Abstract
Sprouting angiogenesis is a major form of neovascularization of tissues suffering from hypoxia and other related stress. Endothelial cells (ECs) undergo proliferation, differentiation, programmed death, and migration during angiogenic sprouting, but the underlying molecular mechanisms regulating ECs in angiogenesis have been incompletely elucidated. Here we report that the transmembrane protein 215 (TMEM215) is involved in angiogenesis by regulating EC survival. The murine TMEM215 gene, which possesses two transcriptional starting sites as determined by 5′‐rapid amplification of complementary DNA (cDNA) ends (RACE), encodes a two‐pass TMEM. The TMEM215 transcripts were detected in ECs in addition to other tissues by quantitative reverse transcription‐polymerase chain reaction. Immunofluorescence showed that TMEM215 was expressed in the vasculature in retina, liver, and tumor, and colocalized with EC markers. We show that knockdown of TMEM215 in ECs induced strong cell death of ECs in vitro without affecting cell proliferation and migration, suggesting that TMEM215 was required for EC survival. Downregulation of TMEM215 expression compromised lumen formation and sprouting capacities of ECs in vitro. Moreover, intravitreous injection of TMEM215 small interfering RNA resulted in delayed and abnormal development of retinal vasculature with poor perfusion. These results identified TMEM215 as a novel molecule involved in angiogenesis by regulating the survival of ECs.
Keywords: angiogenesis, apoptosis, ECs, retina, TMEM215
1. INTRODUCTION
Vascularization is essential for tissue growth and homeostasis. Except for vasculogenesis in early embryonic development, angiogenesis, the outgrowth of new vessels from existing ones, plays a central role in neovascularization under both physiological and pathological conditions, although vasculogenesis has also been suggested in adults in recent years (Adams & Alitalo, 2007). Angiogenesis is largely accomplished by the sprouting of endothelial cells (ECs), which involves cell proliferation, differentiation, adhesion, and guided migration (Gerhardt et al., 2003). Of note, sprouting angiogenesis is triggered by hypoxia and other related stress signals that hold potentials to induce cell death; and remodeling of newly formed vessel plexus requires apoptosis of ECs (Korn & Augustin, 2015; Pugh & Ratcliffe, 2003). Therefore, appropriate regulation of EC survival and apoptosis is critically involved in angiogenic sprouting, but the underlying molecular mechanisms have not been completely elucidated.
A number of molecules and signaling pathways have been identified in regulating EC behavior during angiogenesis. The initial step of angiogenesis involves activation and differentiation of quiescent ECs into the tip and stalk ECs, which navigate hypoxic tissues by long filopodia and elongate vessel sprouts via proliferation, respectively (Jakobsson et al., 2010). Tip cells express a high level of vascular endothelial growth factor receptor 2/3 (VEGFR2/3) to sense vascular endothelial growth factor A (VEGFA), the most important angiogenic cytokine (Blanco & Gerhardt, 2013). On the other hand, stalk cell behavior is suppressed in tip cells by neuropilin 1 (NRP1)‐mediated inhibition of bone morphogenetic protein (BMP)‐activin receptor‐like kinase (ALK) signaling (Aspalter et al., 2015; Fantin et al., 2013). Roundabout homologue 4 (ROBO4)‐uncoordinated 5 homologue B (UNC5B) interaction blocks angiogenesis, and maintains vessel integrity (Koch et al., 2011). In addition, metabolism serves as one of the elements controlling EC behaviors in sprouting. It has been shown that 6‐phosphofructo‐2‐kinase/fructose2, 6‐bisphosphatase‐3 (PFKFB3) enhances glycolysis in tip cells to guarantee the competitive energy supply (De Bock et al., 2013; Schoors et al., 2014). Carnitine palmitoyltransferase 1a (CPT1a)‐controlled fatty acid oxidation regulates nucleotide synthesis and proliferation of ECs in stalk cells (Schoors et al., 2015). MicroRNAs (miRNAs) are novel angiogenic regulators that modulate gene expression by binding to the 3′‐untranslated region (3′‐UTR) of messenger RNAs (mRNAs) with seed sequence (Kane, Thrasher, Angelini & Emanueli, 2014; Landskroner‐Eiger, Moneke, & Sessa, 2013). Knockout of Dicer in ECs results in reduced angiogenic response to limb ischemia and vascular endothelial growth factor (VEGF) stimulation (Suarez et al., 2008). Several miRNAs including miR‐126, miR‐218, miR‐342‐5p, among others, have been implicated in angiogenesis (Fish et al., 2008; Small, Sutherland, Rajagopalan, Wang, & Olson, 2010; Yan et al., 2016).
The Notch signaling pathway represents an evolutionarily highly conserved pathway mediating contact‐dependent communications between neighboring cells (Gridley, 2007). Proper activation of the Notch signal is essential for endothelial sprouting in angiogenesis and maintaining vascular homeostasis (Blanco & Gerhardt, 2013; Hellstrom et al., 2007). Deficiency of recombination binding protein Jκ (RBP‐J), the integrative downstream transcription factor of canonical Notch signaling, leads to excessive sprouting and malformation of vessels, while activation of Notch signaling restricts angiogenesis (Dou et al., 2008). In an attempt to elucidate downstream mechanisms of Notch signaling, we identified the transmembrane protein 215 (TMEM215) in ECs. Here we reported that, although not directly regulated by Notch, TMEM215 is expressed in ECs and likely promotes angiogenic sprouting by maintaining EC survival.
2. MATERIALS AND METHODS
2.1. Human tissues and mice
Human umbilical cord biopsies were obtained from the Department of Obstetrics and Gynecology of Xijing Hospital, Fourth Military Medical University. Informed consent was obtained from individuals who donated their samples. The protocols involving human samples were approved by the Ethics Committee of Xijing Hospital, Fourth Military Medical University.
Mice of the C57BL/6 lineage were maintained under specific pathogen‐free conditions. Transgenic mice with conditionally activated EC‐specific Notch intracellular domain expression (NICeCA) and the control (Ctrl), as well as EC‐specific RBP‐J knockout (RBPj△E) and control mice, were described previously (Yan et al., 2016). For intravitreal injection of small interfering RNA (siRNA), pups of postnatal day (P) 3 were anesthetized by incubating on the ice, and intravitreally injected with 6 μg of siRNA in 0.5 μl phosphate‐buffered saline (PBS) in one eye and an equal amount of control oligonucleotides on the other side. Retinas were collected on P6 or P7, fixed in 4% paraformaldehyde (PFA) at 4°C overnight, and subjected to immunofluorescence staining as described below (Pitulescu, Schmidt, Benedito, & Adams, 2010). The siRNA and control were synthesized as in vivo modified STABLE siRNA by RiboBio (Ribobio, Guangzhou, China). The sequences were as follows (Si‐215‐1, 5′‐CAGGAAGACAUCCAGAUdTdT; Si‐215‐2, 5′‐GCUCAGAUGAGCUGGCUAAdTdT; Si‐Ctrl, 5′‐UUCUCCGAACGUGUCACGUdTdT). All animal experiments were approved and followed the guidelines issued by the Animal Experiment Administration Committee of the Fourth Military Medical University.
2.2. Cell culture and transfection
Primary human umbilical vein endothelial cells (HUVECs) were isolated by collagenase treatment of human umbilical tissues. Single‐cell suspensions were cultured in endothelial cell medium (ECM; ScienCell, San Diego, CA) supplemented with 5% fetal bovine serum (FBS) and endothelial cell growth supplements (ECGS), 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified atmosphere with 5% CO2 at 37°C. Cells between passages 3 and 5 were used in the experiments. Lewis lung carcinoma (LLC) cells, Hepa1–6 hepatic carcinoma cells, bEND.3 cells (lined murine ECs) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) routinely. Mouse primary neural stem cells, mesenchymal stem cells, and macrophages were previously preserved in our lab.
HUVECs were transfected with siRNA or the control oligonucleotides at the concentration of 100 nM in a 6‐well plate with 5 µl Lipofectamine 2000™ reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The medium was replaced 6 hr later with normal growth medium. The γ‐secretase inhibitor (GSI; Alexis Biochemicals, San Diego, CA) was used at the concentrations of 25 μM. For staining with 5‐ethynyl‐2′‐deoxyuridine (EdU), cells were incubated with 50 μM EdU (Ribobio, Guangzhou, China) in the medium for 2 hr, and then fixed with 4% PFA at room temperature for 30 min, followed by staining with Apollo® 567. Images were captured under a fluorescence microscope (BX51; Olympus, Tokyo, Japan). Lactate dehydrogenase (LDH) in the culture medium was determined using a kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
2.3. Immunofluorescence staining
Mouse embryos of embryonic (E) Day 12.5, LLC tumors of Day 21 after subcutaneous inoculation (5 × 106 cells), and normal liver were collected and fixed with 4% PFA, followed by embedding in paraffin according to routine protocol. Deparaffinized tissues were sectioned at 8‐µm thickness and blocked in 5% bovine serum albumin (BSA), and then permeabilized with 0.3% Triton X‐100. Sections were stained with primary antibodies diluted in blocking buffer by incubating at 4°C overnight. After washing, sections were incubated with secondary antibodies for 2 hr, and then for an additional 15 min with 4,6‐diamidino‐2‐phenylindole (DAPI) and mounted with Fluorescent Mounting Medium (Dako, Denmark). Images were taken under a laser scanning confocal microscope (LSM 700; Zeiss, Oberkochen, Germany). Antibodies and related staining reagents include anti‐TMEM215 (1:500, HPA052804; Sigma‐Aldrich, St. Louis, MO), anti‐CD31 (1:400, 102502; BioLegend, San Diego, CA), anti‐Lyve‐1 (1:500, D17; Reliatech, Wolfenbüttel, Germany), Isolectin B4 (IB4, 1:100, FL‐1201, Vector Laboratories, Burlingame, CA), Alexa Fluor 594‐conjugated donkey anti‐Rat (1:400, A‐21209; Invitrogen, Carlsbad, CA), Dylight 488‐conjugated goat anti‐rabbit IgG (H + L) (1:400, A‐23220, Abbkine), and Alexa Fluor 488‐conjugated goat anti‐mouse IgM (1:400, ab150121; Abcam, Cambridge, MA).
For whole‐mount retina staining, eyes from P7 pups were prefixed for 2 hr in 4% PFA at room temperature. Retinas were dissected and postfixed for 12 hr in 4% PFA at 4°C. Then retinas were permeabilized and blocked in PBS containing 1% BSA and 0.5% Triton X‐100 overnight. Samples were incubated with IB4 and anti‐TMEM215 in PBS‐0.5% Triton X‐100 overnight at 4°C, followed by incubation with the secondary antibodies in PBS overnight at 4°C. Each step was followed by three washes in PBS for 10 min. The retinal tissues were then flat mounted on a glass slide under a dissecting microscope and examined under a confocal laser scanning microscope.
Terminal deoxynucleotidyl transferase‐mediated dUTP nick end labeling (TUNEL) was performed by using the DeadEnd™ Fluorometric TUNEL System (Promega Corporation, Madison, CA) following the manufacturer’s instructions. Briefly, samples were fixed with 4% PFA and permeabilized in 0.3% Triton X‐100, and then incubated with the TUNEL reagents containing Cyanine 3 (Cy3)‐dUTP at 37°C for 1 hr. The samples were washed with PBS and counterstained with DAPI and observed under a fluorescence microscope.
2.4. Transmission electron microscopy (TEM)
Cells were fixed in 2.5% glutaraldehyde followed by ferrocyanide‐reduced osmium tetroxide. After uranyl staining en bloc, cells were embedded in epoxy resin according to standard procedures. Ultra‐thin sections were made and observed under an electron microscope (LEO 912AB; Omega) equipped with a camera (SiSVeleta; Olympus) and the iTEM software.
2.5. Lumen formation assay
Matrigel basement membrane matrix (BD Biosciences, Franklin Lakes, NJ) was diluted 1:1 with DMEM and was used to precoat 48‐well plates at 37°C. HUVECs were trypsinized and 8 × 104 cells were seeded in each well in ECM containing 0.5% FBS. Cells were observed and recorded under the microscope at 6 hr. The number of branches and the length of cell cords was measured.
2.6. Fibrin gel beads sprouting assay
Fibrin beads sprouting assay was conducted using a fibrin beads assay kit (Amersham‐Pharmacia Biotech, Piscataway, NJ) as described previously (Yan et al., 2016). Briefly, HUVECs were incubated with the Cytodex 3 microcarrier beads (Sigma‐Aldrich; 400 cells per bead) at 37°C overnight. The beads were then embedded in the fibrinogen (Sigma‐Aldrich) containing 0.625 U/ml thrombin (Sigma‐Aldrich) at a density of 100 beads/ml in a 48‐well plate, and 0.5 ml of EGM‐2 medium (Clonetics, Walkersville, MD) was added with lung fibroblasts (20,000 cells/well). The medium was changed every other day for 2 or 4 days. Images were taken under a microscope (CKX41; Olympus) with a CCD camera (DP70; Olympus), and sprouting was quantified by measuring the number and length of sprouts.
2.7. Aortic ring culture
Mice were killed and aortae were collected as previous (Baker et al., 2011) and transfected with 200 nM siRNA or control oligonucleotides for 12 hr by using Lipofectamine 2000™. The aortae were then embedded in 50 μl Matrigel basement membrane matrix in a 96‐well plate containing 150 μl of opti‐MEM supplemented with 2.5% FBS and 30 ng/ml VEGF (Promega Corporation), and cultured at 37°C in 5% CO2–95% air for 4 days. Samples were then photographed under a microscope and sprouts were measured.
2.8. Cell migration and adhesion assays
Cell migration was measured by wound healing assay as described. In brief, after siRNA transfection in a 6‐well plate, the scratch was created with a pipette tip, and the medium was then replaced by ECM supplemented with 0.5% FBS without ECGS. Wound closure was observed under a microscope (CKX41; Olympus) with a CCD camera (DP70; Olympus) and measured 16 hr after the scratch was made. For cell adhesion assay, 4 × 105 HUVECs were seeded in normal growth medium in a 6‐well plate precoated with 0.2% gelatin. The plates were incubated for 5 min and then shaken for 10 s. After washing, adherent cells were fixed with 4% formaldehyde and stained with crystal violet.
2.9. Reverse transcription‐polymerase chain reaction (RT‐PCR)
Total RNA was prepared from cells or tissues with the TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. Complementary DNA (cDNA) was synthesized using a reverse transcription kit (Takara, Dalian, China). Real‐time polymerase chain reaction (PCR) was performed using an SYBR Premix Ex Taq Kit (Takara) and an ABI PRISM 7500 Real‐time PCR system (Life Technologies, Waltham, MA), with β‐actin as internal controls. The sequences of the primers are as follows: mouse β‐actin, 5′‐CATCCGTAAAGACCTCTATGCCAAC, and 5′‐ATGGAGCCACCGATCCACA; mouse TMEM215, 5′‐TGGAGACAGTGAGAGGTGGC, and 5′‐AGTTTCCCCCTTCATGCCAG; human β‐actin, 5′‐TGGCACCCAGCACAATGAA, and 5′‐CTAAGTCATAGTCCGCCTAGAAGCA; human TMEM215, 5′‐CCCACCTGCCTTGTATCTGATTGATAT, and 5′‐CTGAGCCTGAGGATAGCATCTTCTT.
2.10. Rapid amplification of cDNA ends (RACE)
RNA isolated from mice brain was reverse‐transcribed using the SMARTer RACE cDNA Amplification Kit (Clontech Laboratories, Mountain View, CA), followed by two rounds of PCR amplification of cDNA. The first‐round PCR was performed using the specific primer 5′‐RACE‐TMEM215 or 3′‐RACE‐TMEM215, and the universal primer mix, and the product was diluted and subjected to the second‐round PCR using 5′‐NEST‐RACE‐TMEM215 or 3′‐NEST‐RACE‐TMEM215 primers and nest universal primer (NUP). The sequences of the primers are as follows: 5′‐RACE‐TMEM215, 5′‐CAAGGCACTGTAGGCGAGGGAACTGG; 3′‐RACE‐TMEM215, 5′‐GGGGCATAGTTTAGGGAAGTACGGAGAA; 5′‐NEST‐RACE‐TMEM215, 5′‐TCAGGCCGCATCTTTCACTCCATCC; 5′‐NEST‐RACE‐TMEM215, 5′‐TGCCCCAGGACAGTATCATCGTTTG.
2.11. Western blot analysis
Cells were lysed in RIPA buffer (Beyotime, Shanghai, China) containing 10 mM phenylmethanesulfonyl fluoride (PMSF). Samples (20 μg total proteins) were separated by SDS‐PAGE and blotted onto polyvinylidene fluoride (PVDF) membranes, followed by probing with primary antibodies and HRP‐conjugated secondary antibodies. β‐actin was used as a loading control. Membranes were developed using an enhanced chemiluminescence (ECL) system (Clinx Science Instruments, Shanghai, China).
2.12. Statistics
Statistical analysis was performed with the Image‐Pro Plus 6.0 (Media Cybernetics, MD) and GraphPad Prism 5 (GraphPad Software, CA). All quantitative data were presented as mean ± SEM. Statistical significance was calculated using unpaired or paired Student’s t test. p < 0.05 was considered as significant.
3. RESULTS
3.1. Characterization of the TMEM215 gene
In an attempt to identify Notch downstream molecules involved in angiogenesis using microarray, we found that the expression of TMEM215 was lower in retinal vessels from conditional RBP‐J knockout (RBPj∆E) mice (data not shown). To characterize the TMEM215 gene, we performed the RACE experiment to identify its full‐length transcripts (Figure 1a). Sequencing of the 5′‐RACE products indicated that TMEM215 could have two transcription start sites, with the second transcription starting site located 36 bp downstream to the first one (Figure 1b). Moreover, 3′‐RACE identified a poly(A)‐adding signal AATAAA near the 3′ end of TMEM215 cDNA (Figure 1b). The sequence of transcript 1 and transcript 2 have been submitted to the DDBJ/EMBL/GenBank databases under accessions MH686368 and MH686369, respectively. The amino acid sequence of TMEM215 appeared highly conserved in mammals and contained putative sites for posttranslational modifications, suggesting conserved functions (Figure 1c). Analysis with the TMHMM software (DTU Bioinformatics, Denmark) suggested that TMEM215 was a two‐pass TMEM, with the N and C terminal domains inside the cytoplasm (Figure 1d).
Figure 1.
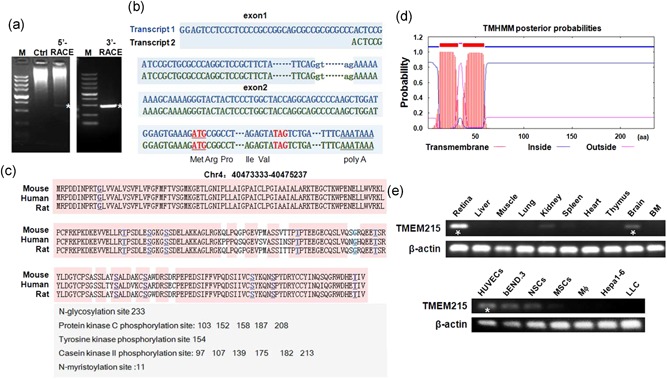
Structure of the TMEM215 gene and tissue distribution of its transcripts. (a) Electrophoresis of PCR products of 5′‐RACE and 3′‐RACE. The 700 bp 5′‐RACE band and the 450 bp 3′‐RACE band were marked by white asterisks. Control (Ctrl), not amplified by TMEM215‐specific primers. M, molecular weight markers (from the top, 5000, 3000, 2000, 1500, 1000, 750, 500, 250, and 100 bp). (b) Transcript 1 and transcript 2 of TMEM215. The transcript 2 starts 36 bp downstream of transcript 1. Exons are in capitals, while the splicing donor and acceptor in the intron are in lowercase. The translation starting codon (ATG) and terminating codon (TAG) are marked in red, and the putative poly(a)‐adding signal (AATAAA) is underlined. Parts of exons and intron sequences are omitted. (c) Alignment of the amino acid sequences of mouse, human, and rat TMEM215. Predicted N‐glycosylation site, protein kinase C phosphorylation sites, tyrosine kinase phosphorylation site, and casein kinase II phosphorylation sites are listed. (d) Prediction of transmembrane domain of TMEM215 using the TMHMM2.0 software (DTU Bioinformatics, Denmark). (e) Detection of TMEM215 transcripts in different types of cells and tissues by qRT‐PCR and electrophoresis, with β‐actin as a reference control. TMEM215 is highly expressed in retina, brain, and ECs, as indicated with white asterisks. ECs: endothelial cells; PCR: polymerase chain reaction; qRT‐PCR: quantitative reverse transcription‐polymerase chain reaction; RACE: 5′‐rapid amplification of complementary DNA ends [Color figure can be viewed at wileyonlinelibrary.com]
We then examined the expression of TMEM215 in different mouse tissues by RT‐PCR. The result showed that the TMEM215 transcript could be readily detected in retina, and brain, and weakly detected in the kidney (Figure 1e, upper). In purified or lined cells, TMEM215 mRNA was expressed at the highest level in ECs, although it could also be detected in neural stem cells (Figure 1e, lower). These data suggested that TMEM215 could be involved in the regulation of ECs.
3.2. TMEM215 was dynamically expressed in ECs
To investigate the role of TMEM215 in ECs, we examined its expression in ECs isolated from tissues under different conditions. Retinas were isolated from P3 and P6 pups, and subjected to immunofluorescence staining with anti‐TEME215 and IB4. The result showed that while few ECs were labeled by anti‐TMEM215 on P3, a considerable proportion of retinal ECs was positive for TMEM215 on P6, especially in areas behind the angiogenic front (Figure 2a). Similarly, anti‐TMEM215 also labeled a part of liver sinusoidal endothelial cells (LSECs) and tumor endothelial cells (TECs) in subcutaneous LLC tumors (Figure 2b,c). Next, retinal ECs were isolated from P3, P5, and P7 pups, and the expression of TMEM215 was determined by qRT‐PCR. The result showed that with the maturation of retinal vasculature, the expression of TMEM215 mRNA increased in retinal ECs (Figure 1d). Then we performed in vitro lumen formation assays using HUVECs and bEND.3 ECs, and compared TMEM215 expression before and after lumen formation. The result showed that the TMEM215 mRNA level increased significantly in ECs in the lumen as compared with ECs under proliferation (Figure 2e). These data suggested that TMEM215 was indeed expressed in ECs, especially in mature vasculature.
Figure 2.
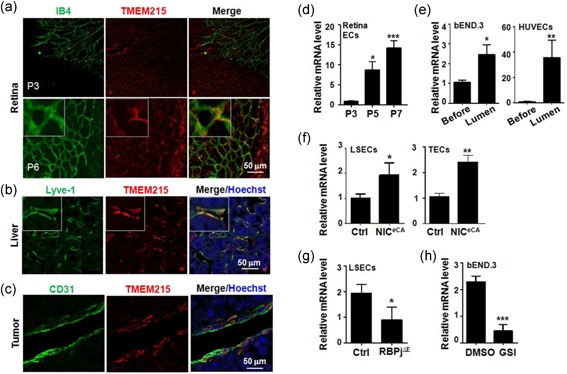
Expression of TMEM215 in ECs. (a) Whole‐mount staining of retina from P3 and P6 pups with IB4 and an anti‐TMEM215 antibody. (b) Liver sections from C57BL/6 mice were stained with anti‐Lyve‐1 and anti‐TMEM215 antibodies. (c) Sections of subcutaneous LLC tumors (21 days after inoculation) were stained with anti‐CD31 and anti‐TMEM215 antibodies. (d) Retinal vasculature was isolated from P3, P5, and P7 pups. The expression of TMEM215 was determined by quantitative RT‐PCR (n = 7). (e) bEND.3 or HUVECs were subjected to in vitro lumen formation assay, and TMEM215 expression in cells before and after lumen formation was determined by qRT‐PCR (n = 7). (f) LSECs and TECs (from subcutaneous LLC tumors) were separated from NICeCA and Ctrl mice, and the expression of TMEM215 was measured by qRT‐PCR (n = 4). (g) LSECs were separated from RBP‐J cKO (RBPj∆E) and Ctrl mice, and the expression of TMEM215 was measured by qRT‐PCR (n = 4). (h) bEND.3 was treated with DMSO or GSI, and the expression of TMEM215 was determined by qRT‐PCR (n = 5). Bar = means ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001. DMSO: dimethyl sulfoxide; ECs: endothelial cells; GSI: γ‐secretase inhibitors; HUVEC: human umbilical vein endothelial cell; IB4: isolectin B4; LLC: Lewis lung carcinoma; LSECs: liver sinusoidal endothelial cells; qRT‐PCR: quantitative reverse transcription‐polymerase chain reaction; SD: standard deviation; TEC: tumor endothelial cells [Color figure can be viewed at wileyonlinelibrary.com]
To evaluate the relationship between TMEM215 and Notch signaling, we isolated LSECs and TECs from mice with EC‐specific Notch activation (NICeCA) and/or Notch blockade (RBPj∆E). qRT‐PCR showed that the TMEM215 mRNA level increased in ECs with activated Notch signaling, but decreased when Notch signal was blocked (Figure 2f,g). Moreover, blocking Notch signaling by treatment with GSI reduced TMEM215 mRNA level (Figure 2h). However, a reporter assay showed that Notch signaling did not influence the transactivation of TMEM215 promoter (data not shown), suggesting that Notch signaling regulated the expression of TMEM215 indirectly.
3.3. TMEM215 was required for survival of ECs
To assess the role of TMEM215 in ECs, we used siRNAs to suppress TMEM215 expression in HUVECs, which was confirmed at both RNA and protein levels (Figure 3a,b). We found that transfection of TMEM215 siRNA (Si‐215) resulted in an obvious reduction of viability of HUVECs as determined by 3‐(4, 5‐dimethylthiazolyl‐2)‐2, 5‐diphenyltetrazolium bromide (MTT) assay (Figure 3c). Furthermore, we examined cell proliferation and apoptosis of HUVECs transfected with Si‐215 using EdU incorporation assay and TUNEL, respectively. The result showed that, while no significant difference was detected in EdU incorporation between HUVECs transfected with Si‐215 and Si‐Ctrl, transfection of Si‐215 resulted in a remarkable increase in apoptotic cells (Figure 3d,e). Ultrastructure of HUVECs transfected with Si‐215 exhibited not only apoptotic bodies but also discontinuous membrane, suggesting coexistence of apoptosis and necrosis (Figure 3f). Indeed, fluorescence‐activated cell sorting (FACS) analysis after staining with Annexin V and propidium iodide (PI) showed that HUVECs transfected with Si‐215 displayed high proportion of AnnexinV+PI− and AnnexinV+PI+ cells, consistent with apoptotic and necrotic populations, respectively (Figure 3g,h). The level of lactate dehydrogenase (LDH) increased in the culture supernatant of HUVECs transfected with Si‐215, suggesting increased membrane fragmentation (Figure 3i). Taken together, these data suggested that knockdown of TMEM215 in ECs led to cell death by increased apoptosis and necrosis.
Figure 3.
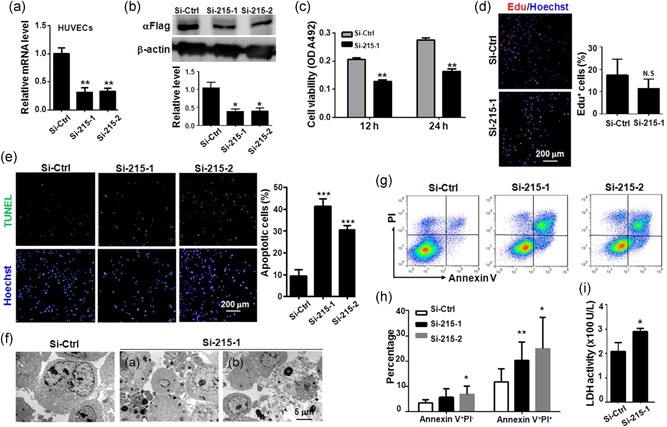
TMEM215 was required for EC survival. (a, b) HUVECs were transfected with Si‐Ctrl, Si‐215‐1, or Si‐215‐2, and mRNA level of TMEM215 was determined by qRT‐PCR. (b) HUVECs expressing a Flag‐tagged TMEM215 were transfected with Si‐Ctrl, Si‐215‐1, or Si‐215‐2, and protein level of TMEM215‐Flag was determined by western blot analysis using anti‐Flag. (c) HUVECs were transfected with Si‐Ctrl or Si‐215‐1, and cell viability was evaluated by MTT assay (n = 6). (d) HUVECs were transfected with Si‐Ctrl or Si‐215‐1, and cell proliferation was evaluated by EdU incorporation assay (n = 7). (e) HUVECs were transfected with Si‐Ctrl, Si‐215‐1, or Si‐215‐2, and cell apoptosis was evaluated by TUNEL (n = 5). (f) HUVECs were transfected with Si‐Ctrl or Si‐215‐1, and cells were observed under TEM. (g, h) HUVECs were transfected with Si‐Ctrl, Si‐215‐,1 or Si‐215‐2, and stained by Annexin V and PI followed by FACS analysis. Percentage of dead cells was determined (n = 5). (i) HUVECs were transfected with Si‐Ctrl or Si‐215‐1, and LDH in the culture medium was determined (n = 5). Bar = means ± SD, *p < 0.05, **p < 0.01, and ***p < 0.001. EC: endothelial cell; EdU: 5‐ethynyl‐2′‐deoxyuridine; FACS: fluorescence‐activated cell sorting; HUVEC: human umbilical vein endothelial cells; LDH: lactate dehydrogenase; N.S.: not significant; mRNA: messenger RNA; MTT: 3‐(4,5‐dimethylthiazolyl‐2)‐2,5‐diphenyltetrazolium bromide; PI: propidium iodide; qRT‐PCR: quantitative reverse transcription‐polymerase chain reaction; SD: standard deviation; TEM: transmission electron microscopy; TUNEL: terminal deoxynucleotidyl transferase‐mediated dUTP nick end labeling [Color figure can be viewed at wileyonlinelibrary.com]
3.4. Knockdown of TMEM215 disrupted angiogenic sprouting in vitro
We then examined the role of TMEM215 in angiogenesis by using in vitro systems. HUVECs were transfected with Si‐215 and Si‐Ctrl, and assayed for lumen formation in vitro. The result showed that knockdown of TMEM215 significantly attenuated lumen formation by HUVECs (Figure 4a). In contrast, cell migration and adhesion appeared to be not influenced by TMEM215 knockdown in HUVECs (Figure 4b).
Figure 4.
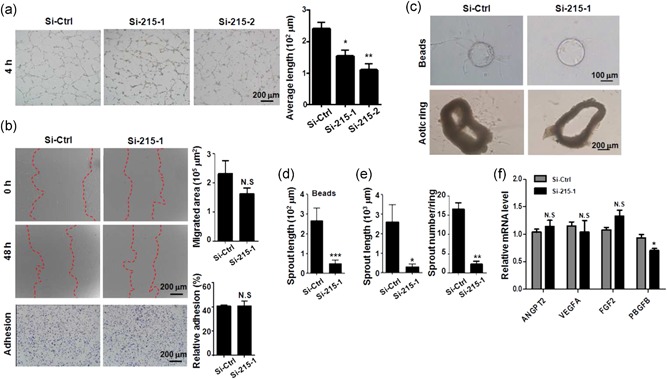
Knockdown of TMEM215 repressed EC angiogenic capacity in vitro. (a) HUVECs were transfected with Si‐Ctrl, Si‐215‐1, or Si‐215‐2, followed by lumen formation assay. The length of cell cords was measured and compared (n = 7). (b) HUVECs were transfected as in (a). Cell migration and adhesion were examined by scratch assay and matrix‐adhesion assay, respectively, (n = 5 for scratch assay and n = 6 for adhesion assay). (c–e) HUVECs were transfected with Si‐Ctrl or Si‐215‐1, and beads‐based sprouting assay was performed (c, upper). Average length of sprouting was determined (d) (n = 10). In lower panels of (c), mouse aortic rings were transfected with Si‐Ctrl or Si‐215‐1, and cultured. Average length and number of sprouts were measured (n = 7). (f) HUVECs were transfected with Si‐Ctrl or Si‐215‐1. The expression of the indicated cytokines was measured. Bar = means ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001. EC: endothelial cells; HUVEC: human umbilical vein endothelial cells; N.S., not significant; SD: standard deviation [Color figure can be viewed at wileyonlinelibrary.com]
Next, HUVECs were transfected with Si‐215 and subjected to fibrin gel beads‐mediated angiogenesis in vitro. The result showed that knockdown of TMEM215 significantly reduced the length of sprouting by HUVECs (Figure 4c, upper,d). Furthermore, we cultured aortic ring from normal mice and transfected cells with Si‐215 or Si‐Ctrl. Consistently with the fibrin gel beads‐mediated angiogenesis, Si‐215 significantly reduced the number and length of angiogenic sprouting from the cultured aortic rings (Figure 4c, lower,e). These results suggested that TMEM215 is necessary for angiogenic sprouting by ECs. Meanwhile, we evaluated the expression of a few angiogenic factors including Angpt2, VEGF, FGF2, and PBGFB, in HUVECs treated with TMEM215 siRNA. The result showed that the mRNA levels of these angiogenic factors appeared not influenced by TMEM215 knockdown in HUVECs, except for PDGFB, which was reduced slightly (Figure 4g). These data suggested that TMEM215 most likely influenced EC survival cell autonomously.
3.5. Suppression of TMEM215 expression resulted in abnormal retinal angiogenesis
To further investigate the role of TMEM215 in angiogenesis in vivo, we performed intravitreal injection of P3 pups with Si‐215 and Si‐Ctrl, and evaluated angiogenesis of retinal vasculature on P7 (Pitulescu et al., 2010). Whole‐mount staining with IB4 showed that transfection with Si‐215 reduced expansion of retinal plexus (Figure 5a,b), and impaired the formation of deep plexus of retinal vasculature (Figure 5c,d). The width of arteries and veins of retinal vasculatures was significantly reduced in Si‐215‐injected eyes (Figure 5e,f). We examined apoptosis of ECs by staining with IB4 and TUNEL. The result showed that transfection with Si‐215 significantly increased TUNEL+ ECs in retinal vasculature (Figure 5g,h). Finally, we evaluated perfusion of retinal vasculature transfected with Si‐215 or Si‐Ctrl by injection Dextran (molecular weight [MW], 2 × 106 Da) through the left ventricle. The result showed that transfection with Si‐215 significantly damaged perfusion of retinal vasculature (Figure 5i,j). Taken together, our data indicated that TMEM215 was required for retinal angiogenesis and insufficient TMEM215 expression could lead to malformation and functional defects of the vasculature.
Figure 5.
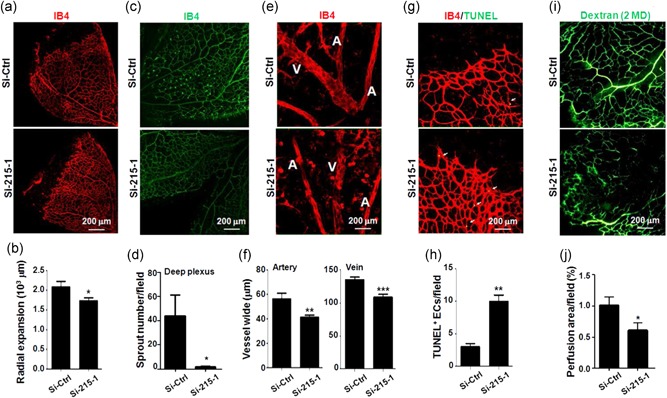
Knockdown of TMEM215 resulted in abnormal retinal vessel development. P3 pups were intravitreally injected with chemically modified Si‐Ctrl or Si‐215‐1. Retinas were dissected on P7 and subjected to whole‐mount immunofluorescence staining. (a, b) Retinas were stained by IB4 (a), and diameter of retinal plexus was measured and compared (b) (n = 7). (c, d) Retinas were stained by IB4 (g), and deep plexus sprouting was counted (h) (n = 3). (e, f) Retinas were stained by IB4 (e), and width of arteries (a) and veins (V) was measured and compared (f) (n = 3). (g, h) Retinas were stained with IB4 and TUNEL (c), and apoptotic ECs were compared (d) (n = 3). (i, j) P3 pups were intravitreally injected with chemically modified Si‐Ctrl or Si‐215‐1. On P7, pups were injected with Dextran (MW, 2 MD) through left ventricle. Retinas were removed 10 min after the injection and photographed. Vessel perfusion was determined (j) (n = 3). Bar = means ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001. EC: endothelial cells; IB4: isolectin B4; MW: molecular weight; TMEM: transmembrane protein; TUNEL: terminal deoxynucleotidyltransferase‐mediated dUTP nick end labeling; SD: standard deviation [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
In the current study, we identified TMEM215 as a novel molecule participating in regulating ECs in angiogenesis. The TMEM215 gene encodes for a two‐pass TMEM with multiple potential posttranslational modification sites. TMEM215 is expressed in ECs derived from different tissues that we have examined including mouse retina, liver, and tumor, as well as in primary and lined ECs cultured in vitro. We noticed that TMEM215 is expressed at a higher level in mature retinal vasculature and in ECs that have formed vessel lumens in vitro, suggesting that it might be involved in late‐angiogenic stages such as vessel remodeling and stabilization. However, the expression of TMEM215 is not limited to vessels. Our data have shown that TMEM215 is also expressed highly in mouse retina, brain, and cultured neural stem cells. Consistently, a recent study has reported that TMEM215 is expressed in a complex subset of cone bipolar cells and amacrine cells in the mature mouse retina, as shown by genetic tracing using the TMEM215‐LacZ transgene (Park, Randazzo, Jones, & Brzezinski, 2017), suggesting that this gene could play a role in bipolar cell subtype fate choice, maturation, and/or physiology. In addition, although we have originally identified TMEM215 as a Notch downstream molecule, and the mRNA level of TMEM215 in ECs was up‐ or downregulated as Notch signaling was activated or disrupted, respectively, our unpublished data showed that Notch signaling did not influence the transactivation of the TMEM215 promoter, suggesting that Notch may regulate TMEM215 expression through indirect mechanism(s).
Our data strongly suggested that TMEM215 participates in angiogenesis by promoting EC survival. Indeed, knockdown of TMEM215 by using a siRNA directly resulted in cell death in ECs with characteristics of both apoptosis and necrosis in vitro, and intravitreal injection of TMEM215 siRNA disturbed angiogenesis with increased apoptosis of ECs during the development of retinal vasculature. Angiogenic process is accomplished by coordinated cellular actions including proliferation, differentiation, guided migration, and quiescence of ECs (Fruttiger, 2007). Controlled cell survival and death of ECs are essential for successful neovascularization by angiogenesis. The initial step of sprouting angiogenesis is typically triggered by hypoxia, which leads to the activation of the Hif‐1α signaling pathway followed by the expression of a large panel of angiogenesis‐related genes (Pugh & Ratcliffe, 2003). However, exposure to severe hypoxia leads to the accumulation of p53, which in turn leads to rapid apoptosis of cells (Banasiak & Haddad, 1998). Hypoxia can also lead to attenuated generation of ATP, which can trigger an unfolded protein response (UPR) when the structure of proteins in the endoplasmic reticulum (ER) could not be maintained, a process known as ER stress. Continuous or uncompensated ER stress could result in apoptosis through the activation of the PERK‐ATF4 axis that leads to the accumulation of the transcription factor CHOP, or the activation of TRAF2‐JNK by IRE1a (Binet & Sapieha, 2015). Moreover, after the outgrowth and full extension of angiogenic sprouts, the newly formed vessels must undergo remodeling to gain efficient tissue perfusion by vessel pruning and regression (Korn & Augustin, 2015). These processes are essentially mediated by EC apoptosis through different mechanisms such as survival factor withdrawal, metabolic changes, and activation of apoptotic pathways. Our in vitro and in vivo data showed that TMEM215 is required for EC survival because suppression of TMEM215 expression leads to EC apoptosis and necrosis. It will be of significance to investigate the role of TMEM215 in vessel pruning and regression. Moreover, it is noteworthy that with siRNAs used in the current study, we had just reduced the expression level of TMEM215 to one third as compared with the control, but resulting in significant increase in cell death in ECs. This could imply that TMEM215 plays a nonredundant role in an important pathway that regulates EC death. However, more molecular experiments with gene knockout mice is required to demonstrate this opinion.
The molecular mechanism underlying TMEM215‐mediated EC survival is currently unknown. TMEM215 is predicted to interact with MAGI‐1, a membrane‐associated guanylate kinase (MAGUK) with inverted domain organization (Luck et al., 2011). MAGI‐1 is present in adherent and tight junctions of all epithelial cell types. MAGI‐1 is important for VE‐cadherin‐dependent Rap1 activation and the adherens junction formation upon cell–cell contact in ECs (Sakurai et al., 2006). In apoptosis, MAGI‐1 is a target of caspase‐mediated cleavage, an impotent step in the disassembly of cell–cell contacts during apoptosis (Gregorc et al., 2007). Whether TMEM215 participates in angiogenesis by regulating MAGI‐1 requires further investigation. Moreover, MAGI‐1 interacts with Delta family Notch ligands including Delta‐like 1 and 4 in mammals and Delta C and D in zebrafish through the C terminal ATEV motif of Delta (Wright, Leslie, Ariza‐McNaughton, & Lewis, 2004). In zebrafish, it was shown that the Delta–MAGI interaction may play some part in the control of neuron migration. Given the critical and multidimensional functions of Notch signaling in angiogenesis, whether this kind of interaction in ECs influences Notch signaling and angiogenesis is another open question.
ACKNOWLEDGMENTS
This study was supported by grants from the National Basic Research Program of China (973 program no. 2015CB553602), the Foundation of Key Laboratory of Modern Separation Science in Shaanxi Province (2010JS104; 11JS098), and the Foundation of Science and Technology in Shaanxi Province (grant numbers 2013SZS18‐Z0, 12018JM7052, and 2015KTCL03‐11) and National Natural Science Foundation of China (grant numbers 31730041, 31671523, 91649106, 31770917, and 31570777).
Liu Y, Zheng Q, He G, et al. Transmembrane protein 215 promotes angiogenesis by maintaining endothelial cell survival. J Cell Physiol. 2019;234:9525–9534. 10.1002/jcp.27641
Contributor Information
Jiankang Liu, Email: j.liu@mail.xjtu.edu.cn.
Liang Liang, Email: lliang2@fmmu.edu.cn.
Hua Han, Email: huahan@fmmu.edu.cn.
References
REFERENCES
- Adams, R. H. , & Alitalo, K. (2007). Molecular regulation of angiogenesis and lymphangiogenesis. Nature Reviews Molecular Cell Biology, 8, 464–478. [DOI] [PubMed] [Google Scholar]
- Aspalter, I. M. , Gordon, E. , Dubrac, A. , Ragab, A. , Narloch, J. , Vizán, P. , … Gerhardt, H. (2015). Alk1 and Alk5 inhibition by Nrp1 controls vascular sprouting downstream of Notch. Nature Communications, 6, 7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, M. , Robinson, S. D. , Lechertier, T. , Barber, P. R. , Tavora, B. , D’Amico, G. , … Hodivala‐Dilke, K. (2011). Use of the mouse aortic ring assay to study angiogenesis. Nature Protocols, 7, 89–104. [DOI] [PubMed] [Google Scholar]
- Banasiak, K. J. , & Haddad, G. G. (1998). Hypoxia‐induced apoptosis: Effect of hypoxic severity and role of p53 in neuronal cell death. Brain Research, 797, 295–304. [DOI] [PubMed] [Google Scholar]
- Binet, F. , & Sapieha, P. (2015). ER stress and angiogenesis. Cell Metabolism, 22, 560–575. [DOI] [PubMed] [Google Scholar]
- Blanco, R. , & Gerhardt, H. (2013). VEGF and Notch in tip and stalk cell selection. Cold Spring Harbor Perspectives in Medicine, 3, a006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bock, K. , Georgiadou, M. , Schoors, S. , Kuchnio, A. , Wong, B. W. , Cantelmo, A. R. , … Carmeliet, P. (2013). Role of PFKFB3‐driven glycolysis in vessel sprouting. Cell, 154, 651–663. [DOI] [PubMed] [Google Scholar]
- Dou, G. R. , Wang, Y. C. , Hu, X. B. , Hou, L. H. , Wang, C. M. , Xu, J. F. , … Han, H. (2008). RBP‐J, the transcription factor downstream of Notch receptors, is essential for the maintenance of vascular homeostasis in adult mice. FASEB Journal, 22, 1606–1617. [DOI] [PubMed] [Google Scholar]
- Fantin, A. , Vieira, J. M. , Plein, A. , Denti, L. , Fruttiger, M. , Pollard, J. W. , & Ruhrberg, C. (2013). NRP1 acts cell autonomously in endothelium to promote tip cell function during sprouting angiogenesis. Blood, 121, 2352–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, J. E. , Santoro, M. M. , Morton, S. U. , Yu, S. , Yeh, R. F. , Wythe, J. D. , … Srivastava, D. (2008). miR‐126 regulates angiogenic signaling and vascular integrity. Developmental Cell, 15, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruttiger, M. (2007). Development of the retinal vasculature. Angiogenesis, 10, 77–88. [DOI] [PubMed] [Google Scholar]
- Gerhardt, H. , Golding, M. , Fruttiger, M. , Ruhrberg, C. , Lundkvist, A. , Abramsson, A. , … Betsholtz, C. (2003). VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. Journal of Cell Biology, 161, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorc, U. , Ivanova, S. , Thomas, M. , Guccione, E. , Glaunsinger, B. , Javier, R. , … Turk, B. (2007). Cleavage of MAGI‐1, a tight junction PDZ protein, by caspases is an important step for cell‐cell detachment in apoptosis. Apoptosis, 12, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley, T. (2007). Notch signaling in vascular development and physiology. Development, 134, 2709–2718. [DOI] [PubMed] [Google Scholar]
- Hellström, M. , Phng, L. K. , Hofmann, J. J. , Wallgard, E. , Coultas, L. , Lindblom, P. , … Betsholtz, C. (2007). Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature, 445, 776–780. [DOI] [PubMed] [Google Scholar]
- Jakobsson, L. , Franco, C. A. , Bentley, K. , Collins, R. T. , Ponsioen, B. , Aspalter, I. M. , … Gerhardt, H. (2010). Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nature Cell Biology, 12, 943–953. [DOI] [PubMed] [Google Scholar]
- Kane, N. M. , Thrasher, A. J. , Angelini, G. D. , & Emanueli, C. (2014). MicroRNAs as modulators of stem cells and angiogenesis. Stem Cells, 32, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Koch, A. W. , Mathivet, T. , Larrivée, B. , Tong, R. K. , Kowalski, J. , Pibouin‐Fragner, L. , … Eichmann, A. (2011). Robo4 maintains vessel integrity and inhibits angiogenesis by interacting with UNC5B. Developmental Cell, 20, 33–46. [DOI] [PubMed] [Google Scholar]
- Korn, C. , & Augustin, H. G. (2015). Mechanisms of vessel pruning and regression. Developmental Cell, 34, 5–17. [DOI] [PubMed] [Google Scholar]
- Landskroner‐Eiger, S., Moneke, I., & Sessa, W. C. (2013). miRNAs as modulators of angiogenesis. Cold Spring Harb Perspect Med 3, a006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, K. , Fournane, S. , Kieffer, B. , Masson, M. , Nominé, Y. , & Travé, G. (2011). Putting into practice domain‐linear motif interaction predictions for exploration of protein networks. PLOS One, 6, e25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K. U. , Randazzo, G. , Jones, K. L. , & Brzezinski, J. A. (2017). Gsg1, Trnp1, and Tmem215 mark subpopulations of bipolar interneurons in the mouse retina. Investigative Ophthalmology and Visual Science, 58, 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu, M. E. , Schmidt, I. , Benedito, R. , & Adams, R. H. (2010). Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nature Protocols, 5, 1518–1534. [DOI] [PubMed] [Google Scholar]
- Pugh, C. W. , & Ratcliffe, P. J. (2003). Regulation of angiogenesis by hypoxia: Role of the HIF system. Nature Medicine, 9, 677–684. [DOI] [PubMed] [Google Scholar]
- Sakurai, A. , Fukuhara, S. , Yamagishi, A. , Sako, K. , Kamioka, Y. , Masuda, M. , … Mochizuki, N. (2006). MAGI‐1 is required for Rap1 activation upon cell‐cell contact and for enhancement of vascular endothelial cadherin‐mediated cell adhesion. Molecular Biology of the Cell, 17, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors, S. , De Bock, K. , Cantelmo, A. R. , Georgiadou, M. , Ghesquière, B. , Cauwenberghs, S. , … Carmeliet, P. (2014). Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metabolism, 19, 37–48. [DOI] [PubMed] [Google Scholar]
- Schoors, S. , Bruning, U. , Missiaen, R. , Queiroz, K. C. S. , Borgers, G. , Elia, I. , … Carmeliet, P. (2015). Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature, 520, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, E. M. , Sutherland, L. B. , Rajagopalan, K. N. , Wang, S. , & Olson, E. N. (2010). MicroRNA‐218 regulates vascular patterning by modulation of Slit‐Robo signaling. Circulation Research, 107, 1336–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, Y. , Fernandez‐Hernando, C. , Yu, J. , Gerber, S. A. , Harrison, K. D. , Pober, J. S. , … Sessa, W. C. (2008). Dicer‐dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proceedings of the National Academy of Sciences of the United States of America, 105, 14082–14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, G. J. , Leslie, J. D. , Ariza‐McNaughton, L. , & Lewis, J. (2004). Delta proteins and MAGI proteins: An interaction of Notch ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development, 131, 5659–5669. [DOI] [PubMed] [Google Scholar]
- Yan, X. C. , Cao, J. , Liang, L. , Wang, L. , Gao, F. , Yang, Z. Y. , … Han, H. (2016). miR‐342‐5p is a Notch downstream molecule and regulates multiple angiogenic pathways including Notch, vascular endothelial growth factor and transforming growth factor beta signaling. Journal of the American Heart Association, 5, e003042. [DOI] [PMC free article] [PubMed] [Google Scholar]


