019;79: 425–432. 10.1002/pros.23748
REFERENCES
- 1.The Danish Data Authority D. Nye Kræfttilfælde i Danmark: Cancerregisteret 2016. 2017.
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 3. Sun L, Gancarczyk K, Paquette EL, et al. Introduction to Department of Defense Center for Prostate Disease Research Multicenter National Prostate Cancer Database, and analysis of changes in the PSA‐era. Urol. Oncol. 2001;6:203–209. [Google Scholar]
- 4. Etzioni R, Penson DF, Legler JM, et al. Overdiagnosis due to prostate‐specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94:981–990. [DOI] [PubMed] [Google Scholar]
- 5. Capitanio U, Briganti A, Gallina A, et al. Predictive models before and after radical prostatectomy. Prostate. 2010;70:1371–1378. [DOI] [PubMed] [Google Scholar]
- 6. Partin AW, Kattan MW, Subong EN, et al. Combination of prostate‐specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi‐institutional update. JAMA J Am Med Assoc. 1997;277:1445–1451. [PubMed] [Google Scholar]
- 7. Boorjian SA, Karnes RJ, Crispen PL, et al. The impact of discordance between biopsy and pathological gleason scores on survival after radical prostatectomy. J Urol. 2009;181:95–104. [DOI] [PubMed] [Google Scholar]
- 8. Melia J, Moseley R, Ball RY, et al. A UK‐based investigation of inter‐ and intra‐observer reproducibility of Gleason grading of prostatic biopsies. Histopathology. 2006;48:644–654. [DOI] [PubMed] [Google Scholar]
- 9. Mian BM, Lehr DJ, Moore CK, et al. Role of prostate biopsy schemes in accurate prediction of Gleason scores. Urology. 2006;67:379–383. [DOI] [PubMed] [Google Scholar]
- 10. D'Amico AV, Whittington R, Malkowicz SB, et al. Pretreatment nomogram for prostate‐specific antigen recurrence after radical prostatectomy or external‐beam radiation therapy for clinically localized prostate cancer. J Clin Oncol. 1999;17:168–172. [DOI] [PubMed] [Google Scholar]
- 11. Klotz L, Vesprini D, Sethukavalan P, et al. Long‐term follow‐up of a large active surveillance cohort of patients with prostate cancer. J Clin Oncol. 2015;33:272–277. [DOI] [PubMed] [Google Scholar]
- 12. Schlomm T, Erbersdobler A, Mirlacher M, Sauter G. Molecular staging of prostate cancer in the year 2007. World J Urol. 2007;25:19–30. [DOI] [PubMed] [Google Scholar]
- 13. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: An overview of nuclear functions. Int J Mol Sci. 2016;17: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez A, Griffiths‐Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. [DOI] [PubMed] [Google Scholar]
- 17. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir‐14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. [DOI] [PubMed] [Google Scholar]
- 19. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egidi MG, Cochetti G, Serva MR, et al. Circulating microRNAs and Kallikreins before and after radical prostatectomy: Are they really prostate cancer markers? Biomed Res Int. 2013;2013:241780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuzaki J, Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: a systematic review. Int J Clin Oncol. 2017;22:413–420. [DOI] [PubMed] [Google Scholar]
- 22. Chun‐Jiao S, Huan C, Li‐Zhong C, Guo‐Mei R, Jian‐Jun G, Qian‐Nan D. The potential of microRNAs as human prostate cancer biomarkers: a meta‐analysis of related studies. J Cell Biochem. 2017;119:2763–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mottet N, Bellmunt J, Briers E, et al. EAU‐ESTRO − ESUR − SIOG guidelines on prostate cancer. Eur Assoc Urol. 2017:16 http://uroweb.org/wp‐content/uploads/09‐Prostate‐Cancer_2017_web.pdf. Accessed July 8, 2018. [Google Scholar]
- 25. Zedan AH, Hansen TF, Jo P. MicroRNA expression in tumour tissue and plasma in patients with newly diagnosed metastatic prostate cancer. Tumor Biol. 2018;40:1–11. [DOI] [PubMed] [Google Scholar]
- 26.Danish Urological Cancer Group. DaProCa (Prostatacancer) guidelines. 2018. http://ducg.dk/daproca‐prostatacancer/. Accessed July 26, 2018.
- 27. Zedan AH, Blavnsfeldt SG, Hansen TF, et al. Heterogeneity of miRNA expression in localized prostate cancer with clinicopathological correlations. PLoS ONE 2017;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andersen CL, Jensen JL, Ørntoft TF, et al. Normalization of real‐time quantitative reverse transcription‐ PCR data: a model‐based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. [DOI] [PubMed] [Google Scholar]
- 29. Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT‐PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Templin T, Paul S, Amundson SA, et al. Radiation‐induced micro‐RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. John‐Aryankalayil M, Palayoor ST, Makinde AY, et al. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leung C‐M, Li S‐C, Chen T‐W, et al. Comprehensive microRNA profiling of prostate cancer cells after ionizing radiation treatment. Oncol Rep. 2014;31:1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng EKO, Chong WWS, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. [DOI] [PubMed] [Google Scholar]
- 35. Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating micrornas as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. [DOI] [PubMed] [Google Scholar]
- 36. Mahn R, Heukamp LC, Rogenhofer S, Von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77:1265.e9–1265.e16. [DOI] [PubMed] [Google Scholar]
- 37. Kelly B, Miller N, Sweeney K, et al. A circulating MicroRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015;4:1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lehmusvaara S, Erkkilä T, Urbanucci A, et al. Goserelin and bicalutamide treatments alter the expression of microRNAs in the prostate. Prostate. 2013;73:101–112. [DOI] [PubMed] [Google Scholar]
- 39.European Medicines Agency. European Medicines Agency. 2006; 2(November 1994). http://www.ema.europa.eu/ema/. Accessed July 26, 2018.
- 40. Mihelich BL, Maranville JC, Nolley R, Peehl DM, Nonn L. Elevated serum microRNA levels associate with absence of high‐grade prostate cancer in a retrospective cohort. PLoS One. 2015;10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moltzahn F, Olshen AB, Baehner L, et al. Microfluidic‐based multiplex qRT‐PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011;71:550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci. 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh PK, Preus L, Hu Q, et al. Serum microRNA expression patterns that predict early treatment failure in prostate cancer patients. Oncotarget. 2014;5:824–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banzhaf‐Strathmann J, Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun Signal. 2014;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen J, Hruby GW, McKiernan JM, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72:1469–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shariat SF, Karakiewicz PI, Roehrborn CG, Kattan MW. An updated catalog of prostate cancer predictive tools. Cancer. 2008;113:3075–3099. [DOI] [PubMed] [Google Scholar]
- 47. Palisaar JR, Noldus J, Löppenberg B, von Bodman C, Sommerer F, Eggert T. Comprehensive report on prostate cancer misclassification by 16 currently used low‐risk and active surveillance criteria. BJU Int. 2012;110:E172–E181. [DOI] [PubMed] [Google Scholar]
- 48. Kattan MW, Shariat SF, Andrews B, et al. The addition of interleukin‐6 soluble receptor and transforming growth factor beta1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J Clin Oncol. 2003;21:3573–3579. [DOI] [PubMed] [Google Scholar]
- 49. Harries LW. MicroRNAs as mediators of the ageing process. Genes (Basel). 2014;5:656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mottet N, Bellmunt J, Briers E, Bolla M, Cornford P, et al. EAU‐ESTRO‐SIOG Guidelines on Prostate Cancer. 2016:1–146. 10.1016/j.eururo.2007.09.002. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Supporting Table S1.
Supporting Table S2.
Supporting Table S3.


