Abstract
Cervical screening aims to identify women with high‐grade squamous intraepithelial lesion/cervical intraepithelial neoplasia 2‐3 (HSIL/CIN2‐3) or invasive cervical cancer (ICC). Identification of women with severe premalignant lesions or ICC (CIN3+) could ensure their rapid treatment and prevent overtreatment. We investigated high‐risk human papillomavirus (hrHPV) detection with genotyping and methylation of FAM19A4/miR124‐2 for detection of CIN3+ in 538 women attending colposcopy for abnormal cytology. All women had an additional cytology with hrHPV testing (GP5+/6+‐PCR‐EIA+), genotyping (HPV16/18, HPV16/18/31/45), and methylation analysis (FAM19A4/miR124‐2) and at least one biopsy. CIN3+ detection was studied overall and in women <30 (n = 171) and ≥30 years (n = 367). Positivity for both rather than just one methylation markers increased in CIN3, and all ICC was positive for both. Overall sensitivity and specificity for CIN3+ were, respectively, 90.3% (95%CI 81.3–95.2) and 31.8% (95%CI 27.7–36.1) for hrHPV, 77.8% (95%CI 66.9–85.8) and 69.3% (95%CI 65.0–73.3) for methylation biomarkers and 93.1% (95%CI 84.8–97.0) and 49.4% (95%CI 44.8–53.9) for combined HPV16/18 and/or methylation positivity. For CIN3, hrHPV was found in 90.9% (95%CI 81.6–95.8), methylation positivity in 75.8% (95%CI 64.2–84.5) and HPV16/18 and/or methylation positivity in 92.4% (95%CI 83.5–96.7). In women aged ≥30, the sensitivity of combined HPV16/18 and methylation was increased (98.2%, 95%CI 90.6–99.7) with a specificity of 46.3% (95%CI 40.8–51.9). Combination of HPV16/18 and methylation analysis was very sensitive and offered improved specificity for CIN3+, opening the possibility of rapid treatment for these women and follow‐up for women with potentially regressive, less advanced, HSIL/CIN2 lesions.
Keywords: Triage, FAM19A4, miR124‐2, human papillomavirus, intraepithelial neoplasia, cervical cancer
Short abstract
What's new?
Reliable triage of women with cervical intraepithelial neoplasia (CIN) is of high priority as not all lesions progress to invasive carcinoma. Here the authors show that combining the methylation status of tumor suppressor genes FAM19A4 and miR124‐2 with genotyping for high‐risk human papillomavirus results in a highly sensitive and moderately specific triage strategy that identifies women with CIN lesions likely to need rapid treatment. The authors recommend clinical evaluation of the strategy in prospective studies.
Abbreviations
- 95% CI
95% confidence interval
- ACTB
b‐actin
- AGC
atypical glandular cells
- ASC‐H
atypical squamous cells cannot exclude HSIL
- ASC‐US
atypical squamous cells of undetermined significance
- CIN
cervical intraepithelial neoplasia
- Cq
quantification cycle
- FAM19A4
family with sequence similarity 19 (chemokine (C–C)‐motif)‐like), member A4
- hrHPV
high‐risk human papillomavirus
- HSIL
high‐grade squamous intraepithelial lesion
- ICC
invasive cervical carcinoma
- LLETZ
large loop excision of the transformation zone
- LSIL
low‐grade squamous intraepithelial lesion
- miR124‐2
micro‐ribonucleic acid 124‐2
- PCR
polymerase chain reaction
- qMSP
quantitative methylation‐specific PCR
Background
The mortality rate from invasive cervical cancer (ICC) has decreased significantly in developed countries through the introduction of organized cervical screening programs, mostly based on conventional cytology.1, 2 Women with abnormal cytology or with a positive result in the testing for high‐risk human papillomavirus (hrHPV) are either referred directly, or after triage, for colposcopy and biopsy to identify high‐grade squamous intraepithelial lesion/cervical intraepithelial neoplasia grade 2 or 3 (HSIL/CIN2‐3) or ICC.3 A biopsy diagnosis of HSIL/CIN2‐3 is the current threshold for excision of a precancerous lesion.4
However, most HSIL/CIN2‐3 will not progress to ICC if not treated.5, 6 HSIL/CIN2‐3 lesions are heterogeneous, especially those graded as CIN2, and include both productive hrHPV infections as well as early and advanced transforming hrHPV lesions. Productive infections and early transforming infections may regress spontaneously and immediate surgical removal may incur harm and increase the risk of adverse outcome for any future pregnancy.7 Preventing such unwanted outcomes would be valuable. In contrast, advanced transforming lesions with a high short‐term risk of progression should be treated without delay. A triage strategy able to provide accurate risk stratification, identifying the different subgroups of women with HSIL/CIN2‐3, could reduce harm, especially in young women of reproductive age.
A number of different molecular markers are currently being investigated to optimize risk stratification for patients with hrHPV infection and cervical lesions. These include hrHPV genotyping, expression of viral genes and methylation of viral genes or human tumor suppressor genes.8 A high proportion of ICC and a large number of HSIL are associated with HPV16/18, even in the presence of multiple HPV infections, and HPV16 is also associated with larger HSIL/CIN3 lesions in younger women.9, 10, 11 Several human tumor suppressor genes have shown hypermethylation of the CpG islands in the promotor regions, which are of value in identifying women with ICC and advanced HSIL/CIN3 defined as long duration of preceding hrHPV infection.12 Hypermethylation of the genes FAM19A4 and miR124‐2 have shown promising results as single triage markers or in combination with other tumor suppressor genes in physician‐taken samples in detecting HSIL/CIN3, particularly in women with HPV infections of long duration.13, 14, 15
The EVAH study (Evaluating the Visual Appearance of Cervical Lesions in Relation its Histological Diagnosis, Human Papillomavirus Genotype and Other Viral Parameters)16 provides a basis for the evaluation of various tests that could be used for risk stratification. It allows comparison of standard clinical and pathological information with new molecular tests and colposcopy follow‐up for up to 2 years and provides a framework for the long‐term outcome of different triage tests.
This study aimed to detect women with HSIL/CIN3 or ICC (CIN3+) in a cytology‐screened (atypical squamous cells of unknown significance or more severe lesions [≥ASC‐US]) referral population by using referral cytological grade, hrHPV testing and genotyping and methylation testing of human tumor suppressor genes FAM19A4 and miR124‐2 performed on physician‐taken smears obtained at colposcopy. Sensitivities, specificities, positive and negative predictive values (PPV and NPV) of the different triage tests for CIN3+ detection were calculated for women <30 and ≥30 years of age.
Methods
The EVAH study is a prospective multicenter observational cohort study16 conducted in Voorburg, The Netherlands, and Barcelona, Spain. Inclusion criteria were (i) an abnormal cytological test result (≥ASC‐US) and (ii) 18 years of age or older. The criteria of exclusion were (i) previous diagnosis of ICC, (ii) history of surgery to the cervix or previous pelvic radiotherapy, (iii) current pregnancy or pregnancy in the previous 3 months, (iv) current breastfeeding or breastfeeding in the previous 3 months and (v) insufficient material for hrHPV testing and methylation analysis.
From each subject, data were collected on ethnicity, relevant medical history, number of sexual partners and contraception, using a standardized questionnaire. Results from the first 601 inclusions from Barcelona were analyzed in this study.
Colposcopy
Women included in the study were seen in the colposcopy clinic, where a physician‐taken smear was collected and digital colposcopy was performed. During colposcopy, at least one cervical biopsy was taken from each patient. Up to four directed biopsies were taken from suspected lesions. If less than four directed biopsies were taken, a random biopsy was also taken from normal appearing tissue. Colposcopy findings were described using the criteria of the International Federation for Cervical Pathology and Colposcopy (IFCPC).17
Physician‐taken smear
A trained physician collected a cervical smear using a Cervex‐Brush (Rovers Medical Devices B.V., Oss, The Netherlands). The Cervex‐Brush was rinsed in 20 mL of ThinPrep medium (Hologic, Marlborough, MA). Samples were stored at room temperature for up to 6 months. An aliquot of 2 mL was sent to DDL Diagnostic Laboratory, Rijswijk, The Netherlands for molecular testing. At arrival at DDL, the samples were stored at 4°C.
Biopsy
Biopsies were fixed in 10% formalin, embedded in paraffin and sectioned for hematoxylin and eosin (H&E) and additional immunohistochemical staining at the local laboratory. p16 immunohistochemical staining was performed and slides were scored as negative, patchy staining, block staining restricted to lower 1/3 the lower 2/3, or extending above the lower 2/3 of the epithelium up to full thickness staining. Sections were reviewed independently by two expert gynecological pathologists and classified as either normal, CIN 1, CIN 2, CIN 3, adenocarcinoma in situ (AIS) or ICC. In case of discrepancies between the two pathologists, a third gynecological expert pathologist was consulted, resulting in a majority consensus diagnosis.
Sample preparation and hrHPV detection
From the physician‐taken smears, DNA was isolated for hrHPV testing. An input volume of 250 μL was used to obtain 100 μL of eluate with the QIAamp MinElute Virus Spin kit (QIAgen Inc., Valencia, CA). hrHPV detection was performed using the GP5+/6+‐PCR‐EIA (Labo Bio‐medical Products, Rijswijk, The Netherlands). The EIA‐positive GP5+/6+ amplimers were genotyped using a strip‐based test by the Genotyping kit HPV GP (Labo Bio‐medical Products, Rijswijk, The Netherlands) genotyping hrHPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68.
Methylation
The residual cervical sample was concentrated at Hospital Clínic and sent to DDL Diagnostic Laboratory for methylation testing.
DNA was isolated using the MagNA Pure 96 (500 μL input, 50 μL eluate) for methylation testing. The level of amplifiable human DNA was assessed using a qPCR of the in‐house reference gene RNaseP with the Phocine herpes virus as an internal control for the absence of PCR‐inhibition.18
All samples with a DNA concentration of 5.5 ng/μL or higher were subjected to bisulfite conversion using the EZ DNA Methylation kit (Zymo Research, Orange, CA). Up to 250 ng/45 μL of DNA was used. A standardized multiplex qMSP (QIAsure), targeting for the promotor regions of FAM19A4 and miR124‐2 (QIAgen Inc., Valencia, CA), was performed on the bisulfite‐converted DNA according to the manufacturer's protocol. The bisulfite‐converted human reference gene beta‐actin (ACTB) was included in the multiplex to determine the total amount of converted human DNA. The samples were scored hypermethylation positive when C T value of the housekeeping gene β‐actin was ≤26.4 and at least one of the marker genes (i.e., FAM19A4 and hsa‐mir124‐2) had a ΔΔC T below the cutoff according to the kit insert. Methylation positivity was then correlated to the histological diagnosis outcome.
Statistical analysis
Results of cytology, hrHPV testing and methylation testing were studied for their performance in the detection of CIN3+ lesions. For analysis cytological outcome was classified as ASC‐US/LSIL (ASC‐US and LSIL) or ASC‐H/HSIL (HSIL, ASC‐H and AGC). Interpretation of the hrHPV testing resulted in hrHPV positive or negative and interpretation of the genotyping resulted in HPV16/18/31/45 or no‐HPV16/18/31/45 and HPV16/18 or no‐HPV16/18. Smears that tested positive for FAM19A and/or miR124‐2 according to the standardized criteria were considered as methylation positive, whereas methylation negative smears were defined as samples negative for both markers. After categorizing women using these test results, different combinations were made to compute sensitivity, specificity, PPV and NPV with corresponding 95% confidence intervals. Diagnostic performance was calculated for the entire cohort and for women aged <30 or ≥30 years. In addition, results are shown for all women with ASC‐US/LSIL referral cytology and women with ASC‐H/HSIL referral cytology.
Statistical analysis was performed using the SPSS software (Version 22.0, SPSS, Inc, Chicago, IL). The results are presented as absolute numbers and percentages or mean and standard deviation. χ 2 tests, independent sample t‐tests and McNemar's tests were used, as appropriate, for comparisons between categorical, between scale variables and for comparison of the performance of diagnostic tests. A p value <0.05 was considered statistically significant.
Results
Characteristics of the EVAH study participants
Of the 601 selected women, 540 (89.8%) had a physician‐taken smear with a DNA concentration ≥5.5 ng/μL and were suitable for methylation testing. Methylation testing resulted in 538 valid results (340, 63.2% negative; 198, 36.8% positive) that were used for further analysis. Two invalid results based on a negative result of the reference gene ACTB were excluded. The flowchart of the women included in the study with the cross‐sectional results of the referral smear, hrHPV testing and genotyping and methylation testing are shown in Figure 1.
Figure 1.
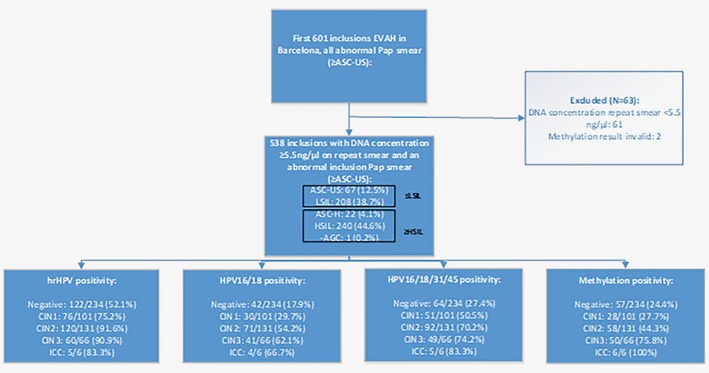
Flowchart of the first 601 women that were included in the EVAH study in Barcelona with cross‐sectional results of the referral smear, hrHPV testing and genotyping and methylation testing. Results are presented as percentages with a positive test result per histological diagnosis category.
Mean age of women at the time of the first visit to the colposcopy clinic was 36.2 years (95% CI 35.3–37.2; range 18–75). Histological diagnoses of the cervical biopsies were normal squamous epithelium in 234 women (43.5%), CIN1 in 101 (18.8%), CIN2 in 131 (24.3%), CIN3 in 66 (12.3%) and ICC (invasive cervical carcinoma) in 6 (1.1%).
hrHPV testing using the clinical test GP5+/6+‐PCR‐EIA, was positive in 383 women (71.2%), and the remaining 155 women (28.8%) were hrHPV negative. Of the hrHPV‐positive samples, 188 (49.1%) tested positive for HPV16/18 and 195 (50.9%) tested positive for non‐HPV16/18 hrHPV genotypes.
Sensitivity and specificity of hrHPV testing and methylation for the detection of CIN3+ in women with abnormal cytology.
Single triage markers
The performance of hrHPV detection, genotyping for HPV16/18 and HPV16/18/31/45 and methylation testing as single triage tests were explored for this population for the outcome of CIN3+ including CIN3 lesions and carcinomas (Table 1) and for CIN3 only (Table 2). hrHPV positivity with GP5+/6+‐PCR‐EIA was found in 65/72 women with CIN3+ and in 148/466 of the women with a ≤CIN2 lesion no hrHPV was found (sensitivity 90.3%, specificity 31.8%). HPV16/18/31/45 was detected in the physician‐taken smear in 54/72 women with CIN3+ lesions and in 259/466 with ≤CIN2 lesions these types were not detected (sensitivity 75.0%, specificity 55.6%). HPV16/18 was present in 45/72 of women with CIN3+ lesions and 323/466 women with ≤CIN2 lesions were HPV16/18 negative (sensitivity 62.5%, specificity 69.3%).
Table 1.
(a) Sensitivity, specificity, PPV and NPV of hrHPV testing, hrHPV genotyping and methylation testing and (b) different combinations (1b) for the detection of CIN3+ (CIN3 and invasive carcinoma) in biopsies of women referred after abnormal cytology
| All women (N = 538) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | 95% CI | Specificity (%) | 95% CI | PPV (%) | 95% CI | NPV (%) | 95% CI | |
| (a) Single triage tests | ||||||||
| hrHPV testing | 90.3 | 81.3–95.2 | 31.8 | 27.7–36.1 | 17.0 | 13.5–21.1 | 95.5 | 91.0–97.8 |
| HPV16/18/31/45 | 75.0 | 63.9–83.6 | 55.6 | 51.0–60.0 | 20.7 | 16.2–26.0 | 93.5 | 90.0–95.9 |
| HPV16/18 | 62.5 | 51.0–72.8 | 69.3 | 65.0–73.3 | 23.9 | 18.4–30.5 | 92.3 | 89.0–94.6 |
| Methylation (FAM19A4/miR124‐2) | 77.8 | 66.9–85.8 | 69.3 | 65.0–73.3 | 36.4 | 29.8–43.6 | 98.3 | 96.1–99.3 |
| (b) Combinations of triage tests | ||||||||
| hrHPV and/or methylation | 98.6 | 92.5–99.8 | 24.3 | 20.6–28.3 | 16.8 | 13.5–20.6 | 99.1 | 95.2–99.8 |
| HPV16/18/31/45 and/or methylation | 95.8 | 88.5–98.6 | 39.7 | 35.4–44.2 | 19.7 | 15.9–24.2 | 98.4 | 95.4–99.5 |
| HPV16/18 and/or methylation | 93.1 | 84.8–97.0 | 49.4 | 44.8–53.9 | 22.1 | 17.8–27.1 | 97.9 | 95.1–99.1 |
Table 2.
(a) Sensitivity, specificity, PPV and NPV of hrHPV testing, hrHPV genotyping and methylation testing and (b) different combinations for the detection of CIN3 in biopsies of women referred after abnormal cytology
| All women (N = 538; total number of women is 538) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | 95% CI | Specificity (%) | 95% CI | PPV (%) | 95% CI | NPV (%) | 95% CI | |
| (a) Single triage tests | ||||||||
| hrHPV testing | 90.9 | 81.6–95.8 | 31.8 | 27.7–36.1 | 15.9 | 12.5–19.9 | 96.1 | 91.8–98.2 |
| HPV16/18/31/45 | 74.2 | 62.6–83.3 | 55.6 | 51.0–60.0 | 19.1 | 14.8–24.4 | 93.8 | 90.4–96.1 |
| HPV16/18 | 62.1 | 50.1–72.9 | 69.3 | 65.0–73.3 | 22.3 | 16.9–28.8 | 92.8 | 89.6–95.1 |
| Methylation (FAM19A4/miR124‐2) | 75.8 | 64.2–84.5 | 69.3 | 65.0–73.3 | 25.9 | 20.2–32.5 | 95.3 | 92.5–97.1 |
| (b) Combinations of triage tests | ||||||||
| hrHPV and/or methylation | 98.5 | 91.9–99.7 | 24.3 | 20.6–28.3 | 15.6 | 12.4–19.3 | 99.1 | 95.2–99.8 |
| HPV16/18/31/45 and/or methylation | 95.5 | 87.5–98.4 | 39.7 | 35.4–44.2 | 18.3 | 14.6–22.7 | 98.4 | 95.4–99.5 |
| HPV16/18 and/or methylation | 92.4 | 83.5–96.7 | 49.4 | 44.8–53.9 | 20.5 | 16.3–25.5 | 97.9 | 95.1–99.1 |
Methylation testing was positive in 56/72 of women CIN3+ lesions and 323/466 of women with ≤CIN2 lesions were methylation negative overall (sensitivity 77.8%, specificity 69.3%). Methylation detected all ICC and 75.8% (50/66) of CIN3. There was a significant difference in mean age between women with a methylation positive CIN3+ lesion (41 years) and women with methylation negative CIN3+ lesions (30 years, p < 0.001). Among the women with ≤CIN2, methylation was positive in 24.4% of women with negative and CIN1 biopsies and in 44.3% of women with CIN2. Again, a significant difference in mean age between methylation positive (41 years) and methylation negative women (34 years) was found (p < 0.001).
FAM19A4 and miR124‐2 positivity in different grades of CIN were assessed separately in order to detect trends in positivity of the individual markers and the combination of the two biomarkers (Fig. 2). FAM19A4 was the commonest individual positive marker. 50/85 (58.8%) biopsies without dysplastic lesions and CIN1 lesions that were methylation positive showed positivity for FAM19A4 only. Positivity for both FAM19A4 and miR124‐2 was seen in most CIN3 lesions and all ICC. We found that 19.7% (13/66) of CIN3 lesions were detected by FAM19A4 only, while miR124‐2 only showed positivity in five low‐grade lesions and one CIN2 lesion but no CIN3+. Recalculation of sensitivity and specificity when assessing FAM19A4 only resulted in an identical sensitivity of 77.8% and a nonsignificantly higher specificity of 70.6%.
Figure 2.
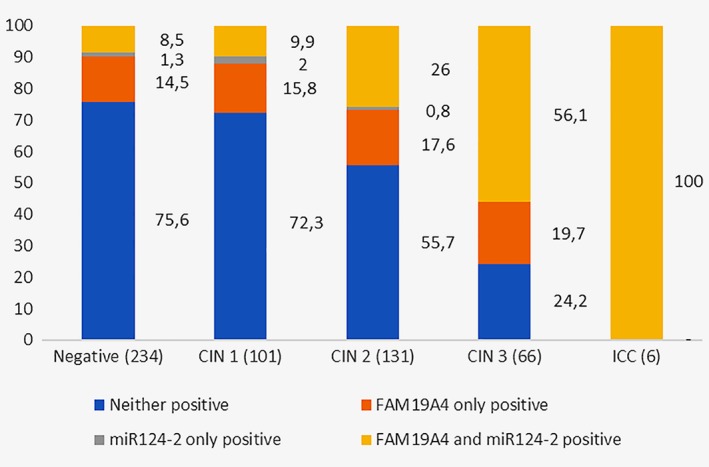
FAM19A4 and miR124‐2 positivity (%) in different grades of CIN in all women.
Combinations of triage markers
Table 1b shows the sensitivity, specificity, PPV and NPV of different combinations of hrHPV testing, hrHPV genotyping and methylation testing for the detection of CIN3+ in biopsies of women referred after abnormal cytology, and Table 2b shows this for the outcome of CIN3 only. Among the different combinations that were made, the combination of HPV16/18 and/or methylation positivity had the highest specificity (49.4%, 95% CI 44.8–53.9), while sensitivity was comparable to that of methylation testing combined with testing for all hrHPV or HPV16/18/31/45.
As the combination of HPV16/18 and/or methylation positivity performed best among the combinations, with the highest specificity and comparable sensitivity, PPV and NPV, this combination was used in further analyses. In these analyses, the combination of HPV16/18 and/or methylation was compared to the single triage markers with the highest sensitivity (hrHPV) and highest specificity and NPV:methylation (Table 1).
Performance of hrHPV testing and methylation for the detection of CIN3+ in women <30 and ≥30 years of age with ASC‐US/LSIL and ASC‐H/HSIL cytology.
Table 3 shows the sensitivity, specificity, PPV and NPV for the detection of CIN3+ by hrHPV testing, hrHPV genotyping and methylation in women <30 and ≥30 years of age with ASC‐US/LSIL and ASC‐H/HSIL referral cytology.
Table 3.
Comparison of sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for the detection of CIN3+ of hrHPV testing and methylation and combined HPV16/18 and methylation testing in women <30 and ≥30 years of age with ASC‐US/LSIL cytology and ASC‐H/HSIL cytology
| Referral cytology ≥ASC‐US | Referral cytology ASC‐US/LSIL | Referral cytology ASC‐H/HSIL | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Triage test | Sensitivity (%) | 95% CI | Specificity (%) | 95% CI | PPV (%) | 95% CI | NPV (%) | 95% CI | Sensitivity (%) | 95% CI | Specificity (%) | 95% CI | PPV (%) | 95% CI | NPV (%) | 95% CI | Sensitivity (%) | 95% CI | Specificity (%) | 95% CI | PPV (%) | 95% CI | NPV (%) | 95% CI |
| Women <30 years of age (N = 171) | Women <30 years of age (N = 102) | Women <30 years of age (N = 69) | ||||||||||||||||||||||
| hrHPV testing | 93.8 | 71.7–98.9 | 26.5 | 20.1–33.9 | 11.6 | 7.2–18.3 | 97.6 | 87.7–99.6 | 33.3 | 6.2–79.2 | 89.9 | 8.2–94.4 | 9.1 | 1.6–37.7 | 97.8 | 92.3–99.4 | 76.9 | 49.7–91.8 | 39.3 | 27.6–52.4 | 22.7 | 12.8–37.0 | 88.0 | 70.0–95.8 |
| Methylation | 37.5 | 18.5–61.4 | 85.8 | 79.4–90.4 | 21.4 | 10.2–39.5 | 93.0 | 87.6–96.2 | 100 | 43.9–100 | 32.3 | 23.9–42.1 | 4.3 | 1.5–11.9 | 100 | 89.3–100 | 38.5 | 17.7–64.5 | 78.6 | 66.2–87.3 | 29.4 | 13.3–53.1 | 84.6 | 72.5–92.0 |
| HPV16/18 and/or methylation | 75.0 | 50.5–89.8 | 55.5 | 47.6–63.1 | 14.8 | 8.7–24.1 | 95.6 | 89.1–98.3 | 66.7 | 20.8–93.9 | 64.7 | 54.8–73.4 | 5.4 | 1.5–17.7 | 98.5 | 91.8–99.7 | 92.3 | 66.7–98.6 | 16.1 | 8.7–27.8 | 20.3 | 12.0–32.3 | 90.0 | 59.6–98.2 |
| Women ≥ 30 years of age (N = 367) | Women ≥ 30 years of age (N = 173) | Women ≥ 30 years of age (N = 194) | ||||||||||||||||||||||
| hrHPV testing | 89.3 | 78.5–95.0 | 34.4 | 29.3–39.9 | 19.7 | 15.3–25.0 | 94.7 | 88.9–97.5 | 66.7 | 20.8–93.9 | 41.2 | 34.1–48.7 | 2.0 | 0.5–6.9 | 98.6 | 92.4–99.8 | 90.6 | 79.8–95.9 | 26.2 | 19.7–34.1 | 31.6 | 24.7–39.5 | 88.1 | 75.0–94.8 |
| Methylation | 89.3 | 78.5–95.0 | 61.1 | 55.6–66.3 | 30.4 | 24.0–37.7 | 98 | 94.9–99.2 | 66.7 | 20.8–93.9 | 71.2 | 64.0–77.5 | 3.9 | 1.1–13.2 | 99.2 | 95.5–99.9 | 90.6 | 79.8–95.9 | 48.9 | 40.8–57.1 | 40.0 | 31.7–48.9 | 93.2 | 85.1–97.1 |
| HPV16/18 and/or methylation | 98.2 | 90.6–99.7 | 46.3 | 40.8–51.9 | 24.8 | 19.6–30.9 | 99.3 | 96.2–99.9 | 100 | 43.9–100 | 58.2 | 50.7–65.4 | 4.1 | 1.4–11.3 | 100 | 96.3–100 | 98.1 | 90.1–99.7 | 31.9 | 24.8–40.0 | 35.1 | 27.9–43.1 | 97.8 | 88.7–99.6 |
Women <30 years of age
The group of women <30 years of age consisted of 171 women, of whom 16 had an HSIL/CIN3 lesion and none had ICC. Age ranged from 18 to 29 with a mean of 25.4 years. In this group, HPV16/18 was found in 68.8% of the women with CIN3+. Sensitivity of hrHPV testing (93.8%, 95% CI 71.7–98.9) was significantly higher than that of methylation testing (37.5%, 95% CI 18.5–61.4) (p = 0.004), whereas the specificity of methylation analysis (85.8%, 95% CI 79.4–90.4) was significantly higher than that of hrHPV testing (26.5%, 95% CI 20.1–33.9) (p < 0.001). The performance of the combination of HPV16/18 and/or methylation positivity provided the optimal balance between sensitivity (75.0%, 95%CI 50.5–89.8) compared to methylation analysis (p = 0.031) and specificity (55.5%, 95% CI 47.6–63.1) compared to hrHPV testing (p < 0.001).
Only 3/16 CIN3+ lesions were found among women with ASC‐US/LSIL referral cytology and, therefore, 95% confidence intervals in this group are wide making the performance of all markers as a triage test for CIN3+ detection comparable. However, the specificity of HPV16/18 and/or methylation (64.75, 95% CI 54.8–73.4) was significantly higher than that of methylation analysis (32.3%, 95% CI 23.9–42.1) (p < 0.001). Most CIN3+ lesions were found in women with ASC‐H/HSIL referral cytology (13/16 CIN3+ lesions). In this group, the sensitivity of HPV16/18 and/or methylation (92.3%, 95% CI 66.7–98.6) was higher than that of methylation analysis (38.5%, 95% CI 17.7–64.5) (p = 0.061), but the specificity of methylation analysis (87.6%, 95%CI 66.2–87.3) was significantly higher than that of HPV16/18 and/or methylation (16.1%, 95% CI 8.8–27.8) (p < 0.001) and hrHPV testing (39.3%, 95% CI 27.6–52.4) (p < 0.001).
Women ≥30 years of age
The group of women ≥30 years (mean 41.3 years, range 30–75) consisted of 367 women, 56 of whom had CIN3+ (50 CIN3, 6 ICC). HPV16/18 was detected in the smear of 60.7% of the women with CIN3+. Sensitivity of hrHPV testing and methylation analysis in this population were the same (89.3%, 95% CI 78.5–95.0), but the specificity of methylation analysis (61.1%, 95% CI 55.6–66.3) was significantly higher than that of hrHPV testing (34.4%, 95% CI 29.3–39.9) (p < 0.001). The combination of HPV16/18 and/or methylation positivity resulted in an increase in sensitivity to 98.2% (p = 0.063) and specificity of 46.3% which was improved compared to hrHPV testing (p < 0.001).
In this group, 3/56 CIN3+ lesions were found in women with ASC‐US/LSIL cytology leading to overlapping 95% confidence intervals of the performance of all three markers. The only woman with ASC‐US cytology and a CIN3 lesion was among these women, smear which was hrHPV negative and methylation positive. 53/56 CIN3+ lesions were found in women with ASC‐H/HSIL. All three tests performed equally well regarding sensitivity, PPV and NPV, but the specificity of methylation analysis (48.9%, 40.8–57.1) was significantly higher than that of HPV16/18 and/or methylation (31.9%, 95% CI 24.8–40.0) (p < 0.001) and hrHPV testing (26.2%, 95% CI 19.7–34.1) (p < 0.001).
Discussion
This study explored different triage tests and combinations for CIN3+ detection in a population of women referred to colposcopy because of abnormal cytology, including women younger and older than 30 years. Many studies have investigated repeated cytology, reflex hrHPV testing and genotyping for HPV16/18 as triage tests in patients referred to colposcopy clinic.19, 20 This study examined, in addition, the value of a test for methylation of two specific tumor suppressor genes (FAM19A4 and miR124‐2). It compared the results with those of hrHPV triage, and evaluated the potential combinations of hrHPV testing, including genotyping, with methylation analysis. A combination of HPV16/18 genotyping and/or methylation positivity performed well, with a high sensitivity (93.1%) and moderate specificity (49.4%) for ≥CIN3. This combination performed particularly well in women ≥30 years of age (sensitivity 98.2% and specificity 46.3%). In women with ≤LSIL, the study offered an adequate balance between sensitivity (83.3%) and specificity (60.6%). The results indicate that the combination of the two biomarkers (HPV16/18 genotyping and/or methylation positivity) could separate women into risk groups more accurately than either HPV genotyping or methylation analysis alone.
The diagnostic performance of the combined testing is based on the importance of HPV16/18 in ICC and in HSIL/CIN3,21, 22 and the presence of methylation in advanced precancers and ICC. Methylation testing with markers FAM19A4 and miR124‐2 on physician‐taken smears had a sensitivity for ASC‐H/HSIL/CIN3 detection of 77.8% and a specificity of 69.3% for the overall population. Interestingly, the sensitivity and specificity of the methylation analysis in women 30 years of age and over were particularly high (89.3% and 61.1%, respectively). This is in line with findings by Luttmer et al., who found an association between age and the performance of FAM19A4 as a triage test in hrHPV‐screened women, making it an attractive marker for women ≥30 years.23 Positivity for miR124‐2 and/or FAM19A4 detected all ICC, and 75.8% of HSIL/CIN3 lesions in the overall population, suggesting that the combination of these biomarkers identifies the most severe precancerous lesions.
The accurate identification of women who have histological HSIL/CIN3, within the subset of women with abnormal smears prior to colposcopy and biopsy, is potentially very important in avoiding overinvestigation and overtreatment. Even among women with cytological HSIL, many will not have HSIL/CIN3 or ICC. In this study, only 25% of these women had histological ASC‐H/HSIL/CIN3. hrHPV testing has shown high sensitivity for HSIL/CIN3 detection24, 25, 26, 27 and in many Western countries, hrHPV testing already has a place in screening, either as a stand‐alone screening test or in combination with cytology, or in triage. However, using the clinically validated GP5+/6+‐PCR‐EIA for triage of women with ≥ASC‐US on screening, hrHPV positivity showed high sensitivity (90.3%) but the lowest specificity (31.8%). This results in potential overtreatment of a large number of women who are not at high short‐term risk of cancer, and who may have regressing lesions.
The results obtained in this study are in keeping with previous evidence showing that older age and longer duration of hrHPV infection are associated with methylation positive HSIL/CIN3+.14, 28, 29 These findings suggest that further investigation of whether the current practice of treating all histologically confirmed ASC‐H/HSIL/CIN2 by LLETZ could be modified to provide more selective individual management of women with screen‐detected cervical abnormalities. Current thresholds for referral to colposcopy and treatment are based on the risk of HSIL/CIN2+.30 This leads to unnecessary referral and overtreatment of women with lesions that have a low short‐term risk of progression and a definite probability of regression.31 Our methylation results demonstrate heterogeneity in HSIL/CIN2 and HSIL/CIN3. It is important to investigate in prospective studies whether combined methylation and HPV16/18 testing could be used in practice to distinguish methylation positive women, who need urgent further investigation and treatment, from women who are methylation‐negative but positive for HPV16/18 and could be followed at appropriate intervals for regression to avoid unnecessary treatment.
This study has a number of limitations. The study was relatively small and only one woman with ASC‐US and five women with LSIL cytology had HSIL/CIN3+. Also, patients were not selected for treatment on the basis of the results of the methylation biomarkers. When the methylation biomarkers were studied individually, FAM19A4 was positive in all methylation‐positive HSIL/CIN3+ lesions, but only miR124‐2 was positive in five women with negative and LSIL/CIN1 biopsies. In the studied population, the additive value of miR124‐2 in the detection of CIN3+ and squamous cell carcinoma seems limited. Its benefit seems more evident in avoiding unnecessary biopsies (i.e., increase negative predictive value) when FAM19A4 is positive and miR124‐2 is negative. Further investigation of the use of these methylation markers individually requires large‐scale clinical trials conducted under careful supervision.
This was a cytology‐screened referral population including women who were screened opportunistically without prior screening. Therefore, the prior probability of cytological and histological abnormalities might have been higher32 and results from this study cannot be directly extrapolated to screening by hrHPV testing alone or co‐testing with cytology. The consistency, however, with other studies of primary HPV screening suggests that the principles identified here could be applied to different screening strategies.33, 34, 35 Colposcopy was used to identify possible lesions. To address the limitations to the accuracy of colposcopy and taking of biopsies,36, 37 multiple biopsies were taken, endocervical curettage, and a random biopsy in every woman who had <4 lesion‐directed biopsies to improve detection of lesions.
This study shows the potential for early accurate identification of HSIL/CIN3+ by testing for hrHPV and methylation of human tumor suppressor genes in routine liquid‐based cervical cytological samples. It suggests that HSIL/CIN3 is a heterogeneous group, consisting of methylation positive lesions and methylation negative lesions. We hypothesize that methylation positive HSIL/CIN3 lesions are advanced transforming lesions and are mostly detected in women ≥30 years of age with a persisting hrHPV infection.28 The methylation negative HSIL/CIN3 lesions may be found in younger women caused by HPV infections of a shorter duration or infections that follow a different pathway.29, 38 Further, larger, follow‐up studies in different screened populations are needed to investigate whether a combination of HPV16/18 typing and methylation testing is feasible for stratifying clinical management.
To evaluate further the heterogeneity of high‐grade precancer lesions, more markers should be evaluated. Such potential markers include methylation of more human genes, methylation of HPV DNA, study of miRNAs, study of somatic and germline SNPs and use of immunohistochemical marker patterns such as p16INK4a and HPV E4 to distinguish transforming and productive elements in CIN lesions. The development and clinical evaluation of these markers in prospective studies opens up the possibility of early and reliable identification of women who need rapid treatment of HSIL/CIN3 at high risk of progression to ICC or have ICC and avoid unnecessary treatment of potentially regressing HSIL/CIN2.
Acknowledgements
The authors thank Nadia Abu‐Lhiga, Agata Rodriguez and Inma Nicolas, Hospital Clínic for their clinical assistance. They thank Lorena Marimon and Esther Barnadas, Hospital Clínic, and Laura van der Hoeven, Stephanie van Zoelen, Peter Lanser and Henk van den Munckhof, DDL Diagnostic Laboratory, for their technical assistance. They also thank Miekel van de Sandt, DDL Diagnostic Laboratory, and Frank Smedts, Reinier de Graaf Gasthuis, for reviewing the biopsy diagnoses. Finally, they wish to thank all clinical and laboratory personnel and all women who participated in the study.
Conflict of Interest: C.J.L.M.M. is minority shareholder of Self‐screen B.V., a spin‐off company of VUmc; Self‐screen B.V. holds patents related to the work (i.e., hrHPV test and methylation markers for cervical screening). C.J.L.M.M. served occasionally on the scientific advisory board (expert meeting) of Qiagen and SPMSD/Merck, and has by occasion been consultant for Qiagen. He has been co‐investigator on a Sanofi Pasteur/MSD sponsored HPV vaccination trial in men of which his institute received research funding; he has small number of shares of Qiagen, and was minority shareholder of Diassay B.V. until April 2016.
References
- 1. de Kok IM, van der Aa MA, van Ballegooijen M, et al. Trends in cervical cancer in The Netherlands until 2007: has the bottom been reached? Int J Cancer 2011;128:2174–81. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 3. Rijkaart DC, Berkhof J, van Kemenade FJ, et al. HPV DNA testing in population‐based cervical screening (VUSA‐Screen study): results and implications. Br J Cancer 2012;106:975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santesso N, Mustafa RA, Schunemann HJ, et al. World Health Organization Guidelines for treatment of cervical intraepithelial neoplasia 2‐3 and screen‐and‐treat strategies to prevent cervical cancer. Int J Gynaecol Obstet 2016;132:252–8. [DOI] [PubMed] [Google Scholar]
- 5. McCredie MR, Sharples KJ, Paul C, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol 2008;9:425–34. [DOI] [PubMed] [Google Scholar]
- 6. Schiffman M, Wentzensen N. From human papillomavirus to cervical cancer. Obstet Gynecol 2010;116:177–85. [DOI] [PubMed] [Google Scholar]
- 7. Kyrgiou M, Mitra A, Arbyn M, et al. Fertility and early pregnancy outcomes after conservative treatment for cervical intraepithelial neoplasia. Cochrane Database Syst Rev 2015;9:CD008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ebisch RM, Siebers AG, Bosgraaf RP, et al. Triage of high‐risk HPV positive women in cervical cancer screening. Expert Rev Anticancer Ther 2016;16:1073–85. [DOI] [PubMed] [Google Scholar]
- 9. Khan MJ, Castle PE, Lorincz AT, et al. The elevated 10‐year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type‐specific HPV testing in clinical practice. J Natl Cancer Inst 2005;97:1072–9. [DOI] [PubMed] [Google Scholar]
- 10. Porras C, Rodriguez AC, Hildesheim A, et al. Human papillomavirus types by age in cervical cancer precursors: predominance of human papillomavirus 16 in young women. Cancer Epidemiol Biomarkers Prev 2009;18:863–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tjalma WA, Fiander A, Reich O, et al. Differences in human papillomavirus type distribution in high‐grade cervical intraepithelial neoplasia and invasive cervical cancer in Europe. Int J Cancer 2013;132:854–67. [DOI] [PubMed] [Google Scholar]
- 12. Hesselink AT, Heideman DA, Steenbergen RD, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high‐risk human papillomavirus DNA‐positive women. Clin Cancer Res 2011;17:2459–65. [DOI] [PubMed] [Google Scholar]
- 13. De Strooper LM, Hesselink AT, Berkhof J, et al. Combined CADM1/MAL methylation and cytology testing for colposcopy triage of high‐risk HPV‐positive women. Cancer Epidemiol Biomarkers Prev 2014;23:1933–7. [DOI] [PubMed] [Google Scholar]
- 14. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014;7:1251–7. [DOI] [PubMed] [Google Scholar]
- 15. Hesselink AT, Heideman DA, Steenbergen RD, et al. Methylation marker analysis of self‐sampled cervico‐vaginal lavage specimens to triage high‐risk HPV‐positive women for colposcopy. Int J Cancer 2014;135:880–6. [DOI] [PubMed] [Google Scholar]
- 16. van der Marel J, van Baars R, Quint WG, et al. The impact of human papillomavirus genotype on colposcopic appearance: a cross‐sectional analysis. BJOG 2014;121:1117–26. [DOI] [PubMed] [Google Scholar]
- 17. Bornstein J, Bentley J, Bosze P, et al. 2011 colposcopic terminology of the International Federation for Cervical Pathology and Colposcopy. Obstet Gynecol 2012;120:166–72. [DOI] [PubMed] [Google Scholar]
- 18. Luo W, Yang H, Rathbun K, et al. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real‐time PCR assay. J Clin Microbiol 2005;43:1851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arbyn M, Roelens J, Simoens C, Buntinx F, Paraskevaidis E, Martin‐Hirsch PP, Prendiville WJ. Human papillomavirus testing versus repeat cytology for triage of minor cytological cervical lesions. Cochrane Database Syst Rev 2013: CD008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McKenna M, McMenamin M, McDowell A. HPV16 and HPV18 genotyping triage in young women with borderline cytology or mild dyskaryosis: effect of age on genotype‐specific risk of high‐grade CIN. Cytopathology 2016;27:261–8. [DOI] [PubMed] [Google Scholar]
- 21. Cogliano V, Baan R, Straif K, et al. Cancer WHOIAfRo. carcinogenicity of human papillomaviruses. Lancet Oncol 2005;6:204. [DOI] [PubMed] [Google Scholar]
- 22. Wheeler CM, Hunt WC, Joste NE, et al. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst 2009;101:475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luttmer R, De Strooper LM, Berkhof J, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high‐risk HPV‐positive women of a gynecologic outpatient population (COMETH study). Int J Cancer 2016;138:992–1002. [DOI] [PubMed] [Google Scholar]
- 24. Bulkmans NW, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5‐year follow‐up of a randomised controlled implementation trial. Lancet 2007;370:1764–72. [DOI] [PubMed] [Google Scholar]
- 25. Castle PE, Stoler MH, Wright TC Jr, et al. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol 2011;12:880–90. [DOI] [PubMed] [Google Scholar]
- 26. Rijkaart DC, Berkhof J, Rozendaal L, et al. Human papillomavirus testing for the detection of high‐grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol 2012;13:78–88. [DOI] [PubMed] [Google Scholar]
- 27. Ronco G, Giorgi‐Rossi P, Carozzi F, et al. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol 2010;11:249–57. [DOI] [PubMed] [Google Scholar]
- 28. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV‐positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–9. [DOI] [PubMed] [Google Scholar]
- 29. Steenbergen RD, Snijders PJ, Heideman DA, et al. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer 2014;14:395–405. [DOI] [PubMed] [Google Scholar]
- 30. Stanczuk GA, Baxter GJ, Currie H, et al. Defining optimal triage strategies for hrHPV screen‐positive women‐an evaluation of HPV 16/18 genotyping, cytology, and p16/Ki‐67 cytoimmunochemistry. Cancer Epidemiol Biomarkers Prev 2017;26:1629–35. [DOI] [PubMed] [Google Scholar]
- 31. McAllum B, Sykes PH, Sadler L, Macnab H, Simcock BJ, Mekhail AK. Is the treatment of CIN 2 always necessary in women under 25 years old? Am J Obstet Gynecol 2011;205:478e1–7. [DOI] [PubMed] [Google Scholar]
- 32. Tranberg M, Larsen MB, Mikkelsen EM, et al. Impact of opportunistic testing in a systematic cervical cancer screening program: a nationwide registry study. BMC Public Health 2015;15:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Strooper LMA, Verhoef VMJ, Berkhof J, et al. Validation of the FAM19A4/mir124‐2 DNA methylation test for both lavage‐ and brush‐based self‐samples to detect cervical (pre)cancer in HPV‐positive women. Gynecol Oncol 2016;141:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dijkstra MG, van Niekerk D, Rijkaart DC, et al. Primary hrHPV DNA testing in cervical cancer screening: how to manage screen‐positive women? A POBASCAM trial substudy. Cancer Epidemiol Biomarkers Prev 2014;23:55–63. [DOI] [PubMed] [Google Scholar]
- 35. Luttmer R, De Strooper LM, Steenbergen RD, et al. Management of high‐risk HPV‐positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn 2016;16:961–74. [DOI] [PubMed] [Google Scholar]
- 36. Hopman EH, Kenemans P, Helmerhorst TJ. Positive predictive rate of colposcopic examination of the cervix uteri: an overview of literature. Obstet Gynecol Surv 1998;53:97–106. [DOI] [PubMed] [Google Scholar]
- 37. Stoler MH, Schiffman M, Atypical Squamous Cells of Undetermined Significance‐Low‐grade Squamous Intraepithelial Lesion Triage Study G . Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS‐LSIL Triage Study. JAMA 2001;285:1500–5. [DOI] [PubMed] [Google Scholar]
- 38. Herfs M, Yamamoto Y, Laury A, et al. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc Natl Acad Sci U S A 2012;109:10516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]


