Abstract
Background
The incidence of squamous cell carcinoma of the oral tongue (SCCOT) is increasing in people under age 40. There is an urgent need to identify prognostic markers that help identify young SCCOT patients with poor prognosis in order to select these for individualized treatment.
Materials and methods
To identify genetic markers that can serve as prognostic markers for young SCCOT patients, we first investigated four young (≤40 years) and five elderly patients (≥50 years) using global RNA sequencing and whole‐exome sequencing. Next, we combined our data with data on SCCOT from the cancer genome atlas (TCGA), giving a total of 16 young and 104 elderly, to explore the correlations between genomic variations and clinical outcomes.
Results
In agreement with previous studies, we found that SCCOT from young and elderly patients was transcriptomically and also genomically similar with no significant differences regarding cancer driver genes, germline predisposition genes, or the burden of somatic single nucleotide variations (SNVs). However, a disparate copy number variation (CNV) was found in young patients with distinct clinical outcome. Combined with data from TCGA, we found that the overall survival was significantly better in young patients with low‐CNV (n = 5) compared to high‐CNV (n = 11) burden (P = 0.044).
Conclusions
Copy number variation burden is a useful single prognostic marker for SCCOT from young, but not elderly, patients. CNV burden thus holds promise to form an important contribution when selecting suitable treatment protocols for young patients with SCCOT.
Keywords: age, copy number variation, prognosis, squamous cell carcinoma of the oral tongue, whole‐exome sequencing
1. INTRODUCTION
Squamous cell carcinoma of the head and neck (SCCHN) is the sixth most common cancer in the world.1 SCCHN occurs predominantly in patients over 50 years old, and similar to most malignancies aging is a pervasive risk factor.2 Epidemiologic studies show an increasing incidence of SCCHN in patients under 40 years of age, especially for SCC of the oral tongue (SCCOT).3, 4, 5 As these patients have not encountered extensive exposure to alcohol/tobacco, the main risk factors for SCCHN, tumor induction in young patients are likely to be etiologically distinct.5 Numerous efforts have been made to identify differences in pathogenesis between the two age‐groups, but no consensus has yet been reached.5 More recent next‐generation sequencing techniques indicate that the genomic profiles and mutations in driver genes are very similar between young and older SCCOT patients,6 suggesting similar mechanisms of tumorigenesis.
The prognosis of SCCHN in young patients is controversial. Many studies have not shown any significant difference in prognosis between young and older patients, whereas some have suggested that young patients have worse outcomes and thus need more aggressive approaches to improve locoregional control and survival.7, 8 Other studies in turn have demonstrated that young patients have a better overall prognosis than older patients.3, 9 Our own results from SCCOT showed recurrences more frequently in young compared to old patients.10 To gain further insight into SCCOT in young patients and enable identification of those with poor prognosis and in need of more individualized treatment, we used next‐generation sequencing to investigate both gene expression profiles and genomic features in tumors from young and elderly patients and evaluated the correlation between molecular pattern and clinical outcome.
2. MATERIALS AND METHODS
2.1. Patients and samples
In order to provide a clear‐cut distinction for age classification, we considered patients aged ≤40 years at diagnosis as young and ≥50 years as elderly. Four young and five elderly patients with varying clinical features of SCCOT that were treated at Norrland's University hospital (NUS) were investigated. One patient (age 42), not grouped as young or elderly, was included for sequencing but not analyzed for age‐related correlations. Tumor tissue, matched tumor‐free tongue tissue from the opposite side of the tumor, and/or blood samples were collected. Patient characteristics and samples are shown in Table 1. The study was approved by the Regional Ethics Review Board, Umeå, Sweden (Dnr 03‐201 and Dnr 08‐003 M) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients.
Table 1.
Clinical data of patients with squamous cell carcinoma of the oral tongue (SCCOT)
| No. | ID | Gender | Age at diagnosis | Smoking history | Locationa | TNM | Follow‐up months | Status | Samples |
|---|---|---|---|---|---|---|---|---|---|
| 1 | p35 | Female | 24 | Non‐smoker | 2 | T2N0M0 | 13 | Dead | T, TF |
| 2 | p82 | Female | 19 | Former Smokerb | 1 | T4aN0M0 | 18 | Dead | T, BL |
| 3 | p98 | Male | 31 | Non‐smoker | 3 | T2N0M0 | 62 | Alive | T, TF,BL |
| 4 | p111 | Female | 31 | Smoker | 2 | T1N0M0 | 53 | Alive | T, TF,BL |
| 5 | p119 | Male | 67 | Non‐smoker | 2 | T2N0M0 | 47 | Alive | T, TF,BL |
| 6 | p124 | Male | 55 | Unknown | 1, 3 | T4N2bM0 | 3 | Dead | T, TF,BL |
| 7 | p137 | Female | 71 | Non‐smoker | 2 | T2N0M0 | 38 | Alive | T, TF,BL |
| 8 | p149 | Female | 69 | Former smokerc | 2 | T1N0M0 | 18 | Deadb | T, TF,BL |
| 9 | p154 | Female | 42 | Unknown | 4 | T1N1M0 | 28 | alive | T, TF,BL |
| 10 | p212 | Male | 52 | Non‐smoker | 1, 5 | T4aN2bM0 | 5 | alive | T, TF,BL |
BL, Blood; T, Tumor; TF, Tumor‐free tongue.
1 = Tongue; 2 = Border of tongue; 3 = Overgrowth into floor of mouth; 4 = Ventral side of tongue; 5 = Base of tongue.
Smoked for 1 y, stopped 3 y before the tumor was diagnosed.
Stopped smoking 20 y before the tumor was diagnosed.
Died from another disease.
2.2. DNA/RNA isolation
Biopsies were fresh frozen in liquid nitrogen and stored at −80°C. AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany) was used to isolate DNA and RNA. For the blood samples, fractions of plasma, buffy coat, and red blood cells were prepared and stored at −80°C. Buffy coat DNA was extracted using the illustra Nucleon Genomic DNA Extraction kit (GE Healthcare, UK). Quantity and purity of DNA/RNA was measured using a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). DNA quality was confirmed by gel electrophoresis and RNA quality by Agilent RNA 6000 Nano kit (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA).
2.3. RNA sequencing (RNA‐Seq) and data analysis
RNA samples from tumor and tumor‐free tongue tissues were subjected to RNA‐Seq at Otogenetics (Otogenetics Corporation, Atlanta, GA, USA) or Novogene (Novogene Bioinformatics Institute, Beijing, China). Sequencing libraries were generated using The Illumina TruSeq Stranded Total RNA Library Preparation Kit (Illumina, San Diego, CA, USA) or the NEBNext Ultra Directional RNA Library Prep Kit (NEB, Ipswich, MA, USA), respectively. RNA‐Seq alignment, assembly, and differential gene expression were carried out using HISAT2, StringTie, and Ballgown.11 Sample similarity in gene expression profiles was compared by performing principal component analysis (PCA). Human papillomavirus (HPV) infection was assessed by HPVDetector using whole‐transcriptome data.12
2.4. Whole‐exome sequencing (WES) and data analysis
DNA samples were subjected to WES at Otogenetics or Novogene, and the Agilent SureSelect Human All Exon kits (V5 or V6, respectively; Santa Clara, CA, USA) were used to capture exome regions. Somatic single nucleotide variations (SNVs) were identified using MuTect (v.1.1.7) and called according to the filter parameter PASS.13 Whenever available, paired blood sample was used as germline reference. Otherwise, the matched tumor‐free tongue sample was used to identify somatic SNVs in the tumor sample. To reduce false‐positive calls, variants were filtered out if coverage <30 and an alternate allele read depth <5. Common variants with minor allele frequency >0.1% reported in the 1000 genomes, dbSNP common, ESP6500, ExAC, or CG46 databases were also removed.
Somatic copy number variation (CNV) analysis was conducted using the EXCAVATOR2 tool.14 Whenever available, paired blood sample was used as germline reference (paired mode). Otherwise, the matched tumor‐free tongue sample (paired mode) was used. Log2R was denoted as the log‐transformed copy number ratio between tumor and control samples. A segmentation log2R value 0 indicates normal diploid status. To measure CNV burden, fraction of copy number‐altered genome (FCA) was calculated by dividing the number of bases in segments with mean log2R >0.2 or <−0.2 by the number of bases in all segments.15
Germline single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were identified using GATK HaplotypeCaller (version 4.0).16 Variants with at least 5 counts of the alternate allele and an alternative allele frequency of at least 20% were identified. Of these, we filtered out variants with >0.05% allele frequency in the 1000 genomes, dbSNP common, ESP6500, ExAC, or CG46 databases. Based on the latest curated list of 152 cancer predisposition genes,17 we further selected for potential cancer predisposing truncation and missense variants.
2.5. The cancer genome atlas (TCGA) data collection and analysis
TCGA has studied 528 patients with SCCHN, including 133 patients with SCCOT. Thirteen SCCOT patients were young (19‐40 years), and 102 were elderly (50‐90 years). Clinical information, somatic SNV, and CNV data (in terms of FCA) were downloaded using cBioPortal.15 In the TCGA data, overall survival time was unknown for one young SCCOT patient and CNV data were not available for three elderly patients. The effect of CNV on overall survival was estimated by Kaplan‐Meier and correlations between CNV and clinical outcome in young and elderly patients were determined by Fisher's exact test using IBM SPSS statistics 23. P < 0.05 was considered significant.
3. RESULTS
3.1. Overview of transcriptomes in tumor and tumor‐free tongue samples
RNA‐Seq data were analyzed to study gene expression profiles (both coding and non‐coding RNAs) in tumor and matched tumor‐free tongue samples. Principal component analysis (PCA) clearly separated tumor‐free and tumor samples (Figure 1). As SCCHN genetic profiles and prognosis differ according to HPV status,18 we investigated HPV infection using HPVDetector. In keeping with previous reports from SCCOT,19 no sign of HPV was seen. Comparing tumors between young and elderly patients, no age‐specific transcriptional profiles were identified.
Figure 1.
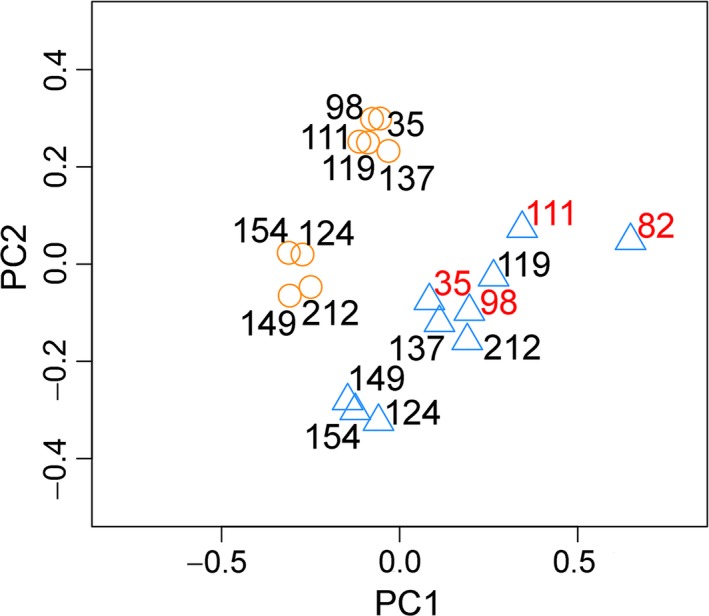
Gene expression profiles in tumor and tumor‐free tongue samples from ten patients with SCCOT. Principal component analysis (PCA) revealed distinct gene expression profiles in tumors (blue triangle) compared to tumor‐free controls (orange circle). Overall, gene expression profiles in tumors from young patients (patient ID in red) were similar to those from older patients
3.2. Somatic single nucleotide variations (SNVs)
Somatic SNVs in tumors from 10 patients were identified by MuTect. The number of identified SNVs in the tumor samples ranged from 97 to 400. No correlation between variant number and age was seen. The highest number of mutations was identified in the youngest patient (p82), a former smoker with a T4aN0M0 tumor and poor outcome. According to the list of cancer driver genes found by 20/20+, TUSON, or MutsigCV,20 several cancer driver mutations were identified (Table 2). Gene ontology analysis showed that these genes were enriched in chromatin organization and negative regulation of cell proliferation. Compared to previous reports and the elderly patients studied here, no driving events were specific for the four young patients, or related to clinical outcome.
Table 2.
Summary of data in patients with SCCOT
| Studya | ID | Age‐groupb | Somatic variants in tumor samples | Germline SNVs in cancer predisposition genes | Clinical outcome | |
|---|---|---|---|---|---|---|
| CNV burden (measured as FCA) | SNVs in cancer driver genes | |||||
| TCGA | TCGA‐CR‐7393 | Young | 0.000 | TP53 | NA | Better |
| TCGA | TCGA‐CR‐7391 | Young | 0.000 | MORC4 | NA | Better |
| NUS | p111 | Young | 0.002 | AMOT, BRWD3, KMT2E, ZNF268 | BRCA1 | Better |
| NUS | p98 | Young | 0.004 | CTNNB1, HERC4, KMT2D | ERCC2, EXT1 | Better |
| TCGA | TCGA‐BA‐A6DB | Young | 0.025 | TP53 | NA | Worse |
| NUS | p154 | 0.05 | ABCA7, DHX15, RIMS2, SPEN, TP53 | RET | Better | |
| TCGA | TCGA‐CN‐A640 | Young | 0.093 | ATRX, CDKN2A, FAT1, TP53, RAC1, BCL9, TARDBP | NA | Worse |
| NUS | p119 | Elderly | 0.102 | ATM, JAK2, KRT15, TP53, UBR5 | ND | Better |
| TCGA | TCGA‐H7‐A6C4 | Young | 0.118 | CDKN2A, TP53, GATA3, USP28, XPO1, BIRC6, WDR33 | NA | Worse |
| NUS | p212 | Elderly | 0.123 | GNPTAB, KDM5C, KMT2D, PIK3CA, TGFBR2, TP53, TRIP12 | ND | NA |
| TCGA | TCGA‐BA‐6873 | Young | 0.143 | CASP8, CDKN2A, IDH1, TP53, CCAR1, EPHA2, NOTCH1, CSMD3 | NA | Worse |
| NUS | p149 | Elderly | 0.165 | INPPL1, MED12 | ND | Worse |
| NUS | p82 | Young | 0.17 | CDKN2A, FAT1, TP53 | ND | Worse |
| TCGA | TCGA‐CV‐7180 | Young | 0.213 | CDKN2A, TP53 | NA | Worse |
| TCGA | TCGA‐CV‐5979 | Young | 0.240 | TP53 | NA | Better |
| TCGA | TCGA‐MT‐A51X | Young | 0.253 | ARID1B, BCLAF1 | NA | Better |
| TCGA | TCGA‐CQ‐7065 | Young | 0.288 | TP53 | NA | Worse |
| NUS | p137 | Elderly | 0.298 | CDKN2A, FLT3, TP53 | RAD51C | Better |
| TCGA | TCGA‐CV‐7255 | Young | 0.303 | CDKN2A, TP53, CHD8, SOX9, BRWD3, CSMD3, HLA‐B, LYN | NA | Worse |
| NUS | p124 | Elderly | 0.32 | CDKN2A, SMAD3, TP53 | ND | Worse |
| TCGA | TCGA‐CV‐A465 | Young | 0.379 | CDKN2A, TP53, NOTCH1 | NA | Worse |
| TCGA | TCGA‐CN‐4737 | Young | 0.512 | TP53, KMT2D | NA | Better |
| NUS | p35 | Young | 0.447c | ARID2, CHRDL1, FGFR1, RB1, TP53 | NA | Worse |
CNV, copy number variation; FCA, fraction of copy number‐altered genome; NA, not available; ND, not detected; SNV, single nucleotide variation.
NUS (10 SCCOT patients treated at Norrland's University Hospital), TCGA (13 young SCCOT patients in the TCGA study).
Young (19‐40 y), Elderly (50‐90 y).
FCA derived from comparison with tumor‐free tongue. Patients are listed according to increasing CNV burden in tumor.
3.3. Somatic copy number variations (CNVs)
Based on segmented WES data by EXCAVATOR2, broad level CNVs (length >50% of a chromosome arm) were found in the tumor samples of two young patients, p35 and p82, including loss of 3p and 8p and gain of 3q, 5p, and 8q, typical for SCCHN,18 whereas no broad level CNV was found in the other two young patients (p98 and p111) (Figure 2). CNV burden, calculated as the fraction of copy number‐altered genome (FCA), showed that only 0.4% and 0.2% of the tumor genomes from p98 and p111 showed changes in copy number, respectively. All tumor samples from elderly patients showed broad level CNVs (Figure 2). CNV burden (measured as FCA) in tumor samples is shown in Table 2.
Figure 2.
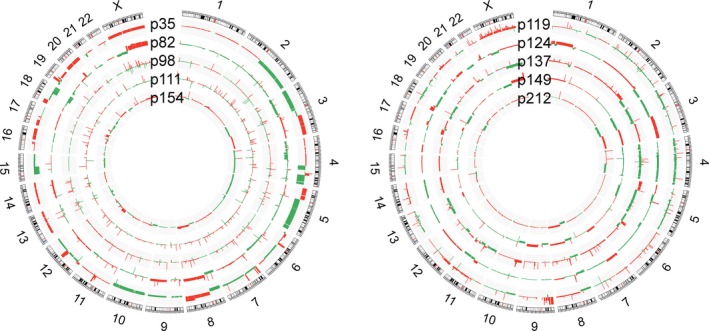
Somatic copy number variations (CNVs) analyzed by EXCAVATOR2. Circos plot showing the segmented data of 10 tumors. Each ring represents one tumor sample with corresponding patient ID. Copy number gains are marked in red, and copy number losses are marked in green. Young patients (≤40 y) (p35, p82, p98 and p111) and one patients aged 42 y (p154) are shown in the left circle. In the right circle, data from the 5 elderly patients (aged ≥ 50 y) (p119, p124, p137, p149, and p212) are shown
3.4. Correlation between CNV and clinical outcome
High levels of CNV were seen in tumors from the two young patients with poor outcome (p35 and p82), whereas low tumor CNV burdens were present in the two patients with better outcome (p98 and p111) (Table 2). Patient 35 died from disease with brain metastasis 13 months after treatment and patient 82 from disease 18 months after treatment. In contrast, p98 and p111 showing low‐CNV levels were alive without any sign of disease at 62 and 53 months of follow–up, respectively. To investigate the correlation between CNV levels and clinical outcome further, data from TCGA were analyzed using the same age criteria (patients aged ≥50 years were considered elderly, and those ≤40 years were classified as young). Based on the estimate that 4.8% of the human genome can be variable,21 an FCA value >0.05 (5% of the copy number‐profiled genome) was defined as high CNV and ≤0.05 as low CNV. First, we looked at the whole group of SCCHN, showing no significant correlations between CNV burden and overall survival using log‐rank test (Figure 3, upper panel). When looking at SCCOT only and combining SCCOT data from TCGA with our SCCOT data (ALL‐SCCOT), significantly better overall survival was seen in young SCCOT patients with low CNV (n = 5) compared to young patients with high CNV (n = 11) (P = 0.044) (Figure 3, lower panel). Even if the total number of young patients analyzed is low, no young patient with low‐CNV levels had died, whereas for elderly patients, no significant difference in overall survival was seen between patients with low‐ and high‐CNV burden. Considering patients alive without tumor after 2 years as having better clinical outcome, we found that low CNV correlated with better clinical outcome in young (P = 0.017, Fisher's exact test), but not elderly patients with SCCOT (P = 0.595).
Figure 3.
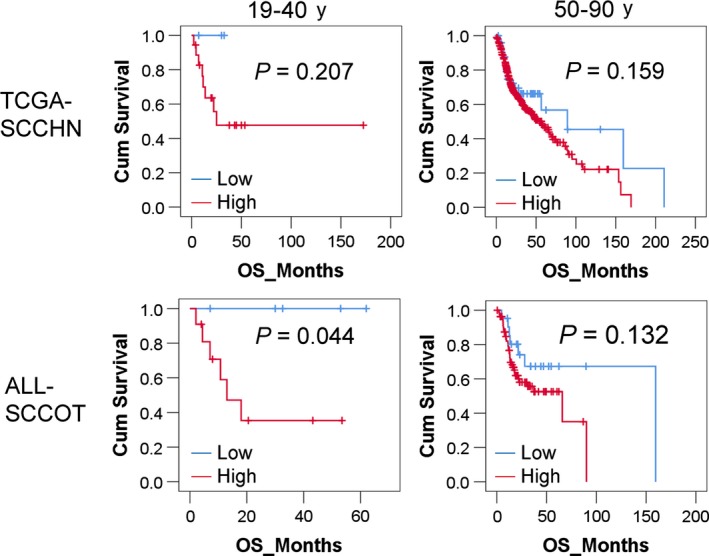
Overall survival analysis by Kaplan‐Meier method. Patients were divided into groups according to age. Overall survival analysis was performed using Kaplan‐Meier method. Log‐rank P values are shown in the plots. The red line represents patients with high‐CNV burden and the blue line patients with low‐CNV burden. According to TCGA data (TCGA‐SCCHN), there was no significant correlation between overall survival and CNV burden for the whole group of patients with SCCHN. The number of young patients (19‐40 y) with low or high CNV being 3 and 18, respectively. Combining TCGA on SCCOT with our data (ALL‐SCCOT), the overall survival rate in young patients with low‐CNV (n = 5) was significantly higher than that in young patients with high‐CNV (n = 11) (log‐rank test P = 0.044)
3.5. Pathogenic germline variants
No blood sample was available from one young patient (p35), and thus, germline variant calling was conducted for nine blood samples. Five pathogenic germline variants were identified in four patients (Table 2). One young patient (p98) harbored missense variants in ERCC2 and EXT1, and a missense variant in BRCA1 was detected in another young patient (p111). Patient 154 (aged 42 years) contained a missense variant in RET and a truncated variant in RAD51C was identified in patient 137, the oldest patient in this study. Of these, EXT1 has been reported as a tumor suppressor22 and RET as a proto‐oncogene.23 The other three genes with germline variants are tumor suppressor genes involved in various DNA repair pathways and the most frequently identified germline variants across different cancer types.24
4. DISCUSSION
SCCHN traditionally arises in older men with a history of smoking and alcohol use. Epidemiological studies demonstrate an increasing incidence in younger patients, particularly for SCCOT.4 In view of their younger age and non‐smoking and none or low alcohol using status, distinct tumorigenic mechanisms are likely involved in young patients, which may include oncogenic viral infection, an altered oral microbiome, changes in dietary constituents, environmental exposure to an unknown mutagen, or an inherited genetic polymorphism that increases risk.25, 26 However, despite decades of research, no conclusive results have been obtained.5 A recent WES study on young (<45 years) and old (>45 years) SCCOT patients showed striking similarities between the two groups, with similar mutations and copy number changes,6 findings in accordance with our present results. The failure to identify a unique cause for SCCOT in the young suggests that it is not a distinct tumor type with a distinct etiology.6 With the aim of seeking a tool for pinpointing young patients with poor prognosis and thus in urgent need of more aggressive and individualized treatment, we here characterized in depth four young patients with varying clinical features, with five elderly patients, also with varying clinical features, as comparison.
Looking at genomic variants across tumor samples, we noticed that two young patients showing high‐level CNV had both died rapidly from their disease, prompting us to investigate whether these genomic characteristics had prognostic impact. Incorporating patient data from TCGA with our findings, we could include another 12 young and 99 elderly patients with survival data and could clearly show that high‐CNV burden predicted poor prognosis in young patients only. Tumor CNV burden as a prognostic factor has previously been suggested in various cancers, including SCCHN,18, 27, 28, 29, 30 but not in connection with patient age. Here, for the first time, we showed that overall tumor CNV burden provides prognostic information specifically for young patients with SCCOT. Even if the total number of young patients is limited, data clearly show all young patients with low‐CNV burden to be alive. The difference in CNV burden and prognosis between young and elderly patients might reflect a combination of aging‐ and tumor‐related mechanisms that regulate genomic alterations. Thus, the impact of type of genomic abnormalities clearly differs between young and elderly patients.
In conclusion, our data have uncovered the transcriptomic and genomic features of young patients in comparison with elderly/traditional SCCOT patients. With additional data from TCGA, we convincingly show that the prognostic value of tumor CNV burden differs between young and elderly patients. The connection between CNV burden and prognosis seen in young patients pinpoints an important feature in young patients affected by SCCOT, with promising clinical impact on the identification of young individuals with poor prognosis that should be considered for more aggressive management and closer follow‐up.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AVAILABILITY OF DATA AND MATERIAL
The raw sequencing data are not publicly available due to it being against Swedish legislation but are available from the corresponding author after consulting the Swedish Data Protection Agency.
AUTHOR CONTRIBUTIONS
X.G designed and performed experiments, analyzed data, and wrote the manuscript. P.J.C analyzed data and wrote the manuscript. L.B performed experiments. L.W and A.K analyzed data. T.H and R.F designed experiments. L.N.S, N.S, and T.W provided medical materials. K.N supervised the project and wrote the manuscript. All authors commented on the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was reviewed and approved by the Regional Ethics Review Board, Umeå, Sweden (Dnr 03‐201 and Dnr 08‐003 M) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients.
PATIENT CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
This research was carried out using the resources of the High Performance Computer Center North (HPC2N) in Sweden.
Gu X, Coates PJ, Boldrup L, et al. Copy number variation: A prognostic marker for young patients with squamous cell carcinoma of the oral tongue. J Oral Pathol Med. 2019;48:24–30. 10.1111/jop.12792
Funding information
This study was supported by Lion's Cancer Research Foundation, Umeå University; the Swedish Cancer Society [contract number 17 0663]; Umeå University; Västerbottens Läns Landsting; the Grant Agency of the Czech Republic [project P206/12/G151]; and the Ministry of Education Youth and Sports in the Czech Republic [project MEYS‐NPSI‐LO1413]. The funding sources had no role other than financial support.
REFERENCES
- 1. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137‐2150. [DOI] [PubMed] [Google Scholar]
- 2. Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26:123‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Annertz K, Anderson H, Biorklund A, et al. Incidence and survival of squamous cell carcinoma of the tongue in Scandinavia, with special reference to young adults. Int J Cancer. 2002;101:95‐99. [DOI] [PubMed] [Google Scholar]
- 4. Tota JE, Anderson WF, Coffey C, et al. Rising incidence of oral tongue cancer among white men and women in the United States, 1973‐2012. Oral Oncol. 2017;67:146‐152. [DOI] [PubMed] [Google Scholar]
- 5. Dos Santos Costa SF, Brennan PA, Gomez RS, et al. Molecular basis of oral squamous cell carcinoma in young patients: is it any different from older patients? J Oral Pathol Med. 2018;47:541‐546. [DOI] [PubMed] [Google Scholar]
- 6. Pickering CR, Zhang J, Neskey DM, et al. Squamous cell carcinoma of the oral tongue in young non‐smokers is genomically similar to tumors in older smokers. Clin Cancer Res. 2014;20:3842‐3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarkaria JN, Harari PM. Oral tongue cancer in young‐adults less‐than 40 years of age ‐ rationale for aggressive therapy. Head Neck‐J Sci Spec. 1994;16:107‐111. [DOI] [PubMed] [Google Scholar]
- 8. Jeon JH, Kim MG, Park JY, et al. Analysis of the outcome of young age tongue squamous cell carcinoma. Maxillofac Plast Reconstr Surg. 2017;39:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldenberg D, Brooksby C, Hollenbeak CS. Age as a determinant of outcomes for patients with oral cancer. Oral Oncol. 2009;45:E57‐E61. [DOI] [PubMed] [Google Scholar]
- 10. Lundqvist L, Stenlund H, Laurell G, Nylander K. The importance of stromal inflammation in squamous cell carcinoma of the tongue. J Oral Pathol Med. 2012;41:379‐383. [DOI] [PubMed] [Google Scholar]
- 11. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript‐level expression analysis of RNA‐seq experiments with HISAT. Nat Protoc. 2016;11:1650‐1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandrani P, Kulkarni V, Iyer P, et al. NGS‐based approach to determine the presence of HPV and their sites of integration in human cancer genome. Br J Cancer. 2015;112:1958‐1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D'Aurizio R, Pippucci T, Tattini L, Giusti B, Pellegrini M, Magi A. Enhanced copy number variants detection from whole‐exome sequencing data using EXCAVATOR2. Nucleic Acids Res. 2016;44:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 2010;20:1297‐1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang KL, Mashl RJ, Wu Y, et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173(355–370):e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas N . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sgaramella N, Coates PJ, Strindlund K, et al. Expression of p16 in squamous cell carcinoma of the mobile tongue is independent of HPV infection despite presence of the HPV‐receptor syndecan‐1. Br J Cancer. 2015;113:321‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tokheim CJ, Papadopoulos N, Kinzler KW, Vogelstein B, Karchin R. Evaluating the evaluation of cancer driver genes. Proc Natl Acad Sci U S A. 2016;113:14330‐14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172‐183. [DOI] [PubMed] [Google Scholar]
- 22. Daakour S, Hajingabo LJ, Kerselidou D, et al. Systematic interactome mapping of acute lymphoblastic leukemia cancer gene products reveals EXT‐1 tumor suppressor as a Notch1 and FBWX7 common interactor. BMC Cancer. 2016;16:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jhiang SM. The RET proto‐oncogene in human cancers. Oncogene. 2000;19:5590‐5597. [DOI] [PubMed] [Google Scholar]
- 24. Lu C, Xie M, Wendl MC, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Llewellyn CD, Johnson NW, Warnakulasuriya KA. Risk factors for squamous cell carcinoma of the oral cavity in young people–a comprehensive literature review. Oral Oncol. 2001;37:401‐418. [DOI] [PubMed] [Google Scholar]
- 26. Toporcov TN, Znaor A, Zhang ZF, et al. Risk factors for head and neck cancer in young adults: a pooled analysis in the INHANCE consortium. Int J Epidemiol. 2015;44:169‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smeets SJ, Brakenhoff RH, Ylstra B, et al. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cell Oncol. 2009;31:291‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zack TI, Schumacher SE, Carter SL, et al. Pan‐cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134‐1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hieronymus H, Schultz N, Gopalan A, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111:11139‐11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wangsa D, Chowdhury SA, Ryott M, et al. Phylogenetic analysis of multiple FISH markers in oral tongue squamous cell carcinoma suggests that a diverse distribution of copy number changes is associated with poor prognosis. Int J Cancer. 2016;138:98‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequencing data are not publicly available due to it being against Swedish legislation but are available from the corresponding author after consulting the Swedish Data Protection Agency.


