Abstract
The cerebral cortex is composed of a large variety of distinct cell‐types including projection neurons, interneurons, and glial cells which emerge from distinct neural stem cell lineages. The vast majority of cortical projection neurons and certain classes of glial cells are generated by radial glial progenitor cells in a highly orchestrated manner. Recent studies employing single cell analysis and clonal lineage tracing suggest that neural stem cell and radial glial progenitor lineage progression are regulated in a profound deterministic manner. In this review we focus on recent advances based mainly on correlative phenotypic data emerging from functional genetic studies in mice. We establish hypotheses to test in future research and outline a conceptual framework how epigenetic cues modulate the generation of cell‐type diversity during cortical development.
Keywords: cell‐type diversity, cerebral cortex, gliogenesis, lineage progression, neurogenesis, radial glial progenitor cells
Abbreviations used
- 5caC
5‐carboxylcytosine
- 5fC
5‐formylcytosine
- 5hmC
5‐hydroxymethylcytosine
- 5mC
5‐methylcytosine
- BAF
Brahma‐associated factor
- bRG
basal radial glial progenitor
- CBP
CREB binding protein
- Cdkn1c
cyclin‐dependent kinase inhibitor 1c
- CHD
chromodomain‐helicase‐DNA‐binding
- cPcdh
clustered protocadherin
- CP
cortical plate
- CTCF
CCCCTC‐binding factor
- Dlk1
delta‐like homologue 1
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- EED
embryonic ectoderm development
- EZH2
enhancer of zeste 2
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- Igf2
insulin‐like growth factor 2
- IP
intermediate progenitor
- IZ
intermediate zone
- lincRNA
long intergenic non‐coding RNA
- lncRNA
long non‐coding RNA
- MADM
Mosaic analysis with double markers
- NESC
neuroepithelial stem cell
- NF1
nuclear factor 1
- NICD
Notch intracellular domain
- Norad
non‐coding RNA activated by DNA damage
- NSC
neural stem cell
- NuRD
nucleosome remodeling deacetylase
- OPC
oligodendrocyte progenitor cell
- oRG
outer radial glia
- PRC
polycomb repressive complex
- PUM
Pumilio
- RCoR
REST co‐repressor
- RGP
radial glial progenitor
- RNA
ribonucleic acid
- SCPN
subcerebral projection neuron
- SUZ12
suppressor of zeste 12
- SVZ
subventricular zone
- TAD
topologically‐associated domains
- TDG
thymine DNA glycosylase
- TET
ten‐eleven translocation protein
- VZ
ventricular zone
- Zac1
zinc finger protein regulating apoptosis and cell cycle arrest
The human cerebral cortex is the seat of our cognitive abilities and composed of an extraordinary number of neurons and glial cells. A remarkable heterogeneity in the cortical projection neuron types has been described (Lein et al. 2017; Luo et al. 2017; Zeng and Sanes 2017), yet the identity and development of the neuronal classes that constitute the cortical microcircuits appears to a large extent genetically hard‐wired (Lodato and Arlotta 2015). During development, the mammalian cerebral cortex derives from the embryonic neuroectoderm. At the end of neurulation and neural tube closure the neuroepithelium is composed of neuroepithelial stem cells (NESCs) from which all subsequent neural progenitor cells and their neuron lineages derive. NESCs initially amplify their pool in fast cell cycle divisions before they transform into radial glial progenitors (RGPs) (Taverna et al. 2014). RGPs have been demonstrated to be the main source in the developing cortex for the vast majority of cortical excitatory neurons, transient amplifying progenitors such as intermediate progenitors (IPs) (Kowalczyk et al. 2009; Vasistha et al. 2014), outer subventricular zone (SVZ) radial glial progenitors (oRGs aka basal RGs or bRGs) (Beattie and Hippenmeyer 2017), a subset of glial lineages and adult SVZ stem cells (Bayraktar et al. 2015). The apical processes of RGPs serve as a scaffold for nascent cortical neurons, which migrate from the ventricular zones (VZ)/SVZ through the intermediate zone, in order to reach the cortical plate (CP) (Evsyukova et al. 2013; Hippenmeyer 2014). Cortical layering occurs in an ‘inside‐out’ fashion whereby earlier born neurons populate deep layers and later born neurons progressively occupy upper layers (Angevine and Sidman 1961). Thus, the sequential generation of discrete cell fates, and concerted migration to correct laminae, is critical for the assembly of the neocortex.
Radial glial progenitors generate cell‐type diversity in the cerebral cortex
The concerted production of the correct number and diversity of neurons and glial cells is essential for intricate cortical circuit assembly and an exquisite balance between RGP proliferation/differentiation must be reached in order to generate a neocortex of appropriate size. To elucidate the precise patterns of RGP division, neuron, and glial cell production, Mosaic analysis with double markers (MADM)‐based quantitative clonal analysis has recently been performed (Zong et al. 2005; Hippenmeyer 2013; Gao et al. 2014). This systematic clonal analysis suggests that the behavior of RGPs is remarkably coherent and predictable across all developmental stages. RGPs in the neurogenic phase do not undergo terminal differentiation in a stochastic manner but rather follow a defined program of cell cycle exit resulting in a unitary output of about 8–9 neurons per individual RGP. The size of asymmetric neurogenic clones is however similar across neocortical areas with distinct functions, providing evidence that the unitary neuronal output is a general property of cortical RGPs. Upon completion of neurogenesis, a defined fraction of individual RGPs proceed to gliogenesis whereby about 1 in 6 neurogenic RGPs produce glia – astrocytes and/or oligodendrocytes – indicating a coupling between gliogenesis and neurogenesis at a predictable rate. While the MADM‐based lineage analysis revealed definitive quantitative ontogeny of neocortical excitatory neurons and glial cells (Gao et al. 2014), the cellular and molecular mechanisms dictating neural progenitor cell lineage progression are not well understood (Beattie and Hippenmeyer 2017). Major progress has been made in classifying cell‐types based on single cell transcriptome analysis (Lein et al. 2017; Luo et al. 2017; Zeng and Sanes 2017), however it remains elusive which neuronal and glial cell types arise from an individual progenitor cell. Furthermore, the regulatory modules and epigenetic cues that furnish RGPs with their precise programs to generate projection neuron and glial cell diversity are poorly defined. In this review we discuss recent progress advancing our conceptual understanding and stimulating new hypotheses that can be tested in future research. Epigenetic signaling cues include specific chemical modifications which modulate chromatin structure and organization. The major biochemical signaling pathways organizing the chromatin architecture include DNA methylation, histone modifications, or expression of long non‐coding RNAs (Di Croce and Helin 2013; Yao et al. 2016). Cells combine these features, defining the epigenetic code, in a cell‐type and temporally specific manner. The code determines whether the chromatin configuration at particular genomic loci exerts an active state, characterized by opening of chromatin allowing access by the transcriptional machinery; or repression, defined by chromatin condensation (Kouzarides 2007; Karlić et al. 2010; Portela and Esteller 2010). Progressive modification of the epigenetic landscape in RGPs and nascent cortical neurons controls transcriptional accessibility of specific target genes (Albert et al. 2017). Epigenetic cues may instruct neural stem cell lineage progression in a number of ways and below we outline three major hypothetical conceptual frameworks: (i) Lineage instruction – an epigenetic factor acts during a specific developmental window to initiate the differentiation into a specific cell‐type; (ii) Lineage pre‐priming – an epigenetic factor is present throughout development but is only instructive at later stages to direct the development of a certain cell‐type e.g. glial cells; (iii) Lineage priming – an epigenetic mark affects the development of an entire lineage and has functional impact on all cell‐types generated within this lineage (Fig. 1). Considering these hypothetical frameworks we focus on specific key questions in light of recent correlative phenotypic data obtained from genetic studies in mice: How do epigenetic regulatory cues modulate the quantitative and qualitative output of a single cortical stem cell? Which signaling pathways are transcriptionally regulated in order to modulate stem cell potential over time? And in a broader context, how do epigenetic factors regulate lineage priming and/or instruction in the course of RGP‐mediated generation of cortical cell‐type diversity?
Figure 1.
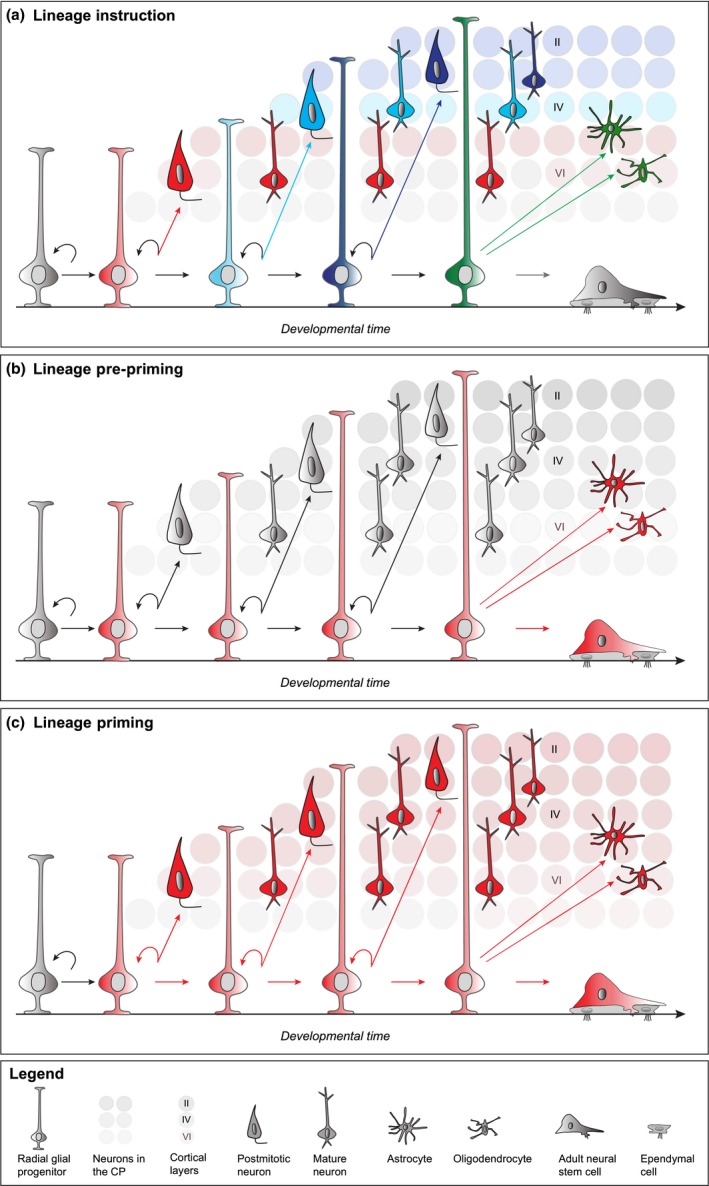
Epigenetic regulation of RGP‐mediated generation of cell‐type diversity. (a) Lineage instruction: epigenetic factors in RGPs together with local factors work in an orchestrated manner to instruct distinct neuronal/glial fates. These factors act only at a specific time window and during a distinct step of cell‐type generation. (b) Lineage pre‐priming: the entire RGP lineage is pre‐primed by a specific epigenetic factor in early RGPs throughout the course of development. However, the functional impact manifests only at a later stage of lineage progression (e.g. during glial differentiation or adult NSC proliferation). (c) Lineage priming: the entire lineage is instructed by the functional impact of an epigenetic factor that is uniformly present throughout all stages of development. In combination with local niche‐derived factors the epigenetic mark exerts its function on progenitor state, RGP proliferation and the entire successive lineage.
DNA methylation and hydroxymethylation during cortical projection neuron development
DNA methylation represents a critical epigenetic mark modifying DNA‐protein interactions and thus controlling transcriptional states and cellular identity. Methylation of cytosine (5‐methylcytosine, 5mC), at CpG dinucleotides modulates core epigenomic processes including gene expression, imprinting, X‐inactivation, silencing of repetitive elements and regulation of heterochromatin (Robertson and Wolffe 2000; Jaenisch and Bird 2003; Bergman and Cedar 2013). DNA methylation most often occurs at CpG dinucleotides, but also marks CpH (H being any other nucleotide than G), particularly in a CpA context (Lister et al. 2013). The methylation pattern during embryogenesis is highly dynamic and exhibits a remarkable degree of tissue and cell‐type specificity. Postmitotic neuron maturation requires accumulation of methylation marks at both, CpG and CpH sites. Lineage specification was shown to correlate with differences in CpA methylation patterns (Sharma et al. 2016).
In mammals, DNA methylation is catalyzed by a family of DNA methyltransferases (DNMTs), including maintenance methyltransferase DNMT1, de novo methyltransferases DNMT3a and 3b, and the catalytically inactive DNMT3L (Lyko 2018). Dnmt1 is expressed throughout cortical development, with increasing expression from progenitor cells to neurons (Hutnick et al. 2009). Dnmt1 controls both, quantitative and qualitative RGP neuron and glia output. Dnmt1–deficient RGPs display upregulation of genes involved in apoptosis and downregulation of genes required for neuronal differentiation and maturation. Consequently, Dnmt1 mutant mice display cortical degeneration, defective neuronal layering, absence of barrel fields and precocious astrocyte generation (Fan et al. 2001, 2005; Golshani et al. 2005; Hutnick et al. 2009). The precise function of DNA methylation in RGP lineage control is currently not known but lineage instruction at the neurogenic to astrocytic transition was analyzed in more detail. During early neurogenesis, CpG sites within promoters of genes regulating gliogenesis, e.g. Gfap, are methylated in a Dnmt1‐dependent manner, thus preventing their expression (Takizawa et al. 2001). At progressively later neurogenic stages, Notch activation in RGPs induces the expression of nuclear factor I, which displaces DNMT1 at the Gfap promoter (Namihira et al. 2009). Therefore methylation of astrogenic gene promoters is selectively abolished (Takizawa et al. 2001), which in turn leads to binding of STAT1/3 heterodimers and the promotion of astrogenic gene expression (Fan et al. 2005; Hatada et al. 2008). In the oligodendrocyte lineage, Dnmt1 regulates oligodendrocyte specification not only by silencing genes involved in oligodendrocyte precursor cell (OPC) proliferation and neuronal differentiation; but also by orchestrating alternative splicing events (Moyon et al. 2016). In mice, the ablation of Dnmt1 results in severe hypomyelination. At the cellular level OPC maturation is impaired because of misfolded proteins and subsequent activation of endoplasmic reticulum stress response (Moyon et al. 2016). Furthermore, based on loss of function data Dnmt1 appears to be required for (olfactory bulb‐destined) neuroblast generation from adult neural stem cells (NSCs) (Noguchi et al. 2016) albeit the precise mechanisms remain elusive and require further studies.
The function of Dnmt3a/b in corticogenesis is even less clear. The expression patterns of Dnmt3a and Dnmt3b are distinct from each other, indicating non‐redundant functions in cortical neurogenesis. Dnmt3b is highly expressed in early progenitors from E10.5 until E13.5 and in differentiated neurons from E17.5 onwards. Dnmt3a is robustly expressed in immature neurons from E13.5 until E17.5 but shows low expression in mature neurons (Feng et al. 2005; Watanabe et al. 2006). Recent studies provide evidence for Dnmt3b‐dependent methylation of the promoters of clustered protocadherin (cPcdh) isoforms, a family of adhesion molecules (Chen and Maniatis 2013), at early stages of neurogenesis. Strikingly, the loss of Dnmt3b results in altered expression of cPcdh isoforms (Toyoda et al. 2014). These findings are intriguing in the context of the possible functional role of cell lineage in modulating the preferential connectivity of clonally related cortical projection neurons (Yu et al. 2009; Li et al. 2012). Indeed, a transient increase of reciprocal connections of clonally related neurons in the somatosensory barrel cortex depends on functional Dnmt3b regulating proper cPcdh isoform expression (Tarusawa et al. 2016). To which extent Dnmt3b activity is required in proliferating RGPs to prime the lineage and thus clonally related progeny remains a key question for future studies.
Hydroxymethylation and in particular 5‐hydroxymethylcytosine (5hmC), which is preferentially detected in intragenic regions, is an abundant epigenetic chemical modification in the brain (Hahn et al. 2013). 5hmC is generated by ten‐eleven translocation protein (TET)‐dependent 5mC oxidation. The TET family includes the three dioxygenases TET1–3 that convert 5mC to 5hmC in a Fe(II)‐ and α‐ketoglutarate‐dependent manner (Tahiliani et al. 2009). In the mammalian brain 5hmC accounts for 1% of all cytosines in cortical DNA (which is equal to ~ 20–25% of total 5mC) and the relative levels of 5mC versus 5hmC are implicated in the regulation of cortical neurogenesis (Jin et al. 2011). TET2 and 3 are highly expressed during cortical neurogenesis, with increasing expression levels from progenitors to neurons. TET2 is most prevalent in outer cortical layers, whereas Tet3 is broadly expressed in all cortical layers (Hahn et al. 2013; Diotel et al. 2017). Accordingly, RGPs in the VZ and young neurons in the intermediate zone contain lower concentration of 5hmC as compared to neurons in the CP. Intragenic 5hmC‐enriched genes are associated with higher transcript levels than others and include many genes critical for neuronal differentiation, migration or axon guidance. Recent evidence suggests that increased TET activity and reduced levels of Polycomb‐mediated repressive histone methylation (discussed in more detail below) work in a synergistic manner to promote neuronal differentiation (Hahn et al. 2013). How TETs regulate lineage priming and qualitative RGP output remains an important unsolved question and requires the analysis of loss and gain‐of TET function at single cell resolution.
5hmC is oxidized to generate 5‐formylcytosine (5fC) and 5‐carboxylcytosine (5caC). Both 5fC and 5caC are recognized and excised by thymine DNA glycosylase (TDG). TDG coupled with base excision repair substitutes 5fC and 5caC by an unmodified cytosine, resulting in DNA demethylation (He et al. 2011; Nabel et al. 2012). During embryonic development, 5hmC and 5caC levels are inversely correlated in different cell types. While RGPs are almost completely devoid of 5caC, this mark accumulates during lineage specification at cell type specific promotors. Accumulation of 5caC at promoters of key glial markers correlates with high transcript levels and glial differentiation. However, it remains unclear whether increased 5caC is a cause or consequence of glial differentiation, or whether a third mechanism could drive both potential responses independently. Experimental evidence from genetic studies suggests that 5caC drives RGP lineage progression towards gliogenesis, since TDG knock‐down results in 5caC retention and enhanced glial differentiation (Wheldon et al. 2014). Future studies will be required to mechanistically dissect the causal link between 5caC levels and astroglial production in more detail.
Role of genomic imprinting in neural stem cell proliferation behavior
Besides global effects on gene expression, differential DNA methylation at imprinting control regions serves as fundamental regulator of genomic imprinting. Imprinting results in parent‐of‐origin specific gene expression where certain genes are expressed solely from the paternally inherited allele and others only from the maternally inherited allele (Barlow and Bartolomei 2014). A key characteristic of imprinted genes is reflected in their cardinal gene‐dosage sensitivity. A number of imprinted genes have been shown to play critical roles in neurogenesis and neuronal differentiation including cyclin‐dependent kinase inhibitor 1c (Cdkn1c), zinc finger protein regulating apoptosis and cell cycle arrest (Zac1), delta‐like homologue 1 (Dlk1) and insulin‐like growth factor 2 (Igf2).
The Cdkn1c gene (aka p57 KIP2) is a member of the CDK interacting protein/kinase inhibitory protein (CIP/KIP) family of cyclin‐dependent kinase inhibitors which regulate G1/S transition by inhibiting cyclin/CDK complexes (Sherr and Roberts 1999). Cdkn1c is maternally expressed in the developing cortex from E11.5 onwards, with highest expression at E14.5 in RGP and IP nuclei. Cdkn1c −/− mice exhibit macrocephaly with disrupted cortical lamination resulting from increased RGP proliferation because of decreased overall cell cycle length and shortening of G1 phase (Mairet‐Coello et al. 2012). Cdkn1c has been shown recently to mark slowly dividing prospective post‐natal precursors which emerge from progenitors located in the ganglionic eminence (Furutachi et al. 2015). Despite the fact that such slowly dividing stem cell precursors have been identified in the developing cortical VZ (Fuentealba et al. 2015) it is not clear whether and how Cdkn1c instructs cortical RGP lineage progression.
The gene encoding Zac1 is expressed from the paternal allele with particular high expression in neuroectodermal stem cells during early development (Valente et al. 2005). Full knockout of Zac1 results in hydrocephaly and decreased brain size, whereas Zac1 overexpression in RGPs triggers premature cell cycle exit because of induction of Cdkn1c expression (Daniel et al. 2015; Rraklli et al. 2016). It is an intriguing hypothetical concept that the expression level of one imprinted gene (Zac1) regulates the expression of a second dosage‐sensitive imprinted gene (Cdkn1c) to modulate unitary RGP output. Furthermore, independent of Cdkn1c, Zac1 negatively controls the neurogenic to astrogenic switch in proliferating RGPs by inducing expression of the JAK/STAT3 signaling inhibitor Socs3 (Schmidt‐Edelkraut et al. 2013).
Dlk1 encodes a transmembrane protein of the Notch/Delta/Serine signaling family. Two different isoforms, a membrane‐bound form and a secreted form have been identified (Smas et al. 1997; Wang and Sul 2006). Dlk1 exhibits paternal specific expression throughout embryonic development (Kobayashi et al. 2000) but allele specific expression of Dlk1 is lost in adult NSCs. Biallelic Dlk1 expression is required for post‐natal SVZ neurogenesis and OB neuron production (Ferrón et al. 2011). However the underlying mechanisms how Dlk1 gene dosage controls stem cell proliferation behavior remain to be determined.
Igf2 encodes a potent growth factor promoting cell survival, proliferation, and differentiation upon binding to insulin‐like growth factor receptors (Nielsen 1992; Daniel et al. 2015). IGF2 binding to IGF1R positively stimulates growth signaling whereas IGF2 binding to IGF2R results in internalization and lysosomal degradation of IGF2, thereby reducing the growth signal (Stewart and Rotwein 1996). In the embryonic brain, Igf2 is expressed from the paternal allele but exhibits biallelic expression shortly after birth and switches to maternal expression in the post‐natal brain (Andergassen et al. 2017). During corticogenesis, IGF2 is secreted from the choroid plexus into the ventricular CSF thereby stimulating the proliferation of RGPs via IGF1R. Igf2 −/− mice display reduced brain size, decreased numbers of dividing progenitors and diminished numbers of upper‐layer neurons (Lehtinen et al. 2011). In future studies it will be important to decipher the precise functional role of Igf2 gene dosage in controlling embryonic RGP proliferation behavior and the generation of the correct number of distinct classes of upper‐layer neurons. Interestingly, during post‐natal neurogenesis, biallelic Igf2 expression is required for adult NSC proliferation (Ferron et al. 2015).
In summary, specific imprinted genes have been shown to regulate RGP and adult NSC proliferation behavior and thus their quantitative and qualitative output. The above cited work also supports the hypothesis that imprinted genes encoding for signaling molecules require biallelic expression in adult NSCs to maintain proper OB neuron generation. The control of imprinted gene expression dosage through epigenetic DNA modification represents an intriguing regulatory module with high potential to regulate the generation of cell‐type diversity during cortical development.
DNA topology controlling RGP lineage progression
The 4D DNA topology orchestrates the ultimate structure and organization of chromatin. Certain genomic regions contact each other in so‐called topologically associated domains (TADs). Cohesin and CCCTC‐binding factor (CTCF) are required for TAD formation and enhancer‐promoter interactions. Association of CTCF to its consensus sequence (three regularly spaced CCCTC repeats) induces cohesin recruitment and formation of a ring‐like structure around distinct sites of the chromosome, thereby inducing DNA looping (Ong and Corces 2014). TAD formation and DNA looping are regulated via modulation of accessibility of CTCF association sites. DNA methylation at CTCF binding sites prevents the interaction of CTCF with DNA (Bell and Felsenfeld 2000; Wang et al. 2012), thus excluding the formation of TAD boundaries at methylated DNA sequences. > 77 000 CTCF binding sites widely distributed throughout the genome have been mapped so far (Chen et al. 2012; Wang et al. 2012). CTCF is highly expressed during neocortical development (Sams et al. 2016) and modulates RGP output by maintaining the progenitor state (Watson et al. 2014). Qualitatively, CTCF instructs cortical cell‐type diversity by promoting fate specification of post‐mitotic neurons through regulation of genes involved in cell adhesion, 58% of those being cPcdh genes. Almost all promoters of stochastically expressed cPcdh isoforms contain a CTCF‐binding site and Ctcf deletion in post‐mitotic neurons leads to misexpression of cPcdh genes and concomitant absence of barrel structures despite layer IV presence (Hirayama et al. 2012). Intriguingly, Dnmt3b deletion also leads to altered cPcdh expression (Toyoda et al. 2014), providing evidence for a potential functional link between DNMT3B and the accessibility of CTCF binding sites which in turn may control cPcdh expression.
The role of histone modifications in RGP proliferation behavior
N‐terminal histone tails are targets for a variety of post‐translational modifications including the reversible covalent attachment of methyl‐, acetyl‐, phospho‐ or ubiquitin groups to distinct lysine (K) or arginine (R) residues (Yao et al. 2016). Such histone modifications activate or repress gene expression (Kouzarides 2007; Karlić et al. 2010; Portela and Esteller 2010).
Methylation of histones is catalyzed by histone methyltransferases and reversed by histone demethylases. The most extensively studied histone methylation sites include histone H3 lysine 4 (H3K4), H3K9, H3K27, H3K36, H3K79 and H4K20. Methylation of each of these distinct lysine residues influences the accessibility of chromatin in a different manner. Generally, H3K4me serves as an active mark, whereas H3K9me2/me3 and H3K27me3 are associated with transcriptional repression (Hyun et al. 2017).
Repressive histone marks
H3K27me3 is catalyzed by the multisubunit Polycomb repressive complex (PRC)2, which consists of three core subunits: enhancer of zeste 2 (EZH2) or its homolog EZH1, embryonic ectoderm development, and suppressor of zeste 12 (Di Croce and Helin 2013). Both, EZH2 and EZH1, contain a conserved SET domain catalyzing the mono‐, di‐, and tri‐methylation of H3K27. PRC1 binds to H3K27me3 and catalyzes the mono‐ubiquitinylation of lysine 119 of histone H2A (H2AK119ub) through a homolog of Drosophila RING protein, thereby ultimately inducing transcriptional silencing. Methylation of H3K27 is reversible, with the two proteins JMJD3 and UTX acting as H3K27 demethylases (Di Croce and Helin 2013).
Ezh2 and Ring1B show high expression in RGPs up to E14.5 and have been proposed to regulate RGP identity and proliferation behavior, as well as RGP‐to‐glial‐progenitor transition (Hirabayashi et al. 2009; Pereira et al. 2010). Ablation of Ezh2 and thus H3K27me3 in RGPs correlates with premature RGP differentiation, increased generation of lower‐layer neurons, decreased upper‐layer neuron production, and precocious astrocyte generation (Pereira et al. 2010; Hahn et al. 2013). Ezh2‐mediated repression of gene expression in cortical RGPs is therefore essential for controlling lineage progression and appropriate neuron and glia output. In a complementary experiment with specific deletion of the SET domain of Ezh2 during mid neurogenesis, RGPs fail to downregulate proneurogenic Ngn1 signaling, which leads to the suppression of glial cell generation (Hirabayashi et al. 2009). Deletion of Ring1B during mid neurogenesis does not alter RGP maintenance, but results in alterations of timed production of specific projection neuron populations such as sustained production of CTIP2+ layer V neurons (Morimoto‐Suzki et al. 2014) and BRN2+ upper‐layer neurons (Hirabayashi et al. 2009). Similar to deletion of the Ezh2 SET domain, Ring1B‐deficient cortices display defective RGP lineage progression from the neurogenic to the gliogenic state, presumably by failing to suppress proneurogenic genes (Hirabayashi et al. 2009). The precise mechanisms by which PRC instructs RGP proliferation behavior are unknown. It is however an attractive hypothesis that PRC association with target genes is differentially regulated in progenitors at distinct stages and post‐mitotic cells, respectively. Key questions that require in‐depth analysis in the future are: (i) How do PRC complexes recognize their target genes? (ii) Which co‐factors regulate PRC recruitment? (iii) How is PRC activity modulated at distinct neurogenic and gliogenic stages? A functionally relevant group of PRC co‐factors in RGPs are chromodomain‐helicase‐DNA‐binding proteins (CHDs) that will be discussed in more detail below.
Repressive H3K9me2/me3 marks are established by the methyltransferases SETDB1, SUV39H1, G9a and G9a‐like protein. H3K9me3 binds heterochromatin protein 1 for transcriptional repression leading to formation and maintenance of heterochromatin. Similar to H3K27me, H3K9me is a reversible mark (Hyun et al. 2017). Setdb1 is highly expressed in proliferating NESCs in the VZ at E9.5 but its expression declines at E15.5 and is not detectable at E17.5. While deletion of Setdb1 does not affect RGP numbers, it leads to increased upper‐layer neuron production at the expense of deep‐layer neurons. Furthermore, ablation of Setdb1 causes accelerated astrogliogenesis, demonstrating that Setdb1 not only regulates the timing of late neurogenic events, but also neurogenic RGP‐to‐astrogenic‐progenitor transition (Tan et al. 2012). At the molecular level, SETDB1 catalyzes H3K9 methylation at promoters of glial differentiation genes (e.g. Sox9 and Gfap), resulting in their repression during neurogenic stages. Taken together, repressive H3K27me3 and H3K9me3 marks correlate with inhibition of precocious neuronal differentiation and controlled timing of gliogenesis. Future studies should aim at identifying functionally‐relevant SETDB1 targets and how these regulate RGP proliferation behavior and lineage progression.
Activating histone marks
Transcriptionally active loci are associated with acetylation of histone lysines, e.g. H3K27ac, mediated by histone acetyl transferases and reversed by histone deacetylases (HDACs) (Wang et al. 2009). Both types of enzymes are recruited to their target promoters through interaction with sequence‐specific transcription factors.
The gene encoding the histone acetylase cAMP‐response element binding protein binding protein (Cbp) is expressed in proliferating RGPs and post‐mitotic neurons during corticogenesis and induces acetylation of H3K9, H3K14 and H3K27 within target gene promoters, such as α1‐tubulin (acetylation peak at E13‐E16), Gfap (peaking at E16‐P3) and Mbp (peaking at post‐natal stages). Cbp knockdown or haploinsufficiency diminishes the acetylation levels at those promoters and concurrently leads to reduced production of late‐born upper‐layer neurons from RGPs, as well as decreased transition to glial progenitors (Wang et al. 2010). How CBP targeting specificity is achieved by temporally controlled expression of binding partners represents an important line of future research. A key candidate in this regard is NGN1 which prevents interaction of CBP with STAT proteins and subsequent activation of astrogenic gene expression (Sun et al. 2001).
Chromatin remodeling complexes controlling RGP lineage progression
Chromatin remodeling is mediated by multi‐subunit protein complexes including the nucleosome remodeling deacetylase (NuRD) and the Brahma‐associated factors (BAF) complex. The NuRD complex consists of lysine‐specific histone demethylase 1A (LSD1), HDAC1/2, the histone binding proteins RBAP46 and 48, metastasis‐associated protein, methyl‐CpG‐binding domain protein 3, and a CHD protein (Lai and Wade 2011). The BAF complex consists of BRG1, BRM and several distinct BAF proteins (Kadoch et al. 2013). Both complexes exhibit alternative subunit composition with temporally regulated expression during development (Son and Crabtree 2014; Nitarska et al. 2016).
NuRD complex
The demethylase LSD1 is expressed in RGPs and post‐mitotic neurons populating the developing CP. LSD1 specifically removes activating H3K4me2 marks from promoters of either neurogenic differentiation‐inducing genes (Zhang et al. 2014) or progenitor‐maintaining genes (Wang et al. 2016). Targeting of specific promoters by LSD1 is mediated by co‐factors, such as REST corepressor (RCoR)2 (Qureshi et al. 2010). The promoters of several RCoR2 target genes including dorso‐ventral CNS specification genes such as Dlx2, Dlx5, Shh and Ascl1, are transcriptionally repressed by removal of activating H3K4me marks through LSD1. Thus genes maintaining RGP stem cell state, e.g. Sonic Hedgehog (SHH) pathway components are upregulated, whereas genes positively involved in neurogenesis, such as Emx1, Tbr2, Trnp1, Foxg1 and Reln, are downregulated in Rcor2 knock‐out mice (Wang et al. 2016).
HDAC1 and 2 are both expressed in RGPs throughout embryonic development. Together LSD1 and HDACs interact with RCoR1/2 (Qureshi et al. 2010). Conditional HDAC1/2 double knockout mice recapitulate the phenotype observed in RCoR1/2 double knockouts, characterized by microcephaly caused by a massive block of projection neuron and oligodendrocyte production (Monaghan et al. 2017), severe laminar disorganization, and accompanied by a global increase in histone acetylation marks (Montgomery et al. 2009; Hagelkruys et al. 2014). HDAC1/2 appears to regulate lineage priming by orchestrating neurogenesis at the level of both, RGP cell fate maintenance and specification of distinct neuronal subtypes. As such, removal of histone acetylation promotes layer II/III callosal projection neuron development by inhibiting subcerebral projection neuron fate specification. HDACs are recruited by LHX2, SATB2 and SKI to mediate NuRD complex‐dependent silencing of Fezf2, Ctip2 and Sox11 subcerebral projection neuron specification genes (Alcamo et al. 2008; Britanova et al. 2008; Baranek et al. 2012; Muralidharan et al. 2017). In OPCs HDACs compete with β‐catenin for TCF7L2 interaction. While the β‐catenin‐TCF complex activates the negative oligodendrocyte differentiation regulator Id2, TCF‐HDAC suppresses Id2 transcription and thus allows oligodendrocyte production (Ye et al. 2009). In summary, the interaction of LSD1, HDAC1/2 and RCoR1/2 activates critical temporal gene expression programs that may impact on lineage priming and lineage instruction in RGPs.
The CHD family is characterized by tandem chromodomains and a SNF2‐like ATPase domain (Murawska and Brehm 2011). CHDs exhibit subunit‐specific functions and display mutually exclusive occupancy within the NuRD complex at different stages of corticogenesis (Nitarska et al. 2016). Thus CHD proteins have been implicated in modulating the overall output of proliferating RGPs. Indeed, based on loss of function studies, CHD2 and CHD7 have been proposed to regulate self‐amplification of RGPs and prevent precocious cell cycle exit (Micucci et al. 2013; Shen et al. 2015; Ohta et al. 2016). In contrast, CHD3 controls the timing of upper‐layer neuron specification (Nitarska et al. 2016) and CHD4 maintains neurogenic RGP fate in an Ezh2‐dependent fashion (Sparmann et al. 2013). These findings indicate that CHDs interact with PRC and regulate H3K27me3 deposition at target promoters, a hypothesis further supported by recent studies on CHD5 and CHD8. Chd5 is expressed in neurons throughout cortical development and promotes SATB2+ upper‐layer projection neuron production. CHD5 is required to activate expression of genes essential in neuron production, migration and differentiation (such as Tubb3, NeuN and Ncam), but at the same time to also induce PRC‐mediated silencing of a small cohort of genes involved in development of non‐neuronal lineages (Egan et al. 2013). Chd8 is strongly expressed around the transition from symmetric proliferative to asymmetric neurogenic RGP division (Sugathan et al. 2014) and promotes the expression of PRC2 components EZH2 and suppressor of zeste 12. Similar to Ezh2 deletion (Pereira et al. 2010; Hahn et al. 2013), knockdown of Chd8 results in premature depletion of RGPs and impaired neurogenesis (Durak et al. 2016). In contrast, twofold reduction of CHD8 protein by Chd8 haploinsufficiency (deletion of exon 5) or Chd8 heterozygous loss‐of‐function mutations in humans causes macrocephaly by increasing proliferation of neural progenitors (Katayama et al. 2016; Gompers et al. 2017; Platt et al. 2017). At first glance the results obtained by the knockdown and haploinsufficiency studies appear contradictory. However one may hypothesize that differential gene dosage of CHD8 results in distinct RGP proliferation dynamics. Whereas substantial depletion of CHD8 drastically impairs RGP lineage progression, twofold protein reduction might just delay activation of neuronal differentiation programs. Thus, determining the precise function of CHD8 in controlling RGP proliferation behavior and unitary neuron output remains an important task for further studies.
BAF complex
In the developing neocortex, distinct BAF subunits are expressed in a temporal and cell‐type specific manner. Proliferating RGPs and post‐mitotic neurons contain BAF complexes with distinct subunit composition, with the RGP BAF complex containing BAF45a and BAF53a, and neuron BAF complex including BAF45b, BAF45c and BAF53b (Lessard et al. 2007). BAF45a promotes progenitor cell proliferation and transition from neurogenic to gliogenic RGP cell fate in a BRG‐dependent manner. Knockdown of progenitor BAF components in RGPs results in slow‐down of the cell cycle and overall decrease of proliferating RGPs, thereby strongly reducing the numbers of IPs and upper‐layer neurons (Lessard et al. 2007; Matsumoto et al. 2016). Brg1‐deficiency in embryonic RGPs inhibits the neurogenic to gliogenic switch but E16.5 cortical cultures lacking Brg1 are not impaired in astrocyte generation (Lessard et al. 2007). These findings suggest that niche‐derived signals determine the fate of Brg1‐deficient RGPs in vivo. Progenitor BAF complex controls RGP proliferation and maintenance on different mechanistic levels: (i) by activating transcription of stem cell differentiation inhibitor Mash1 (Matsumoto et al. 2006, 2016); (ii) by stimulating expression of Notch‐dependent proliferation‐promoting signals and (iii) by repressing SHH‐dependent differentiation‐promoting signals (Lessard et al. 2007). During RGP lineage progression, BRG1 also controls OPC specification and oligodendrocyte formation by suppressing precocious Olig2 transcription (Matsumoto et al. 2016). Intriguingly, progenitor BAF displays mutually exclusive incorporation of either BAF170 or BAF155 at distinct developmental stages. Conditional BAF155/170 double mutants display reduced numbers of proliferative RGPs, dramatic thinning of the cortical SVZ and extensive loss of projection neurons, emphasizing a crucial function of BAF complexes in corticogenesis (Narayanan et al. 2015). Conditional BAF155/170 deletion is accompanied by a global shift from activating H3K9ac to repressive H3K27me2/me3 marks (Nguyen et al. 2016). What are the exclusive functions of BAF170 and BAF155, respectively, during RGP‐mediated neurogenesis? Neural progenitor BAF complexes harbor BAF170 until E14.5 to repress IP generation by inhibiting the expression of many genes typically activated by PAX6 during upper‐layer neuron development (e.g. Tbr2, Cux1 and Tle1) in a BRM‐dependent manner. BAF170 and PAX6 recruit the REST repressor complex to the promoters of target genes, which induces transcriptional silencing of genes involved in late neurogenic events. Between E14.5 and E15.5, BAF170 is replaced by BAF155, which activates expression of IP‐inducing PAX6 target genes in RGPs via association with the H3K27 demethylases JMJD3 and UTX (Lee et al. 2007; Tuoc et al. 2013; Narayanan et al. 2015). Taken together, expression of distinct BAF subunits correlates with the timed generation of cortical projection neuron subtypes; and the interaction of PAX6 with progenitor BAF complexes plays a role in the maintenance of the neurogenic fate of adult NSC‐derived neuroblasts. Upon deletion of either Pax6 or Brg1 from adult NSCs, neuroblasts located outside of the neurogenic niche differentiate to glial lineages, especially OPCs (Ninkovic et al. 2013). Progenitor BAF complexes thus generally regulate cell‐type diversity by promoting neurogenic fate.
Control of RGP proliferation behavior by long non‐coding RNAs
Long non‐coding RNAs (lncRNAs) are untranslated transcripts longer than 200 nucleotides modulating chromatin organization, gene transcription, pre‐mRNA metabolism, and RNA turnover (Grammatikakis and Gorospe 2016). The mammalian genome encodes for thousands of lncRNA, most of which are expressed in the brain (Aprea and Calegari 2015). Recent RNA‐seq experiments using human samples at distinct developmental stages revealed that only a few lncRNAs are abundantly expressed in all cortical cell types (e.g. Norad and Brn1b), whereas the majority of lncRNAs display highly cell type specific expression (e.g. Pnky and LOC646329 in RGPs) (Liu et al. 2016). Several lncRNAs have been shown to play important regulatory functions in cortical development in vivo. For instance the nuclear lncRNA Pinky (Pnky) has been implicated in the promotion of RGP stem cell maintenance, presumably by interacting with the RNA splicing factor PTBP1 albeit the precise mechanism how Pnky controls RGP proliferation behavior remains to be elucidated (Ramos et al. 2015). The long intergenic ncRNA (lincRNA) Brn1b (aka Dali in humans) is expressed in the developing brain from E13.5 until E18.5 and modulates RGP turnover by promoting expression of the neighboring Brn1 gene. Deletion of linc‐Brn1b suppresses IP generation, leading to abnormal cortical lamination particularly affecting upper‐layer neurons and barrel cortex organization (Sauvageau et al. 2013). Since the corresponding human gene product Dali interacts with DNMTs (Chalei et al. 2014), an appealing hypothesis may propose that linc‐Brn1b regulates barrel cortex structures through DNA‐methylation dependent cPcdh expression in upper‐layer neurons. Similar to linc‐Brn1b and Brn1b, many lncRNAs share identical expression patterns with specific neurogenic genes, suggesting that distinct lncRNAs exert a general regulatory function in cell fate (Aprea et al. 2013; Liu et al. 2016). The cytoplasmic non‐coding RNA activated by DNA damage (Norad) is highly expressed in neuronal tissues (Tichon et al. 2016) and antagonizes the activity of the RNA binding proteins Pumilio 2 and 3, which are negative regulators of mRNA translation (Lee et al. 2016). Cortical RGPs are transcriptionally primed to generate diverse types of neurons by simultaneously expressing mRNA of transcriptional regulators of both deep and superficial layer neurons. As such, the Pum2/E4‐T complex promotes translational repression of deep‐layer fate in upper‐layer neurons, thereby controlling correct temporal specification of newborn upper‐layer neurons (Zahr et al. 2018). It will be important in future studies to further elaborate whether or how Norad contributes to Pum2 target recognition and which exact role Norad exerts in generating cortical cell‐type diversity.
Conclusions and perspectives
The mammalian cerebral cortex consists of an extraordinary diversity of neurons and glial cells. However, the complete picture of cortical cell‐type diversity is just emerging. While cortical laminar position enables a rough classification, many other criteria ranging from morphological and physiological to transcriptomic and epigenetic fingerprinting have also been employed. In particular, single cell RNA sequencing has greatly transformed our understanding of cell‐type diversity in the developing and adult cerebral cortex (Poulin et al. 2016; Lein et al. 2017; Zeng and Sanes 2017). While single cell transcriptomes and methylomes (Luo et al. 2017) represent a robust measure to classify cell types, the mechanistic principles controlling their generation by RGPs in vivo remain mostly unclear. In Fig. 2, we summarize the most important epigenetic modulators and their proposed function in distinct steps of RGP lineage progression. RGPs display silencing of genes mediating post‐mitotic cell fates by maintaining repressive DNA methylation, H3K9me, and H3K27me marks. Successive temporally controlled production of neuronal and glial subtypes requires selective removal of those repressive modifications and the addition of activating H3K4me or acetylation marks at specific target loci. At the same time, genes conferring alternative cell fates need to be silenced. Target specificity can be mediated by expression of mutually exclusive subunits of large epigenetic complexes or by distinct co‐factors serving as recruitment hubs for specific epigenetic modulators.
Figure 2.
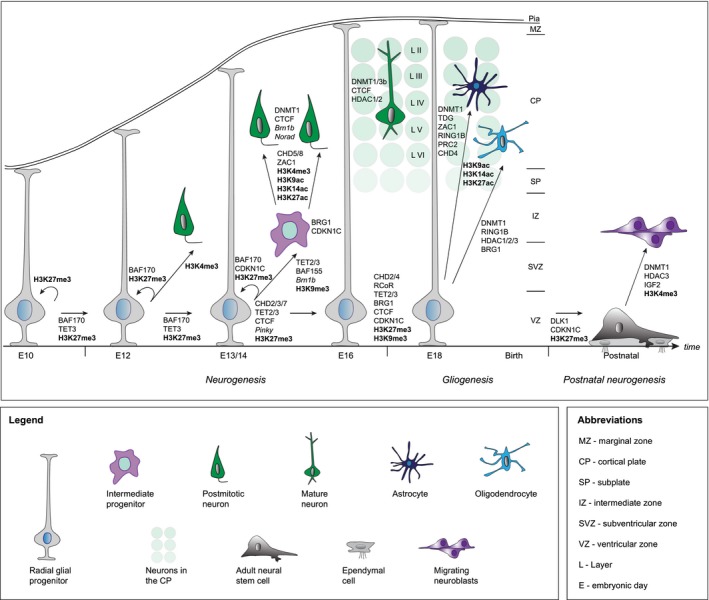
Epigenetic factors and modifications regulating the generation of cell‐type diversity during RGP lineage progression. High content of repressive H3K9me2/me3 and H3K27me3 marks and the presence of chromatin regulators such as BAF170, BRG1, CHD2, Pnky, TET1/3 and CTCF regulate stem cell maintenance and RGP self‐renewal while suppressing genes involved in neuron differentiation. For neuron production, repressive marks are replaced by active marks such as H3K4me3 or histone acetylation to promote expression of proneural genes mediating neuronal differentiation and maturation. Transition from repressive to activating epigenetic regulation is mediated through BAF155, BRN1, CHD5/8 and CBP. PRC and histone acetylation are essential for mediating the neurogenic to gliogenic transition. Adult NSCs display high levels of H3K27me3 and require the accumulation of H3K4me3 and expression of DNMT1, HDAC3, and CDKN1C for the faithful generation of OB inhibitory neurons. BAF, Brahma‐associated factor; Cdkn1c, cyclin‐dependent kinase inhibitor 1c; CHD, chromodomain‐helicase‐DNA‐binding; CTCF, CCCCTC‐binding factor; DNMT, DNA methyltransferase; HDAC, histone deacetylase, NSCs, neural stem cell; PRC, polycomb repressive complex; RGP, radial glial progenitor; TET, ten‐eleven translocation protein.
Recent single cell lineage tracing approaches (Woodworth et al. 2017) including MADM‐based experimental paradigms (Hippenmeyer et al. 2010, 2013; Gao et al. 2014; Beattie et al. 2017) have revealed a rough inaugural quantitative framework of RGP lineage progression. Over the last years, it became apparent from genetic studies that RGP lineage progression is modulated by epigenetic components. We propose three major hypothetical conceptual frameworks how epigenetic cues may control RGP‐mediated generation of cortical cell‐type diversity: (i) direct but progressive distinct RGP‐mediated lineage instruction at the time of neuron/glia production; (ii) epigenetic pre‐priming of RGPs which functionally only precipitates at a later stage in the lineage and (iii) priming of an entire successive RGP lineage at a defined developmental stage (Fig. 1). Many past studies focused on the analysis of global knockdown or genetic loss of function of epigenetic regulators and therefore little is known about the functional epigenetic mechanisms at the single RGP level. In order to probe the function of genes encoding epigenetic regulators at single cell level in vivo, MADM technology may offer a promising approach for future analysis. Despite that epigenetic processes regulate the expression of downstream target genes it is currently not clear how the precise epigenetic state of a proliferating RGP correlates with its neuron/glia output. The epigenetic landscape is highly dynamic and even during distinct phases in the RGP cell cycle crucial transcriptional changes associated with differences in the epigenetic marks may be required for correct lineage progression and/or priming. It remains a substantial challenge to rigorously analyze transcriptome and epigenome fingerprints in real time and at the single cell level to address the following questions in more detail: What are the precise cell‐autonomous mechanisms regulating RGP output and what are the essential non‐autonomous signals elicited by the stem cell niche? Which epigenetically controlled signaling molecules contribute to RGP lineage progression? How do epigenetic cues contribute to the regulatory process to instruct whether RGPs progress into either astrocyte progenitors or OPCs? In light of the emerging evidence that DNA methylation can affect the modification states on accompanying histones and vice versa (Vaissière et al. 2008; Rose and Klose 2014; Nishiyama et al. 2016) it will be important to determine whether such interactions play an instructive role in RGP lineage progression and/or post‐mitotic fate specification. The function and impact of distinct histone modifications, in general, on RGP proliferation behavior and beyond requires also more investigation in the future. Furthermore, it will be essential to comprehensively analyze the precise molecular and biochemical function of the various epigenetic protein complexes, described in the above sections, in RGP lineage progression at single cell and high temporal resolution. It will be revealing to more precisely categorize specific epigenetic modulators (Fig. 2) with regard to functional requirement in lineage instruction, lineage pre‐priming or lineage priming (Fig. 1). Lastly, most functional analyses of epigenetic regulators that contributed to our current understanding of RGP lineage progression were reliant on mouse genetic approaches. How are the proliferative RGP potential and the generation of cell‐type diversity regulated in different species? It will be particularly important to analyze epigenetic mechanisms also in higher mammalian brains. Interestingly, neural stem cells in ferret and human were recently shown to express the histone methyltransferase Prdm16 (Baizabal et al. 2018) although the precise role of Prdm16 in stem cell lineage progression remains somewhat elusive since it was mainly addressed by loss of gene function in mice. Experimental access to the human embryonic brain is extremely limited. Yet, recent advances in pluripotent stem cell technology now enable the generation of cerebral organoids that at least recapitulate some aspects of early‐to mid‐fetal human cortical development (Lancaster and Knoblich 2014; Suzuki and Vanderhaeghen 2015; Quadrato and Arlotta 2017). Therefore, future studies with the goal to contribute to our understanding of the epigenetic mechanisms controlling the generation of cortical neuron and glial cell diversity in distinct species and humans may also help to build a potential foundation for prospective reprogramming and/or stem cell‐based approaches in regenerative medicine.
Acknowledgments and conflict of interest disclosure
We thank all members of the Hippenmeyer laboratory for discussion and comments on the manuscript. This work was supported by IST Austria institutional funds; NÖ Forschung und Bildung n[f+b] (C13‐002) to SH; a program grant from the Human Frontiers Science Program (RGP0053/2014) to SH; the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (FP7/2007‐2013) under REA grant agreement No 618444 to SH, and the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (grant agreement No 725780 LinPro) to SH. The authors have no conflicts of interest to disclose.
References
- Albert M., Kalebic N., Florio M., Lakshmanaperumal N., Haffner C., Brandl H., Henry I. and Huttner W. B. (2017) Epigenome profiling and editing of neocortical progenitor cells during development. EMBO J. 36, 2642–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamo E. A., Chirivella L., Dautzenberg M., Dobreva G., Farinas I., Grosschedl R. and McConnell S. K. (2008) Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377. [DOI] [PubMed] [Google Scholar]
- Andergassen D., Dotter C. P., Wenzel D. et al (2017) Mapping the mouse Allelome reveals tissue‐specific regulation of allelic expression. Elife 6, pii: e25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angevine J. B. and Sidman R. L. (1961) Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768. [DOI] [PubMed] [Google Scholar]
- Aprea J. and Calegari F. (2015) Long non‐coding RNAs in corticogenesis: deciphering the non‐coding code of the brain. EMBO J. 34, 2865–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J., Prenninger S., Dori M., Ghosh T., Monasor L. S., Wessendorf E., Zocher S., Massalini S., Alexopoulou D. and Lesche M. (2013) Transcriptome sequencing during mouse brain development identifies long non‐coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baizabal J.‐M., Mistry M., García M. T., Gómez N., Olukoya O., Tran D., Johnson M. B., Walsh C. A. and Harwell C. C. (2018) The epigenetic state of PRDM16‐regulated enhancers in radial glia controls cortical neuron position. Neuron 98, 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranek C., Dittrich M., Parthasarathy S., Bonnon C. G., Britanova O., Lanshakov D., Boukhtouche F., Sommer J. E., Colmenares C. and Tarabykin V. (2012) Protooncogene Ski cooperates with the chromatin‐remodeling factor Satb2 in specifying callosal neurons. Proc. Natl Acad. Sci. 109, 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. P. and Bartolomei M. S. (2014) Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O. A., Fuentealba L. C., Alvarez‐Buylla A. and Rowitch D. H. (2015) Astrocyte development and heterogeneity. Cold Spring Harb. Perspect. Biol. 7, a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie R. and Hippenmeyer S. (2017) Mechanisms of radial glia progenitor cell lineage progression. FEBS Lett. 591, 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie R., Postiglione M. P., Burnett L. E., Laukoter S., Streicher C., Pauler F. M., Xiao G., Klezovitch O., Vasioukhin V. and Ghashghaei T. H. (2017) Mosaic analysis with double markers reveals distinct sequential functions of Lgl1 in neural stem cells. Neuron 94, 517–533. [DOI] [PubMed] [Google Scholar]
- Bell A. C. and Felsenfeld G. (2000) Methylation of a CTCF‐dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482. [DOI] [PubMed] [Google Scholar]
- Bergman Y. and Cedar H. (2013) DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 20, 274–281. [DOI] [PubMed] [Google Scholar]
- Britanova O., de Juan Romero C., Cheung A., Kwan K. Y., Schwark M., Gyorgy A., Vogel T., Akopov S., Mitkovski M. and Agoston D. (2008) Satb2 is a postmitotic determinant for upper‐layer neuron specification in the neocortex. Neuron 57, 378–392. [DOI] [PubMed] [Google Scholar]
- Chalei V., Sansom S. N., Kong L., Lee S., Montiel J. F., Vance K. W. and Ponting C. P. (2014) The long non‐coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3, e04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. V. and Maniatis T. (2013) Clustered Protocadherins. Development 140, 3297–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Tian Y., Shu W., Bo X. and Wang S. (2012) Comprehensive identification and annotation of cell type‐specific and ubiquitous CTCF‐binding sites in the human genome. PLoS ONE 7, e41374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel G., Schmidt‐Edelkraut U., Spengler D. and Hoffmann A. (2015) Imprinted Zac1 in neural stem cells. World J. Stem Cells 7, 300–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L. and Helin K. (2013) Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Diotel N., Mérot Y., Coumailleau P., Gueguen M. M., Sérandour A. A., Salbert G. and Kah O. (2017) 5‐hydroxymethylcytosine marks postmitotic neural cells in the adult and developing vertebrate central nervous system. J. Comp. Neurol. 525, 478–497. [DOI] [PubMed] [Google Scholar]
- Durak O., Gao F., Kaeser‐Woo Y. J., Rueda R., Martorell A. J., Nott A., Liu C. Y., Watson L. A. and Tsai L. H. (2016) Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 19, 1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C. M., Nyman U., Skotte J. et al (2013) CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell 26, 223–236. [DOI] [PubMed] [Google Scholar]
- Evsyukova I., Plestant C. and Anton E. S. (2013) Integrative mechanisms of oriented neuronal migration in the developing brain. Annu. Rev. Cell Dev. Biol. 29, 299–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Beard C., Chen R. Z., Csankovszki G., Sun Y., Siniaia M., Biniszkiewicz D., Bates B., Lee P. P. and Kühn R. (2001) DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Martinowich K., Chin M. H., He F., Fouse S. D., Hutnick L., Hattori D., Ge W., Shen Y. and Wu H. (2005) DNA methylation controls the timing of astrogliogenesis through regulation of JAK‐STAT signaling. Development 132, 3345–3356. [DOI] [PubMed] [Google Scholar]
- Feng J., Chang H., Li E. and Fan G. (2005) Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746. [DOI] [PubMed] [Google Scholar]
- Ferron S. R., Radford E. J., Domingo‐Muelas A. et al (2015) Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat. Commun. 6, 8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrón S. R., Charalambous M., Radford E., McEwen K., Wildner H., Hind E., Morante‐Redolat J. M., Laborda J., Guillemot F. and Bauer S. R. (2011) Postnatal loss of Dlk1 imprinting in stem cells and niche‐astrocytes regulates neurogenesis. Nature 475, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L. C., Rompani S. B., Parraguez J. I., Obernier K., Romero R., Cepko C. L. and Alvarez‐Buylla A. (2015) Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K. I. and Hirabayashi Y. (2015) Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18, 657–665. [DOI] [PubMed] [Google Scholar]
- Gao P., Postiglione M. P., Krieger T. G. et al (2014) Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P., Hutnick L., Schweizer F. and Fan G. (2005) Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long‐term potentiation. Thalamus Relat. Syst. 3, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompers A. L., Su‐Feher L., Ellegood J. et al (2017) Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 20, 1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I. and Gorospe M. (2016) Identification of neural stem cell differentiation repressor complex Pnky‐PTBP1. Stem Cell Investig. 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelkruys A., Lagger S., Krahmer J. et al (2014) A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development 141, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. A., Qiu R., Wu X. et al (2013) Dynamics of 5‐hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 3, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada I., Namihira M., Morita S., Kimura M., Horii T. and Nakashima K. (2008) Astrocyte‐specific genes are generally demethylated in neural precursor cells prior to astrocytic differentiation. PLoS ONE 3, e3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.‐F., Li B.‐Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z. and Li L. (2011) Tet‐mediated formation of 5‐carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S. (2013) Dissection of gene function at clonal level using mosaic analysis with double markers. Front. Biol. 8, 557–568. [Google Scholar]
- Hippenmeyer S. (2014) Molecular pathways controlling the sequential steps of cortical projection neuron migration, in Cellular and Molecular Control of Neuronal Migration (Laurent Nguyen, and Simon Hippenmeyer, ed.), pp. 1–24. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S., Youn Y. H., Moon H. M., Miyamichi K., Zong H., Wynshaw‐Boris A. and Luo L. (2010) Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S., Johnson R. L. and Luo L. (2013) Mosaic analysis with double markers reveals cell‐type‐specific paternal growth dominance. Cell Rep. 3, 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., Koseki H., Vidal M. and Gotoh Y. (2009) Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613. [DOI] [PubMed] [Google Scholar]
- Hirayama T., Tarusawa E., Yoshimura Y., Galjart N. and Yagi T. (2012) CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2, 345–357. [DOI] [PubMed] [Google Scholar]
- Hutnick L. K., Golshani P., Namihira M., Xue Z., Matynia A., Yang X. W., Silva A. J., Schweizer F. E. and Fan G. (2009) DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 18, 2875–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun K., Jeon J., Park K. and Kim J. (2017) Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. and Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245. [DOI] [PubMed] [Google Scholar]
- Jin S.‐G., Wu X., Li A. X. and Pfeifer G. P. (2011) Genomic mapping of 5‐hydroxymethylcytosine in the human brain. Nucleic Acids Res. 39, 5015–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J. and Crabtree G. R. (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlić R., Chung H.‐R., Lasserre J., Vlahoviček K. and Vingron M. (2010) Histone modification levels are predictive for gene expression. Proc. Natl Acad. Sci. 107, 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Nishiyama M., Shoji H., Ohkawa Y., Kawamura A., Sato T., Suyama M., Takumi T., Miyakawa T. and Nakayama K. I. (2016) CHD8 haploinsufficiency results in autistic‐like phenotypes in mice. Nature 537, 675. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Wagatsuma H., Ono R., Ichikawa H., Yamazaki M., Tashiro H., Aisaka K., Miyoshi N., Kohda T. and Ogura A. (2000) Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells 5, 1029–1037. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T., Pontious A., Englund C., Daza R. A. M., Bedogni F., Hodge R., Attardo A., Bell C., Huttner W. B. and Hevner R. F. (2009) Intermediate neuronal progenitors (basal progenitors) produce pyramidal–projection Neurons for all layers of cerebral cortex. Cereb. Cortex 19, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. Y. and Wade P. A. (2011) NuRD: a multi‐faceted chromatin remodeling complex in regulating cancer biology. Nat. Rev. Cancer 11, 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A. and Knoblich J. A. (2014) Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125. [DOI] [PubMed] [Google Scholar]
- Lee M. G., Villa R., Trojer P., Norman J., Yan K.‐P., Reinberg D., Di Croce L. and Shiekhattar R. (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318, 447–450. [DOI] [PubMed] [Google Scholar]
- Lee S., Kopp F., Chang T.‐C., Sataluri A., Chen B., Sivakumar S., Yu H., Xie Y. and Mendell J. T. (2016) Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K., Zappaterra M. W., Chen X. et al (2011) The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E. S., Belgard T. G., Hawrylycz M. and Molnár Z. (2017) Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu. Rev. Neurosci. 40, 629–652. [DOI] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A. and Crabtree G. R. (2007) An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu H., Cheng P.‐L., Ge S., Xu H., Shi S.‐H. and Dan Y. (2012) Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Mukamel E. A., Nery J. R., Urich M., Puddifoot C. A., Johnson N. D., Lucero J., Huang Y., Dwork A. J. and Schultz M. D. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. J., Nowakowski T. J., Pollen A. A., Lui J. H., Horlbeck M. A., Attenello F. J., He D., Weissman J. S., Kriegstein A. R. and Diaz A. A. (2016) Single‐cell analysis of long non‐coding RNAs in the developing human neocortex. Genome Biol. 17, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S. and Arlotta P. (2015) Generating neuronal diversity in the mammalian cerebral cortex. Annu. Rev. Cell Dev. Biol. 31, 699–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Keown C. L., Kurihara L., Zhou J., He Y., Li J., Castanon R., Lucero J., Nery J. R. and Sandoval J. P. (2017) Single‐cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. (2018) The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92. [DOI] [PubMed] [Google Scholar]
- Mairet‐Coello G., Tury A., Van Buskirk E., Robinson K., Genestine M. and DiCicco‐Bloom E. (2012) p57(KIP2) regulates radial glia and intermediate precursor cell cycle dynamics and lower layer neurogenesis in developing cerebral cortex. Development 139, 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S., Banine F., Struve J., Xing R., Adams C., Liu Y., Metzger D., Chambon P., Rao M. S. and Sherman L. S. (2006) Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 289, 372–383. [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Banine F., Feistel K., Foster S., Xing R., Struve J. and Sherman L. S. (2016) Brg1 directly regulates Olig2 transcription and is required for oligodendrocyte progenitor cell specification. Dev. Biol. 413, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micucci J. A., Layman W. S., Hurd E. A., Sperry E. D., Frank S. F., Durham M. A., Swiderski D. L., Skidmore J. M., Scacheri P. C. and Raphael Y. (2013) CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet. 23, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan C. E., Nechiporuk T., Jeng S., McWeeney S. K., Wang J., Rosenfeld M. G. and Mandel G. (2017) REST corepressors RCOR1 and RCOR2 and the repressor INSM1 regulate the proliferation–differentiation balance in the developing brain. Proc. Natl Acad. Sci. 114, E406–E415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R. L., Hsieh J., Barbosa A. C., Richardson J. A. and Olson E. N. (2009) Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl Acad. Sci. USA 106, 7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto‐Suzki N., Hirabayashi Y., Tyssowski K., Shinga J., Vidal M., Koseki H. and Gotoh Y. (2014) The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development 141, 4343–4353. [DOI] [PubMed] [Google Scholar]
- Moyon S., Huynh J. L., Dutta D., Zhang F., Ma D., Yoo S., Lawrence R., Wegner M., John G. R. and Emery B. (2016) Functional characterization of DNA methylation in the oligodendrocyte lineage. Cell Rep. 15, 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan B., Khatri Z., Maheshwari U., Gupta R., Roy B., Pradhan S. J., Karmodiya K., Padmanabhan H., Shetty A. S. and Balaji C. (2017) LHX2 interacts with the NuRD complex and regulates cortical neuron subtype determinants Fezf2 and Sox11. J. Neurosci. 37, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M. and Brehm A. (2011) CHD chromatin remodelers and the transcription cycle. Transcription 2, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel C. S., Jia H., Ye Y., Shen L., Goldschmidt H. L., Stivers J. T., Zhang Y. and Kohli R. M. (2012) AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T. and Nakashima K. (2009) Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245–255. [DOI] [PubMed] [Google Scholar]
- Narayanan R., Pirouz M., Kerimoglu C. et al (2015) Loss of BAF (mSWI/SNF) complexes causes global transcriptional and chromatin state changes in forebrain development. Cell Rep. 13, 1842–1854. [DOI] [PubMed] [Google Scholar]
- Nguyen H., Sokpor G., Pham L., Rosenbusch J., Stoykova A., Staiger J. F. and Tuoc T. (2016) Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle 15, 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen F. C. (1992) The molecular and cellular biology of insulin‐like growth factor II. Prog. Growth Factor Res. 4, 257–290. [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Steiner‐Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., Von Holst A., Beckers J. and Lie C. D. (2013) The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross‐regulatory transcriptional network. Cell Stem Cell 13, 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Yamaguchi L. and Nakanishi M. (2016) Regulation of maintenance DNA methylation via histone ubiquitylation. J. Biochem. 159, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitarska J., Smith J. G., Sherlock W. T., Hillege M. M. G., Nott A., Barshop W. D., Vashisht A. A., Wohlschlegel J. A., Mitter R. and Riccio A. (2016) A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 17, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H., Kimura A., Murao N., Namihira M. and Nakashima K. (2016) Prenatal deletion of DNA methyltransferase 1 in neural stem cells impairs neurogenesis and causes anxiety‐like behavior in adulthood. Neurogenesis 3, e1232679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Yaguchi T., Okuno H., Chneiweiss H., Kawakami Y. and Okano H. (2016) CHD7 promotes proliferation of neural stem cells mediated by MIF. Mol. Brain 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.‐T. and Corces V. G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. D., Sansom S. N., Smith J., Dobenecker M. W., Tarakhovsky A. and Livesey F. J. (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self‐renewal and differentiation in the cerebral cortex. Proc. Natl Acad. Sci. USA 107, 15957–15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R. J., Zhou Y., Slaymaker I. M., Shetty A. S., Weisbach N. R., Kim J.‐A., Sharma J., Desai M., Sood S. and Kempton H. R. (2017) Chd8 mutation leads to autistic‐like behaviors and impaired striatal circuits. Cell Rep. 19, 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A. and Esteller M. (2010) Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Poulin J.‐F., Tasic B., Hjerling‐Leffler J., Trimarchi J. M. and Awatramani R. (2016) Disentangling neural cell diversity using single‐cell transcriptomics. Nat. Neurosci. 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Quadrato G. and Arlotta P. (2017) Present and future of modeling human brain development in 3D organoids. Curr. Opin. Cell Biol. 49, 47–52. [DOI] [PubMed] [Google Scholar]
- Qureshi I. A., Gokhan S. and Mehler M. F. (2010) REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle 9, 4477–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. D., Andersen R. E., Liu S. J., Nowakowski T. J., Hong S. J., Gertz C. C., Salinas R. D., Zarabi H., Kriegstein A. R. and Lim D. A. (2015) The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. and Wolffe A. P. (2000) DNA methylation in health and disease. Nat. Rev. Genet. 1, 11. [DOI] [PubMed] [Google Scholar]
- Rose N. R. and Klose R. J. (2014) Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta 1839, 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rraklli V., Södersten E., Nyman U., Hagey D. W. and Holmberg J. (2016) Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem Cell Res. 16, 1–9. [DOI] [PubMed] [Google Scholar]
- Sams D. S., Nardone S., Getselter D., Raz D., Tal M., Rayi P. R., Kaphzan H., Hakim O. and Elliott E. (2016) Neuronal CTCF is necessary for basal and experience‐dependent gene regulation, memory formation, and genomic structure of BDNF and arc. Cell Rep. 17, 2418–2430. [DOI] [PubMed] [Google Scholar]
- Sauvageau M., Goff L. A., Lodato S., Bonev B., Groff A. F., Gerhardinger C., Sanchez‐Gomez D. B., Hacisuleyman E., Li E. and Spence M. (2013) Multiple Knockout Mouse Models Reveal LincRNAs are Required for Life and Brain Development. ELife 2, e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt‐Edelkraut U., Hoffmann A., Daniel G. and Spengler D. (2013) Zac1 regulates astroglial differentiation of neural stem cells through Socs3. Stem Cells 31, 1621–1632. [DOI] [PubMed] [Google Scholar]
- Sharma A., Klein S. L., Barboza L., Lodhi N. and Toth M. (2016) Principles governing DNA methylation during neuronal lineage and subtype specification. J. Neurosci. 36, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T., Ji F., Yuan Z. and Jiao J. (2015) CHD2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells 33, 1794–1806. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. and Roberts J. M. (1999) CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Smas C. M., Chen L. and Sul H. S. (1997) Cleavage of membrane‐associated pref‐1 generates a soluble inhibitor of adipocyte differentiation. Mol. Cell. Biol. 17, 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E. Y. and Crabtree G. R. (2014) The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A., Xie Y., Verhoeven E., Vermeulen M., Lancini C., Gargiulo G., Hulsman D., Mann M., Knoblich J. A. and van Lohuizen M. (2013) The chromodomain helicase Chd4 is required for Polycomb‐mediated inhibition of astroglial differentiation. EMBO J. 32, 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. E. and Rotwein P. (1996) Growth, differentiation, and survival: multiple physiological functions for insulin‐like growth factors. Physiol. Rev. 76, 1005–1026. [DOI] [PubMed] [Google Scholar]
- Sugathan A., Biagioli M., Golzio C. et al (2014) CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl Acad. Sci. USA 111, E4468–E4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nadal‐Vicens M., Misono S., Lin M. Z., Zubiaga A., Hua X., Fan G. and Greenberg M. E. (2001) Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. [DOI] [PubMed] [Google Scholar]
- Suzuki I. K. and Vanderhaeghen P. (2015) Is this a brain which I see before me? Modeling human neural development with pluripotent stem cells. Development 142, 3138–3150. [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R. and Aravind L. (2009) Conversion of 5‐methylcytosine to 5‐hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M. and Taga T. (2001) DNA methylation is a critical cell‐intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1, 749–758. [DOI] [PubMed] [Google Scholar]
- Tan S.‐L., Nishi M., Ohtsuka T., Matsui T., Takemoto K., Kamio‐Miura A., Aburatani H., Shinkai Y. and Kageyama R. (2012) Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 139, 3806–3816. [DOI] [PubMed] [Google Scholar]
- Tarusawa E., Sanbo M., Okayama A., Miyashita T., Kitsukawa T., Hirayama T., Hirabayashi T., Hasegawa S., Kaneko R. and Toyoda S. (2016) Establishment of high reciprocal connectivity between clonal cortical neurons is regulated by the Dnmt3b DNA methyltransferase and clustered protocadherins. BMC Biol. 14, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E., Götz M. and Huttner W. B. (2014) The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. [DOI] [PubMed] [Google Scholar]
- Tichon A., Gil N., Lubelsky Y., Solomon T. H., Lemze D., Itzkovitz S., Stern‐Ginossar N. and Ulitsky I. (2016) A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 7, 12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda S., Kawaguchi M., Kobayashi T., Tarusawa E., Toyama T., Okano M., Oda M., Nakauchi H., Yoshimura Y. and Sanbo M. (2014) Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82, 94–108. [DOI] [PubMed] [Google Scholar]
- Tuoc T. C., Boretius S., Sansom S. N., Pitulescu M. E., Frahm J., Livesey F. J. and Stoykova A. (2013) Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell 25, 256–269. [DOI] [PubMed] [Google Scholar]
- Vaissière T., Sawan C. and Herceg Z. (2008) Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 659, 40–48. [DOI] [PubMed] [Google Scholar]
- Valente T., Junyent F. and Auladell C. (2005) Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1‐expressing cells during development. Dev. Dyn. 233, 667–679. [DOI] [PubMed] [Google Scholar]
- Vasistha N. A., García‐Moreno F., Arora S., Cheung A. F. P., Arnold S. J., Robertson E. J. and Molnár Z. (2014) Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cereb. Cortex 25, 3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. and Sul H. S. (2006) Ectodomain shedding of preadipocyte factor 1 (Pref‐1) by tumor necrosis factor alpha converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol. Cell. Biol. 26, 5421–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W. and Zhao K. (2009) Genome‐wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Weaver I. C., Gauthier‐Fisher A., Wang H., He L., Yeomans J., Wondisford F., Kaplan D. R. and Miller F. D. (2010) CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein‐Taybi syndrome brain. Dev. Cell 18, 114–125. [DOI] [PubMed] [Google Scholar]
- Wang H., Maurano M. T., Qu H., Varley K. E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M. and Sandstrom R. (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu Q., Yang P., Wang C., Liu J., Ding W., Liu W., Bai Y., Yang Y. and Wang H. (2016) LSD1 co‐repressor Rcor2 orchestrates neurogenesis in the developing mouse brain. Nat. Commun. 7, 10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D., Uchiyama K. and Hanaoka K. (2006) Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience 142, 727–737. [DOI] [PubMed] [Google Scholar]
- Watson L. A., Wang X., Elbert A., Kernohan K. D., Galjart N. and Berube N. G. (2014) Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J. Neurosci. 34, 2860–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheldon L. M., Abakir A., Ferjentsik Z., Dudnakova T., Strohbuecker S., Christie D., Dai N., Guan S., Foster J. M. and Corrêa I. R. (2014) Transient accumulation of 5‐carboxylcytosine indicates involvement of active demethylation in lineage specification of neural stem cells. Cell Rep. 7, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Woodworth M. B., Girskis K. M. and Walsh C. A. (2017) Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet. 18, 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Christian K. M., He C., Jin P., Ming G.‐L. and Song H. (2016) Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Chen Y., Hoang T., Montgomery R. L., Zhao X.‐H., Bu H., Hu T., Taketo M. M., Van Es J. H. and Clevers H. (2009) HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the β‐catenin–TCF interaction. Nat. Neurosci. 12, 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.‐C., Bultje R. S., Wang X. and Shi S.‐H. (2009) Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr S. K., Yang G., Kazan H., Borrett M. J., Yuzwa S. A., Voronova A., Kaplan D. R. and Miller F. D. (2018) A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520–537. [DOI] [PubMed] [Google Scholar]
- Zeng H. and Sanes J. R. (2017) Neuronal cell‐type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530. [DOI] [PubMed] [Google Scholar]
- Zhang F., Xu D., Yuan L., Sun Y. and Xu Z. (2014) Epigenetic regulation of Atrophin1 by lysine‐specific demethylase 1 is required for cortical progenitor maintenance. Nat. Commun. 5, 5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H., Espinosa J. S., Su H. H., Muzumdar M. D. and Luo L. (2005) Mosaic analysis with double markers in mice. Cell 121, 479–492. [DOI] [PubMed] [Google Scholar]


