Summary
Cupressaceae subfamily Callitroideae has been an important exemplar for vicariance biogeography, but its history is more than just disjunctions resulting from continental drift. We combine fossil and molecular data to better assess its extinction and, sometimes, rediversification after past global change.
Key fossils were reassessed and their phylogenetic placement for calibration was determined using trait mapping and Bayes Factors. Five vicariance hypotheses were tested by comparing molecular divergence times with the timing of tectonic rifting. The role of adaptation to fire (serotiny) in its spread across a drying Australia was tested for Callitris.
Our findings suggest that three transoceanic disjunctions within the Callitroideae probably arose from long‐distance dispersal. A signature of extinction, centred on the end‐Eocene global climatic chilling and drying, is evident in lineages‐through‐time plots and in the fossil record. Callitris, the most diverse extant callitroid genus, suffered extinctions but surviving lineages adapted and re‐radiated into dry, fire‐prone biomes that expanded in the Neogene. Serotiny, a key adaptation to fire, likely evolved in Callitris coincident with the biome shift.
Both extinction and adaptive shifts have probably played major roles in this chronicle of turnover and renewal, but better understanding of biogeographical history requires improved taxonomy of fossils.
Keywords: biome shift, Callitris, conifers, extinction, fossils, long‐distance dispersal, serotiny, vicariance
Introduction
The cypress family of conifers (Cupressaceae) has a rich and ancient fossil record going back > 200 Myr to Pangaea. Today it is widespread across both the northern and southern continents and therefore is an excellent model for testing biogeographical hypotheses about the relative roles of continental drift and long‐distance dispersal and establishment (LDDE) in explaining transoceanic disjunctions (Crisp et al., 2009). The cypress fossil record is exceptional (Hill & Brodribb, 1999; Stockey et al., 2005), better than for most plant families, providing tangible evidence for the presence of lineages in different places at different times. This record also provides multiple calibration points for molecular clock analyses.
A recent biogeographical modelling study (Mao et al., 2012) explained the distribution of cypresses by overland migration across the supercontinents (Pangaea, Laurasia and Gondwana) before they rifted apart and broke up. That is, they concluded that phylogeny and distribution of the family reflect patterns consistent with vicariance rather than LDDE. However, there were assumptions and omissions in their analysis that might have affected the accuracy of their analysis of the Southern Hemisphere subfamily Callitroideae (callitroids). For example, they treated Australia and Zealandia as a single landmass until the present, whereas they rifted apart c. 80–65 million yr ago (Ma) (McLoughlin, 2001; Mortimer et al., 2017), well before two inferred transoceanic disjunctions in the callitroids. Also, they omitted some key callitroid fossils that are relevant to molecular clock calibration and biogeography, and they sampled fewer than half of the living species of Callitris s.l., which is the largest callitroid group. Callitris is defined broadly herein to include Actinostrobus and Neocallitropsis, which have been formally synonymized (Piggin & Bruhl, 2010; Byng, 2015).
During the Oligocene, following initiation of global climate change at the end of the Eocene, there was extensive extinction of conifers including callitroids, as indicated by both fossil (Niklas, 1997) and phylogenetic evidence (Crisp & Cook, 2011). This extinction particularly impacted Australia, where most of the fire‐sensitive, ever‐wet adapted Southern Hemisphere cypresses had disappeared from the fossil record by the early Miocene (Hill & Brodribb, 1999; Hill, 2004; Hill & Brodribb, 2006; see also Fig. 2) as the continent dried and became dominated by fire. Exceptionally, Callitris survived and rediversified into multiple habitats throughout Australia, including the arid zone, and into New Caledonia (NC). Callitris is sensitive to crown fire but its seed cones have the morphological characteristics of serotiny, a syndrome which is hypothesized to be adaptive to crown‐fire regimes (Lamont et al., 1991). However, it is unclear whether serotiny in Callitris evolved in response to some other selective pressure (exaptation: Bradshaw et al., 2011) or was a direct response to pyric selection after transition to a fire‐dominated biome (adaptation: Keeley et al., 2011; Lamont & He, 2017).
In the present paper we re‐evaluate published hypotheses on cypress biogeography, particularly the model for the whole family by Mao et al. (2012), as well as the hypothetical scenario of Crisp et al. (2011) that posits that multiple transoceanic disjunctions are the product of vicariance followed by differential extinction. Our main focus is the Southern Hemisphere subfamily Callitroideae using a much more comprehensive taxon sample and better area definitions than in previous studies. We use the first fully sampled molecular phylogeny of Callitris species to investigate the Cenozoic revival and spread of this genus across Australia and NC, including the role of adaptive shifts into fire‐dominated communities. Central to our approach is a re‐evaluation of the fossil record of the callitroids, focussing on the seed‐cone fossils of ‘Callitris octothamna’ (mid‐Cretaceous) and Libocedrus mesibovii (late Oligocene), which have not been used previously for calibration of molecular phylogenies. (The name of ‘C. octothamna’ is placed in quotes because it has not been formally published.) Fossils provide the only direct evidence for our hypothesis that differential extinction was responsible for at least some of the multiple transoceanic disjunctions in the extant genera (cf. Crisp et al., 2011). Fossils also provide direct evidence of turnover and rediversification in Callitris.
We test five hypotheses explaining transoceanic divergences (A to E in Fig. 1) by comparing our new molecular divergence‐time estimates with published tectonic rifting times. Mao et al. (2012) inferred four of these to be vicariance and one to be LDDE. Also, we test the fossil‐based extinction–rediversification hypothesis by examining lineages‐through‐time (LTT) plots for Callitris for signatures of these processes (Crisp & Cook, 2009), relative to its callitroid sister taxa, which suffered extinctions in Australia but did not rediversify there. Finally, we ask whether serotiny in Callitris could be an adaptation to fire by testing the expectation that the shift to a fire‐prone biome predated the origin of cone morphology associated with serotiny.
Figure 1.
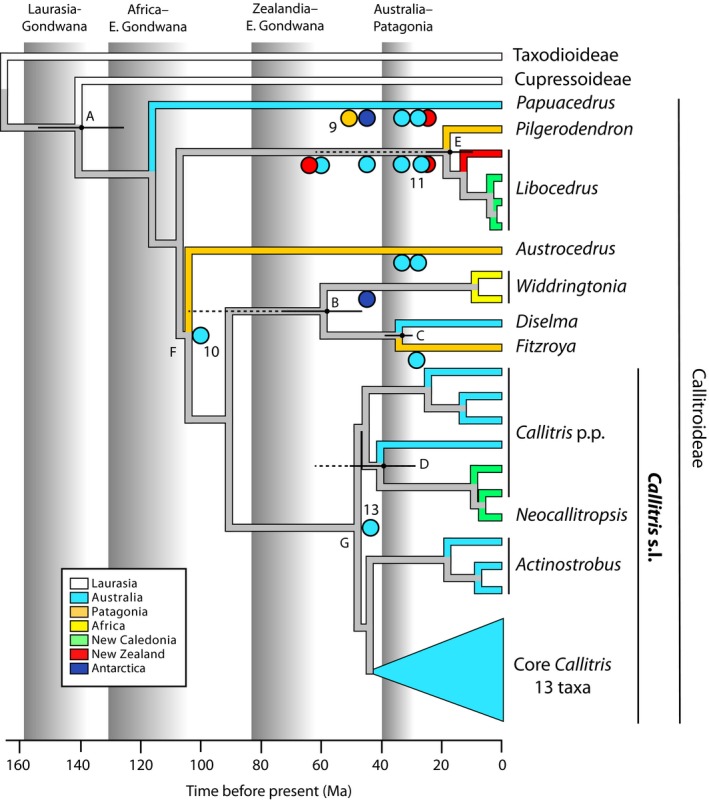
Simplified time tree of Cupressaceae estimated from the combined cpDNA + nDNA dataset, using a partitioned Beast analysis and calibrated using lognormal priors. Terminal branches and circles (indicating fossils) are coloured by geographical occurrence, as in the key. Fossils used to calibrate the Beast analysis are numbered as in Supporting Information Table S3 and Fig. S3. Fossil 10 (‘Callitris octothamna’) is at the position (node F) preferred by Bayes Factors. Fossil 11 (Libocedrus mesibovii) could be a Pilgerodendron (see text). Broad grey vertical bars show events in the breakup of Pangaea and Gondwana, graded from commencement to cessation of rifting. Black horizontal bars at nodes A to E show the 95% HPD of estimated divergence times (Table 2) between extant taxa in now‐separated parts of Laurasia and/or Gondwana. Dotted lines show gaps between these HPDs and the rifting event that putatively caused vicariance (B, D and E). Node G is the crown of Callitris s.l. The complete tree is shown in Fig. S11. Ma, million yr ago.
Materials and Methods
Taxon sampling
The taxon sample for the phylogeny (Supporting Information Table S1) comprised all 23 extant species of Callitris based on the taxonomy of Hill (1998), with the addition of the four species previously recognized as Actinostrobus (Western Australia) and Neocallitropsis (NC). Additionally, all three subspecies of C. oblonga (Hill, 1998) were sampled. To test the monophyly of Callitris s.l. and to estimate a well‐calibrated time‐tree, we also sampled 36 taxa across the rest of the family Cupressaceae, including all genera and most species of subfamily Callitroideae. We included two samples per species from Callitris and, where possible, the rest of Callitroideae. We could not obtain data for two of the four Widdringtonia species and one of the five Libocedrus species.
DNA extraction, sequencing and alignment
Genomic DNA was extracted from c. 1.0 cm of dried foliage tissue using a slightly modified hexadecyltrimethylammonium bromide (CTAB) method (Murray & Thompson, 1980; Doyle, 1991). We sequenced c. 6 kb of standard loci from the chloroplast genome, including gene‐coding regions rbcL, matK and trnL‐trnF, as well as noncoding regions. We also sequenced three nuclear loci (c257 = inorganic pyrophosphatase; c22306 = erd1 and c36749 = alpha‐glucan protein synthase) using primers designed from an EST library of C. intratropica (Sakaguchi et al., 2011). Details of all loci and primer sequences, including references, are in Table S2. GenBank accession numbers for sequences are in Table S1. PCR amplification of all loci was carried out following the standard protocol from the Qiagen multiplex PCR kit (Qiagen, Hilden, Germany). PCR products were sequenced using both forward and reverse primers with an ABI prism bigdye terminator cycle‐sequencing ready‐reaction kit v.3.1 (Applied Biosystems, Waltham, MA, USA) and electrophoresed on an ABI prism 3100 genetic analyser. Sequences were edited using bioedit v.7.0.8.0 (Hall, 1999), aligned initially using mafft (Katoh & Toh, 2010) in cipres (Miller et al., 2010) and manually corrected in geneious ® v.10.2.2, with exons adjusted to the open reading frame. Some hypervariable regions were excluded from the alignment and the final aligned lengths were: cpDNA = 6658, c257 = 282, c22306 = 729, c36759 = 273, total = 7942. The three nuclear loci could be sequenced successfully only within Callitroideae.
Phylogenetic analysis
Maximum‐likelihood (ML) trees were generated using RaxML v.7.4.2 (Stamatakis et al., 2008) in cipres. DNA sequences were partitioned with substitution models comprising a SRD06 codon‐based model for the rbcL exon and separate general time‐reversible (GTR) + Γ models for the matK exon, the noncoding cpDNA and the three nuclear loci (c257, c22306 and c36749) combined into a single partition. Separate analyses of cpDNA (Fig. S1) and the three nuclear loci combined (Fig. S2) found no conflict between the two datasets in nodes with strong bootstrap support (BS > 80), so all loci were concatenated for subsequent analysis. The RaxML tree inferred from the combined data (Fig. S3) was then used as a starting tree for the Beast analyses.
Time‐calibrated phylogenies were inferred using Beast v.1.8.4 (Drummond et al., 2012) with an uncorrelated lognormal (UCLN) clock model (both Cupressaceae and Callitris datasets) and a random local clocks (RLC) model for Callitris only (Markov chain Monte Carlo (MCMC) chain stationarity could not be achieved using the RLC model with the full Cupressaceae dataset). Each partition (defined as for RaxML) was given a separate substitution and clock model in Beast. Relative fit of the alternative clock models (RLC and UCLN) in Callitris alone was evaluated using Bayes Factors (BF; Raftery, 1996) calculated from marginal likelihoods obtained by path (PS) and stepping‐stone (SS) sampling (Baele et al., 2012, 2013). This test indicated ‘very strong’ support (BF > 10) for the UCLN clock (Table S4), which was used in all subsequent Beast analyses. To model the speciation process, a birth–death model, corrected for incomplete sampling (Stadler, 2009), was used because species sampling was sparse in the non‐callitroid outgroups. To ensure stationarity and convergence of the Bayesian MCMC chains, three parallel chains were run and tracer v.1.6 (Rambaut et al., 2013) was used to check that the effective sample sizes in the combined logs (after a 10% burnin) were > 200 for all parameters. The MCMC chain length was set to a minimum of 100 million generations (full Cupressaceae dataset) and was increased to 200 million for clock model tests in Callitris to achieve stationarity and convergence. To display the results, the maximum clade credibility tree of Callitris was annotated in figtree v.1.4.3 (Rambaut, 2016), as recommended by the author.
Fossil‐based calibration
For the time‐scaled (Beast) phylogenetic analysis of Cupressaceae, we used the fossil‐based calibration points listed in Table S3, including several from previous studies and some new ones, detailed in the next section. Calibration priors were given lognormal distributions, as recommended in theory (Morrison, 2008; Ho & Phillips, 2009). By contrast, Mao et al. (2012) used mostly uniform minimum‐age priors, whose flat distribution makes all ages equally probable between a hard minimum and maximum. But uniform priors increase temporal uncertainty in the calibrations and tend to bias estimates towards slower clock rates and older ages over the tree (Dornburg et al., 2012). A lognormal prior also sets a hard‐minimum age (i.e. the ‘offset’, which is usually set at the age of the fossil) but the lognormal tail limits regress of the age estimate into the past with diminishing probability (Ho & Phillips, 2009). In any case, we tested the fit of lognormal vs uniform prior distributions with otherwise matched datasets. Lognormal prior distributions were preferred very strongly by BF calculated from the Beast posterior marginal likelihoods (PS = 27.7, SS = 31.4; Table S4) and used in all subsequent Beast analyses.
Assessment and model‐testing of previously unused fossils for calibration
Callitroideae has a rich fossil record (Fig. 1; Hill & Brodribb, 1999; Stockey et al., 2005; Wilf et al., 2009) and we evaluated some of its fossil taxa that have not been used previously for calibration. We chose these particular fossils because their age and phylogenetic placement potentially affected the biogeographical reconstruction of Callitroideae and Callitris. Fossils are typically fragmentary, limiting the number of characters available for their identification and phylogenetic assignment. One source of error has been the assignment of fossils to extant genera based on ancestral character states that are shared among multiple genera (symplesiomorphies), for example in Libocedrus (Whang & Hill, 1999). Therefore, to assess whether the fossils discussed below have unique defining characters (synapomorphies) for any lineages within Callitroideae, we scored potential characters (Table S5) and mapped them on the molecular phylogeny using mesquite v.3.4 (Maddison & Maddison, 2018) with parsimony and the ML model Mk1 (Lewis, 2001). Characters were scored for the fossil C. leaensis from the detailed description and photographic plate in Paull & Hill (2010).
‘Callitris octothamna’ M.D. Peters (1985). These seed cone fossils are similar to cones of extant Callitris and potentially could greatly increase the age estimate of the genus, being from mid‐Cretaceous (99.6 Ma) sediments near Winton, Queensland. We assigned the calibration alternately to (a) the stem node of Callitris, being the most recent common ancestor (MRCA) of Callitris, Fitzroya, Diselma and Widdringtonia (hereafter, CFDW clade) and (b) its parent node. These assignments were based on mapping the characters phyllotaxis (Fig. S4), leaf dimorphism (Fig. S5) and cone‐scale arrangement (Fig. S6) on a tree. The fit of these calibrations relative to exclusion of the fossil, was assessed using BF.
Libocedrus: A number of Libocedrus fossils have been described from Australia and New Zealand (NZ) (Hill & Carpenter, 1989; Pole, 1998, 2007a,b; Whang & Hill, 1999) but none was used for calibration by Mao et al. (2012) even though they are relevant to biogeography. Extant Libocedrus is endemic to Zealandia (NC + NZ; Mortimer et al., 2017). We used the only known seed cone fossil, L. mesibovii (Hill & Carpenter, 1989), from the late Oligocene–early Miocene (c. 24 Ma), to calibrate the MRCA of sister genera Libocedrus and Pilgerodendron, and assessed its fit relative to its exclusion, using BF.
Assessment of these fossils is explained in more detail in Methods S1.
Testing biogeographical hypotheses
Hypotheses proposed to explain transoceanic divergences between extant lineages of the callitroids (Crisp et al., 2011; Mao et al., 2012) were tested by comparing divergence times inferred from the molecular phylogeny with the timing of rifting events in the break‐up of Gondwana. The tests followed the protocol of Crisp et al. (2011: Box 2). This approach tests the prediction of a vicariance hypothesis that sister taxa on either side of a barrier should have diverged at the same time as the barrier originated. The test is two‐tailed, that is, vicariance is rejected if the divergence between the taxa either postdates or pre‐dates the origin of the barrier. The test also should take account of uncertainty in timing of both the lineage divergence times and the geological rifting. The test of vicariance is explicit because it addresses a specific divergence (node) in the phylogeny, which is hypothesized to be caused by the origin of a particular barrier. A rejection of a particular vicariance event does not generalize to a statement that vicariance does not explain the distribution of the whole taxon. It rejects only the specific hypothesis that ‘vicariance event X explains node Y’.
Diversification of Callitris
The trend of Callitris lineages through the Cenozoic was plotted using mesquite. This plot was based on the Beast tree calibrated with the fossil ‘Callitris octothamna’ at the MRCA of Austrocedrus and Callitris (Fig. S7), with the addition of the fossil C. leaensis, which was inserted by hand mid‐way along the stem‐lineage of Callitris (cf. Figs S8, S9), diverging at 68 Ma and terminating at c. 34 Ma (= age of the fossil).
Evolution of fire‐related traits
The seed‐cones of most Callitris species are woody and serotinous (Lamont et al., 1991; Ladd et al., 2013). That is, mature cones containing ripe seeds are held unopened in the canopy for one or more seasons, so that multiple generations of unopened cones are distributed along branches at any one time. Wildfires that burn tree canopies usually kill serotinous individuals, which then release their seeds that germinate in the ash‐bed and re‐establish the population. Serotiny is considered an adaptation to wildfire (Keeley et al., 2011; Lamont & He, 2017). Some species of Callitris, and most other callitroid genera, do not have serotinous cones but release their seeds as soon as they are ripe, generally within a year of pollination. If serotiny truly is an adaptation to wildfire (as opposed to an exaptation), it should have arisen by selection following the advent of recurrent fire into communities in which ancestral, nonserotinous species occurred – or the migration of an ancestral nonserotinous population into a new, fire‐prone habitat (Keeley et al., 2011; Lamont & He, 2017). This leads to the prediction that evolutionary transitions from nonserotiny to serotiny should correlate with transitions from nonfire‐prone to fire‐prone habitats, with the adaptive shift following the change of habitat, that is of the selective regime (Keeley et al., 2011). We tested this prediction using the data in Table S5 with the Pagel94 ML module (Pagel, 1994) in mesquite. All species of Callitris and many of the outgroups were visited in the field, where both traits were scored from direct observation. The remainder were scored from the literature, for example from Farjon (2005). Given the prediction above, the correlation model treated the serotiny variable (Y) as dependent upon the habitat flammability variable (X). As the Pagel94 model does not accept dimorphic traits, we scored alternative states for each of the duplicate tree‐tip samples in two dimorphic species (Table S5). Each trait was dimorphic in some species but no species was dimorphic for both traits; that is, duplicated tips were sufficient to score known combinations of the two traits.
Results
Phylogeny
The Beast MCMC chains were stationary and converged after 200 million generations and the marginal likelihood tests very strongly preferred the UCLN clock model to the RLC model (BF: PS = 17.6, SS = 24.4; Table S4). Nonetheless, identical topologies were found from the combined data under both clocks and the age estimates were similar, with overlapping 95% HPDs (highest posterior densities). Hereafter, we use only the results from the preferred UCLN model. The RaxML (Figs S1–S3) and Beast (Figs S7, S10, S11) analyses of all Cupressaceae gave topologies consistent with Mao et al. (2012), except in the position of Austrocedrus, as noted below. Within Callitris, our phylogeny was more resolved, with higher nodal support, than that of Mao et al., who sampled only half (12) of the species. Hereafter the label ‘core Callitris’ (e.g. Figs 1, 2) is used for a clade that received maximum support (BS = 100 or PP = 1.0) in all trees. This clade includes 13 of the 23 species, and excludes some Callitris and all species previously assigned to other genera (Actinostrobus and Neocallitropsis, e.g. as in Farjon, 2005).
Figure 2.
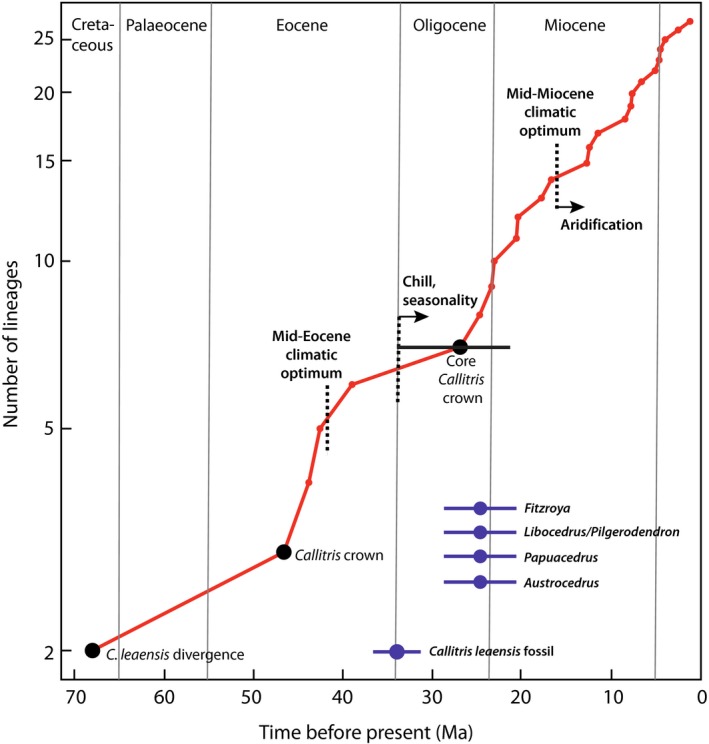
Lineages‐through‐time (LTT) plot for Callitris. Based on the Beast tree calibrated with the fossil ‘Callitris octothamna’ at the most recent common ancestor (MRCA) of Austrocedrus and Callitris (Supporting Information Fig. S7). The fossil C. leaensis was inserted by hand mid‐way along the stem‐lineage of Callitris (cf. Figs S8, S9), diverging at 68 million yr ago (Ma) and terminating at 34 Ma (= age of the fossil). Black filled circles are highlighted nodes from the tree. The core Callitris crown node marks the end of the plateau and likely extinction time, and the horizontal black bar is the 95% HPD of the age estimate for this node. Blue filled circles represent the most recent Australian fossils for callitroid genera, with bars representing uncertainty in their stratigraphic age. The C. leaensis fossil is not the most recent for the genus but clearly represents an ever‐wet habitat.
In the ML analyses, the cpDNA dataset (Fig. S1) placed Austrocedrus as sister to the CFDW clade without strong support (BS = 70). By contrast, the nuclear dataset (Fig. S2) placed it as sister to CFDW plus Libocedrus and Pilgerodendron (BS = 81). The combined data (Fig. S3) gave the same topology as cpDNA (BS = 72). Additionally, in trees from all three datasets, and in both ML and Beast analyses, the branch subtending Austrocedrus and its sister group was always very short. Given this uncertainty, the Beast analysis of the combined data was alternately constrained to give the nDNA topology (Fig. S10) and the result compared with the unconstrained analysis (Figs S7). Both topologies were compared in alternate downstream analyses where they could have made a difference to key results. However, the results were virtually identical in divergence times relevant to biogeography (Table 1), fossil trait reconstructions for calibration placement (Table 2), and mapping of fire‐related traits (not shown).
Table 1.
Estimated divergence times from Beast for key nodes for testing biogeographic hypotheses
| Nodes | Not calibrated with ‘Callitris octothamna’ | ‘C. octothamna’ (calibration 10) at parent of Callitris stem node | ‘C. octothamna’ at Callitris stem node | ||
|---|---|---|---|---|---|
| Unconstrained | Constrained | Unconstrained | Constrained | Unconstrained | |
| A. Callitroideae–Cupressoideae | 127.0 (105–148) | 127.0 (106–150) | 139.7 (125–154) | 139.6 (126–156) | 148.4 (134–163) |
| B. Widdringtonia–(Diselma, Fitzroya) | 52.5 (41–66) | 52.3 (41–66) | 58.3 (45–73) | 58.7 (45–73) | 64.6 (47–82) |
| C. Diselma–Fitzroya | 32.8 (30–38) | 32.8 (30–38) | 33.2 (30–39) | 33.2 (31–39) | 33.5 (30–41) |
| D. Callitris NC–C. macleayana | 34.8 (25–46) | 35.3 (25–46) | 39.3 (28–50) | 39.4 (28–51) | 43.0 (31–56) |
| E. Pilgerodendron–Libocedrus | 15.6 (9–24) | 15.7 (9–24) | 17.3 (10–26) | 17.3 (10–27) | 18.5 (11–28) |
| F. CFDW stem | 87.2 (71–105) | 87.6 (71–105) | 103.0 (100–110) | 103.1 (100–110) | 113.0 (106–112) |
| G. Callitris crown | 41.6 (33–51) | 42.0 (33–52) | 46.9 (38–57) | 47.3 (38–57) | 51.5 (41–62) |
Node labels (A–G) refer to Fig. 1. Primary columns represent different calibration settings for ‘Callitris octothamna’, including its exclusion. Sub‐columns differ in whether monophyly of the CFDW–Libodedrus–Pilgerodendron clade was constrained, to the exclusion of Austrocedrus. All these results are from the combined cpDNA + nDNA dataset. The unconstrained topology reflects that from cpDNA alone and places Libocedrus + Pilgerodendron as sister to CFDW (Supporting Information Fig. S1). The constrained topology reflects that from nDNA alone (Fig. S2). Values are median age estimates in million yr ago (Ma) (95% HPD). NC, New Caledonia; CFDW, Callitris, Fitzroya, Diselma and Widdringtonia.
Table 2.
Mapping of fossil characters to guide calibration placement on the molecular phylogeny: proportional likelihoods at candidate nodes
| Characters and states | Tree: Austrocedrus in | Tree: Austrocedrus out | ||||
|---|---|---|---|---|---|---|
| Node B (CFDW stem) | Node A (Callitris stem) | Callitris crown | Node B (CFDW stem) | Node A (Callitris stem) | Callitris crown | |
| Phyllotaxis ternate | 0.03 | 0.51* | 1.00* | 0.03 | 0.33* | 1.00* |
| Phyllotaxis decussate | 0.96* | 0.48* | 0.00 | 0.97* | 0.65* | 0.00 |
| Leaves monomorphic | 0.16 | 0.85 | 1.00* | 0.20 | 0.85 | 1.00* |
| Leaves dimorphic | 0.83 | 0.14 | 0.00 | 0.80 | 0.15 | 0.00 |
| Cone‐scales two whorls of three | 0.02 | 0.26* | 1.00* | 0.01 | 0.16* | 1.00* |
| Cone‐scales decussate | 0.98* | 0.74* | 0.00 | 0.99* | 0.84* | 0.00 |
Proportional likelihoods under the Mk1 model (Lewis, 2001) were sampled at candidate nodes on alternative topologies: (a) with Austrocedrus sister to the CFDW clade (‘Austrocedrus in’), as supported by cpDNA and the combined data, and (b) with Libocedrus + Pilgerodendron constrained to be sister to CFDW (‘Austrocedrus out’), as supported by the nDNA alone. For each character, the plesiomorphic (ancestral) state is listed after the derived state. The transition between the plesiomorphic and derived state for each character is reconstructed as occurring between node B and the Callitris crown node, possibly in the Callitris stem (node A). The node labels refer to Supporting Information Figs S4–S6. CFDW, Callitris, Fitzroya, Diselma and Widdringtonia.
*States judged ‘best’ (likelihoods are not significantly different at that node if both states are asterisked).
Assessment and model‐testing of previously unused fossils for calibration
Three models were compared with respect to the assignment of ‘Callitris octothamna’ as a calibration and ranked by their marginal Loge likelihoods (Table S4). The first‐ranked model, with ‘C. octothamna’ placed at the MRCA of Callitris and Austrocedrus, was positively preferred (BF: PS = 4.7, SS = 5.2) to the third‐placed model, with ‘C. octhothamna’ placed at the Callitris stem node. The second‐ranked model (‘C. octothamna’ calibration omitted) differed minimally from the first‐ranked model (BF: PS = 0.84, SS = 0.22; Table S4), at a level considered by Raftery (1996) as ‘not worth more than a bare mention’ (Raftery, 1996). Consequently, both best models were used in downstream analyses. Divergence times (Table 1) estimated using the best model, with placement of ‘C. octothamna’ at the CFDW stem (Fig. S7), were older than those estimated without this calibration (Fig. S11), although the 95% HPDs overlapped. The largest difference (15.9 Ma) was at the calibrated node itself.
Exclusion of the calibration using Libocedrus mesibovii was very strongly preferred to its inclusion (BF: PS = 33, SS = 600; Table S4), so it was thereafter omitted.
Testing biogeographical hypotheses
The divergence between the Cupressoideae and Callitroideae (node A in Fig. 1) is consistent with vicariance (Fig. 1) under all calibration models (Table 1). The 95% HPDs of the divergence‐time estimates overlap the period (c. 160–140 Ma) when Laurasia and Gondwana finally separated as Pangaea broke up (McLoughlin, 2001). Likewise, all five divergence‐time estimates for Diselma and Fitzroya are consistent with the separation of Australia, South America and Antarctica c. 40–34 Ma (Fig. 1; Table 1). All other divergences (B, D, E) relevant to biogeography are too recent to be explained by vicariance because the upper (older) 95% HPDs are > 10 Ma younger than the completion of rifting between the respective landmasses (Fig. 1; Table 1). These are: B, between Widdringtonia in West Gondwana (Africa) and Diselma + Fitzroya in East Gondwana (c. 130–105 Ma); D, between Callitris in NC and the C. baileyi clade in Australia (c. 80–65 Ma); and E, between Pilgerodendron (Patagonia) and Libocedrus in Australia and NZ (c. 80–65 Ma). Details of biogeographical hypothesis tests for all callitroid genera follow.
Papuacedrus was once extant in South America, Antarctica, southern Australia and NZ (Fig. 1; Wilf et al., 2009) but is now extinct in all those places. According to the reconstruction by Mao et al., its range expanded from South America to area U (Australasia, including New Guinea) between 130 and 52 Ma, followed by vicariance separating the South American fossil from the extant New Guinea lineage. This inference was based on the fossil record alone because, with a single living species in New Guinea, there is no molecular divergence time against which this hypothesis can be tested.
Libocedrus and possibly also Pilgerodendron (see Methods S1) have an extensive fossil record between 60 and 22 Ma, exclusively from Australia and NZ (Fig. 1). Mao et al. (their Fig. S4) postulate that the ancestor of Libocedrus + Pilgerodendron spread from South America to Australasia (their area U) after c. 105 Ma and then c. 35 Ma, diverged by vicariance into these two genera. However, the latter event is much too young to be explained by the tectonic separation of Zealandia and South America (85–65 Ma). Our most conservative dating (Fig. 1; Table 1, column 5) of the divergence between these genera was 11–28 Ma, supporting a hypothesis of trans‐Pacific dispersal. The dispersal could have gone in either direction but, given the extensive fossil record for the clade in Australasia during that period (reviewed in Wilf et al., 2009), perhaps it was the source. There is no known fossil record of either genus from South America (Wilf et al., 2009).
Austrocedrus fossils are known from Australia at the right time (36–30 Ma) for the genus to be vicariant in South America and Australia, followed by extinction of the Australian population. Consistently with this, Mao et al. estimated the divergence at c. 30 Ma.
We estimate that Widdringtonia (southern Africa) diverged from Diselma + Fitzroya (East Gondwana) 73–41 Ma (Fig. 1; Table 1), long after these land masses finished separating (c. 105 Ma; McLoughlin, 2001), so this appears to have been a transoceanic dispersal. Mao et al. (2012) also reconstructed a dispersal to Africa around the Cretaceous–Palaeogene boundary but from a source in South America. However, fossil wood has been tentatively identified as Widdringtonia from Miocene to Quaternary sediments on Kerguelen Island, which lies in the southern Indian Ocean (= Widdringtonioxylon antarcticum; Philippe et al., 1998). This small island is part of the much more extensive Kerguelen Plateau, which is mostly submerged now but supported terrestrial flora in the early Late Cretaceous (McLoughlin, 2001). Therefore, Kerguelen could have been a stepping stone if the ancestor of Widdringtonia actually dispersed from Antarctica (when part of East Gondwana) to Africa.
Fitzroya has a single species endemic to Patagonia and is sister to the monotypic Diselma in Tasmania. Mao et al. reconstructed the MRCA of these genera as present in East Gondwana from c. 110 Ma, then diverging (nonvicariant, i.e. within East Gondwana) c. 45 Ma, followed by vicariant speciation within Fitzroya c. 35 Ma when Australia, Antarctica and South America rifted apart. This was followed by extinction of the Australian Fitzroya population. Our dating linked the Fitzroya–Diselma divergence to that rifting event, but given the error bars (Fig. 1), either that or the divergence between Australia and South America within Fitzroya could be related to it – but not both.
Multiple genera were more widespread in the past and all callitroid genera except Widdringtonia are represented in Australia by either fossils or living species (Fig. 1). The relevance of fossils to tests of biogeographical hypotheses is explained under ‘Fossils combined with molecular evidence reveal the extent of turnover’ in the Discussion section.
Diversification and substitution rates
The log‐linear plot (Fig. 2) depicting the accumulation of Callitris lineages‐through‐time (LTT) in the ‘C. octothamna’‐calibrated tree rises steeply until it plateaus between c. 40 and 28 Ma, possibly indicating an extinction event in the late Oligocene (Crisp & Cook, 2009). Thereafter, the LTT graph rises steeply and linearly to the present, indicating a more or less constant rate of diversification. The estimated ages of the last known Australian fossils of at least four callitroid genera overlap the plateau in the Callitris LTT plot (Fig. 2). The alternative tree with no ‘C. octothamna’ calibration gave a very similar plot (not shown), shifted c. 5 Ma towards the present.
Evolution of fire‐related traits
A significant correlation (P = 0.04) in which serotiny (Y) depends upon habitat flammability (X) was found using Pagel's (1994) test of correlated (discrete‐state) character evolution from 100 simulations on the tree (Figs S8, S9). Using the alternative tree with no ‘C. octothamna’ calibration (Fig. S11), the correlation also was significant (P = 0.01).
Mapping these traits onto the phylogeny indicated a possible time‐lag between transitions, with the habitat shifts occurring before the origin of serotiny, at least within Callitris. In parsimony mapping, the MRCA of all Callitris was reconstructed as having a fire‐prone habitat (Fig. S9), whereas the same node was reconstructed indecisively – as either serotinous or not (Fig. S8). Similarly, ML mapping indicated decisively that the transition to fire‐prone habitat had occurred by the crown node of Callitris (proportional likelihood ≥ 0.90*, which is significant) on both trees (with and without calibration using ‘C. octothamna’) (Table 3). By contrast, the proportional likelihood of serotiny did not exceed the decisive value of 0.90 until two nodes later (core Callitris crown node). These traits are homoplasious, with multiple gains and/or losses in both the Cupressoideae (Cupressus, Juniperus and Tetraclinis) and the Callitroideae (Callitris and Widdringtonia) (Figs S8, S9). Within Callitris, absences of serotiny reconstructed as reversals in the MRCA of the C. macleayana‐NC clade and the MRCA of the C. columellaris s.l. clade. Callitris baileyi also lacks serotiny but the trait reconstruction was equivocal, that is serotiny could have been lost in C. baileyi or it could have originated independently in its sister species (C. roei and C. drummondii).
Table 3.
Fire‐related trait transitions: proportional likelihoods at nodes
| Node | Tree with ‘C. octothamna’ calibration at CFDW stem node | Tree with no ‘C. octothamna’ calibration | ||||
|---|---|---|---|---|---|---|
| Age (Ma) | X = habitat flammability | Y = cone serotiny | Age (Ma) | X = habitat flammability | Y = cone serotiny | |
| Widdringtonia – Callitris | 90 | 0.52 | 0.40 | 76 | 0.51 | 0.38 |
| Callitris crown – C. leaensis | 72 | 0.57 | 0.44 | 59 | 0.56 | 0.40 |
| Callitris crown (= C. roei – C. oblonga) | 47 | 0.90* | 0.70 | 42 | 0.90* | 0.68 |
| Actinostrobus – core Callitris clade | 43 | 0.92* | 0.82 | 39 | 0.92* | 0.81 |
| Core Callitris crown (= C. canescens – C. oblonga) | 27 | 0.99* | 0.99* | 24 | 0.99 | 0.99* |
Proportional likelihoods under the Mk1 model (Lewis, 2001) were sampled at successive nodes (top row is nearest the root). Alternative Beast trees differ in node ages, depending on whether they were calibrated with ‘C. octothamna’. As the characters are binary, only the proportional likelihood for the derived state is shown. Correlations between these traits, where Y depends upon X, were assessed using the Pagel94 test. The node references are to the most recent common ancestor (MRCA) of the named taxa (see Supporting Information Figs S8, S9). Ages are median estimates for that node. CFDW, Callitris, Fitzroya, Diselma and Widdringtonia.
*States judged ‘best’ (likelihoods are not significantly different at that node if both states are asterisked). Ma, million yr ago.
Recently, serotiny was reported in Protodammara, a putative Cupressaceae fossil from mid‐Cretaceous NZ (Mays et al., 2017). The authors suggest an affinity to members of Cupressaceae that are not closely related to Callitroideae, so serotiny is likely to be independently evolved in Protodammara and the callitroids. The reference by Mays et al. to serotiny in a mid‐Cretaceous fossil of Widdringtonia (McIver, 2001) is based on a likely misidentification of this fossil (Stockey et al., 2005; Crisp & Cook, 2011).
Discussion
Biogeography: post‐Gondwanan transoceanic dispersals
The callitroid cypresses have an ancient fossil record, tracing back to the separation of Gondwana and Laurasia > 150 Ma (Hill & Brodribb, 1999; Wilf et al., 2009). Using the most comprehensive phylogeny of the callitroids (Fig. 1), we have confirmed that they spread widely across Gondwana before it broke up (Crisp et al., 2011; Leslie et al., 2012; Mao et al., 2012). At least some divergences are consistent in timing and location with vicariance hypotheses, notably Cupressoideae–Callitroideae (Laurasia–Gondwana) and Diselma–Fitzroya (Australia–South America), because the 95% HPD intervals of their divergence‐time estimates overlap the relevant tectonic rifting periods (Fig. 1; Table 1).
By contrast, our tests of biogeographical hypotheses found three post‐Gondwanan transoceanic divergences that were too young to be explained by vicariance, even from our oldest estimates (Table 1, last column), and therefore likely resulted from long‐distance dispersal and establishment. These were the divergences between (a) Pilgerodendron (Patagonia) and Libocedrus (Zealandia), (b) Callitris macleayana (Australia) and the NC clade, and (c) Widdringtonia (southern Africa) and Diselma (Tasmania) + Fitzroya (Patagonia). Of these, (a) and (b) were not inferred as transoceanic dispersals by Mao et al. (2012) because those authors lumped Zealandia into Australia (their area ‘U’), yet Zealandia rifted away from Australia (then part of East Gondwana) in the Cretaceous, long before the trans‐Pacific divergences in these two clades (Fig. 1; Table 1). By treating Australia and Zealandia as separate regions, we have been able to more accurately reconstruct their biogeography.
In a large modelling analysis across all conifers, Condamine et al. (2017) concluded that islands, especially those of Zealandia, were refugia and centres of diversification, from where they dispersed back to continents such as Australia. The authors inferred that conifer diversification was faster on islands than on continents, due to lower speciation and higher extinction rates in the latter. This is not supported by our data for the callitroids, except that there was extensive extinction in Australia during the Cenozoic (Figs 1, 2). Both callitroid clades in Zealandia appear to have dispersed across the Tasman Sea from Australia during the Neogene, with no evidence for dispersal in the opposite direction. Within Australia, Callitris diversified rapidly into multiple habitats following cooling and aridification (Fig. 2). Today, more than two‐thirds of the callitroids (25 of 38 spp.) are restricted to Australia but all are Callitris species, except Diselma archeri. Condamine et al. sampled barely half the callitroid species and most of the missing taxa are Australian Callitris (14 of 24 spp.). Therefore, their analysis would have underestimated the extent and speed of the Australian (i.e. continental) radiation.
Fossils combined with molecular evidence reveal the extent of turnover
The present‐day distribution of callitroids reflects long persistence in the southern continents, but the biogeographical history is obscured by extensive extinctions following global climatic change at the end of the Eocene, especially in Australia (Hill, 2004; Hill & Brodribb, 2006). Fossils of many extant genera have been found in places where they no longer exist, especially in Australia (Figs 1, 2). This is not surprising because, in general, extinct species vastly outnumber living species, even if only the Cenozoic period is considered (Marshall, 2017). How does knowledge of extinctions change the historical interpretation of the biogeographical pattern of extant callitroids (Crisp et al., 2011)? Our critical examination of significant but hitherto neglected fossils has revealed more extensive past distributions than used previously for biogeography, and these suggested alternative biogeographical reconstructions, for example Pilgerodendron and Widdringtonia.
Some fossils identified as Fitzroya from late Oligocene to Miocene sediments in Tasmania (Hill & Paull, 2003; Paull & Hill, 2010) appear to postdate the inferred vicariant divergence between Diselma in Australia and the single extant species of Fitzroya in Patagonia (Table 1). Did Fitzroya diverge from Diselma sympatrically in Tasmania and then go extinct there, or simply differentiate into Diselma? The latter possibility seems unlikely because the oldest (Early Oligocene) Fitzroya fossils, from the Lea River in Tasmania, include a female cone in which the cone‐scale phyllotaxis is ternate (Paull & Hill, 2010). This character state is derived relative to the decussate cone‐scale phyllotaxis of Diselma (Fig. S9).
A similar conundrum is the possible co‐occurrence of Pilgerodendron (= Libocedrus mesibovii, fossil 11 in Fig. 1) with Libocedrus jacksonii in Tasmania in the late Oligocene (c. 25 Ma) (Hill & Carpenter, 1989), about the same time as the trans‐Pacific divergence between these genera (10–26 Ma; column 3 in Table 1). We suggest that L. mesibovii could be a Pilgerodendron because it shares a putative synapomorphy (asymmetric cones; see Methods S1) with the extant Patagonian population. All other fossils assigned to Libocedrus are vegetative only, so they might have had asymmetric cones before this feature was lost after the extant lineage established in Zealandia.
Callitris fossils are unknown outside Australia, and even there the record is minimal (Peters, 1985; Hill & Brodribb, 1999; Paull & Hill, 2010). ‘Callitris octothamna’ (Peters, 1985) is by far the oldest known callitroid fossil (mid‐Cretaceous) and so is important for molecular clock calibration. However, our phylogenetic mapping of morphological traits (Methods S1) indicates that it was not a Callitris but more likely an early member of the Callitroideae. Callitris leaensis, which is represented by a good range of leaf material, clearly shows features such as exposed stomata scattered over the abaxial surface, which characterize extant species from ever‐wet habitats (Paull & Hill, 2010). The age of this fossil coincides with the global chilling and cooling triggered by the opening of the Southern Ocean (Zachos et al., 2001). This led to contraction of the Eocene rainforests and expansion of seasonally dry, fire‐dominated sclerophyll communities Australia‐wide (Hill, 2004; Hill & Brodribb, 2006; Crisp & Cook, 2013). A plateau in the Lineages‐through‐time plot (Fig. 2) indicates that Callitris suffered a pulse of extinction at this time and then rediversified through the Neogene and until the present. The same period marks the last known occurrence of four (or five) other callitroid genera in Australia (Fig. 2). That is, ever‐wet taxa went extinct as their habitats disappeared, but Callitris rediversified because some species adapted and moved into dry, fire‐prone biomes.
Fire‐related adaptation
Serotiny is a trait hypothesized to enable a population of fire‐sensitive woody plants to survive despite being killed by recurrent crown fires. It is found in communities with intermediate fire frequency, relative to the generation time of the plants. If fire recurrence is very infrequent (e.g. in rainforest), there is no effective selection for serotiny, and if recurrence is so frequent that a population cannot re‐establish between fires (e.g. in savannah), it will go extinct (Lamont & Enright, 2000; Ladd et al., 2013) unless there are adaptations to resist (thick bark) or vegetatively recover (stem or basal sprouts) from fire (Bowman et al., 2017). Our correlation tests indicate that serotiny in callitroids and cupressoids has a statistically significant dependence upon the putative selective agent, habitat flammability. Note that Pagel's (1994) test does not rely upon reconstruction of ancestral states. Although correlation alone cannot indicate causation, this result is consistent with the hypothesis that shifts to serotiny in Callitris are adaptations resulting from shifts into a crown‐fire regime (Keeley et al., 2011).
However, ancestral state reconstruction indicated that serotinous cones originated up to 20 Ma later than the shifts from nonfire‐prone to fire‐prone forest and scrub habitats (Table 3; Figs S8, S9). This time‐lag suggests that serotiny was not the trait enabling survival of Callitris in fire‐prone communities, at least initially. Alternative traits, such as those described below, might have enabled survival under wildfire. Such traits would be considered exaptations if they were ancestral, having been selected by factors other than wildfire (Bradshaw et al., 2011). Serotiny, originating later, might have been an adaptation resulting from a change in fire regime, for example towards crown fires with greater intensity, as climatic drying increased later in the Oligocene.
Two groups within Callitris lack serotiny. One is the clade including C. columellaris, C. intratropica and C. glaucophylla and reconstructs as having lost serotiny secondarily (Fig. S8). These three species inhabit sclerophyll or savannah communities subject to frequent surface fires, rather than crown fires. They release their seeds as soon as they mature and depend upon the soil seed‐bank for regeneration (Lamont et al., 1991). Moreover, these species are community dominants, forming extensive stands, and it has been suggested that they survive partly by suppressing low‐intensity fire where the stands are sufficiently dense, and partly in fire refugia such as rocky areas or patches that are unburned by chance (Cohn et al., 2011; Trauernicht et al., 2012). They have thick bark and even seedlings can withstand surface fires with low flame heights (Bowman et al., 2017). These traits are widely present in nonflammable communities and thus are likely exaptations for fire.
The second nonserotinous group occurs in wet habitats that burn infrequently (C. baileyi, C. macleayana and the three NC species). They belong to a clade that also includes C. roei and C. drummondii, both of which are serotinous and occur in communities prone to recurrent crown fires. The most recent common ancestor (MRCA) of the inclusive clade reconstructs as having a flammable habitat (Fig. S9) but could have been either serotinous or not (Fig. S8). If it was serotinous, then serotiny was lost in C. baileyi and the C. macleayana‐NC clade. If the MRCA was not serotinous, then serotiny was gained in C. roei and C. drummondii.
Conclusion
The Gondwanan subfamily Callitroideae of the cypresses has a long history (150 Ma) in Australia: five of the eight extant genera were present in ever‐wet rainforests by the early Oligocene. However, global climate change through the Oligocene resulted in contraction of rainforest and expansion of sclerophyll communities, and all the genera except Callitris were extinguished in Australia by the mid‐Miocene. Callitris survived and rediversified through to the present by adaptively shifting into seasonally dry environments, some of which were prone to crown fires, and where serotiny might have been a selective advantage. We have shown how fossils can be integrated with molecular phylogenies to illuminate how species turnover and renewal have shaped the biogeography of southern cypresses.
Author contributions
MDC led the project including design of the research, data analysis, interpretation of results and writing the manuscript; the project was originally conceived by DMJSB, LGC, MDC and YI, and LGC and MC were involved in design of the research; all authors contributed to the field sampling; SS led the laboratory work, including loci selection, sequencing and sequence assembly and alignment; LGC and DMJSB contributed to interpretation of results and manuscript writing.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Maximum‐likelihood phylogeny of the cpDNA dataset, estimated using a partitioned RaxML analysis.
Fig. S2 Maximum‐likelihood phylogeny of the nDNA dataset, estimated using an unpartitioned RaxML analysis.
Fig. S3 Maximum‐likelihood phylogeny of combined cpDNA and nDNA datasets, using a partitioned RaxML analysis.
Fig. S4 Leaf phyllotaxis reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S5 Leaf dimorphism and monomorphism reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S6 Cone‐scale phyllotaxis reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S7 Time tree of Cupressaceae estimated from the combined cpDNA‐nDNA dataset, calibrated with ‘C. octothamna’ but not constrained to nDNA topology.
Fig. S8 Parsimony reconstruction of the fire‐adaptive trait cone serotiny.
Fig. S9 Parsimony reconstruction of fire‐prone habitat.
Fig. S10 Time tree of Cupressaceae estimated in Beast from the combined cpDNA‐nDNA dataset, calibrated with ‘C. octothamna’ and constrained to nDNA topology.
Fig. S11 Time tree of Cupressaceae estimated from the combined cpDNA‐nDNA dataset, not calibrated with ‘C. octothamna’ and not constrained to nDNA topology.
Methods S1 Further details on assessment of previously unused fossils for calibration.
Notes S1 References for Supporting Information.
Table S1 Taxa sampled, sample sources, their geographic origin and GenBank accession numbers for sequences included in this study.
Table S2 Loci sequenced, primers used for PCR and their design sources.
Table S3 Molecular‐clock calibrations.
Table S4 Model comparisons using Bayes factors calculated from marginal likelihoods in Beast.
Table S5 Trait data: fossil morphology and fire‐adaptive traits.
Acknowledgements
We are grateful to the Curators of the Australian National Herbarium and the Queensland Herbarium for access to the collections. We thank Lynda Prior for field collection of samples and data, general advice and suggestions for improving the manuscript. We also thank Greg Jordan and two anonymous referees for their useful suggestions. Tim Brodribb kindly donated leaf tissue samples from plants cultivated at the University of Tasmania.
References
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molecular Biology and Evolution 29: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Molecular Biology and Evolution 30: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman DMJS, Haverkamp C, Rann KD, Prior LD, Edwards D. 2017. Differential demographic filtering by surface fires: how fuel type and fuel load affect sapling mortality of an obligate seeder savanna tree. Journal of Ecology 106: 1010–1022. [Google Scholar]
- Bradshaw SD, Dixon KW, Hopper SD, Lambers H, Turner SR. 2011. Little evidence for fire‐adapted plant traits in Mediterranean climate regions. Trends in Plant Science 16: 69–76. [DOI] [PubMed] [Google Scholar]
- Byng JW. 2015. The gymnosperms handbook: a practical guide to extant families and genera of the world. Hertford, UK: Plant Gateway Ltd. [Google Scholar]
- Cohn JS, Lunt ID, Ross KA, Bradstock RA. 2011. How do slow‐growing, fire‐sensitive conifers survive in flammable eucalypt woodlands? Journal of Vegetation Science 22: 425–435. [Google Scholar]
- Condamine FL, Leslie AB, Antonelli A. 2017. Ancient islands acted as refugia and pumps for conifer diversity. Cladistics 33: 69–92. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Arroyo MTK, Cook LG, Gandolfo MA, Jordan GJ, McGlone MS, Weston PH, Westoby M, Wilf P, Linder HP. 2009. Phylogenetic biome conservatism on a global scale. Nature 458: 754–756. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2009. Explosive radiation or mass extinction? Interpreting signatures in molecular phylogenies. Evolution 63: 2257–2265. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2011. Cenozoic extinctions account for low diversity of extant gymnosperms compared with angiosperms. New Phytologist 192: 997–1009. [DOI] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2013. How was the Australian flora assembled over the last 65 million years? A molecular phylogenetic perspective. Annual Review of Ecology, Evolution, and Systematics 44: 303–324. [Google Scholar]
- Crisp MD, Trewick SA, Cook LG. 2011. Hypothesis testing in biogeography. Trends in Ecology and Evolution 26: 66–72. [DOI] [PubMed] [Google Scholar]
- Dornburg A, Brandley MC, McGowen MR, Near TJ. 2012. Relaxed clocks and inferences of heterogeneous patterns of nucleotide substitution and divergence time estimates across whales and dolphins (Mammalia: Cetacea). Molecular Biology and Evolution 29: 721–736. [DOI] [PubMed] [Google Scholar]
- Doyle JJ. 1991. DNA protocols for plants In: Hewitt GM, Johnston AWB, Young JPW, eds. Molecular techniques in taxonomy. Berlin, Germany: Springer, 283–293. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjon A. 2005. A monograph of Cupressaceae and Sciadopitys. Kew, UK: Royal Botanic Gardens. [Google Scholar]
- Hall TA. 1999. BIOEDIT: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hill KD. 1998. Pinophyta In: McCarthy PM, ed. Flora of Australia, vol. 48. Ferns, gymnosperms, and allied groups. Melbourne, Australia: CSIRO Publishing, 545–596. [Google Scholar]
- Hill RS. 2004. Origins of the southeastern Australian vegetation. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 359: 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RS, Brodribb TJ. 1999. Southern conifers in time and space. Australian Journal of Botany 47: 639–696. [Google Scholar]
- Hill RS, Brodribb TJ. 2006. The evolution of Australia's living biota In: Attiwill P, Wilson B, eds. Ecology: an Australian perspective. Melbourne, Australia: Oxford University Press, 19–40. [Google Scholar]
- Hill RS, Carpenter RJ. 1989. Tertiary gymnosperms from Tasmania: Cupressaceae. Alcheringa 13: 89–102. [Google Scholar]
- Hill RS, Paull R. 2003. Fitzroya (Cupressaceae) macrofossils from Cenozoic sediments in Tasmania, Australia. Review of Palaeobotany & Palynology 126: 145–152. [Google Scholar]
- Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology 58: 367–380. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley JE, Pausas JG, Rundel PW, Bond WJ, Bradstock RA. 2011. Fire as an evolutionary pressure shaping plant traits. Trends in Plant Science 16: 406–411. [DOI] [PubMed] [Google Scholar]
- Ladd PG, Midgley JJ, Nield AP. 2013. Serotiny in southern hemisphere conifers. Australian Journal of Botany 61: 486–496. [Google Scholar]
- Lamont BB, Enright NJ. 2000. Adaptive advantages of aerial seed banks. Plant Species Biology 15: 157–166. [Google Scholar]
- Lamont BB, He TH. 2017. Fire‐proneness as a prerequisite for the evolution of fire‐adapted traits. Trends in Plant Science 22: 278–288. [DOI] [PubMed] [Google Scholar]
- Lamont BB, Lemaitre DC, Cowling RM, Enright NJ. 1991. Canopy seed storage in woody plants. Botanical Review 57: 277–317. [Google Scholar]
- Leslie AB, Beaulieu JM, Rai HS, Crane PR, Donoghue MJ, Mathews S. 2012. Hemisphere‐scale differences in conifer evolutionary dynamics. Proceedings of the National Academy of Sciences, USA 109: 16217–16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. 2018. Mesquite: a modular system for evolutionary analysis. Version 3.40. [WWW document] URL http://mesquiteproject.org [accessed 24 April 2018].
- Mao K, Milne RI, Zhang L, Penga Y, Liu J, Thomas P, Mill RR, Renner SS. 2012. The distribution of living Cupressaceae reflects the breakup of Pangea. Proceedings of the National Academy of Sciences, USA 109: 7793–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR. 2017. Five palaeobiological laws needed to understand the evolution of the living biota. Nature Ecology and Evolution 1: 0165. [DOI] [PubMed] [Google Scholar]
- Mays C, Cantrill DJ, Bevitt JJ. 2017. Polar wildfires and conifer serotiny during the Cretaceous global hothouse. Geology 45: 1119–1122. [Google Scholar]
- McIver EE. 2001. Cretaceous Widdringtonia Endl. (Cupressaceae) from North Americ. International Journal of Plant Sciences 162: 937–961. [Google Scholar]
- McLoughlin S. 2001. The breakup history of Gondwana and its impact on pre‐Cenozoic floristic provincialism. Australian Journal of Botany 49: 271–300. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers, 1–8. [Google Scholar]
- Morrison DA. 2008. How to summarize estimates of ancestral divergence times. Evolutionary Bioinformatics 4: 75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer N, Campbell HJ, Tulloch AJ, King PR, Stagpoole VM, Wood RA, Rattenbury MS, Sutherland R, Adams CJ, Collot J et al 2017. Zealandia: earth's hidden continent. GSA Today 27: 27–35. [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ. 1997. The evolutionary biology of plants. Chicago, IL, USA: University of Chicago Press. [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies—a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society of London. Series B: Biological Sciences 255: 37–45. [Google Scholar]
- Paull R, Hill RS. 2010. Early Oligocene Callitris and Fitzroya (Cupressaceae) from Tasmania. American Journal of Botany 97: 809–820. [DOI] [PubMed] [Google Scholar]
- Peters MD. 1985. A taxonomic analysis of a middle Cretaceous megafossil plant assemblage from Queensland, Australia. PhD thesis, University of Adelaide, Adelaide, SA, Australia. [Google Scholar]
- Philippe M, Giret A, Jordan GJ. 1998. Tertiary and Quaternary fossil wood from Kerguelen (southern Indian Ocean). Comptes Rendus L'Académie de Sciences. Série 2, Sciences de la Terre et des Planetes 326: 901–906. [Google Scholar]
- Piggin J, Bruhl JJ. 2010. Phylogeny reconstruction of Callitris Vent. (Cupressaceae) and its allies leads to inclusion of Actinostrobus within Callitris . Australian Systematic Botany 23: 69–93. [Google Scholar]
- Pole M. 1998. Paleocene gymnosperms from Mount Somers, New Zealand. Journal of the Royal Society of New Zealand 28: 375–403. [Google Scholar]
- Pole M. 2007a. Conifer and cycad distribution in the Miocene of southern New Zealand. Australian Journal of Botany 55: 143–167. [Google Scholar]
- Pole M. 2007b. Early Eocene dispersed cuticles and mangrove to rainforest vegetation at Strahan‐Regatta Point, Tasmania. Palaeontologia Electronica 10(15A): 66. [Google Scholar]
- Raftery AE. 1996. Approximate Bayes factors and accounting for model uncertainty in generalised linear models. Biometrika 83: 251–266. [Google Scholar]
- Rambaut A. 2016. FigTree v1.4.3. [WWW document] URL http://tree.bio.ed.ac.uk/software/figtree/ [accessed 24 April 2018].
- Rambaut A, Suchard MA, Drummond A. 2013. Tracer v1.6. [WWW document] URL http://tree.bio.ed.ac.uk/software/tracer/ [accessed 24 April 2018].
- Sakaguchi S, Uchiyama K, Ueno S, Ujino‐Ihara T, Tsumura Y, Prior LD, Bowman DMJS, Crisp MD, Isagi Y. 2011. Isolation and characterization of 52 polymorphic EST‐SSR markers for Callitris columellaris (Cupressaceae). American Journal of Botany 98: e363–e368. [DOI] [PubMed] [Google Scholar]
- Stadler T. 2009. On incomplete sampling under birth‐death models and connections to the sampling‐based coalescent. Journal of Theoretical Biology 261: 58–66. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stockey RA, Kvacek J, Hill RS, Rothwell GW, Kvacek Z. 2005. The fossil record of Cupressaceae s. lat In: Farjon A, ed. A monograph of Cupressaceae and Sciadopitys. Kew, UK: Royal Botanic Gardens, 54–68. [Google Scholar]
- Trauernicht C, Murphy BP, Portner TE, Bowman DMJS. 2012. Tree cover‐fire interactions promote the persistence of a fire‐sensitive conifer in a highly flammable savanna. Journal of Ecology 100: 958–968. [Google Scholar]
- Whang SS, Hill RS. 1999. Late Palaeocene Cupressaceae macrofossils at Lake Bungarby, New South Wales. Australian Systematic Botany 12: 241–254. [Google Scholar]
- Wilf P, Little SA, Iglesias A, Zamaloa MD, Gandolfo MA, Cuneo NR, Johnson KR. 2009. Papuacedrus (Cupressaceae) in Eocene Patagonia: a new fossil link to Australasian rainforests. American Journal of Botany 96: 2031–2047. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Maximum‐likelihood phylogeny of the cpDNA dataset, estimated using a partitioned RaxML analysis.
Fig. S2 Maximum‐likelihood phylogeny of the nDNA dataset, estimated using an unpartitioned RaxML analysis.
Fig. S3 Maximum‐likelihood phylogeny of combined cpDNA and nDNA datasets, using a partitioned RaxML analysis.
Fig. S4 Leaf phyllotaxis reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S5 Leaf dimorphism and monomorphism reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S6 Cone‐scale phyllotaxis reconstructed on the tree, to guide placement of ‘C. octothamna’ fossil for calibration.
Fig. S7 Time tree of Cupressaceae estimated from the combined cpDNA‐nDNA dataset, calibrated with ‘C. octothamna’ but not constrained to nDNA topology.
Fig. S8 Parsimony reconstruction of the fire‐adaptive trait cone serotiny.
Fig. S9 Parsimony reconstruction of fire‐prone habitat.
Fig. S10 Time tree of Cupressaceae estimated in Beast from the combined cpDNA‐nDNA dataset, calibrated with ‘C. octothamna’ and constrained to nDNA topology.
Fig. S11 Time tree of Cupressaceae estimated from the combined cpDNA‐nDNA dataset, not calibrated with ‘C. octothamna’ and not constrained to nDNA topology.
Methods S1 Further details on assessment of previously unused fossils for calibration.
Notes S1 References for Supporting Information.
Table S1 Taxa sampled, sample sources, their geographic origin and GenBank accession numbers for sequences included in this study.
Table S2 Loci sequenced, primers used for PCR and their design sources.
Table S3 Molecular‐clock calibrations.
Table S4 Model comparisons using Bayes factors calculated from marginal likelihoods in Beast.
Table S5 Trait data: fossil morphology and fire‐adaptive traits.


