Abstract
To reduce required capital and time investment in development of new pharmaceutical agents, there is an urgent need for preclinical drug testing models that are predictive of drug response in human tissues or organs. Despite tremendous advancements and rigorous multistage screening of drug candidates involving computational models, traditional cell culture platforms, animal models and most recently humanized animals, there is still a large deficit in our ability to predict drug response in patient groups and overall attrition rates from phase 1 through phase 4 of clinical studies remain well above 90%. Organ-on-a-chip (OOC) platforms have proven potential in providing tremendous flexibility and robustness in drug screening and development by employing engineering techniques and materials. More importantly, in recent years there is a clear upward trend in studies that utilize human-induced pluripotent stem cell (hiPSC) to developed personalized tissue or organ models. Additionally, integrated multiple organs on the single chip with increasingly more sophisticated representation of absorption, distribution, metabolism, excretion and toxicity (ADMET) process are being utilized to better understand drug interaction mechanisms in the human body and thus show great potential to better predict drug efficacy and safety. In this review, we summarize these advances, highlighting studies that took the next step to clinical trials and research areas with the utmost potential and discuss the role of the OOCs in overall drug discovery process at preclinical and clinical stage, as well as outline remaining challenges.
Keywords: Organ-on-a-chip, microfluidic technology, drug development, personalized medicine, human-derived induced pluripotent stem cells, tissue engineering
1. Introduction
The inevitable multiple phases of drug assessment from research and development (pre-human and clinical studies) to post-clinical evaluations have mandated the drug development process to be a slow-paced and over-priced procedure. The average capitalized cost for a new drug has been estimated at $2.5 billion in the R&D stage and a total of $2.8 billion post-approval (1). Formulating new drugs for various disease categories such as cancer, orphan diseases and neurological disorders becomes even more challenging and time-consuming when difficulties such as drug resistance and incomplete understanding of disorder pathophysiology come into play (2–7). For instance, a recent study on colorectal, breast and non–small cell lung cancers found that the mean for drug development duration is 8.9, 6.7 and 6.6 years for each cancer type with unsustainably high attrition rates for the breast cancer (83.9%), colorectal cancer (87.0%), and non–small cell lung cancer (92.0%) (8). In addition, according to a report by Tufts Center for the study of drug development, orphan drug development requires 15.1 years to reach product launch from the patent filing stage. As such, searching for makeshift methods to replace or expedite current drug development methods has gained large attention over the past two decades.
Another challenge facing the current drug development routes is in the achievement of high reliability and predictability in the outcome of drug treatment to ensure that the unforeseen side effects are minimized (9). This has been quite challenging with only 59% of the drugs entering phase II trials and only 21% reaching phase III (1). The use of animal models in drug development and evaluation is considered a key apparatus in medicine for the purpose of studying new therapies and evaluating new drugs. Despite the ubiquity of these models, the success rate of the subsequent drugs in human clinical trials has been noticeably low (10, 11). This degree of failure can be attributed to the major evolutionary differences between human and animal models which result in higher structural and biological complexity of human tissues or organs (12–15). For instance, a survey of 150 compounds resulting in human toxicity events during clinical development summarized and compared drug toxicity in humans and animal models and determined major deficits as rodent-only models were predictive of human toxicity in 43% and non-rodent only were predictive of 63%, with a combined true positive human toxicity concordance rate of 71% (16). As such, the intricate nature of human diseases makes it objectionable to be fully recapitulated by animal models, and the anticipated response and recovery can vastly diverge from the triggered reaction in humans. Failed trials such as Vioxx for rheumatoid arthritis (17, 18), TGN1412 for immunotherapy (19), HIV vaccine (20, 21) and parkinson’s disease treatments CEP-1347 (22) and Cogane (23, 24) are just a few examples.
One method to address such shortcomings is the use of genetically modified animals, especially mice. In this manner specific differences between the species can be addressed to achieve more clinically relevant models with increased representation of targeted physiology or pathways in humans. An example is the established difference between expression patterns of CYP2B gene in mice and humans, where several functional members of the CYP2B family are expressed in mice while in humans, CYP2B6 is the only functional member. To address this difference, investigators have created “CYP2B6 humanized mice” that expresses the CYP2B transgene in the liver, but has suppressed expression of all mouse CYP2B genes. Another species difference addressed in this manner is drug metabolization by UDP-glucuronosyltransferase (UGT) enzymes. Humanized UGT1 mice were established in 2010 by crossing UGT1-null mice with human UGT1 transgenic mice, (25) and a recent summary of toxicity and metabolism studies in hUGT1 mice is presented by Fujiwara et al. (26). Other examples include the use of human and mouse artificial chromosome vectors, (27) as well as chimeric mice with humanized liver (28, 29). Moreover, newer generations of humanized mice have been improved to possess functional and predictive human immune system response (30, 31). However, to realize their full potential, some limitations associated with these models have to be overcome; this includes interference from residual mouse immune system. Additionally, even in transgenic mice with cytokines derived from human genes, residual murine cytokines still show affinity for target sites, and thus may bond but not signal, interfering with critical pathways (32). Other limitations include excessively high costs, complex system-wide drug interactions that hinder mechanistic studies, limited accessibility of targeted sites for manipulation, sensing and imaging.
Moreover, there are other complications in the way of drug development; For instance, consideration of individual genetic variations is a relatively new phenomenon in development of cancer drugs. While epigenetic and environmental factors can be tremendously impactful in a patient’s response to treatment, these effects remain largely under-investigated and unknown. Additionally, intricate system-level drug-drug interactions add another level of complexity that is frequently neglected especially leading up to release of new pharmaceuticals and only after discovery of adverse effects in patients do they come into consideration. Therefore, there is an undeniable and urgent need to refine the process of drug discovery and development to account for genetic and epigenetic variations as well as environmental and circumstantial factors that alter treatment outcomes.
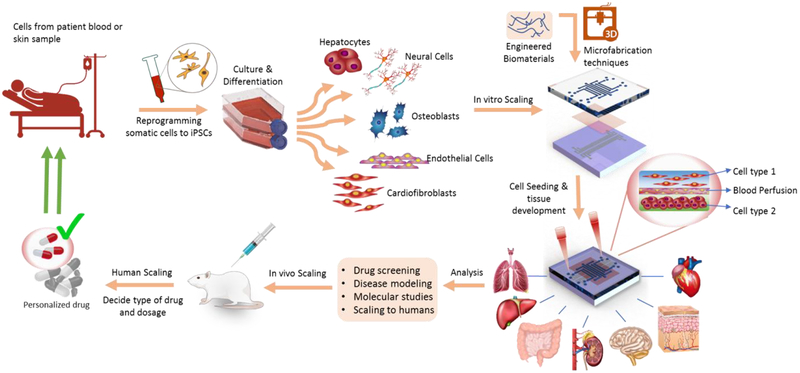
Recently, organ-on-a-chip (OOC) technology has drawn attention to be used in drug development and clinical drug testing model by reflecting the genetic characteristics of cells in each patient. OOC is an engineered assembly of a controlled compartment to study, measure, and control cell behavior and response to various drug stimuli by replicating the cellular behavior of the target tissue microenvironment. OOC has evolved from a combination of various engineering platforms such as microfluidic systems, engineered biomimetic tissues, and non-invasive monitoring system to address the difficulties of conventional drug testing models. Owing to recent advances in engineered biomaterials, it is now possible to design organoids with two-dimensional (2D) and three-dimensional (3D) scaffolds equipped with suitable extracellular matrix (ECM) to closely mimic human cell adhesion, migration, differentiation and function in vitro system. Many types of human and animal stem cells have been used to generate organoids for the OOCs. Especially, using human-induced pluripotent stem cells (hiPSCs), which are obtained from patient’s skin tissue or be directly harvested as pathogenic cells from patients, can be used to engineer personalized tissue constructs or disease models. Therefore, hiPSC-integrated OOCs provide a useful tool to establish personalized drug testing platforms that can mimic human physiology tuned for specific patient groups and individuals. Restrictions such as availability of patient-specific human cells, which used to limit the potential of the OOCs only a decade ago, have been largely lifted recently through the utilization of disease-specific cell lines, primary cells, and hiPSCs. The unlimited renewability and potency of hiPSCs to differentiate into major cell types to create various types of tissues or organoids enable OOCs to be a powerful tool for capturing complex drug interactions within multiple organ systems (Figure 1).
Figure 1.
Schematic of the cycle used in OOCs for personalized medicine. The cells are derived from patient and cultured and reprogrammed to different cell types. The device is fabricated using various microfabrication and 3D printing techniques. Next, the printed cells are seeded and cultured on the device. The target drug candidates are tested and analyzed using the OOC model followed by in vivo test. Next, the drug dosage and type are decided based on the responses received from the in vivo and OOC device and are later scaled to achieve the personalized drug for the patient.
Consequently, OOCs are particularly suitable for patient-specific drug development, owing to their remarkably higher throughputs, additional multimodal functionalities such as precise control of cellular microenvironments as well as ability to provide mechanical and electrical stimuli and recapitulate interactions between different functional units. Other advantages compared with conventional drug testing platforms include higher efficiency in screening time, lower cost, chemical/biological gradient screening (33), and reduced consumption in costly cell lines and chemical/biological reagents (34).
2. Design of the microfluidic OOCs.
The OOC design consists of an array of microfluidic channels which recurrently perfuse biological fluids such as culture media that contains nutrients and oxygen as well as biological agents and drugs in a controllable manner. Microfluidic possesses unique properties different from those of typical fluids. Specifically, Reynolds number is a characteristic (Re = ρvd/μ, where ρ=fluid density (g/cm3), v = fluid velocity (cm/s), d = channel diameter (cm), μ = dynamic viscosity (g/cm·s)), which is defined as the ratio of the inertial force causing turbulent flow to the viscosity causing laminar flow under 2,300. In the microfluidic systems, since the flows are often laminar, encountering two fluids can create stable concentration gradients which are only mixed at the contact interface through diffusion. This gradient can be effectively employed to separate proteins and cells in the microfluidic chips (35). Having uniform laminar characteristics, microfluidic systems can be easily controlled and directed to achieve high reproducibility, particularly in manipulating cell flow interactions (35, 36). The OOC controls the flow of microfluidics at the micro-level to measure and analyze the interactions with a target object (37, 38).
Furthermore, the OOC is combined with a microdevice-based non-invasive monitoring component (e.g., sensors and miniaturized microscopes) with an automated microfluidic control system to conduct sophisticated fluid manipulation and allow for repeated measurements and labor-free testing, reducing human errors and long-term operations. Integration of a biochemical analytical system into the OOCs enables the evaluation of various interactions occurring in the metabolite changes of the organoids under drugs treatment in a real-time manner. Moreover, miniaturization of the analytical system reduces the use of expensive reagents and sampling volume, as well as improving the analytical efficiency to evaluate organoids so that the overall effects of the experimental variables can be maximized (39). A chamber for controlling the microfluidic device, a biofilter and a channel for transferring and controlling analytes, as well as various types of sensors can be integrated with the microfluidic chips through micro-electromechanical systems (MEMS) fabrication techniques. Photolithography and soft lithography are some of the main techniques used. Excellent optical characteristics of these techniques can produce a chip with sophisticated patterns of cells and flow by spreading photoresists (PR) on a physiochemically stable silicon board, forming patterns by applying UV irradiation through a patterned mask, and removing PR. Polydimethylsiloxane (PDMS)-based microfluidics chip produced by soft lithography technique is an approach that provides excellent flexibility, biocompatibility, and oxygen permeability, thereby maintaining a high cell or tissue viability and allowing observation of biochemical responses inefficient manner (40). It is the most commonly used technique in the production of OOCs, being more economical than other existing methods (37, 41, 42). To facilitate the prototyping procedure, some studies have used poly (methyl methacrylate) (PMMA) in the fabrication of OOCs as an alternative to PDMS, as it can be easily etched with laser or thermally carved (43–46). Furthermore, the PMMA-based microfluidic chips can reduce drug, protein, or small molecule absorption/adsorption compared to the PDMS-based devices and can improve the robustness of the microfluidic chips during long operations.
Various types of tissues or organs can be cultured on a 2D or 3D scaffold in the microchamber while the nutrients, biological reagents and drugs can be continuously supplied by the microchannel. Since the residence time and physiological parameters are controllable, drug effects and toxicity can also be evaluated in various metabolic environments (39, 47). ECMs surround cells in the complex in vivo microenvironment, and they interact with cells near the ECMs. Therefore, development of engineered ECM to mimic physical and biological properties of the native ECM is considered the core of creating functional tissue models in vitro. Furthermore, various microfabrication technologies can be developed to resemble the architecture of specific tissues or organs (48). Chang and Robert et al. created a hydrogel-based 3D micro-organ via direct cell-writing automated printing (49). They demonstrated the strengths of this technique, which allows for the control of the spatial arrangement of cells using a hydrogel-based direct cell deposition technique as well as culturing of various cell types. The OOCs can mimic the structure-function relationship (hierarchical tissue such as cardiac muscle, hepatic cord) and mechanical force (shear stress, cyclic pressure, tensile) of each organ very well and show vascularization associated with cell viability and tissue function maintenance. It is also possible to apply electrical stimuli to the cells to study their electrophysiological behavior. Therefore, these features enable toxicity assessment as well as the mechanism of action for various drugs and substances on OOCs.
3. Cell resources for developing human-derived or personalized OOCs
To increase the resemblance of OOCs to human tissues, proper selection of biological resources is a key factor. Immortalized human cell lines and primary human cells have been commonly used in the OOC research as they are economical, easy to culture, and similar in biological characteristics to their in vivo counterpart. Immortalized cell lines with genetic variations can be continuously sub-cultured without any genotypic or phenotypic variations. These cell lines proliferate rapidly under simple culture conditions and can be useful in optimizing various parameters at the early stages of OOC development. However, these cell lines are insufficient to accurately predict human physiology such as metabolic drug activity, efficacy, and toxicity (50). Primary cells can be isolated and obtained from human biopsy or discarded tissues. Applying primary cells to the OOC for a target organ can produce pharmacologically reliable results regarding the drug response and toxicity to the primary cell (50). Although the primary cells provide a much improved model of the human physiology compared to the immortalized cell lines by replicating the cellular mechanisms and toxicity, engineering biomimetic tissues at organ level is yet unattainable due to the lack of cell proliferation and human tissue sources (51).
To overcome the issues of immortalized cell lines and primary cells, biomimetic tissue models have targeted the use of human stem cells to accurately predict human responses to drug treatments. Capable of self-regeneration and controllable differentiation to various types of cells or tissues under specific microenvironments, stem cells can provide a powerful gadget for engineering human tissue models. Using hiPSCs, human embryonic stem cells (hESCs), and human mesenchymal stem cells (hMSCs) in the field of the OOCs has recently drawn attention. With the ground-breaking work of Takahashi and Yamanaka in 2006, hiPSCs have ushered in an era of new hope for precision and personalized medicine (52). hiPSCs are created by introducing specific transcription factors (Oct4, Nanog, c-Myc, and Sox2) into somatic cells derived from an adult tissue to differentiate mature cells to immature cells, thereby giving the cells an ability to differentiate into various cell types such as cardiomyocytes, adipocytes, skeletal muscle progenitor cells, neural cells, pancreatic β-cells, and hematopoietic progenitor cells, etc. Moreover, as these cells are derived from normal somatic cells, they elude ethical issues that have limited the tremendous potential of hESCs. Yet, to realize the true impact of these cells in the preclinical stage several issues need to be addressed. For instance, controlled differentiation is challenging in 2D or 3D scaffolds and thus the goal of truly personalized disease models are still extremely cost prohibitive (53). In addition, it is strongly essential to increase the hiPSC differentiation efficiency and reproducibility. To solve these issues, generation of cell stocks from human leukocyte antigens homozygous donors that represent a specific population is a more attainable goal and can greatly reduce the associated costs to build personalized tissue model. Futures research efforts should be further devoted to addressing the mentioned issues.
Consequently, hiPSCs have drawn attention in the field of the personalized OOC platforms because the conversion of patients’ somatic cells to hiPSCs that provide customized healthy and disease models for personalized drug screening platform that recapitulate individual patient’s physiology much closer than animal models (54, 55). Recently, drastic advances in the development of the human OOCs using hiPSCs have been made with the aim of enabling efficient drug screening through the sophisticated control of physiological and chemical microenvironments in drug studies on human diseases (56). Safety and toxicity testing of drug candidates, reducing preclinical research time, and the possibility of developing drugs specific for individual patient genes and diseases are important reasons why hiPSCs must be introduced into the field of human OOC research for the pharmaceutical industry. Introduction of hiPSCs into the human OOCs will allow for a comparison of individual patients’ physiological responses to drugs. Despite the developmental potential of hiPSCs, there are limitations with respect to accurate mimicking of human organs by an OOC developed from hiPSCs. hiPSCs cannot accurately reproduce the complex cell–cell interactions and the interactive mechanisms between the tissues that comprise the cells and the chip due to the confounding variables including the genetic variability of humans (55). For this reason, research on OOC using patient-specific iPSCs is still limited to animal experiments and in vitro cell line assays. It is, therefore, necessary to establish hiPSC-based systematic experimental methods specific for each patient and disease model to study the individual cellular response to external physical stimuli, and changes in ECM during drug screening.
4. The evolution of the human-derived OOCs
The following section is dedicated to the recently developed OOCs for specific target organ and disease models based on human cells or iPSCs cultured on 2D or 3D biocompatible scaffolds. The functionality of each OOC was additionally examined for various drug assays, the summary of which is available in Table 1.
Table 1.
OOCs developed for drug screening and development studies
| OOC | Cell types used | Target disease or condition | Drugs tested | Functionality tested | Ref. |
|---|---|---|---|---|---|
| Lung on a chip | Cell line | Lung Cancer (NSCLC) | Tyrosine kinase inhibitor | Mechanical Strain, transfer across epithelial-endothelial tissue-tissue interface | (122) |
| Liver on a chip | Primary hepatocytes | Non-alcoholic fatty liver disease (NAFLD) | Pioglitazone, metformin | Oxygen gradient, Cell phenotyping, fat accumulation, metabolic capacity | (59) |
| Primary hepatocytes | Potential drug toxic effects | Acetaminophen (APAP) | Configuration, arrangement of capillary layers, inflammatory reaction towards enhanced cellular stress, expression of genes | (60) | |
| Primary hepatocytes | Drug-induced liver injury (DILI) | Troglitazone | Efflux media collection, compatibility for microfluidic coupling, clearance rates for drugs | (123) | |
| Cell line (HepG2/C3A) | Mitochondrial dysfunction | Troglitazone, rotenone | 3D cell aggregates, real-time oxygen measurement, mitochondrial Stress | (61) | |
| Kidney on a chip | hiPSCs | Albuminuria | Adriamycin | Cyclic mechanical strain, urinary filtrate, regulated vacuum, kidney glomerular capillary wall | (67) |
| Primary cell | CysA-induced damage | Cyclosporine A | Renal proximal tubule (PT) composed of a perfusable open lumen that possesses a programmable architecture | (68) | |
| Primary cell | Chronic kidney disease, urothelial cancer | Aristolochic acid I | Organ-organ interactions | (69) | |
| Gut on a chip | Cell line | Drug absorption | SN-38 (7-ethyl-10-hydroxycamptothecin) | Barrier function, microvilli expression, permeability coefficient | (124) |
| Cell line | Gut radiation injury | DMOG (dimethyl oxaloylglycin) | Villus differentiation, paracellular permeability, radio-protective effects of drug | (72) | |
| hiPSCs | Biologically responsive to exogenous stimuli | Tumor necrosis factor-α, IFN-γ | Epithelial-immune cell interactions, permeability | (125) | |
| CNS and PNS on a chip | Human neural progenitor cell line | Molecular toxicology | Acetaminophen, 5-fluorouracil, retinoic acid, doxorubicin, pitavastatin | Various cell states, protein expression | (75) |
| hiPSC-derived neurons | Familial Alzheimer’s disease | β-secretase inhibitor | Fluidic isolation, separation of axons from the soma | (76) | |
| Primary cell, hiPSC | Motor neuron disease | Motor neuron progenitor | Effect of microvascular network perfusion on neural activity | (126) | |
| BBB on a chip | hiPSCs | Central neural system disorders | Caffeine, cimetidine, doxorubicin | Physiologically relevant perfusion rates, medium recirculation, shear stress | (78) |
| Primary cell (hBMVEC) | Inflammatory stimulation | Lipopolysaccharide | Co-differentiating astrocytes, transport of inflammatory signals | (127) | |
| hiPSCs | Brain tumor | Paclitaxel, bortezomib | Barrier integrity with tight junction protein expression, higher TEER, verified the barrier functions of our BBB | (79) | |
| Heart on a chip | hiPSCs | Dilated cardiomyopathy | Isoproterenol, E-4031, verapamil, and metoprolol | Structural sarcomeric abnormalities, electrical function, beat rate & mechanical motion | (85) |
| hiPS-CMs | Low blood pressure, heart failure | Norepinephrine | Electrical stimulation, electrophysiology measurement | (82) | |
| hiPSCs | Cardiac cell outgrowth correlation of pharmacological compounds | Doxorubicin, endothelin-1, isoproterenol, phenylephrine, amiodarone | Fluidized space for the cell spheroids, geometric format suitable, quantitative measurement | (81) | |
| Muscle on a chip | Primary cell, biopsy | Duchenne muscular dystrophy | Stem cell therapy | Dystrophin expression, mechanical and topological properties | (88) |
| Biopsy | Multi-organ toxicity | Doxorubicin, atorvastatin, valproic Acid, acetaminophen, N-acetyl-m-aminophenol. | Culturing of cells in compartmentalized chambers with controlled interactions | (128) | |
| Primary cell | Cardiovascular diseases | Hemodynamic force, mechanical stimuli | Fluid velocity, shear stress, direct cell–cell contact, immunohistochemical analyses | (89) |
4.1. Liver-on-a-chip
Early studies on liver-on-a-chips focused on the functional activities related to the 3D perfusion culture of liver cell aggregates (57). Lee et al. developed a continuous, slow flow using an osmotic pressure pump to develop a hemispheric microfluidic chip that created an environment resembling the native liver environment (Figure 2A) (58). It was shown that the paracrine effects of hepatic stellate cells would improve the liver function without any autocrine effects caused by direct contact during the cultivation of hepatocytes spheroids (Figure 2B). Moreover, the slow flow created by the osmotic pressure pump maintained high survival rates to allow for long-term culturing of liver tissues.
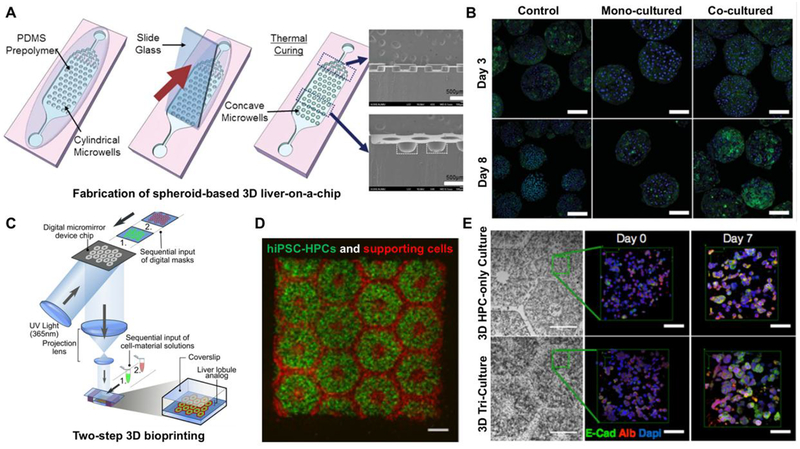
Figure 2.
Schematic showing microwell array PDMS plate based liver-on-a-chip device. (B) Generated 3D spheroids (mono-culture and co-cultured) on day 3 and 8. Reproduced from Lee et al (58) with permission from The Royal Society of Chemistry. (C) Hydrogel-based 3D bioprinted hepatic construct. hiPSC-HPCs and the support cells were patterned using two-step 3D bioprinting technique. (D) Fluorescent image is showing patterns of hiPSC-HPCs (green) and supporting cells (red). (E) Fluorescent images of albumin, E-cadherin, and nucleus staining of hiPSC-HPCs without supporting cells and in 3D triculture constructs. Reproduced from Ma et al. (63) with permission from Proceedings of the National Academy of Sciences of the United States of America.
The liver plays a crucial part in drug metabolism and detoxification and is, therefore, a key organ to study in development of novel drugs regardless of the type of disease. As such, much research has been conducted on the liver functions associated with hepatotoxicity. Essential functional components of liver-on-chips include not only drug metabolism, but also liver zonation (i.e., identifying the major toxic areas within the tissues and balancing them with detoxification) and transporter expression (i.e., localization upon drug exposure). Liver-on-chips with these components are applied to liver disease models namely, liver failure and hepatitis C models for drug screening and optimization. For instance, Kostrzewski et al. used primary human hepatocytes (PHHs) cultured on a 3D perfusable substrate to model the non-alcoholic fatty liver disease in vitro (59). They found that liver cells cultured under 3D conditions are more viable, functional, and consistently steroidal in compared to those cultured under 2D conditions. Effects of anti-steroid compounds were measured to validate the effectiveness of the disease model for drug screening. Freyer et al. used a perfused microscale 3D liver chip to confirm the dose-dependent toxicity of acetaminophen (60). Based on the finding that mitochondrial dysfunction plays an important role in pharmaceutical toxicity, Bavli et al. monitored the metabolic function of 3D aggregates of HepG2/C3A cell in the liver-on-a-chip (61).
In one study by Ware et al., in vitro platforms for assessment of drug-induced liver injuries were developed using iPSC-derived human hepatocyte-like cells (iPSC-HHs). Using industry-standard 96 well plates, these micropatterned cultures are capable of high-throughput drug screening and personalized medicine (62). Along with controls such as micropatterned PHHs and traditional monolayer cultures of iPSC-HHs and PHHs, iPSC-HH-based micropatterned co-cultures (iMPCCs) were treated with 37 different drugs including aspirin, buspirone, and dexamethasone, etc. for 6 days, and their toxicity was assessed by measuring albumin, ammonia, and adenosine triphosphate (ATP) levels. The liver toxicity and metabolic activity of the drugs showed higher sensitivity and specificity under the micropatterned iPSC-HHs and PHHs compared to monolayer controls. In another study, Ma et al. developed a 3D tri-culture model of hiPSC-derived hepatic progenitor cells (hiPSC-HPCs) by combining organ engineering and 3D bioprinting technologies (Figure 2C, D, and E) (63). To generate 3D patterns that approximate dimensions and structure of human liver lobules in vivo, a digital micromirror device chip was used to project a digitally created pattern and photopolymerize hydrogel matrices such as gelatin methacrylate (GelMA) and Glycidyl methacryloyl-hyaluronic acid that encapsulate hiPSC-derived hepatic cells, endothelial and mesenchymal-originated cells for support.
4.2. Kidney-on-a-chip
The kidney, which is the major site for hemofiltration, maintains homeostasis; it is involved in the excretion of waste materials from the cardiovascular system, in maintenance of the acid–base balance and total body fluid, and in electrolyte metabolism. Nephrons which filter and reabsorb substances are the basic unit of the kidney. Each nephron consists of a glomerulus, Bowman capsule, and nephric tubules. In kidney-on-a-chip research, mimicking the tubular structure of nephrons, establishing the complex culture conditions of kidney cells such as podocytes, and sophisticated control of the physical parameters that are applied to cells in microenvironments such as shear stress and flow are important (64). Ingber’s group was the first to study the proximal tubule-on-a-chip made of human cells to mimic the kidney (65). Primary human proximal tubule epithelial cells were used to analyze the morphology and function of cells when a flow that is adequate for an environment resembling the proximal tubules of nephrons is applied. In studies on the human tubule-on-chips resembling the human kidney, the fluidic model was found to recover rapidly due to the expression of Oct-2 inhibitors, in contrast to static models when cells are damaged by cisplatin. In addition, nephron-mimicking chips were produced and observed to reproduce epithelial and vascular tissues. Drug absorption through microfluidics resembling blood flow and the subsequent physiological responses could be observed.
Musah et al. developed a human kidney glomerulus model by inducing differentiation of hiPSC-derived podocytes that serve as markers for mature phenotypes (nephrin+, podocin+, WT1+, PAX2−) (66, 67). Specifically, hiPSC-derived intermediate mesoderm cells were differentiated into podocytes on one side of the channel and primary human glomerular microvascular endothelial cells on the other side, and physical microfluidic flows in the two channels was employed to create a model to mimic the renal glomerulus consisting of urinary and capillary parts. Furthermore, the capillaries of the glomerulus were found to contain macromolecules such as albumin and to perform kidney functions namely, excretion of small molecules. When a microfluidic glomerulus chip with this basic composition was treated with Adriamycin® (Doxorubicin) an anti-cancer drug increased albumin uptake by hiPSC-derived podocytes was observed within the microfluidic chip due to podocyte delamination and albumin loss from the vascular channels (67). Consequently, the possibility of development of a hiPSC-baed disease model and a glomerulus chip was assessed in this study. The applications of this kidney-on-a-chip was further explored in personalized medicine based on the function of the glomerulus in adriamycin-induced albuminuria. In another approach, Homan et al. used the bioprinting method to create 3D renal proximal tubules in a perfusable in vitro platform to quantitatively measure the extent of epithelial wall destruction in the proximal tubule caused by cyclosporine A, a known nephrotoxin (68). Furthermore, by connecting a kidney-on-a-chip and a liver-on-a-chip (69), Chang et al. exhibited that drug metabolism actively takes place in the liver and kidney and reported the bioactivation and transport mechanisms of aristolochic acid I.
4.3. Gut-on-a-chip
The small intestine performs important functions associated with drug absorption and immunity. There are epithelial cells with large surface area needed for nutrient absorption, as well as bacteria and intestinal microflora that transport nutrients on the intestinal surface. In many cases, the adverse effects of drugs affect the gastrointestinal tract, and it can be difficult to understand the causes and pathogenesis of many gut diseases. Therefore, it is essential to gather enough understanding of the relation between the gut and drug pharmacodynamics for effective drug screening. In the same way, proper recapitulation of the mechanical, structural, and physiological characteristics of the gut surface is considered important due to the structural variations in the gut.
Gut-on-a-chip can be a means for analysis of the gut pathophysiology to which the microbiome contributes. In vitro or animal models may be used to analyze disease mechanisms in an uncontrollable manner. For instance, Kim et al. developed a gut-on-a-chip coated with ECM consisting of two microfluidic channels composed of human intestinal epithelial (Caco-2) cells (70). This device created low shear stress on the microchannel and cyclic strain, mimicking the gut movement, thereby causing the fluids to flow at a slow rate, reproducing the microenvironment within the small intestine. It was observed that the cylindrical cells were polarized and spontaneously grew into folds that resembled the gut villi. Moreover, a co-culture of the gut microflora and Lactobacillus rhamnoses CG was done under conditions that did not decrease the survival of epithelial cells. When cells cultured by the transwell method under the same cellular conditions were compared with the above model, cells from the gut-on-a-chip were found to resemble the gut epithelial cells of healthy humans better than cells cultured in the static model. After human intestinal epithelial cells and symbiotic microorganisms were co-cultured for over 1 week, the independent effect of mechanical variations associated with microorganisms, inflammatory cells, and peristalsis were analyzed. It was found that probiotic and antibiotic therapies mostly prevented pathogenic bacteria from damaging the villi.
Furthermore, it was found that bacterial overgrowth was induced due to the lack of variation in epithelial cells similar to that observed in patients with Enterocleisis or Enteritis when the perfusion is maintained and movements such as peristalsis are halted (71). On the other hand, Jalili et al. conducted a study on intestinal damage in humans caused by acute γ-ray exposure, in which they studied cytotoxicity, apoptosis, and deoxyribonucleic acid (DNA) fragmentation that occurred on a gut-on-a-chip after γ-ray exposure and observed an inhibitory response to dimethyloxalylglycine (DMOG), which is a potential prophylactic radiation countermeasure drug (72). Workman et al. investigated an intestine chip by converging human iPSC-derived intestinal organoids and microengineering techniques (72). In this model, a consistent flow of media at 30 μL/h was applied to control the microenvironment within the chip and was then exposed to microbes to enhance cell differentiation. After 14 days of differentiation, cells expressing the intestine markers such as lysozyme+, MUC2+, chromogranin A+, and FABP2+ cells could be observed in the intestine chip. In an assessment of cell phenotypes and functions after exposure to the IFN-γ cytokine, which induces inflammatory bowel disease, expression of indoleamine 2,3-dioxygenase 1 and guanylate-binding protein 1 was observed. This study demonstrates the potential of intestine-on-chip for personalized medicine studies on intestinal cells.
4.4. Central nervous system (CNS) and peripheral nervous system (PNS)-on-a-Chip
The CNS refers to the skull-enclosed nervous tissues and the nerves within the vertebrae. The PNS refers to the nerves outside the skull and vertebrae and is connected to the CNS. The PNS consists of bundles of afferent fibers that carry sensory stimuli to the spinal cord or brain and efferent fibers that send signals from the brain to muscles. When these nerves are damaged, the stimuli sent from them are lost, and the muscles in which these nerves are distributed weaken due to inactivity. The nervous system consists of a variety of cell types; neurons that constitute the conducting tissues of the nervous system, and neuroglia, which are connective tissue cells and support or protect the nervous tissues. Different populations of neuroglia support various CNS tissues and are composed of astrocytes, oligodendrocytes, microglia, and ependymal cells. Furthermore, the PNS consists of schwann and satellite cells. The complexity of the nervous system makes it difficult to fully reproduce all mechanisms involved in different functions of the system. However, a recently developed microplatform enabled a more appropriate control of microbial environments, stimuli, and structures, and improved reproduction of cells in their native environment. Guo et al. used the neuromuscular junction (NMJ), which is a type of synapse in the PNS that is commonly used to study synaptic physiology and pharmacodynamics of a NMJ-on-a-chip that mimics such gaps within the body (73). Taylor et al. polarized axons within a microfluidics culture system and performed a biochemical analysis of a pure axonal fraction to address the challenge of axonal damage in neurodegenerative diseases (74). Increased growth of axons was observed in a chamber treated with neurotrophins, (e.g., neurotrophin-3), and the conditions which induce axonal growth were investigated.
It is necessary to fully understand the physiological conditions of the target area such as the tissue composition when developing CNS- and PNS-on-a-chip. For this reason, the chips must be able to mimic neurovascular units to reproduce the in vivo conditions of nervous-system diseases. Other studies on disease and drug screening using CNS- and PNS-on-a-chip, Nierode et al. observed toxicity differences among various compounds on a chip for human neural progenitor cell lines (75). Woodruff et al. used a microfluidic system made of hiPSC-derived neurons and found that neuron-specific impairment, due to the genetic influence of Alzheimer’s disease was associated with synaptic maintenance (76). This information suggests that OOC technologies are currently being used to understand further disease mechanisms and physiological pathways that affect the human brain, as well as providing a background for future studies on drug toxicity and development.
4.5. Blood brain barrier (BBB)-on-a-chip
The BBB is a cerebrovascular structure to prevent the blood in circulation from diffusion into the brain tissue and its extracellular fluid. It controls the transport of nutrients, and protects the brain from various toxic compounds and pathogens that may exist in the circulating blood. While water, oxygen, and energy sources, which are essential for survival, can easily move across the BBB by diffusion and active transport, potentially dangerous toxic substances or drugs cannot cross BBB. In this manner the BBB protects the brain from infection and harmful effects of different substances. However, such inhibitory effects also make it difficult for drugs to cross the BBB and reach the brain tissue, thus reducing drug efficiency. A BBB-on-a-chip was developed for screening drug penetration with a platform that can realistically mimic the complex function and structure of the BBB. The transwell migration assay, in which a fluorescent probe is used to check the permeability of a confluent monolayer of endothelial cells formed on a porous supporting structure, cannot exactly mimic the blood flow within actual blood vessels. Booth and Kim et al. inserted a porous membrane into a microfluidics chip to develop a BBB-on-a-chip that mimics cell–cell interactions and the flow of fluids (77). Cell–cell interactions were analyzed by culturing cerebrovascular cells (bEnd.3) and astrocytes (C8-D1A) on the porous membrane and characterized the microfluidic flow by measuring transendothelial electrical resistance (TEER). In addition, Wang et al. co-cultured hiPSC-derived cerebrovascular cells and astrocytes and reported higher TEER of membranes for co-culture compared to stand-alone culture and demonstrated the importance of cell co-culture in the reproduction of a BBB model (Figure 3A, B, and C) (78). Regarding studies on disease and drug screening using the BBB-on-a-chip, Wang et. al. (78) produced a microfluidic platform consisting of hiPSC-derived brain microvascular endothelial cells and analyzed drug response. Qi et al. used a hiPSC-based microchip to evaluate the role of the BBB in the presence of anti-brain tumor drugs (Figure 3D and E) (79). Improving upon traditional in vitro models, the main goal of the BBB-on-a-chip is to reproduce the microenvironment of the BBB more precisely through microfluidics technology.
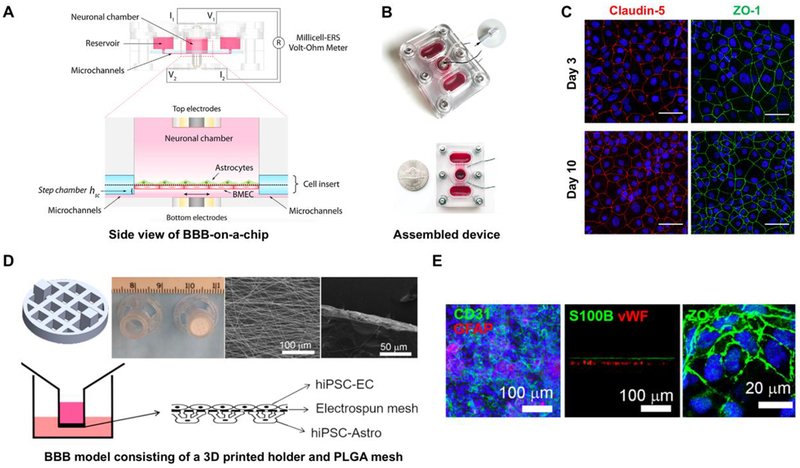
Figure 3.
(A) Side view of designed BBB-on-a-chip showing the fluid pathway, electrode wiring and BBB co-culture orientation. (B) Image is showing the actual assembled device with or without the lid. (C) Verification of BBB characteristic barrier integrity after co-cultures of hiPSC-derived BMECs and astrocytes for 10 days. Reproduced from Wang et al. (78) with permission from Wiley. (D) Schematic and image of a insert of a 3D printed holder and PLGA mesh developed for a BBB model (Top). Schematic showing the co-culture of hiPSC-ECs and hiPSC-Astro on the PLGA mesh. (E) Verification of astrocyte marker (GFAP) and EC junction marker (CD31), and glycoprotein (vWF) along with the tight junction protein after 7 days of culture. Reproduced from Qi et al. (79) with permission from ACS Publications.
4.6. Heart-on-a-chip
In one of the earliest heart-on-a-chip designs developed by Grossberg et al (80), cardiac muscle cells were cultured on a thin PDMS membrane to produce muscular thin films, then, electrical stimuli were applied to quantify the correlation between an electrical stimulus and the level of heart contraction by measuring the angle at which the epimysium was bent. To mimic the anisotropic physiology of the heart ventricles, ESCs were used to micromachine the fibronectin layers, and inducing the expression of cardiomyocytes which successfully demonstrated that cardiomyocytes stimulated in a 2D structure can create a contractile force. Christoffersson et al. used a cardiac-spheroid-on-a-chip to perform a high-throughput cardiac cell outgrowth assay to test 6 compounds and proposed the strengths of this technique in high-throughput analyses (81). Qian et al. developed a platform that can record tissue adhesion, electrophysiology, and contractility on a single chip and collect data regarding the cardiac electrophysiology and contractibility upon drug-induced stimulation (82).
In 2015, Mathur et al., cultured hiPSC-derived cardiomyocytes (hiPSC-CMs) in a biomimetic cardiovascular system to produce a heart-on-a-chip with an endothelial barrier that was subjected to the same shear stress conditions as those found in vivo (83). Cellular toxicity and efficacy of four drugs (Isoproterenol, E-4031, Verapamil, and Metoprolol) was tested in the hiPSC-derived cardiac-micro-physiological system (83). In a 3D biomimetic cardiac muscle tissue that features microcirculation similar to native cardiac muscles, hiPSCs performed ventricular muscle movements with uniform functions and delivered drugs to cells that were consistently exchanging nutrients and oxygen. This experiment confirmed that a drug screening assay can produce predictions of a 3D cell system that mimic the in vivo microenvironment more efficiently than a 2D cell system. In 2017, Ellis et al. co-cultured hiPSC-CMs and hiPSC-derived endothelial cells to reproduce the cardiac and blood vessel tissues by means of a myocardium-on-a-chip (84). In addition, Tzatzalos et al. reported that the hiPSC-CMs can represent an unlimited potential for healthy and disease-specific CMs to assess the efficacy of drugs for dilated cardiomyopathy (85). A drug screening study on a heart-on-a-chip using the hiPSC-derived cardiac-micro-physiological system reported that the heart-on-a-chip will allow researchers to efficiently select drugs that have effective functions and are nontoxic from a group of novel drugs during the early stages of cardiac drug development. These advances in drug development are of major importance for cardiovascular tissue because cardiotoxicity is commonly seen during drug trials and it is often one of the main reasons why clinical trials are suspended or drugs are withdrawn from the market.
4.7. Muscle-on-a-chip
Muscle diseases such as sarcopenia can lead to disabilities, lesions, dependence and overall lower lifestyle quality. Muscle-on-a-chip platforms have been used in mechanistic studies to better understand the human skeletal muscles and evaluate the effect and toxicity of drugs. It is also used to predict and prevent adverse effects of hypoglycemic agents in muscles. Madden et al. collected and cultured human muscle cells and developed a self-organized artificial muscle (86). This self-organized muscle contracted when brain signals that are transmitted to contract muscles were mimicked using electrical stimuli and chemicals. In other words, the muscle-on-a-chip behaved similarly to human muscles, thus mimicking the native conditions. McCain et al. developed a gelatin network with cells around two bioactive hydrogels to allow them to induce cell rearrangement and maturation, thereby characterizing the muscle tissue shape and transformation (87). Serena et al. used myotubes in patients with Duchenne muscular dystrophy to quantitatively assess the restorative ability of dystrophin for reducing muscle damage (88). They observed that mesoangioblasts produced dystrophin more efficiently than myoblasts and suggested that this phenomenon can be used to predict the effect of novel drugs and treatments that are aimed at improving dystrophin accumulation. Van Engeland et al. reported that stimuli affect molecular signaling and may modulate diseases that are hemodynamically associated with vascular smooth muscle cells (Figure 4 D–G) (89).
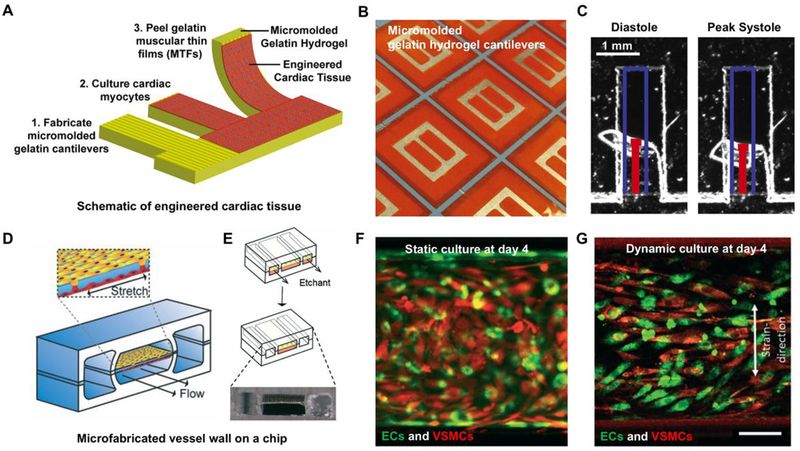
Figure 4.
Fabrication of biomimetic muscle-on-a-chip to study the effect and toxicity of drugs in muscles. (A) and (B) Gelatin hydrogel based muscular thin films (MTFs) were developed and cardiac tissues were cultured on gelatin hydrogel cantilevers. (C) Human iPS-derived cardiac myocytes were cultured on microfabricated MTFs and images were taken at diastole and systole. Reproduced from McCain et al. (87) with permission from Elsevier. (D) and (E) Schematic of vessel wall on a chip capable of creating physiological arterial strain and shear stress from the blood flow. (F) and (G) VSMCs were aligned and were more elongated compared to the cells cultured in the static condition. Reproduced from van England et al. (89) with permission from The Royal Society of Chemistry.
4.8. Multi organs-on-a-chip
OOC devices enable complex cell-cell and cell-ECM interactions of a specific tissue type or functional unit, but also lay the foundation to recapitulate circulation enabled interplay of various organs akin to the in vivo situation. As such, systemic effects of reagents and those involving endocrine or metabolic mechanisms may be elicited. The predictive capacity of these human-on-a-chip systems facilitate investigation of important in vivo effects, such as metabolic functions of hepatic tissue that are at times neglected in in vitro and co-culture studies (Table 2). Such strategies are posed to alleviate the previously mentioned inefficiencies and risks for drug developers by supplementing and possibly supplanting animal testing and are intensely pursued by regulatory and scientific agencies. Recognizing this tremendous potential, Defense Advanced Research Projects Agency, National Institute of Health and Food and Drug Administration have collaboratively supported advancement of tissue chip technology since 2012 with the launch of tissue chip for drug screening program. Ever since, efforts have involved linking individual organ chips to develop human multi-organ model systems (2014) recapitulating aspects of immune, nervous, respiratory, hepatic, gastrointestinal, circulatory, reproductive and urinary systems and in collaboration with Center for the Advancement of Science in Space testing of these systems in microgravity conditions aboard international space stations (2016).
Table 2.
Co-cultured tissues and organs within the scope of 6 multi-organ studies
| Co-Cultured Tissues | References |
|---|---|
| Liver, pancreas, skin, lung, intestine, bone marrow, heart, Fat tissue, spleen, kidney, brain, muscle, adrenal gland | (129) |
| Liver, pancreas | (130) |
| Intestinal epithelium, liver | (131) |
| Liver, colorectal tumors | (132) |
| Liver, brain, muscle, heart | (133) |
| Cervix, fallopian tube, uterus | (134),(135) |
5. Current state on acquiring approval for OOC devices in the preclinical and clinical trials
Preclinical and clinical studies account for 33% and 63% of the overall cost of drug development respectively (90). In every 10,000 new chemical entities (NCE), only one achieves market approval. The first step towards approval is target identification, which involves the identification of a specific potential target (cell receptors, proteins, enzymes, DNA, RNA and ribosomal proteins, among others), which could hold responsibile for the disease (91). Once identified, the target must be validated through experimental data by assessing the effects on certain pathologies, molecular and physiological inhibition as well as modulation activities supporting the possible therapeutic objectives of the target. Possible biologic effects and toxicity of chemical or drug candidates can be predicted via computer simulation to narrow down the options to the most promising molecules to be tested in vivo. For instance, computer aided drug design can be used in predicting receptor binding sites for a potential drug molecul.; structure-activity relationship software can predict the biologic activity of a drug candidate, and Quantitative SAR can analyze the relationship between the physicochemical properties and biological activity. Futhermore, the drug candidates can be evaluated and predicted in terms of carcinogenicity and mutagenicity using computer softwares. At this stage, cost-effectiveness, viability of producing the lead drug, and the drug delivery methods are also evaluated (92).
The main considerations during the preclinical stage drug development fall into the two categories of pharmacokinetics (PK) and pharmacodynamics (PD) studies. PK, determines the fate of an administered substance during the four stages of absorption, distribution, metabolism, excretion and toxicity (ADMET) and PD studies investigate the physiologic effect of the substance and its products on the organism to determine efficacy, toxicity and dose. These studies must be done in at least two different species in order to suggest that they could be safe to proceed to human clinical trials (93). Implementation of in vitro micro-physiological platforms able to determine the ADMET of drug candidates during preclinical stages could accelerate the drug development process.
In addition to their contribution to improving the prediction and performance in terms of drug functionality and effects on human models, the OOC technologies could also be employed as evaluation tools for the already market-approved drugs to study their interaction under certain rare disease conditions, varied populations or drug-drug interactions (94). On the other hand, in cases where a drug is withdrawn from the market – often a major concern for pharmaceutical companies—OOC systems could potentially target such drugs for further studies to increase the probability of future approvals.
Among the first steps that should be taken in order to achieve mass adoption of OOC technology for drug screening is to validate it as a reliable, fast, economical and standardized method. Theoretical schemes have been proposed to achieve such validation and proof of concept for drug screening has been provided by studies that assessed the damage on different tissues after drug administration (92). According to a report by Abaci and Schuler et al., two different platforms can be strategically employed to make reliable OOC and body-on-chip technologies at various stages of the drug development and screening (95). The first proposes a platform suitable for the preclinical stage, while the latter proposes a comprehensive platform for initial human trials as well as later phases of drug development. Currently, there is only one registered clinical trial planning to collect human samples that will be used to establish biomimetic human OOC technologies for disease modeling and for studying the role of the microbiome in the pathogenesis of human gastrointestinal diseases (96). However, there are currently no registered clinical trials using the OOC for drug screening nor validating the use of the OOC technologies for this purpose. The pharmaceutical industry’s major complications regarding approved drugs include failed phase 4 clinical trials, post-marketing failures and black-box warnings (i.e., strictest warning in the labeling of drugs and products given by the Food and Drug Administration) (97). Many commercially available drugs can cause hepatotoxic and cardiotoxic reactions, cardiotoxicity being a major cause of withdrawal (98). Recently, a number of studies have been directed to design proof-of-concept OOC models to perform drug screening on market-approved drugs to assess the damage produced on different tissues after drug administration. Examples of such studies include, (i) liver-on-a-chip platforms targeting acetaminophen hepatotoxicity (99), (ii) kidney-on-a-chip models to study drug-induced kidney damage caused by cisplatin, gentamicin, and doxorubicin (100), and (iii) skin-on-a-chip models for substance penetration studies (101). Additionally, a number of OOC studies have compared the interactions of different market-approved drugs on multiple organs (i.e., liver, lung and heart) to evaluate particular drug interactions such as propranol, acetaminophen and bleomycin (102).
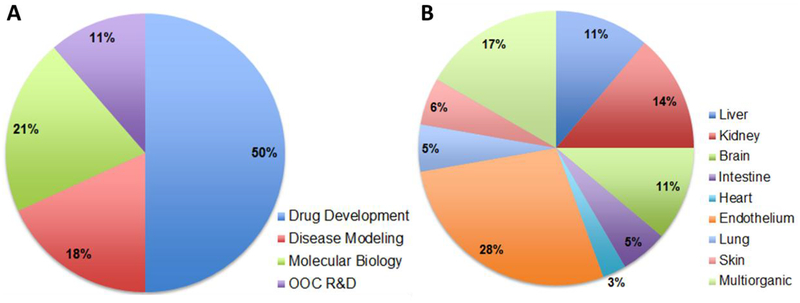
Since the early 2010s, multiple companies have pursued commercialization of various aspects of OOC technology. Examples include TissUse, based in Germany with commercialized 2-organ-chip and 4-organ-chips, Emulate Inc. with liver-chip, lung-chip and intestine-chip products, AxoSim Technologies LLC with a nerve-on-a-chip platform®, and UK based CN Bio Innovations and Hμrel® Corporation with assays covering several species. A total of 44 studies on microfluidic OOC devices were published between 2015 and 2018 in which the pharmaceutical and OOC manufacturing companies listed in (Table 3) were acknowledged or were sponsors. Some of those studies were posted in the publications section of the company’s websites as links to scientific journals. Consistent with the fact that the 44 reviewed publications were related or published by OOC and drug manufacturing companies and their collaborators, the main research area of the publications was related to drug development followed by OOC research and development (Figure 5A). Among the 44 studies, 36 were experimental, presenting endothelium and liver among the most studied single OOC (Figure 5B) by those companies in that timeframe, which is consistent with the use of OOCs for ADMET studies since endothelium and liver play essential roles on drug testing studies (103, 104). On the other hand, multi-OOC studies also represent an important field of study for this technology in which there have been models from co-cultures of 2 organs and up to 14 different tissue chambers (Table 2) (105). This information suggests that OOC technologies are currently being investigated and further developed.
Table 3.
Publications from 2015 to 2018 which acknowledged or were sponsored by companies involved in OOC technologies.
| Company | References |
|---|---|
| Astra Zeneca | (136–138) |
| Johnson & Johnson | (138) |
| Emulate | (137), (134, 139–141) |
| Mimetas | (130, 142–153) |
| SynVivo | (154–159) |
| Roche | (140) |
| GlaxoSmithkline | (160, 161) |
| Tissuse Gmbh | (130, 147, 162–167) |
| Nortis | (168) |
| Insphero | (132, 169, 170) |
| Hesperos | (78, 129, 131, 133, 137, 171–174) |
| Kirkstall | (175, 176) |
Figure 5.
Research on OOCs within Companies. A) Main topics of the publications acknowledged by companies on OOC technologies from 2015 to 2018. B) Most studied single organs for OOC technologies. Co-cultured organs (multi-organ systems) are listed in table 2.
6. Challenges and outlook
A major advantage coming with personalized medicine is the transformation of drug development from a generic “panacea” to a more stratified and individualized path. As such, the variety of drug is also reduced by categorizing comparable cases into finer subgroups with higher condition similarities. Although the use of personalized medicine and individualized drugs can improve the treatment accuracy and drug efficacy, there are still obstructions on the commercialization of these drugs; firstly, there exists an uncertainty in where exactly the line should be drawn when classifying the patients into similar groups. It is often hard to define the benchmarks for choosing the patients who are more credible to benefit from the treatment over the group who are not (106). In many cases, particularly in cancer medication, some drugs are not effective for the patient, but are still prescribed. This condition- often referred to as drug futility- can result in unfavorable effects while interfering with other medications (107–109). For instance, a study report by Fede et al. revealed that omeprazole, a drug prescribed to treat gastrointestinal diseases or as an agent complimenting the anti-inflammatory medication, had been prescribed to 11 of 20 of terminally ill cancer patients with no history of the disease or condition (110). As such, defining the border for medical prescription induces further complexity into the personalized OOC development.
Another barrier in developing personalized medicine is the cost and inadequacy of acquired human materials with respect to the high patient variability as well as deficiency of expertise in working with human tissues which demands time-consuming optimization and characterization steps for all experimental parameters including the culture conditions (111). The technical challenges on the fabrication process of the OOC devices need to be addressed in order to create a robust platform for long-term drug testing. Some challenges include possible microbial contamination in the chambers and throughout the microfluidic pathway, regulating the interactions between the different cell types, as well as maximizing the ECM similarity between the native tissue and the engineered biomaterial.
Moreover, the limitations in sensitivity of the conventional biochemical analytical systems impede the cell response measurements due to the small chamber size and low cell density. It has been suggested that the improvements in MEMS and nanofabrication techniques could contribute to development of highly sensitive nano- and microsensors which would function with considerably lower amounts of input biomarker sample (< pg/ml) (112). Furthermore, to reach large-scale industrial production, the fabrication process developed in the laboratory needs to be fully automated, a challenge which remains unaddressed due to the technical difficulties in material preparation. For instance, the PDMS chambers developed in the proof-of-concept models, often require highly delicate manual preparation lacking standard fully automated preparation protocol (112).
Although creating minimum levels of organ functionality may be an optimum solution for the OOC-based research, some studies still require thoroughly functional organ replication of all cell types, tissues and ECM, such as those focusing on expansive drug toxicity throughout the whole body. In these systems, animal and in vivo studies still represent a more reliable resource. However, current OOC studies can be coupled with in vivo animal studies to reinforce and complement the analysis. Advancements in designing multi-OOC systems could also open new pathways towards comprehensive drug analysis (112). Nonetheless, developing multi-OOCs brings new challenges and complexities to the system. For instance, optimizing the common culture media for multi-OOCs is necessary to achieve an adequate growth and differentiation of the human tissues to mimic their physiologic conditions. The use of some growth factors and differentiation reagents required for a particular cell type might adversely affect other cells during the common media circulation. An example of such interaction could be observed during the co-culture of hepatocytes and human umbilical vein endothelial cells (HUVEC), in which certain hepatocyte culture media can interfere with the metabolic activity of HUVEC cells and their production of other factors, like von willebrand factor (vWF) and metabolites such as lactate, thus, altering their ability to mimic their native conditions (113). This issue has been addressed in the latest designed multi-organ on chips by separating the media or by optimizing the co-culture ratio (114). However, to completely recapitulate the multi-organic tissue interactions, drug and disease effects a single media circulation system would be useful (115).
An important consideration for developing reliable and physiologically relevant OOC systems and to realize their true potential as predictive PK/PD models, at the minimum there should exist functional equivalent sub-systems that can accurately replicate each stage of the complex ADME process. This may be realized by integration of at least intestine-on-a-chip, Inter-organ interactions that are representative of blood or lymph circulation, liver-on-a-chip, and kidney-on-a-chip.
Moreover, it is imperative that every stage of design, test, conclusion and especially in vitro to in vivo extrapolation in such systems are guided by sensible and proven scaling principles. To achieve this, different approaches have been pursued. Allometric scaling which describes the relationship between organ size and function among different species provides a good starting point. Other scaling methods have also been proposed and are actively pursued, such as scaling each organ based on the residence time of blood (116, 117), scaling based on metabolic function and cell number (118) and computational PK/PD models (119). Using such scaling schemes, parameters such as appropriate fluid flow, effective drug dose, tissue masses and volumes, cell population, functional and metabolic activity and safety data can be determined and extrapolated for further investigation in animal models. After completion of studies investigating proper drug dose, efficacy and safety with animal models, the result should be scaled and extrapolated again to determine starting dose in first-in-human trials. In this manner, proper scaling methods bridge studies in conventional cell culture platforms, OOCs, animal models and phase 1 clinical trial. However, even advanced scaling methods impose some uncertainty, as in some cases the scaled effective drug dosage derived from studies in small model animals does not necessarily predict efficacy in humans.
Although the OOC technology is challenging in several aspects and requires clinical and technical modifications before turning into a viable commercialized drug testing resource, the scientific interest and rate of research progress is evident through the outburst of publications and industrial start-ups. Advancement in development of engineering techniques and computational modeling empowers OOCs for physiological characterization of drug screening culture models prior to clinical trials and animal models. Using these methods, it is possible to predict the in vivo drug behavior in humans with higher robustness, owing to the recent improvements in vascularized OOC systems. Prantil-Baun et al describe how Physiologically Based Pharmacokinetic (PBPK) modeling in vascularized OOCs enable the prediction of PK parameters from in vitro data and thus provide a potential solution to the scaling problem (120). Moreover, investing research on improvement of personalized OOCs can be a major investment in clinical testing as these chips can reduce the number of necessary clinical trials through individualizing drug dosage thus lowering the rate of clinical failures (55). Furthermore, the manufacturing process can be designed to streamline drug screening and preclinical testing with higher reliability by increasing repeatability and reproducibility in drug testing procedures. A future plan for OOC-based drug testing involves optimization of highly interactive multi-OOC systems by considering organ-organ interactions through which the process of drug development can be considerably revolutionized by better modeling the dose response and toxicity and prediction of drug resistance (121).
7. Conclusion
The process of drug approval is a long and costly procedure which is often practiced through animal models in vivo or 2D culture dishes in vitro. Those models, however, fail to fully recapitulate and predict the complexities of human tissues and diseases, resulting in probable drug failures for human patients. OOC technology offers a novel method to replace or complement the current drug study methods and expedite the drug assessment procedure by providing a more accurate replication of the 3D ECM of the native tissues and various cell-cell interactions using engineered biomaterials and microfluidic techniques. The present paper suggests the application of hiPSC based OOC devices as a powerful tool for personalized medicine, particularly in the development of drugs for rare and neurodegenerative diseases as well as a tool to minimize the probabilities of drug withdrawal or failure through designing multi-organs-on-a-chip technology.
8. Acknowledgements
This paper was funded by the National Institutes of Health (R01AR074234 and R21EB026824), the Qatar national Research Fund (a part of Qatar Foundation, NPRP9–144-3–021), and by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MSIP) (No. 2015R1A5A1009701). S.R.S. would like to recognize and thank Brigham and Women’s Hospital President Betsy Nabel, MD, and the Reny family, for the Stepping Strong Innovator Award through their generous funding.
10. References
- 1.DiMasi JA, Grabowski HG, Hansen RW JJohe. Innovation in the pharmaceutical industry: new estimates of R&D costs. 2016;47:20–33. [DOI] [PubMed] [Google Scholar]
- 2.Smith TJ. Challenges in Orphan Drug Development: Identification of Effective Therapy for Thyroid-Associated Ophthalmopathy. Annual review of pharmacology and toxicology. 2018. 2018/07//. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta D, Jackson R, Paul G, Shi J, Sabbagh M. Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015., Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015. Expert Opin Investig Drugs. 2017. 2017/06//;26, 26(6, 6):735,-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry-Kravis EM, Lindemann L, Jønch AE, Apostol G, Bear MF, Carpenter RL, et al. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nature Reviews Drug Discovery. 2018. 2018/04//;17(4):280–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley CG, Ellis LM. Raise standards for preclinical cancer research. Nature. 2012. 2012/03/28/;483:531. [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian HM, Prat F, Steensma DP, Kurzrock R, Stewart DJ, Sekeres MA, et al. Cancer research in the United States: A critical review of current status and proposal for alternative models. Cancer. 2018. 2018/07/15/;124(14):2881–9. [DOI] [PubMed] [Google Scholar]
- 7.Kantarjian H, Patel Y. High cancer drug prices 4 years later—Progress and prospects. Cancer. 2017. 2017/04/15/;123(8):1292–7. [DOI] [PubMed] [Google Scholar]
- 8.Nixon NA, Khan OF, Imam H, Tang PA, Monzon J, Li H, et al. Drug development for breast, colorectal, and non–small cell lung cancers from 1979 to 2014. Cancer. 2017. 2017/12/01/;123(23):4672–9. [DOI] [PubMed] [Google Scholar]
- 9.Stewart DJ, Stewart AA, Wheatley-Price P, Batist G, Kantarjian HM, Schiller J, et al. The importance of greater speed in drug development for advanced malignancies. Cancer Medicine. 2018. 2018/05/01/;7(5):1824–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark M, Steger-Hartmann T. A big data approach to the concordance of the toxicity of pharmaceuticals in animals and humans. Regulatory Toxicology and Pharmacology. 2018. 2018/07/01/;96:94–105. [DOI] [PubMed] [Google Scholar]
- 11.Arrowsmith J, Miller P. Phase II and Phase III attrition rates 2011–2012. Nature Reviews Drug Discovery. 2013. 2013/08/01/;12:569. [DOI] [PubMed] [Google Scholar]
- 12.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. 2013;110(9):3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mestas J, Hughes CC JTJoI. Of mice and not men: differences between mouse and human immunology. 2004;172(5):2731–8. [DOI] [PubMed] [Google Scholar]
- 14.Cabrera O, Berman DM, Kenyon NS, Ricordi C, Berggren P-O, Caicedo A JPotNAoS The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. 2006;103(7):2334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masjosthusmann S, Becker D, Petzuch B, Klose J, Siebert C, Deenen R, et al. A transcriptome comparison of time-matched developing human, mouse and rat neural progenitor cells reveals human uniqueness. Toxicol Appl Pharmacol. 2018. 2018/09/01/;354:40–55. [DOI] [PubMed] [Google Scholar]
- 16.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000. 2000/08//;32(1):56–67. [DOI] [PubMed] [Google Scholar]
- 17.Waxman HA. The Lessons of Vioxx — Drug Safety and Sales. New England Journal of Medicine. 2005. 2005/06/23/;352(25):2576–8. [DOI] [PubMed] [Google Scholar]
- 18.Horton R. Vioxx, the implosion of Merck, and aftershocks at the FDA. The Lancet. 2004. 2004/12/04/;364(9450):1995–6. [DOI] [PubMed] [Google Scholar]
- 19.Attarwala H JJoypJ. TGN1412: from discovery to disaster. 2010;2(3):332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayak TR, Andersen H, Makam VS, Khaw C, Bae S, Xu X, et al. Graphene for controlled and accelerated osteogenic differentiation of human mesenchymal stem cells. ACS Nano. 2011. June 28;5(6):4670–8. [DOI] [PubMed] [Google Scholar]
- 21.Benmira S, Bhattacharya V, Schmid ML. An effective HIV vaccine: A combination of humoral and cellular immunity? Curr HIV Res. 2010. 2010/09//;8(6):441–9. [DOI] [PubMed] [Google Scholar]
- 22.Neurology PSGPIJ. Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease. 2007;69(15):1480–90. [DOI] [PubMed] [Google Scholar]
- 23.Bai H, Li C, Shi G. Functional Composite Materials Based on Chemically Converted Graphene. Adv Mater. 2011. January 7;23:1089–115. [DOI] [PubMed] [Google Scholar]
- 24.Safety and Tolerability Study of Cogane™ in Healthy Volunteers and Parkinson’s Disease Patients - Full Text View - ClinicalTrials.gov.
- 25.Fujiwara R, Nguyen N, Chen S, Tukey RH. Developmental hyperbilirubinemia and CNS toxicity in mice humanized with the UDP glucuronosyltransferase 1 (UGT1) locus. PNAS. 2010. 2010/03/16/;107(11):5024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara S Humanized mice: A brief overview on their diverse applications in biomedical research. Journal of Cellular Physiology. 2018. 2018/04/01/;233(4):2889–901. [DOI] [PubMed] [Google Scholar]
- 27.Human and mouse artificial chromosome technologies for studies of pharmacokinetics and toxicokinetics. Drug Metabolism and Pharmacokinetics. 2018. 2018/02/01/;33(1):17–30. [DOI] [PubMed] [Google Scholar]
- 28.Chimeric mice with humanized liver: Application in drug metabolism and pharmacokinetics studies for drug discovery. Drug Metabolism and Pharmacokinetics. 2018. 2018/02/01/;33(1):31–9. [DOI] [PubMed] [Google Scholar]
- 29.Devoy A, Bunton-Stasyshyn RKA, Tybulewicz VLJ, Smith AJH, Fisher EMC. Genomically humanized mice: technologies and promises. Nature Reviews Genetics. 2012. 2012/01//;13(1):14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brehm MA, Shultz LD, Luban J, Greiner DL. Overcoming Current Limitations in Humanized Mouse Research. J Infect Dis. 2013. 2013/11/15/;208(Suppl 2):S125–S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akkina R JV. New generation humanized mice for virus research: comparative aspects and future prospects. 2013;435(1):14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh N, Kenney L, Jangalwe S, Aryee K-E, Greiner DL, Brehm MA, et al. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017. 2017/01/24/;12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagunas A, Martínez E, Samitier J. Surface-Bound Molecular Gradients for the High-Throughput Screening of Cell Responses. Front Bioeng Biotechnol. 2015. 2015/08/31/;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura H, Sakai Y, Fujii T JDm, pharmacokinetics. Organ/body-on-a-chip based on microfluidic technology for drug discovery. 2017. [DOI] [PubMed] [Google Scholar]
- 35.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nature Reviews Drug Discovery. 2006. 03/01/online;5:210. [DOI] [PubMed] [Google Scholar]
- 36.Kang L, Chung BG, Langer R, Khademhosseini A. Microfluidics for drug discovery and development: From target selection to product lifecycle management. Drug Discovery Today. 2008. 2008/01/01/;13(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014. August;32(8):760–72. [DOI] [PubMed] [Google Scholar]
- 38.Huh D, Torisawa Y-s, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: Organs-on-Chips. Lab on a Chip. 2012;12(12):2156–64. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Lee H, Selimović Š, Gauvin R, Bae H JDS. Organ-On-A-Chip: Development and Clinical Prospects Toward Toxicity Assessment with an Emphasis on Bone Marrow. 2015. May 01;38(5):409–18. [DOI] [PubMed] [Google Scholar]
- 40.Whitesides GM. The origins and the future of microfluidics. Nature. 2006. 07/26/online;442:368. [DOI] [PubMed] [Google Scholar]
- 41.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Analytical Chemistry. 1998. 1998/12/01;70(23):4974–84. [DOI] [PubMed] [Google Scholar]
- 42.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014. 03/12/online;507:181. [DOI] [PubMed] [Google Scholar]
- 43.Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. 2017:201612906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Łopacińska JM, Emnéus J, Dufva M JPO. Poly (dimethylsiloxane)(PDMS) affects gene expression in PC12 cells differentiating into neuronal-like cells. 2013;8(1):e53107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong T-F, Ju W-J, Wu M-C, Tai C-H, Tsai C-H, Fu L-M JM, et al. Rapid prototyping of PMMA microfluidic chips utilizing a CO 2 laser. 2010;9(6):1125–33. [Google Scholar]
- 46.Ma L, Zhou C, Lin B, Li W JBm. A porous 3D cell culture micro device for cell migration study. 2010;12(4):753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung JH, Esch MB, Prot J-M, Long CJ, Smith A, Hickman JJ, et al. Microfabricated mammalian organ systems and their integration into models of whole animals and humans. Lab on a Chip. 2013;13(7):1201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SH, Shim KY, Kim B, Sung JH. Hydrogel-based three-dimensional cell culture for organ-on-a-chip applications. Biotechnology Progress. 2017. 2017/05/01;33(3):580–9. [DOI] [PubMed] [Google Scholar]
- 49.Chang R, Nam J, Sun W. Direct cell writing of 3D microorgan for in vitro pharmacokinetic model. Tissue Eng Part C Methods. 2008. June;14(2):157–66. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Lee H, Selimovic S, Gauvin R, Bae H. Organ-on-a-chip: development and clinical prospects toward toxicity assessment with an emphasis on bone marrow. Drug Saf. 2015. May;38(5):409–18. [DOI] [PubMed] [Google Scholar]
- 51.Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol. 2014. February;25:45–50. [DOI] [PubMed] [Google Scholar]
- 52.Sayed N, Liu C, Wu JC JJotACoC. Translation of human-induced pluripotent stem cells: from clinical trial in a dish to precision medicine. 2016;67(18):2161–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka S JC. iPS Cells 10 Years Later. 2016;166. [DOI] [PubMed] [Google Scholar]
- 54.Zhao T, Zhang Z-N, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011. 2011/05/13/;474(7350):212–5. [DOI] [PubMed] [Google Scholar]
- 55.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nature Reviews Drug Discovery. 2015. 03/20/online;14:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott CW, Peters MF, Dragan YP. Human induced pluripotent stem cells and their use in drug discovery for toxicity testing. Toxicology Letters. 2013. 2013/05/10/;219(1):49–58. [DOI] [PubMed] [Google Scholar]
- 57.Ho CT, Lin RZ, Chang WY, Chang HY, Liu CH. Rapid heterogeneous liver-cell on-chip patterning via the enhanced field-induced dielectrophoresis trap. Lab Chip. 2006. June;6(6):724–34. [DOI] [PubMed] [Google Scholar]
- 58.Lee SA, No da Y, Kang E, Ju J, Kim DS, Lee SH. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte-hepatic stellate cell interactions and flow effects. Lab Chip. 2013. September 21;13(18):3529–37. [DOI] [PubMed] [Google Scholar]
- 59.Kostrzewski T, Cornforth T, Snow SA, Ouro-Gnao L, Rowe C, Large EM, et al. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J Gastroenterol. 2017. January 14;23(2):204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freyer N, Greuel S, Knospel F, Gerstmann F, Storch L, Damm G, et al. Microscale 3D Liver Bioreactor for In Vitro Hepatotoxicity Testing under Perfusion Conditions. Bioengineering (Basel). 2018. March 15;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bavli D, Prill S, Ezra E, Levy G, Cohen M, Vinken M, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2016. April 19;113(16):E2231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ware BR, Berger DR, Khetani SR. Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes. Toxicol Sci. 2015. June;145(2):252–62. [DOI] [PubMed] [Google Scholar]
- 63.Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A. 2016. February 23;113(8):2206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paoli R, Samitier J. Mimicking the Kidney: A Key Role in Organ-on-Chip Development. Micromachines. 2016;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jang K-J, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh K-Y, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integrative Biology. 2013;5(9):1119–29. [DOI] [PubMed] [Google Scholar]
- 66.Musah S, Dimitrakakis N, Camacho DM, Church GM, Ingber DE. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nature Protocols. 2018. 2018/07/01;13(7):1662–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng. 2017;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, et al. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci Rep. 2016. October 11;6:34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang SY, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight. 2017. November 16;2(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a Chip. 2012;12(12):2165–74. [DOI] [PubMed] [Google Scholar]
- 71.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016. January 5;113(1):E7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jalili-Firoozinezhad S, Prantil-Baun R, Jiang A, Potla R, Mammoto T, Weaver JC, et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018. February 14;9(2):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo X, Das M, Rumsey J, Gonzalez M, Stancescu M, Hickman J. Neuromuscular Junction Formation Between Human Stem-Cell-Derived Motoneurons and Rat Skeletal Muscle in a Defined System. Tissue Engineering Part C: Methods. 2010. 2010/12/01;16(6):1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2005. August;2(8):599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nierode GJ, Perea BC, McFarland SK, Pascoal JF, Clark DS, Schaffer DV, et al. High-Throughput Toxicity and Phenotypic Screening of 3D Human Neural Progenitor Cell Cultures on a Microarray Chip Platform. Stem Cell Reports. 2016. November 8;7(5):970–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodruff G, Reyna SM, Dunlap M, Van Der Kant R, Callender JA, Young JE, et al. Defective Transcytosis of APP and Lipoproteins in Human iPSC-Derived Neurons with Familial Alzheimer’s Disease Mutations. Cell Rep. 2016. October 11;17(3):759–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB). Lab Chip. 2012. April 24;12(10):1784–92. [DOI] [PubMed] [Google Scholar]
- 78.Wang YI, Abaci HE, Shuler ML. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng. 2017. January;114(1):184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi D, Wu S, Lin H, Kuss MA, Lei Y, Krasnoslobodtsev A, et al. Establishment of a Human iPSC- and Nanofiber-Based Microphysiological Blood–Brain Barrier System. ACS Applied Materials & Interfaces. 2018. 2018/07/05;10(26):21825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grosberg A, Alford PW, McCain ML, Parker KK. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip. 2011. December 21;11(24):4165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Christoffersson J, Meier F, Kempf H, Schwanke K, Coffee M, Beilmann M, et al. A Cardiac Cell Outgrowth Assay for Evaluating Drug Compounds Using a Cardiac Spheroid-on-a-Chip Device. Bioengineering. 2018;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian F, Huang C, Lin YD, Ivanovskaya AN, O’Hara TJ, Booth RH, et al. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip. 2017. May 16;17(10):1732–9. [DOI] [PubMed] [Google Scholar]
- 83.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015. March 9;5:8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellis BW, Acun A, Can UI, Zorlutuna P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics. 2017. March;11(2):024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tzatzalos E, Abilez OJ, Shukla P, Wu JC. Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies. Adv Drug Deliv Rev. 2016. January 15;96:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Madden L, Juhas M, Kraus WE, Truskey GA, Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife. 2015. January 9;4:e04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCain ML, Agarwal A, Nesmith HW, Nesmith AP, Parker KK. Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues. Biomaterials. 2014. July;35(21):5462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serena E, Zatti S, Zoso A, Lo Verso F, Tedesco FS, Cossu G, et al. Skeletal Muscle Differentiation on a Chip Shows Human Donor Mesoangioblasts’ Efficiency in Restoring Dystrophin in a Duchenne Muscular Dystrophy Model. STEM CELLS Translational Medicine. 2016. 2016/12/01;5(12):1676–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van Engeland NCA, Pollet AMAO, den Toonder JMJ, Bouten CVC, Stassen OMJA, Sahlgren CM. A biomimetic microfluidic model to study signalling between endothelial and vascular smooth muscle cells under hemodynamic conditions. Lab on a Chip. 2018;18(11):1607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schuhmacher A, Gassmann O, Hinder M JJotm Changing R&D models in research-based pharmaceutical companies. 2016;14(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drews J JS. Drug discovery: a historical perspective. 2000;287(5460):1960–4. [DOI] [PubMed] [Google Scholar]
- 92.Giri S, Bader A JDdt. A low-cost, high-quality new drug discovery process using patient-derived induced pluripotent stem cells. 2015;20(1):37–49. [DOI] [PubMed] [Google Scholar]
- 93.Hajba L, Guttman A JJoFC. Continuous-flow-based microfluidic systems for therapeutic monoclonal antibody production and organ-on-a-chip drug testing. 2017;7(3–4):118–23. [Google Scholar]
- 94.Materne E-M, Ramme AP, Terrasso AP, Serra M, Alves PM, Brito C, et al. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. 2015;205:36–46. [DOI] [PubMed] [Google Scholar]
- 95.Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol (Camb). 2015. 2015/04//;7(4):383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Collection of Gastrointestinal Malignant and Non-malignant Human Samples - Full Text View - ClinicalTrials.gov.
- 97.Mol PG, Arnardottir AH, Motola D, Vrijlandt PJ, Duijnhoven RG, Haaijer-Ruskamp FM, et al. Post-approval safety issues with innovative drugs: a European cohort study. 2013;36(11):1105–15. [DOI] [PubMed] [Google Scholar]
- 98.Minotti G Cardiotoxicity of non-cardiovascular drugs: John Wiley & Sons; 2010. [Google Scholar]
- 99.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. 2016;8(1):014101. [DOI] [PubMed] [Google Scholar]
- 100.Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R JTib. Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. 2016;34(2):156–70. [DOI] [PubMed] [Google Scholar]
- 101.Schimek K, Hsu H-H, Boehme M, Kornet JJ, Marx U, Lauster R, et al. Bioengineering of a full-thickness skin equivalent in a 96-well insert format for substance permeation studies and organ-on-a-chip applications. 2018;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. 2017;7(1):8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khetani SR, Berger DR, Ballinger KR, Davidson MD, Lin C, Ware BR JJola. Microengineered liver tissues for drug testing. 2015;20(3):216–50. [DOI] [PubMed] [Google Scholar]
- 104.Jung Y, Ji H, Chen Z, Chan HF, Atchison L, Klitzman B, et al. Scaffold-free, human mesenchymal stem cell-based tissue engineered blood vessels. 2015;5:15116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller PG, Shuler ML JB, bioengineering. Design and demonstration of a pumpless 14 compartment microphysiological system. 2016;113(10):2213–27. [DOI] [PubMed] [Google Scholar]
- 106.Kimmelman J, Tannock I JNRCO. The paradox of precision medicine. 2018;15(6):341. [DOI] [PubMed] [Google Scholar]
- 107.Corcoran M JCcjotMCC Polypharmacy in the older patient with cancer. 1997;4(5):419–28. [PubMed] [Google Scholar]
- 108.Guthrie B, Makubate B, Hernandez-Santiago V, Dreischulte T JBm. The rising tide of polypharmacy and drug-drug interactions: population database analysis 1995–2010. 2015;13(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Riechelmann RP, Zimmermann C, Chin SN, Wang L, O’Carroll A, Zarinehbaf S, et al. Potential drug interactions in cancer patients receiving supportive care exclusively. 2008;35(5):535–43. [DOI] [PubMed] [Google Scholar]
- 110.Fede A, Miranda M, Antonangelo D, Trevizan L, Schaffhausser H, Hamermesz B, et al. Use of unnecessary medications by patients with advanced cancer: cross-sectional survey. 2011;19(9):1313–8. [DOI] [PubMed] [Google Scholar]
- 111.Holmes A, Bonner F, Jones D JNRDD. Assessing drug safety in human tissues—what are the barriers? 2015;14(8):585. [DOI] [PubMed] [Google Scholar]
- 112.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov. 2015. 2015/04//;14(4):248–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Freyer N, Greuel S, Knöspel F, Strahl N, Amini L, Jacobs F, et al. Effects of Co-culture media on hepatic differentiation of hiPSC with or without HUVEC Co-Culture. 2017;18(8):1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Calejo I, Costa-Almeida R, Reis R, Gomes M, editors. OPTIMIZATION AND ESTABLISHMENT OF A CO-CULTURE MODEL TO STUDY CELLULAR INTERACTIONS IN TENDON-TO-BONE INTERFACE Orthopaedic Proceedings; 2018: The British Editorial Society of Bone & Joint Surgery. [Google Scholar]
- 115.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. 2013;13(18):3496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab on a Chip. 2009. 2009/05/21/;9(10):1385–94. [DOI] [PubMed] [Google Scholar]
- 117.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab on a Chip. 2010. 2010/02/21/;10(4):446–55. [DOI] [PubMed] [Google Scholar]
- 118.Moraes C, Labuz JM, Leung BM, Inoue M, Chun T-H, Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr Biol (Camb). 2013. 2013/09//;5(9):1149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab on a chip. 2013. 2013/09/21/;13(18):3496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prantil-Baun R, Novak R, Das D, Somayaji MR, Przekwas A, Ingber DE JArop, et al. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. 2018;58:37–64. [DOI] [PubMed] [Google Scholar]
- 121.Bovard D, Iskandar A, Luettich K, Hoeng J, Peitsch MC JTR, Application. Organs-on-a-chip: A new paradigm for toxicological assessment and preclinical drug development. 2017;1:2397847317726351. [Google Scholar]
- 122.Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, et al. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. 2017;21(2):508–16. [DOI] [PubMed] [Google Scholar]
- 123.Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Ying Shun T, Gough A, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Experimental Biology and Medicine. 2015. 2016/01/01;241(1):101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pocock K, Delon L, Bala V, Rao S, Priest C, Prestidge C, et al. Intestine-on-a-Chip Microfluidic Model for Efficient in Vitro Screening of Oral Chemotherapeutic Uptake. ACS Biomaterials Science & Engineering. 2017. 2017/06/12;3(6):951–9. [DOI] [PubMed] [Google Scholar]
- 125.Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, et al. Enhanced Utilization of Induced Pluripotent Stem Cell–Derived Human Intestinal Organoids Using Microengineered Chips. Cellular and Molecular Gastroenterology and Hepatology. 2018. 2018/01/01/;5(4):669–77.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Osaki T, Sivathanu V, Kamm RD. Engineered 3D vascular and neuronal networks in a microfluidic platform. Scientific Reports. 2018. 2018/03/26;8(1):5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brown JA, Codreanu SG, Shi M, Sherrod SD, Markov DA, Neely MD, et al. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. Journal of Neuroinflammation. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oleaga C, Bernabini C, Smith AST, Srinivasan B, Jackson M, McLamb W, et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Scientific Reports. 2016. 02/03/online;6:20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnology and bioengineering. 2016;113(10):2213–27. [DOI] [PubMed] [Google Scholar]
- 130.Bauer S, Huldt CW, Kanebratt KP, Durieux I, Gunne D, Andersson S, et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Scientific Reports. 2017;7(1):14620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Esch MB, Ueno H, Applegate DR, Shuler ML JLoaC. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. 2016;16(14):2719–29. [DOI] [PubMed] [Google Scholar]
- 132.Kim J-Y, Fluri DA, Marchan R, Boonen K, Mohanty S, Singh P, et al. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. Journal of biotechnology. 2015;205:24–35. [DOI] [PubMed] [Google Scholar]
- 133.Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Scientific reports. 2016;6:20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nawroth J, Rogal J, Weiss M, Brucker SY, Loskill P JAhm. Organ- on- a- chip Systems for Women’s Health Applications. 2018;7(2):1700550. [DOI] [PubMed] [Google Scholar]
- 135.Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. 2017;8:14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andersson TB JB, pharmacology c, toxicology. Evolution of novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. 2017;121(4):234–8. [DOI] [PubMed] [Google Scholar]
- 137.Ewart L, Dehne E-M, Fabre K, Gibbs S, Hickman J, Hornberg E, et al. Application of microphysiological systems to enhance safety assessment in drug discovery. 2018;58:65–82. [DOI] [PubMed] [Google Scholar]
- 138.Lal MA, Young KW, Andag U. Targeting the podocyte to treat glomerular kidney disease. Drug discovery today. 2015;20(10):1228–34. [DOI] [PubMed] [Google Scholar]
- 139.Barrile R, van der Meer AD, Park H, Fraser JP, Simic D, Teng F, et al. Organ- on- Chip Recapitulates Thrombosis Induced by an anti- CD154 Monoclonal Antibody: Translational Potential of Advanced Microengineered Systems. 2018. [DOI] [PubMed]
- 140.Ilic D JRm. Latest developments in the field of stem cell research and regenerative medicine compiled from publicly available information and press releases from nonacademic institutions 1 January–28 February 28 2018. 2018;13(4):361–70. [DOI] [PubMed] [Google Scholar]
- 141.Jain A, Barrile R, van der Meer AD, Mammoto A, Mammoto T, De Ceunynck K, et al. Primary Human Lung Alveolus- on- a- chip Model of Intravascular Thrombosis for Assessment of Therapeutics. 2018;103(2):332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vv Duinen, Heuvel A, Trietsch S, Lanz H, Gils J, Zonneveld A, et al. 96 perfusable blood vessels to study vascular permeability in vitro. Scientific reports. 2017;7(1):18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lanz HL, Saleh A, Kramer B, Cairns J, Ng CP, Yu J, et al. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC cancer. 2017;17(1):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nature communications. 2017;8(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Junaid A, Mashaghi A, Hankemeier T, Vulto P. An end-user perspective on Organ-on-a-Chip: Assays and usability aspects. Current Opinion in Biomedical Engineering. 2017;1:15–22. [Google Scholar]
- 146.Wevers NR, Van Vught R, Wilschut KJ, Nicolas A, Chiang C, Lanz HL, et al. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Scientific reports. 2016;6:38856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, et al. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. 2016;33(3):272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R. Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. Trends in biotechnology. 2016;34(2):156–70. [DOI] [PubMed] [Google Scholar]
- 149.Moreno EL, Hachi S, Hemmer K, Trietsch SJ, Baumuratov AS, Hankemeier T, et al. Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab on a Chip. 2015;15(11):2419–28. [DOI] [PubMed] [Google Scholar]
- 150.van Duinen V, Trietsch SJ, Joore J, Vulto P, Hankemeier T. Microfluidic 3D cell culture: from tools to tissue models. Current opinion in biotechnology. 2015;35:118–26. [DOI] [PubMed] [Google Scholar]
- 151.Jang M, Manz A, Volk T, Kleber A. Study of melatonin-mediated effects on various hepatic inflammatory responses stimulated by IL-6 in a new HepG2-on-a-chip platform. Biomedical microdevices. 2018;20(3):54. [DOI] [PubMed] [Google Scholar]
- 152.Jang M, Neuzil P, Volk T, Manz A, Kleber A. On-chip three-dimensional cell culture in phaseguides improves hepatocyte functions in vitro. Biomicrofluidics. 2015;9(3):034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koo Y, Hawkins BT, Yun Y. Three-dimensional (3D) tetra-culture brain on chip platform for organophosphate toxicity screening. Scientific reports. 2018;8(1):2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pradhan S, Smith AM, Garson CJ, Hassani I, Seeto WJ, Pant K, et al. A Microvascularized Tumor-mimetic Platform for Assessing Anti-cancer Drug Efficacy. 2018;8(1):3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tang Y, Soroush F, Sheffield JB, Wang B, Prabhakarpandian B, Kiani MF JSr. A biomimetic microfluidic tumor microenvironment platform mimicking the EPR effect for rapid screening of drug delivery systems. 2017;7(1):9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Terrell-Hall TB, Ammer AG, Griffith JI, Lockman PR JF, CNS Bot. Permeability across a novel microfluidic blood-tumor barrier model. 2017;14(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Soroush F, Zhang T, King DJ, Tang Y, Deosarkar S, Prabhakarpandian B, et al. A novel microfluidic assay reveals a key role for protein kinase C δ in regulating human neutrophil–endothelium interaction. 2016;100(5):1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deosarkar SP, Prabhakarpandian B, Wang B, Sheffield JB, Krynska B, Kiani MF JPo. A novel dynamic neonatal blood-brain barrier on a chip. 2015;10(11):e0142725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Prabhakarpandian B, Shen M-C, Nichols JB, Garson CJ, Mills IR, Matar MM, et al. Synthetic tumor networks for screening drug delivery systems. 2015;201:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grosberg A, Nesmith AP, Goss JA, Brigham MD, McCain ML, Parker KK JJop, et al. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. 2012;65(3):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martz L JS-Be. Pulmonary edema on a chip. 2012;5(48):1251-. [Google Scholar]
- 162.Sieber S, Wirth L, Cavak N, Koenigsmark M, Marx U, Lauster R, et al. Bone marrow- on- a- chip: Long- term culture of human haematopoietic stem cells in a three- dimensional microfluidic environment. Journal of tissue engineering and regenerative medicine. 2018;12(2):479–89. [DOI] [PubMed] [Google Scholar]
- 163.Schimek K, Hsu H-H, Boehme M, Kornet J, Marx U, Lauster R, et al. Bioengineering of a full-thickness skin equivalent in a 96-well insert format for substance permeation studies and organ-on-a-chip applications. Bioengineering. 2018;5(2):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Materne E-M, Ramme AP, Terrasso AP, Serra M, Alves PM, Brito C, et al. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. Journal of biotechnology. 2015;205:36–46. [DOI] [PubMed] [Google Scholar]
- 165.Materne E-M, Maschmeyer I, Lorenz AK, Horland R, Schimek KM, Busek M, et al. The multi-organ chip-a microfluidic platform for long-term multi-tissue coculture. Journal of visualized experiments: JoVE. 2015. (98). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hasenberg T, Mühleder S, Dotzler A, Bauer S, Labuda K, Holnthoner W, et al. Emulating human microcapillaries in a multi-organ-chip platform. Journal of biotechnology. 2015;216:1–10. [DOI] [PubMed] [Google Scholar]
- 167.Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab on a Chip. 2015;15(12):2688–99. [DOI] [PubMed] [Google Scholar]
- 168.Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Ying Shun T, Gough A, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Experimental biology and medicine. 2016;241(1):101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim J-Y, Fluri DA, Kelm JM, Hierlemann A, Frey O. 96-well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. Journal of laboratory automation. 2015;20(3):274–82. [DOI] [PubMed] [Google Scholar]
- 170.Frey O, Misun PM, Fluri DA, Hengstler JG, Hierlemann A. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nature communications. 2014;5:4250. [DOI] [PubMed] [Google Scholar]
- 171.Wang YI, Oleaga C, Long CJ, Esch MB, McAleer CW, Miller PG, et al. Self-contained, low-cost Body-on-a-Chip systems for drug development. Experimental Biology and Medicine. 2017;242(17):1701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang YI, Carmona C, Hickman JJ, Shuler ML. Multiorgan Microphysiological Systems for Drug Development: Strategies, Advances, and Challenges. Advanced healthcare materials. 2018;7(2):1701000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abaci HE, Gledhill K, Guo Z, Christiano AM, Shuler ML. Pumpless microfluidic platform for drug testing on human skin equivalents. Lab on a Chip. 2015;15(3):882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Esch MB, Prot J-M, Wang YI, Miller P, Llamas-Vidales JR, Naughton BA, et al. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab on a Chip. 2015;15(10):2269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chandorkar P, Posch W, Zaderer V, Blatzer M, Steger M, Ammann C, et al. Fast-track development of an in vitro 3D lung/immune cell model to study Aspergillus infections. Scientific reports. 2017;7(1):11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Martin KC, Yuan X, Stimac G, Bannerman K, Anderson J, Roy C, et al. Symmetry- breaking in branching epithelia: cells on micro- patterns under flow challenge the hypothesis of positive feedback by a secreted autocrine inhibitor of motility. Journal of anatomy. 2017;230(6):766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]