Abstract
In Euglena gracilis, wax ester fermentation produces ATP during anaerobiosis. Here, we report that anaerobic wax ester production is suppressed when the mitochondrial electron transport chain complex I is inhibited by rotenone, whereas it is increased by the uncoupler carbonyl cyanide m‐chlorophenylhydrazone (CCCP). The ADP/ATP ratio in anaerobic cells is elevated by treatment with either rotenone or CCCP. Gene silencing experiments indicate that acyl‐CoA dehydrogenase, electron transfer flavoprotein (ETF), and rhodoquinone (RQ) participate in wax ester production. These results suggest that fatty acids are synthesized in mitochondria by the reversal of β‐oxidation, where trans‐2‐enoyl‐CoA is reduced mainly by acyl‐CoA dehydrogenase using the electrons provided by NADH via the electron transport chain complex I, RQ, and ETF, and that ATP production is highly supported by anaerobic respiration utilizing trans‐2‐enoyl‐CoA as a terminal electron acceptor.
Keywords: anaerobic respiration, Euglena gracilis, mitochondrial electron transfer system, mitochondrial fatty acid synthesis, wax ester fermentation
Abbreviations
CCCP, carbonyl cyanide m‐chlorophenylhydrazone
ETF, electron transfer flavoprotein
RQ, rhodoquinone
UQ, ubiquinone
Most eukaryotes, when under anaerobic conditions, obtain ATP by fermentation, where glucose is catabolized to pyruvate through the glycolytic pathway. This yields a net gain of two ATP molecules from substrate‐level phosphorylation with the concomitant formation of two molecules of NADH per molecule of glucose 1, 2. Pyruvate is further metabolized to reoxidize NADH, which is necessary to sustain metabolic flux through glycolysis, and a variety of anaerobic end products, such as lactate and ethanol, are formed. ATP can also be produced by anaerobic respiration, where NADH is oxidized by transferring electrons through the mitochondrial electron transport chain to a terminal electron acceptor other than O2 and ATP is then synthesized by utilizing the proton motive force developed by the electron transport 1, 2, 3. In certain eukaryotes, fumarate of endogenous origin is used as a terminal electron acceptor, resulting in the formation of succinate.
Euglena gracilis, a photosynthetic unicellular protist, has a unique anaerobic energy generating system called wax ester fermentation, in which wax esters are synthesized as end products from paramylon, a storage polysaccharide (β‐1,3‐glucan) 4, 5. Anaerobically synthesized wax esters are coupled with saturated fatty acids and alcohols, mainly myristic acid and myristyl alcohol, and accumulated in cells 6, 7. During wax ester fermentation, fatty acids are synthesized using acetyl‐CoA as both primer and C2 donor in the mitochondria, essentially by the reversal of β‐oxidation 8. It had long been believed that NAD(P)H‐dependent trans‐2‐enoyl‐CoA reductase (acyl‐CoA:NAD(P)+ trans‐2‐oxidoreductase) functions in the fatty acid synthesis, instead of acyl‐CoA dehydrogenase linked to the electron transfer flavoprotein (ETF) (acyl‐CoA:ETF 2,3‐oxidoreductase) that is utilized in the β‐oxidation 8, 9. However, details of mitochondrial fatty acid synthesis, in particular the reduction in trans‐2‐enoyl‐CoA, remain to be clarified. In addition, the physiological significance of why Euglena cells produce wax esters as an anaerobic end product instead of the more common lactate or ethanol is not well understood.
In the present paper, we report that the production of wax esters is suppressed in anaerobic Euglena cells when the mitochondrial electron transport chain complex I is inhibited by rotenone, whereas an uncoupler, carbonyl cyanide m‐chlorophenylhydrazone (CCCP), induces an increase in anaerobic wax ester production. On the basis of results obtained by gene silencing experiments, we suggest that trans‐2‐enoyl‐CoA is reduced mainly by acyl‐CoA dehydrogenase in mitochondrial fatty acid synthesis, and that electrons for this reduction are provided by NADH via complex I, rhodoquinone (RQ), and ETF. In addition, we also suggest that ATP is synthesized by utilizing the proton motive force developed from the electron transport through complex I.
Materials and methods
Organism and culture
Euglena gracilis SM‐ZK, a nonphotosynthetic mutant strain 10, was cultured in Koren–Hutner medium containing glucose as the major carbon source 11 with aeration at 27 °C for 3 or 4 days until the late logarithmic or early stationary phase of growth. Anaerobic treatment was performed as follows: an aliquot (2 mL) of the aerobic cell culture was dispensed into 5 mL plastic tube and bubbled with argon gas for 1 min. Rotenone (10 mm) or CCCP (2 mm) dissolved in methanol was added to a final concentration of 50 μm or 10 μm, respectively. Tubes were then tightly capped and allowed to stand under anaerobic conditions for 24 h. Cells in the culture were counted with a CDA‐1000 particle counter (Sysmex, Kobe, Japan).
RNA interference
Double strand RNA (dsRNA) was synthesized and purified using a MEGAscript RNAi kit (Thermo Fisher Scientific, Waltham, MA, USA), and total RNA extracted from Euglena cells was reverse transcribed with SuperScript III reverse transcriptase (Thermo Fisher Scientific) according to a method described previously 7. Template DNA containing T7 promoter at both ends was amplified from Euglena cDNA by PCR using the primers listed in Table 1. Sequences of mRNA encoding target genes were obtained using a TBLASTN search against a E. gracilis RNA‐Seq database (DRA accession number: SRP060591) 12. These genes included ETF alpha (etfalpha; accession ID GDJR01012930.1) and beta (etfbeta; GDJR01013070.1) subunit genes, RQ biosynthesis gene A (rquA; GDJR01043002.1), and acyl‐CoA dehydrogenase 1 gene (acd1; GDJR01042416.1). In addition, previously reported cDNA sequence data were used in NAD(P)H‐dependent trans‐2‐enoyl‐CoA reductase (ter; AY741582) 9.
Table 1.
Oligonucleotide primers for the synthesis of dsRNA used in RNA interference experiments
| Name of dsRNA | Target gene | Sequencea | |
| ETF‐dsRNA‐1 | etfalpha | Forward | ctaatacgactcactatagggagaTTCTGCAGAATATGCAAATTTG |
| Reverse | ctaatacgactcactatagggagaTTCCGGTCTGTCCAACCTGT | ||
| ETF‐dsRNA‐2 | etfbeta | Forward | ctaatacgactcactatagggagaTTGATGAGATCGCTGTTGAAC |
| Reverse | ctaatacgactcactatagggagaTTCAATCGGAGATCACAGGACAG | ||
| RQUA‐dsRNA | rquA | Forward | ctaatacgactcactatagggagaTTGGGGATGCATGCCTGATGGAGAAC |
| Reverse | ctaatacgactcactatagggagaTTGAACTGCTTTCGCAGCTCCGCGTC | ||
| ACD1‐dsRNA | acd1 | Forward | ctaatacgactcactatagggagaTTGGACTCTGGACACAGTTAAGACTGA |
| Reverse | ctaatacgactcactatagggagaATCCCATCGTGTGGCTTGCCAATCAG | ||
| TER‐dsRNA‐1 | ter | Forward | ctaatacgactcactatagggagaTGCCTGTGCGTGGCAACGGTA |
| Reverse | ctaatacgactcactatagggagaAAGCCACGAATCTTGGGCTGGA | ||
| TER‐dsRNA‐2 | ter | Forward | ctaatacgactcactatagggagaGGTATTGTTGGCGACTGGATCCAA |
| Reverse | ctaatacgactcactatagggagaAGAAACGCCACGGTGTTGTACCAGC |
Lower case letters show T7 promoter sequence.
Euglena cells suspended in phosphate‐buffered saline (4 × 106 cells/400 μL) in a 0.2 cm gap cuvette were electroporated with 30 μg of dsRNA using an ECM630 (BTX, Holliston, MA, USA) at 0.5 kV and 200 μF. After electroporation, the cells were cultured in Koren–Hutner medium under aerobic conditions for 3 or 4 days. The extent of silencing was examined by semiquantitative RT‐PCR (Table S1).
Analyses of wax esters
As described previously 7, lipids were extracted from Euglena cells and directly subjected to gas–liquid chromatography to determine the wax ester content. The lipid extract dissolved in hexane was separated on a silica gel column (Silica gel 60, Merck, Darmstadt, Germany; 6 × 75 mm) equilibrated with hexane, and wax esters were eluted with diethylether‐hexane (1 : 99, by vol.). After saponification, the constituent fatty acids and alcohols of wax esters were analyzed by gas–liquid chromatography 7.
Determination of ADP/ATP ratio
Into cell suspension, 70% perchloric acid was added at a final concentration of 3.5%, homogenized by sonication and neutralized with 3 m K2CO3 at 4 °C. After centrifugation for the removal of precipitate, the ratio of ADP/ATP in the supernatant was determined using a commercial kit (ADP/ATP Ratio Assay Kit (Bioluminescent), Abcam, Cambridge, UK).
Quinone analysis
Quinone content was determined using an HPLC‐electrochemical detection system according to the method described by Shiobara et al. 13. Cells incubated under anaerobic conditions for 24 h were harvested by centrifugation, and 2‐propanol was added (20 μL·mg−1 wet weight of cells) to extract quinones and quinols. In total, 1 μL of 120 mm FeCl3 solution dissolved in 2‐propanol was added to 100 μL of the extract to oxidize quinols prior to analysis 14, and this was used as a sample for measurement. Rhodoquinone‐9 (RQ9) was prepared from Ascaris suum 15 and was used as a standard for HPLC analysis in addition to ubiquinone‐10 (UQ10), which was purchased from Wako Chemicals (Osaka, Japan).
Results
Anaerobic wax ester production in the presence of rotenone and CCCP
When aerobically grown Euglena cells were transferred to anaerobic conditions, wax ester fermentation took place and amounts of wax esters (approximately 40 μg/106 cells) were produced during anaerobic incubation for 24 h. To explore whether the mitochondrial electron transport chain participates in anaerobic wax ester production, rotenone was added at 50 μm to inhibit mitochondrial complex I. In the presence of rotenone, the level of anaerobically produced wax esters reduced to <50% of that in control cells (Fig. 1). This result suggests that mitochondrial complex I is involved in wax ester fermentation. In contrast, an uncoupler, CCCP (10 μm), stimulated anaerobic wax ester production, and the amount of wax esters accumulated increased 1.7‐fold compared to the control.
Figure 1.
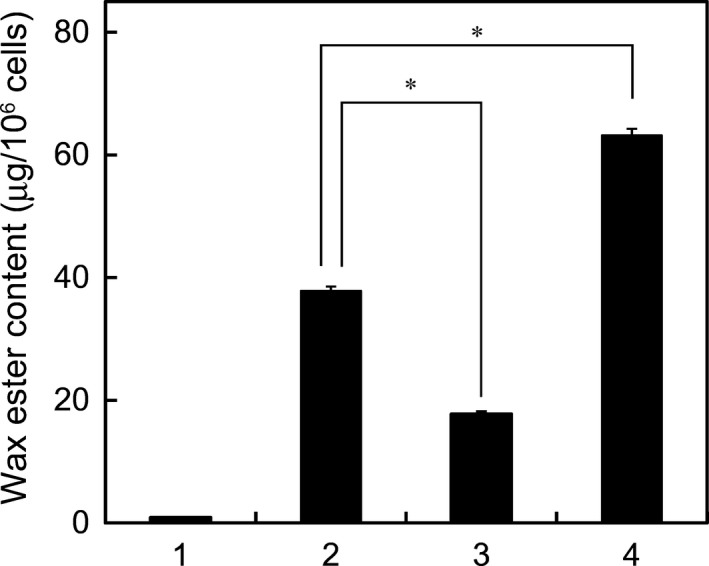
Effects of rotenone and CCCP on anaerobic production of wax esters. Cells cultured aerobically for 4 days were transferred to anaerobic conditions, and wax ester content was determined before (bar 1) or after (bars 2, 3, 4) anaerobic incubation for 24 h in the absence (bar 2) or presence of rotenone (bar 3) or CCCP (bar 4). Data are presented as means ± SD (n = 3). *P < 0.05.
When the constituent fatty acids and alcohols of wax esters produced by anaerobic Euglena cells were analyzed after saponification, odd‐numbered fatty acids and alcohols were detected in large quantities (40% and 35% of total, respectively), as has been reported previously 6, 7. As shown in Fig. 2, when CCCP was added during anaerobic incubation, the proportions of odd‐numbered fatty acids and alcohols decreased both to <15%. In addition, the average carbon chain lengths of the constituent fatty acids and alcohols became slightly longer in the presence of either rotenone or CCCP. Propionyl‐CoA is used as a primer for the synthesis of odd‐numbered fatty acids instead of acetyl‐CoA 16. In E. gracilis, propionyl‐CoA has been reported to be synthesized from phosphoenolpyruvate through oxaloacetate, fumarate, succinate, and methylmalonyl‐CoA, and fumarate reductase is thought to be involved in the conversion from fumarate to succinate in this pathway 17. Hence, it is expected that propionyl‐CoA synthesis is influenced by rotenone and CCCP because mitochondrial complex I participates in the electron transfer from NADH to fumarate during the fumarate reductase reaction. However, it remains unclear why CCCP (but not rotenone) induced large decreases in the proportions of odd‐numbered fatty acids and alcohols in wax esters produced in anaerobic Euglena cells.
Figure 2.
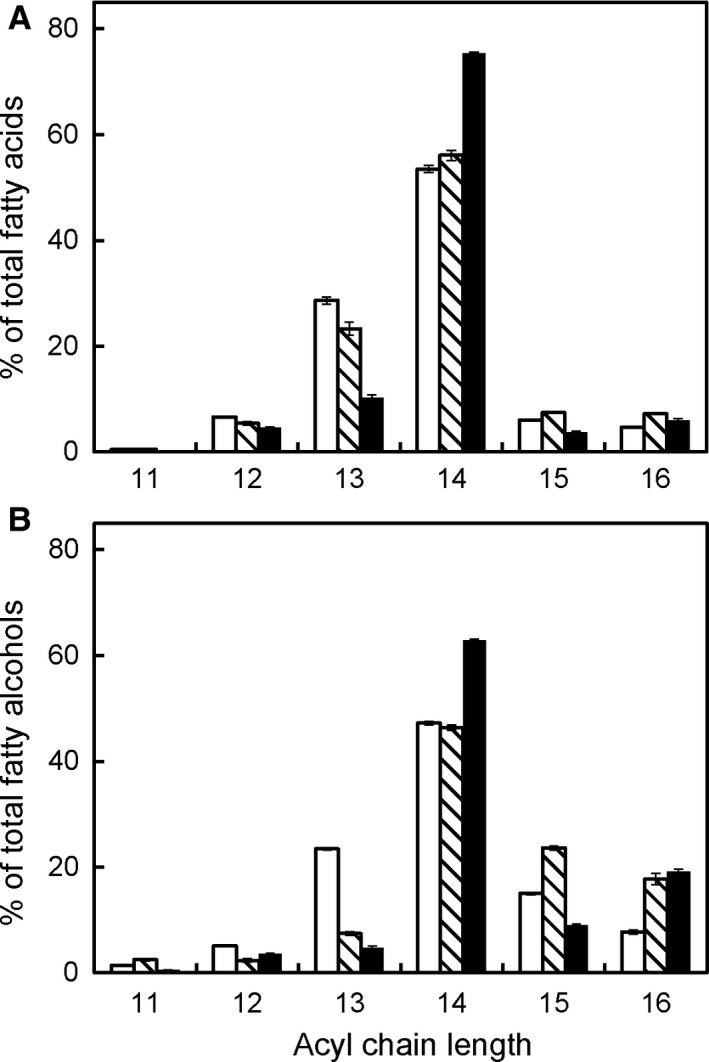
Fatty acid and alcohol moieties composed of anaerobically produced wax esters in the presence of rotenone and CCCP. Cells cultured aerobically for 4 days were incubated under anaerobic conditions for 24 h in the absence (open bars) or presence of rotenone (shaded bars) or CCCP (solid bars). Lipids were extracted from the cells, wax esters purified from the lipid extract were hydrolyzed, and the fatty acids (A) and alcohols (B) were analyzed by gas–liquid chromatography. Data are presented as means ± SD (n = 3).
ADP/ATP ratio in the anaerobic cells in the presence of rotenone and CCCP
The ratio of ADP/ATP in Euglena cells did not change significantly under anaerobic incubation for 24 h (Fig. 3), confirming that ATP can be produced by wax ester fermentation at a sufficient level in these cells. However, the ADP/ATP ratio greatly increased in cells exposed to anaerobic conditions in the presence of rotenone. A further increase in the ADP/ATP ratio was observed in the presence of CCCP. These results suggest that, in anaerobic Euglena cells, ATP is synthesized by mitochondrial complex V (ATP synthase) using a proton motive force developed by the electron flow through complex I.
Figure 3.
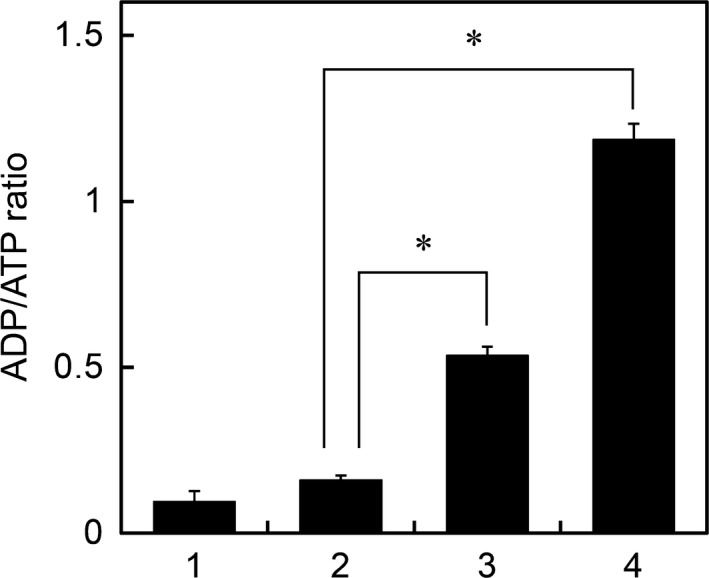
The ADP/ATP ratio in the cells incubated under anaerobic conditions in the presence of rotenone or CCCP. Cells cultured aerobically for 4 days were transferred to anaerobic conditions, and ADP/ATP ratio in the cells was determined before (bar 1) or after (bars 2, 3, 4) anaerobic incubation for 24 h in the absence (bar 2) or presence of rotenone (bar 3) or CCCP (bar 4). Data are presented as means ± SD (n = 3). *P < 0.05.
Participation of ETF, RQ, and acyl‐CoA dehydrogenase in anaerobic wax ester production
To further clarify the function of the mitochondrial electron transport chain in wax ester fermentation, a gene silencing experiment was performed that targeted ETF, which links acyl‐CoA dehydrogenase to the electron transport chain. Gene silencing of either alpha or beta subunit of ETF, which was confirmed by semiquantitative RT‐PCR (Fig. S1A), did not show any significant effects on the growth of Euglena cells under aerobic conditions (data not shown). However, the production of wax esters was significantly suppressed in both of these gene‐silenced cell lines when they were incubated anaerobically (Fig. 4A). These results suggest that ETF, probably together with acyl‐CoA dehydrogenase, participates in mitochondrial fatty acid synthesis in anaerobic Euglena cells.
Figure 4.
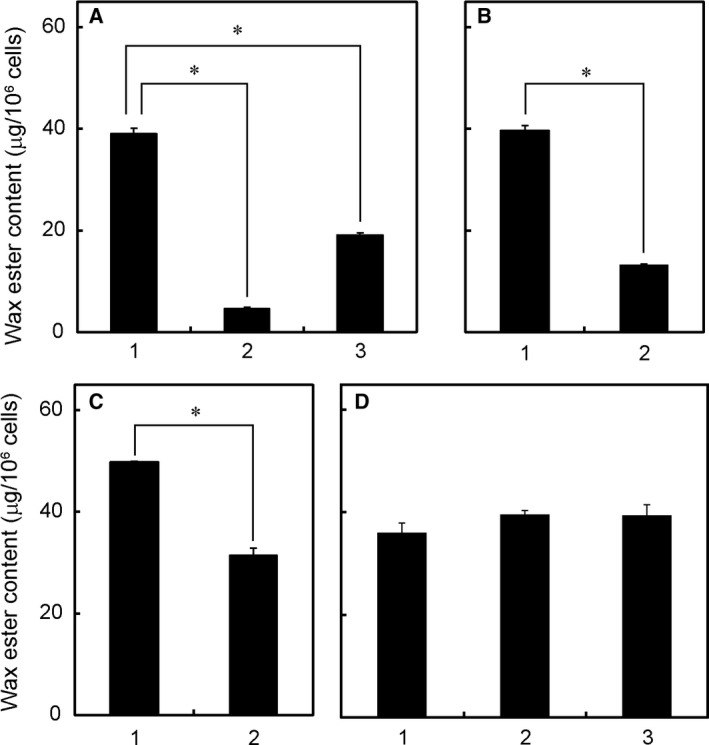
Participation of ETF, RQ, and acyl‐CoA dehydrogenase in anaerobic wax ester synthesis. A: ETF alpha (etfalpha) or beta (etfbeta) subunit gene was silenced by introducing the corresponding dsRNA (ETF‐dsRNA‐1 and ETF‐dsRNA‐2, respectively) into cells. The etfalpha‐ (bar 2) or etfbeta‐ (bar 3) silenced or control (bar 1) cells were cultured aerobically for 4 days and then incubated under anaerobic conditions for 24 h, and wax ester content was determined. B: The RQ biosynthesis gene A (rquA) was silenced by introducing the corresponding dsRNA (RQUA‐dsRNA) into cells. The rquA‐silenced (bar 2) or control (bar 1) cells were cultured under aerobic conditions for 4 days, incubated under anaerobic conditions for 24 h, and wax ester content was determined. C: The acyl‐CoA dehydrogenase gene (acd1) was silenced by introducing the corresponding dsRNA (ACD1‐dsRNA) into cells. The acd1‐silenced (bar 2) or control (bar 1) cells were cultured under aerobic conditions for 3 days, incubated under anaerobic conditions for 24 h, and wax ester content was determined. D: The trans‐2‐enoyl‐CoA reductase gene (ter) was silenced by introducing the corresponding dsRNA (TER‐dsRNA‐1 or TER‐dsRNA‐2) into cells. The ter‐silenced cells with TER‐dsRNA‐1 (bar 2) or with TER‐dsRNA‐2 (bar 3) or control cells (bar 1) were cultured under aerobic conditions for 3 days, incubated under anaerobic conditions for 24 h, and wax ester content was determined. Data are presented as means ± SD (n = 3). *P < 0.05.
It is well known that RQ, which has a more negative redox potential than UQ, is used to transfer electrons from mitochondrial complex I to fumarate reductase during anaerobic respiration 1, 2, 3, 17. In addition, it is also known that Euglena cells contain RQ9 together with UQ9 18. Previously, a gene (rquA) encoding putative methyltransferase involved in the synthesis of RQ from UQ was identified in Rhodospirillum rubrum 19, and its ortholog was found in Euglena RNA‐Seq database 12. When Euglena rquA was silenced, total RQ9 content (sum of quinone and quinol forms) was greatly reduced, whereas total UQ9 content was slightly increased (Table 2). This is the first direct evidence showing that the eukaryotic rhodobacter homologs of rquA actually participate in the synthesis of RQ. The anaerobic production of wax esters in rquA‐silenced cells was attenuated to one‐third of the control cells (Fig. 4B). These results suggest that RQ9 (rather than UQ9) mainly functions to mediate electron transfer between complex I and ETF in mitochondrial fatty acid synthesis in anaerobic Euglena cells.
Table 2.
Ubiquinone‐9 (UQ9) and rhodoquinone‐9 (RQ9) contents in the rquA‐silenced cells. Data are presented as means ± SD (n = 3)
| Total UQ9 content | Total RQ9 content | |
|---|---|---|
| (pmol/106 cells) | ||
| Control | 86.7 ± 3.44 | 51.6 ± 3.44 |
| rquA‐silenced | 104 ± 2.42 | 1.10 ± 0.364 |
The Euglena RNA‐Seq database 12 suggests that there are at least seven ETF‐linked acyl‐CoA dehydrogenase orthologs. One of these (named acd1), which has a mitochondrial targeting sequence and is expressed the most abundantly, was silenced by introducing its dsRNA in Euglena cells to confirm that acyl‐CoA dehydrogenase is involved in mitochondrial fatty acid synthesis. As shown in Fig. 4C, anaerobic wax ester production in acd1‐silenced cells was lowered to 60% than that in the control. In contrast, when NAD(P)H‐dependent trans‐2‐enoyl‐CoA reductase that can utilize crotonyl‐CoA and trans‐2‐hexenoyl‐CoA 9 was knocked down by gene silencing, anaerobic wax ester production was not affected (Fig. 4D) although the enzyme protein, which was examined by western blotting, disappeared almost completely in the gene‐silenced cells (Fig. S2). Recently, trans‐2‐enoyl‐CoA reductase has been shown to be important in chloroplast development under aerobic conditions rather than in anaerobic wax ester synthesis (T. Tomiyama, K. Goto, Y. Tanaka, T. Maruta, T. Ogawa, Y. Sawa, T. Ito, T. Ishikawa, unpublished data), although this enzyme is reportedly localized in the mitochondria 9.
Discussion
The data presented here show that acyl‐CoA dehydrogenase mainly functions to reduce trans‐2‐enoyl‐CoA during mitochondrial fatty acid synthesis in E. gracilis under anaerobic conditions and that electrons for reduction are provided by NADH via complex I, RQ and ETF (Fig. 5). These data also strongly suggest that during wax ester fermentation, Euglena cells produce ATP by anaerobic respiration coupled with mitochondrial fatty acid synthesis, in addition to substrate‐level phosphorylation in the cytosolic glycolytic pathway. Therefore, more amounts of ATP can be obtained via wax ester fermentation compared with that via common lactate or ethanol fermentation.
Figure 5.
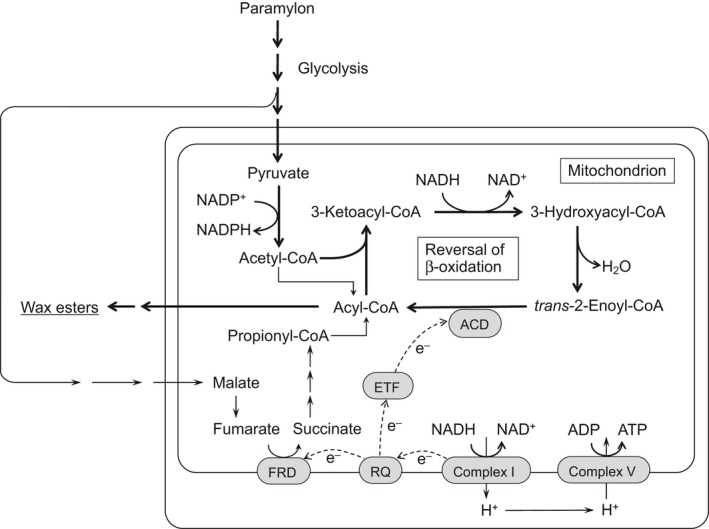
Mitochondrial fatty acid synthesis by the reversal of β‐oxidation linking with ATP synthesis by anaerobic respiration in Euglena gracilis. Abbreviations: ACD, acyl‐CoA dehydrogenase; ETF, electron transfer flavoprotein; FRD, fumarate reductase; RQ, rhodoquinone.
The anaerobic respiration occurring in E. gracilis seems to resemble that reported in Ascaris (giant roundworm) 2, 20, 21. In Ascaris under anaerobic conditions, acetyl‐CoA and propionyl‐CoA (or two molecules of propionyl‐CoA) are condensed and subsequently reduced in mitochondria, and 2‐methylbutanoate (or 2‐methylpentanoate) is synthesized as an end product. In this process, ATP is produced by anaerobic respiration linked to the reduction in 2‐methyl branched‐chain enoyl‐CoA, in which electrons are provided by NADH via mitochondrial complex I, RQ, and ETF. However, in contrast to Ascaris mitochondria in which the anaerobic end products, 2‐methylbutanoate and 2‐methylpentanoate, are short‐chain fatty acids, in Euglena mitochondria, long‐chain fatty acids, mainly myristic acid, are synthesized. Therefore, to the best of our knowledge, this is the first report showing that anaerobic respiration occurs coupled with long‐chain fatty acid synthesis by the reversal of β‐oxidation.
Algal biofuel is considered to be an attractive alternative to fossil fuels. Biofuel produced from Euglena wax esters exhibits good cold flow properties with high oxidative stability and is proposed to be suitable to use as a drop‐in jet fuel 5, 7. For industrial applications, however, it will be necessary to improve the productivity of wax esters from anaerobic Euglena cells. In this context, the results obtained in the present study will be valuable; in particular, mitochondrial uncoupling reagents, such as CCCP, could be useful tools for improving productivity. Anaerobic Euglena cells produce wax esters from paramylon to obtain ATP; therefore, the rate of wax ester production should be controlled by the amount of ATP required in cells. CCCP acts as an ionophore, causes a decrease in proton motive force developed in the mitochondrial inner membrane, and reduces ATP synthesis by anaerobic respiration, in contrast to rotenone, which directly inhibits mitochondrial complex I. Therefore, mitochondrial fatty acid synthesis would not be suppressed even in the presence of CCCP because electron flow through complex I is not inhibited. To compensate for a decrease in ATP synthesized by anaerobic respiration, glycolysis may accelerate to increase ATP synthesis by substrate‐level phosphorylation. Consequently, the anaerobic production of wax esters should be greatly accelerated in the presence of CCCP, providing a possible route toward unraveling the potential of this organism as a renewable energy resource.
Author contributions
All members conceived and designed the experiments, and discussed the results for the completion of the manuscript. MN, HA, AN, and TO performed the experiments and analyzed the data. KS performed quinone analysis. MN and HI wrote the manuscript.
Supporting information
Fig S1. Semiquantitative RT‐PCR analysis to determine the degree of gene silencing.
Fig S2. Immunodetection of trans‐2‐enoyl‐CoA reductase protein. C, without dsRNA; 1, TER‐dsRNA‐1 introduced; 2, TER‐dsRNA‐2 introduced.
Table S1. Oligonucleotide primers and amplification cycle numbers for semiquantitative RT‐PCR.
Acknowledgements
This work was supported by the Core Research of Evolutional Science and Technology program (CREST) from the Japan Science and Technology Agency (JST) (for MN, TI, and HI), PRESTO program from JST (for MN), and JSPS KAKENHI Grant number 16K18828 (for MN).
Edited by Peter Brzezinski
References
- 1. Ginger ML, Fritz‐Laylin LK, Fulton C, Cande WZ and Dawson SC (2010) Intermediary metabolism in protist: a sequence‐based view of facultative anaerobic metabolism in evolutionarily diverse eukaryotes. Protist 161, 642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu R‐Y, van der Giezen M, Tielens AGM and Martin WF (2012) Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76, 444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tielens AGM, Rotte C, van Hellemond JJ and Martin W (2002) Mitochondria as we don't know them. Trends Biochem Sci 27, 564–572. [DOI] [PubMed] [Google Scholar]
- 4. Inui H, Miyatake K, Nakano Y and Kitaoka S (1982) Wax ester fermentation in Euglena gracilis . FEBS Lett 150, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inui H, Ishikawa T and Tamoi M (2017) Wax ester fermentation and its application for biofuel production. Adv Exp Med Biol 979, 269–283. [DOI] [PubMed] [Google Scholar]
- 6. Inui H, Miyatake K, Nakano Y and Kitaoka S (1983) Production and composition of wax esters by fermentation of Euglena gracilis . Agric Biol Chem 47, 2669–2671. [Google Scholar]
- 7. Nakazawa M, Andoh H, Koyama K, Watanabe Y, Nakai T, Ueda M, Sakamoto T, Inui H, Nakano Y and Miyatake K (2015) Alteration of wax ester content and composition in Euglena with gene silencing of 3‐ketoacyl‐CoA thiolase isozymes. Lipids 50, 483–492. [DOI] [PubMed] [Google Scholar]
- 8. Inui H, Miyatake K, Nakano Y and Kitaoka S (1984) Fatty acid synthesis in mitochondria of Euglena gracilis . Eur J Biochem 142, 121–126. [DOI] [PubMed] [Google Scholar]
- 9. Hoffmeister M, Piotrowski M, Nowitzki U and Martin W (2005) Mitochondrial trans‐2‐enoyl‐CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J Biol Chem 280, 4329–4338. [DOI] [PubMed] [Google Scholar]
- 10. Nakazawa M, Hayashi R, Takenaka S, Inui H, Ishikawa T, Ueda M, Sakamoto T, Nakano Y and Miyatake K (2017) Physiological functions of pyruvate:NADP+ oxidoreductase and 2‐oxoglutrate decarboxylase in Euglena gracilis under aerobic and anaerobic conditions. Biosci Biotechnol Biochem 81, 1386–1393. [DOI] [PubMed] [Google Scholar]
- 11. Koren LE and Hutner SH (1967) High‐yield media for photosynthesizing Euglena gracilis Z. J Protozool 14 (Suppl), 17. [Google Scholar]
- 12. Yoshida Y, Tomiyama T, Maruta T, Tomita M, Ishikawa T and Arakawa K (2016) De novo assembly and comparative transcriptome analysis of Euglena gracilis in response to anaerobic conditions. BMC Genom 17, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiobara Y, Harada C, Shiota T, Sakamoto K, Kita K, Tanaka S, Tabata K, Sekie K, Yamamoto Y and Sugiyama T (2015) Knockdown of the coenzyme Q synthesis gene Smed‐dlp1 affects planarian regeneration and tissue homeostasis. Redox Biol 6, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimada H, Kodjabachian D and Ishida M (2007) Specific and rapid analysis of ubiquinones using Craven's reaction and HPLC with postcolumn derivatization. J Lipid Res 48, 2079–2085. [DOI] [PubMed] [Google Scholar]
- 15. Sato M and Ozawa H (1969) Occurrence of ubiquinone and rhodoquinone in parasitic nematodes, Metastrongylus elongatus and Ascaris lumbricoides var. suis . J Biochem 65, 861–867. [DOI] [PubMed] [Google Scholar]
- 16. Nagai J, Ohta T and Saito E (1971) Incorporation of propionate into wax esters by etiolated Euglena . Biochem Biophys Res Commun 42, 523–529. [DOI] [PubMed] [Google Scholar]
- 17. Tucci S, Vacula R, Krajcovic J, Proksch P and Martin W (2010) Variability of wax ester ferementation in natural and bleached Euglena gracilis strain in response to oxygen and the elongase inhibitor flufenacet. J Eukaryot Microbiol 57, 63–69. [DOI] [PubMed] [Google Scholar]
- 18. Threlfall DR (1972) Incorporation of L‐[Me‐3H2]methionine into isoprenoid quinones and related compounds by Euglena gracilis . Biochim Biochem Acta 280, 472–480. [DOI] [PubMed] [Google Scholar]
- 19. Lonjers ZT, Dickson EL, Chu T‐PT, Kreutz JE, Neacsu FA, Anders KR and Shepherd JN (2012) Identification of a new gene required for the biosynthesis of rhodoquinone in Rhodospirillum rubrum . J Bacteriol 194, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komuniecki R, McCrury J, Thissen J and Rubin N (1989) Electron‐transfer flavoprotein from anaerobic Ascaris suum mitochondria and its role in NADH‐dependent 2‐methyl branched‐chain enoyl‐CoA reductase. Biochim Biophys Acta 975, 127–131. [DOI] [PubMed] [Google Scholar]
- 21. Ma Y‐C, Funk M, Dunham WR and Komuniecki R (1993) Purification and characterization of electron‐transfer flavoprotein:rhodoquinone oxidireductase from anaerobic mitochondria of the adult parasitic nematoda, Ascaris suum . J Biol Chem 269, 20360–20365. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Semiquantitative RT‐PCR analysis to determine the degree of gene silencing.
Fig S2. Immunodetection of trans‐2‐enoyl‐CoA reductase protein. C, without dsRNA; 1, TER‐dsRNA‐1 introduced; 2, TER‐dsRNA‐2 introduced.
Table S1. Oligonucleotide primers and amplification cycle numbers for semiquantitative RT‐PCR.


