Abstract
The KCNQ1 channel is a voltage-dependent potassium channel and is ubiquitously expressed throughout the human body including the heart, lung, kidney, pancreas, intestine and inner ear. Gating properties of the KCNQ1 channel are modulated by KCNE auxiliary subunits. For example, the KCNQ1-KCNE1 channel produces a slowly-activating potassium current, while KCNE3 makes KCNQ1 a voltage-independent, constitutively open channel. Thus, physiological functions of KCNQ1 channels are greatly dependent on the type of KCNE protein that is co-expressed in that organ. It has long been debated how the similar single transmembrane KCNE proteins produce quite different gating behaviors. Recent applications of voltage-clamp fluorometry (VCF) for the KCNQ1 channel have shed light on this question. The VCF is a quite sensitive method to detect structural changes of membrane proteins and is especially suitable for tracking the voltage sensor domains of voltage-gated ion channels. In this short review, I will introduce how the VCF technique can be applied to detect structural changes and what have been revealed by the recent VCF applications to the gating modulation of KCNQ1 channels by KCNE proteins.
Keywords: ion channel, potassium channel, auxiliary subunit, voltage sensor, electrophysiology
Significance.
Voltage clamp fluorometry (VCF) is a powerful tool to track conformational rearrangement of the voltage sensor domain (VSD) of voltage-gated ion channels. The KCNQ1 channel is a voltage-gated potassium channel and its gating properties are largely dependent on which types of auxiliary KCNE proteins bind. Recent applications of VCF to KCNQ1 channel have started to reveal the mechanisms in which KCNE proteins modulate the VSD movement and the gating properties of the KCNQ1 channels.
The heart is an organ for pumping blood through the entire body. The atria and ventricles alternately contract in each cardiac cycle and that is how the heart works as an efficient and rhythmic pump. These rhythmic activities are produced by electrical activity (i.e., action potentials), and each action potential is shaped by voltage-gated ion channels including Na+ channels, Ca2+ channels and K+ channels. Ventricular myocytes are depolarized by Na+ channels and Ca2+ channels, leading to contraction. Depolarization is typically maintained for 200–300 ms in the human heart. K+ channels subsequently repolarize the myocytes, leading to relaxation. Therefore, any defect of these ion channels or their associated proteins can cause prolongation (or shortening) of ventricular depolarization. As a result, the QT interval of an electrocardiogram is prolonged (or shortened) and is diagnosed as long (short) QT syndrome (LQTS, SQTS). LQTS (or SQTS) increases the risk of atrial and ventricular arrhythmias and is potentially lethal. Therefore, it is important that these ion channels work at proper timings in cardiac cycles.
Structure and function of KCNQ1 channel and KCNE proteins
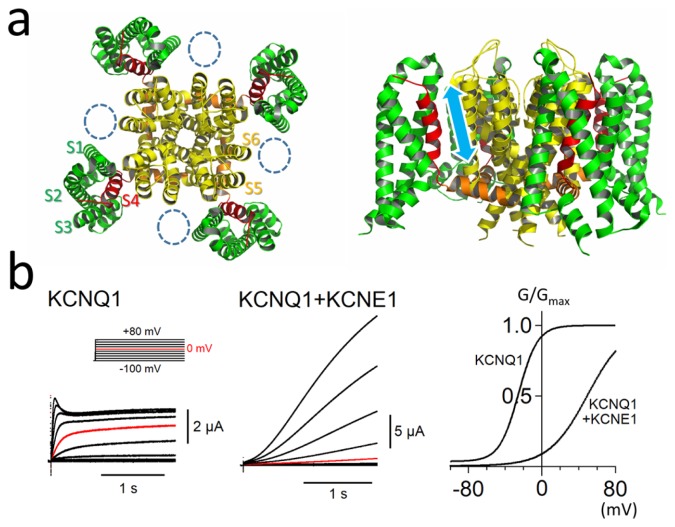
The KCNQ1 channel is a member of the voltage-gated K+ channel family. As KCNQ1 is known as a causative gene for long QT syndrome (LQTS type 1 or LQT1), it is obviously important for shaping cardiac action potentials. KCNQ1 α subunit is a membrane protein having six transmembrane segments (S1–S6), and four α subunits form a single KCNQ1 channel (tetramer) like other voltage-gated K+ channels. The first four transmembrane segments (S1–S4) form a voltage-sensing domain (VSD) and the remaining two segments (S5–S6) form a pore domain (PD) (Fig. 1a). The S4 segments (red helices in Fig. 1a), which possess positively-charged amino acid residues, are known to move up and down voltage-dependently by sensing membrane potential, and the upward movement of the S4 segment leads to channel opening [1]. In addition, KCNQ1 is known to be modulated by auxiliary KCNE subunits. Five KCNE genes (KCNE1-5) have been identified in the human genome. Gating properties of KCNQ1 channel are largely dependent on which types of KCNE proteins bind. KCNE1, for example, binds to a KCNQ1 channel and slows its activation and deactivation kinetics by 100 times [2,3]. KCNE1 also shifts the voltage-dependence (G-V) curve of KCNQ1 channel by 40 mV in a positive direction. In other words, KCNE1 makes the KCNQ1 channel much more difficult to be activated (Fig. 1b). Since KCNE1 has also been identified as a causative gene for LQTS (LQT5), both KCNQ1 and KCNE1 subunits are indispensable for proper rhythmic activity of the heart. The stoichiometry of the KCNQ1-KCNE1 channel has been a matter of debate for quite a while. It has been reported that two KCNE1 subunits bind to a KCNQ1 tetramer (4:2 stoichiometry) [4–6]. We and other groups, on the other hand, showed that multiple stoichiometry (4:1~4:4 stoichiometry) is possible at least in vitro and human embryonic stem cell derived cardiomyocytes [7–10]. Further study, especially in vivo experiments, may be necessary to determine which stoichiometries are predominant in the human heart.
Figure 1.
Structure and ionic currents of the KCNQ1 channel with KCNE1. (a) Top view (left) and side view (right) of the tetrameric KCNQ1 structures [37]. S1–S3, S4, S4–S5 linker, and S5–S6 are colored in green, red, orange and yellow, respectively. Dotted circles indicate putative KCNE1 sites. S4 segments (red helices) voltage-dependently move up and down as indicated by a cyan arrow in the right figure. (b) Representative currents and G-V curves of the KCNQ1 channel in Xenopus oocytes with or without KCNE1 co-expression. G/Gmax indicates apparent open probability of the channel. Co-expression of KCNE1 slows the activation of the channel and shifts the G-V curve in the positive direction (The channel requires higher membrane potential to be activated.). Inset shows the voltage clamp protocol. The currents at 0 mV are colored in red for comparison. Figures and the data presented are adapted from [26,40].
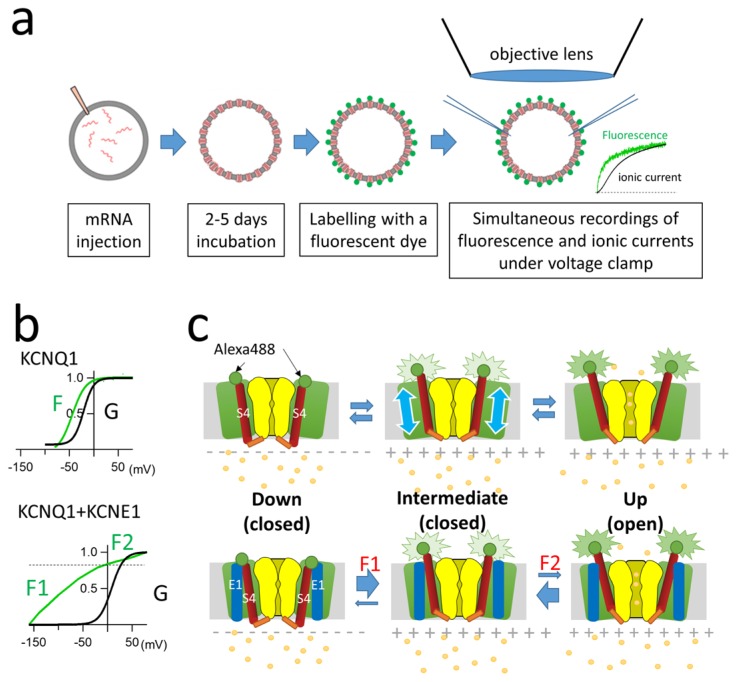
Modulation of KCNQ1 voltage sensor movement by KCNE1
It has also long been debated how KCNE1 changes the KCNQ1 gating. Initial reports suggested that KCNE1 directly interacts with the PD of KCNQ1 and prevents it from opening [11–14]. We and other groups showed that VSD movement (or state transition) was affected by co-expression of KCNE1 by utilizing the substituted cysteine accessibility method (SCAM) [15–17]. Those studies led to works focusing on the VSD movement of KCNQ1 channels, and works in which voltage-clamp fluorometry (VCF) was used to track the KCNQ1 VSD revealed the detailed mechanisms by which KCNE1 regulates KCNQ1 gating via the VSD [18–20]. In VCF experiments, a cysteine residue was introduced at the extracellular side of the S4 segment (S3–S4 linker) by site-directed mutagenesis. Fluorescent dye such as tetra-methylrhodamine maleimide or Alexa488 maleimide was then attached to the site of the introduced cysteine residue to track voltage-dependent conformational rearrangement by detecting change of fluorescence intensity under a voltage-clamp (Fig. 2a) [21–23]. VCF experiments clearly showed that KCNQ1 VSD movement was greatly changed by KCNE1, though the change was far more complicated than expected [18,20]. In KCNQ1, the fluorescence-voltage (F-V) relationship (representing the VSD movement) and the G-V relationship (representing the pore opening) are mostly overlapped (Fig. 2b upper panel) [19]. This is because each individual VSD movement contributes sequentially to channel conductance in KCNQ1 channel [24,25], not like other shaker-type potassium channels in which all four VSD must be in the up state before opening. On the other hand, KCNE1 shifts the G-V relationship in the positive direction, while the F-V relationship is split into two components, a steep component F1 and a shallow component F2 (Fig. 2b bottom panel). F1 reflects a non-conducting state(s) from −160 to −50 mV and F2 nearly overlaps the G-V relationship. It is assumed that there are three states in KCNQ1 gating: closed state (S4 segments in the down state and the PD closed), intermediate state (S4 segments in the up state and the PD closed), and open state (S4 segments in the “full” up state and the PD open). The results of VCF studies indicated that KCNE1 may stabilize the intermediate state and that is why the F-V relationship is split into two (Fig. 2c). Since the intermediate state is non-conducting, channel opening requires higher and longer depolarization. Conversely, the channel requires much larger hyperpolarization (−200 mV) to enter the “fully” down state [18,20]. Since membrane potential of cardiomyocytes rarely reaches −200 mV, most of the KCNQ1 channels with KCNE1 subunits should be in the intermediate state.
Figure 2.
KCNE1 stabilizes the intermediate state of the KCNQ1 channel. (a) Preparation of fluorescence-labelled ion channels expressed in Xenopus oocytes for VCF experiments. Fluorescence changes are acquired by a photomultiplier tube (or a photodiode) while ionic currents are simultaneously recorded under two-electrode voltage clamp. (b) G-V (black; related to pore opening) and F-V (green; related to VSD movement) relationships of KCNQ1 alone and the KCNQ1-KCNE1 channel. The F-V relationship of KCNQ1-KCNE1 is split into two components, F1 and F2 [20]. (c) Model of three states of KCNQ1 channel gating without KCNE1 (upper row) and with KCNE1 (bottom row). Green circles indicate attached fluorophores (e.g., Alexa Fluor 488) for the VCF experiments. S4 segments (red) voltage-dependently move up and down (cyan arrows in upper row). KCNE1 stabilizes the intermediate state. Figures and the data presented are adapted from [26,40].
Steric hindrance between the VSD and PD of KCNQ1 induced by KCNE1
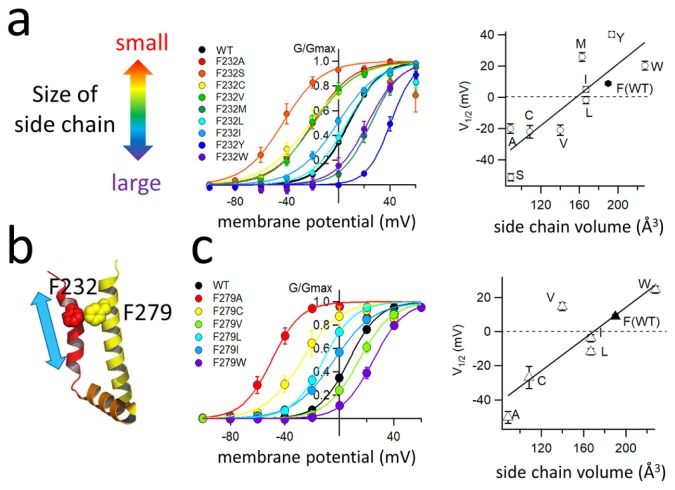
As discussed in the previous section, co-expression of KCNE1 dramatically changes the voltage-dependence of the KCNQ1 VSD and that is at least one of the mechanisms by which KCNE1 slows KCNQ1 gating kinetics. However, it still remains largely unknown how KCNE1 changes the voltage-dependence of KCNQ1. We previously made a series of cysteine mutants of the KCNQ1 S3–S4 linker and the first half of the S4 segment (G219C-Q234C) to identify the best site for SCAM study [15], and we noticed that the F232C mutant did not show the characteristic positive G-V shift induced by KCNE1. Phe232 (F232) is located next to the second positive charge (R231) of the S4 segment. We therefore made several other Phe232 mutants (mostly hydrophobic amino acid residues) to explore the role of Phe232 in the positive G-V shift induced by KCNE1. Remarkably, the G-V curve shifts induced by KCNE1 were dependent on the bulkiness of the amino acid side chain (Fig. 3a). Interestingly, KCNQ1 mutants without KCNE1 did not show bulkiness-dependence in the G-V curves. Phe232 affects the G-V curve only in the presence of KCNE1 [26]. According to the closed state and open state of KCNQ1-KCNE1 structural models by Sanders’s group [27], Phe232 on the S4 segment of the VSD faces the S5 segment of the PD of an adjacent subunit, and Phe232 seems to have to hurdle another bulky Phe279 on the S5 segment during the transition from the closed state to the open state. Therefore, we hypothesized that Phe232 collides with Phe279 during upward movement of the S4 segment (Fig. 3b). It turned out that was probably the case because the size of the Phe279 side chain affected the G-V relationship as the size of the Phe232 side chain did. The larger the introduced side chain at Phe279 was, the greater was the shift in the G-V relationship in the positive direction and, again, the G-V relationship was shifted only in the presence of KCNE1 (Fig. 3c). We concluded that the presence of KCNE1 created a steric hindrance between two phenylalanine residues of the S4 and S5 segments, and this could be the reason why activation of the KCNQ1-KCNE1 channel becomes more difficult than that of the KCNQ1 channel.
Figure 3.
Bulky amino acid residues on S4 and S5 prevent the KCNQ1-KCNE1 channel from opening. (a) Side chain dependence of G-V curves (left) and voltage of half activation (V1/2, right) in KCNQ1 F232 mutant series. Mutation of a larger amino acid residue tends to require higher membrane potential to be activated. (b) Locations of F232 on S4 (red) and F279 on S5 (yellow) in the cryo-EM structure of the KCNQ1 channel [37]. S4 moves up and down in the direction of the cyan arrow. (c) Side chain dependence of G-V curves (left) and voltage of half activation (V1/2, right) in KCNQ1 F279 mutant series. As observed in the F232 mutants, mutation of a larger amino acid residue tends to require higher membrane potential to be activated. Figures and the data presented are adapted from [26,40].
We next applied VCF to reveal which gating step was mostly affected by steric hindrance of the two phenylalanine residues. By attaching Alexa 488 maleimide to G219C mutation on the S3–S4 linker, ionic currents and fluorescence intensity were simultaneously recorded from wild-type and F232A and F279A mutants of KCNQ1 co-expressed with KCNE1 (Fig. 4a and b). In the wild-type KCNQ1-KCNE1 channel, an apparent delay of ionic currents (PD opening) after the increase of fluorescence (upward movement of the VSD) was observed. On the other hand, the delay was substantially diminished or almost disappeared in F232A or F279A mutants (Fig. 4a). In addition, the ratio of a shallow (presumably F2) component was increased in the F-V relationships of the mutants, indicating the transition from the intermediate state to the open state requires less depolarization in the mutants (Fig. 4b). Therefore, it was concluded that the steric hindrance mostly affects the gating transition from the intermediate state to the open state and prevents the channel from opening.
Figure 4.
F232 and F279 create a delay of channel opening after upward VSD movement. (a) Representative ionic currents (black traces) and fluorescence intensity (green traces) of KCNQ1 WT, F232A and F279A in the presence of KCNE1. Membrane potential was +40 mV from the holding potential of −90 mV. (b) F-V (green) and G-V relationships (black) of WT, F232A and F279A in the presence of KCNE1. (c) S4 segment (red bar) easily moves upward during depolarization in the absence of KCNE1. In the presence of KCNE1, the distance between S4 and S5 (yellow bar) becomes smaller and that may cause steric hindrance between the two phenylalanine residues. Figures and the data presented are adapted from [26,40].
Why does steric hindrance occur between Phe232 and Phe279 in the KCNQ1 channel only when KCNE1 is co-expressed? In movies of MD simulation of the voltage-gated K+ channel gating provided by Jensen et al. in 2012, four VSDs are moving around slightly and have a loose coupling with the PD [1]. We hence propose that the distance between the S4 and S5 may become smaller due to the interaction of KCNE1 with both the VSD and PD [28]. Only when KCNE1 interacts with the KCNQ1 channel, Phe232 on S4 and Phe279 on S5 become close enough to collide with each other during gating, and that would prevent the channel from entering the open state (Fig. 4c).
F279I, a novel mutation of KCNQ1, was recently reported and shown to be responsible for short QT syndrome by electrophysiology and simulation study [29]. F279I was also examined in our study. Isoleucine is slightly smaller than phenylalanine, and the G-V curve of the F279I mutant is therefore shifted in the negative direction (easier to be activated) by 10 mV (Fig. 3c, blue curve). This 10-mV shift may be large enough to cause short QT syndrome, which is a result of upregulation of the potassium current and shortened cardiac action potential.
Comparison to KCNE3 and other recent advances revealed by VCF
KCNQ1 can also be modulated by other types of KCNE proteins. KCNE3 is one of them and is known to make the KCNQ1 channel constitutively open. The KCNQ1-KCNE3 complex is known to be expressed in epithelial cells of the intestine and trachea to assist Cl− transport [30–32]. We and other groups have shown by mutagenesis and SCAM experiments that KCNE3 might lock the S4 segment in the up state and that is probably the mechanism by which the channel becomes constitutively open [15,16]. Recent VCF experiments have directly confirmed that VSDs are stabilized in the up state in KCNQ1-KCNE3 channels [33]. The VSD is actually not locked in the up state: the G-V curve is shifted in the negative direction by 100 mV, and the VSD therefore requires large hyperpolarization (lower than −100 mV) to start entering the down state. This means that KCNQ1-KCNE3 channels virtually stay in the open state in the physiological range of membrane potentials.
The KCNQ1 channel is known to require phosphatidylinositol-4,5-bisphosphate (PIP2) for activation [34]. Recent VCF experiments have revealed that the VSD still shows voltage-dependent movement without PIP2. However, the channel loses functional coupling between the VSD and PD in the absence of PIP2 and that is why KCNQ1 is unable to open the pore [35]. PIP2 may directly interact with the S4–S5 linker to control coupling of the VSD and PD of the KCNQ1 channel. Therefore, it is possible to functionally separate the VSD from the PD by abolishing PIP2. VCF experiments were performed in the absence of PIP2 to reveal which part(s) of the KCNQ1 channels was (were) affected by KCNE proteins [36]. The results suggested that KCNE3 functionally interacted only with the VSD and does not need an interaction with the PD to make the channel constitutively open. On the other hand, KCNE1 functionally interacted with both the VSD and PD. While the VSD is stabilized at the intermediate state, KCNE1 also directly affects the PD [36].
Future direction of KCNQ1 study
It still remains unknown how KCNE1 (and KCNE3) interacts with KCNQ1 and modulates the gating. Recently, Sun and MacKinnon reported the first tetrameric KCNQ1 structure determined by cryo-EM (Fig. 1a) [37]. Because the structure lacks PIP2, there is no coupling between the PD and VSD, and the VSD is therefore in the up state (at 0 mV) and the PD is closed. Because it is a homo tetrameric structure, it does not offer any information on how and where KCNE proteins interact. Structures of the KCNQ1-KCNE complex are, therefore, eagerly awaited. On the other hand, as I discussed in this short review, the KCNQ1 channel goes back and forth among many states to produce a voltage-dependent (or independent) potassium current with KCNE proteins. Electrophysiological experiments including current recording and VCF should remain powerful tools for studying state transitions of ion channels.
Applications of VCF to other membrane proteins
Although VCF is most suitable for tracking voltage-dependent conformational change of voltage-gated ion channels, applications of VCF to other membrane proteins such as pumps and transporters have also been reported [38,39]. For example, conformational changes of the Na+/K+-ATPase induced by extracellular potassium are detectable by VCF [38]. Thanks to the recent advancement of cryo-EM, structural information of membrane proteins has been explosively increasing. This simple yet powerful tool for tracking real-time structural changes will be applicable to more diverse membrane proteins by utilizing these new structural information.
Acknowledgements
I thank Professor Yoshihiro Kubo (National Institute for Physiological Sciences) for his continuous support in conducting experiments during my time in Kubo lab. This work was supported by KAKENHI (24590292, 25136724, 17K08552).
Footnotes
Conflict of Interest
The author declares no conflict of interests.
Author contributions
K. N. wrote the manuscript.
References
- 1.Jensen MØ, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 2.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and lsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, et al. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Kim LA, Rajan S, Xu S, Goldstein SA. Charybdotoxin binding in the IKs pore demonstrates two MinK subunits in each channel complex. Neuron. 2003;40:15–23. doi: 10.1016/s0896-6273(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 5.Morin TJ, Kobertz WR. Counting membrane-embedded KCNE beta-subunits in functioning K+ channel complexes. Proc Natl Acad Sci USA. 2008;105:1478–1482. doi: 10.1073/pnas.0710366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plant LD, Xiong D, Dai H, Goldstein SA. Individual IKs channels at the surface of mammalian cells contain two KCNE1 accessory subunits. Proc Natl Acad Sci USA. 2014;111:E1438–1446. doi: 10.1073/pnas.1323548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Xia J, Kass RS. MinK-KvLQT1 fusion proteins, evidence for multiple stoichiometries of the assembled IsK channel. J Biol Chem. 1998;273:34069–34074. doi: 10.1074/jbc.273.51.34069. [DOI] [PubMed] [Google Scholar]
- 8.Nakajo K, Ulbrich MH, Kubo Y, Isacoff EY. Stoichiometry of the KCNQ1 - KCNE1 ion channel complex. Proc Natl Acad Sci USA. 2010;107:18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Terrenoire C, Sampson KJ, Iyer V, Osteen JD, Lu J, et al. Biophysical properties of slow potassium channels in human embryonic stem cell derived cardiomyocytes implicate subunit stoichiometry. J Physiol. 2011;589:6093–6104. doi: 10.1113/jphysiol.2011.220863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray CI, Westhoff M, Eldstrom J, Thompson E, Emes R, Fedida D. Unnatural amino acid photo-crosslinking of the IKs channel complex demonstrates a KCNE1:KCNQ1 stoichiometry of up to 4:4. Elife. 2016;5:e11815. doi: 10.7554/eLife.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KW, Tai KK, Goldstein SA. MinK residues line a potassium channel pore. Neuron. 1996;16:571–577. doi: 10.1016/s0896-6273(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 12.Tai KK, Goldstein SA. The conduction pore of a cardiac potassium channel. Nature. 1998;391:605–608. doi: 10.1038/35416. [DOI] [PubMed] [Google Scholar]
- 13.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Panaghie G, Tai KK, Abbott GW. Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J Physiol. 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakajo K, Kubo Y. KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J Gen Physiol. 2007;130:269–281. doi: 10.1085/jgp.200709805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocheleau JM, Kobertz WR. KCNE peptides differently affect voltage sensor equilibrium and equilibration rates in KCNQ1 K+ channels. J Gen Physiol. 2008;131:59–68. doi: 10.1085/jgp.200709816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, Delaloye K, Zaydman MA, Nekouzadeh A, Rudy Y, Cui J. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J Gen Physiol. 2010;135:595–606. doi: 10.1085/jgp.201010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osteen JD, Gonzalez C, Sampson KJ, Iyer V, Rebolledo S, Larsson HP, et al. KCNE1 alters the voltage sensor movements necessary to open the KCNQ1 channel gate. Proc Natl Acad Sci USA. 2010;107:22710–22715. doi: 10.1073/pnas.1016300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osteen JD, Barro-Soria R, Robey S, Sampson KJ, Kass RS, Larsson HP. Allosteric gating mechanism underlies the flexible gating of KCNQ1 potassium channels. Proc Natl Acad Sci USA. 2012;109:7103–7108. doi: 10.1073/pnas.1201582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barro-Soria R, Rebolledo S, Liin SI, Perez ME, Sampson KJ, Kass RS, et al. KCNE1 divides the voltage sensor movement in KCNQ1/KCNE1 channels into two steps. Nat Commun. 2014;5:3750. doi: 10.1038/ncomms4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 22.Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 23.Cha A, Bezanilla F. Structural implications of fluorescence quenching in the Shaker K+ channel. J Gen Physiol. 1998;112:391–408. doi: 10.1085/jgp.112.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meisel E, Dvir M, Haitin Y, Giladi M, Peretz A, Attali B. KCNQ1 channels do not undergo concerted but sequential gating transitions in both the absence and the presence of KCNE1 protein. J Biol Chem. 2012;287:34212–34224. doi: 10.1074/jbc.M112.364901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westhoff M, Eldstrom J, Murray CI, Thompson E, Fedida D. IKs ion-channel pore conductance can result from individual voltage sensor movements. Proc Natl Acad Sci USA. 2019;116:7879–7888. doi: 10.1073/pnas.1811623116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajo K, Kubo Y. Steric hindrance between S4 and S5 of the KCNQ1/KCNE1 channel hampers pore opening. Nat Commun. 2014;5:4100. doi: 10.1038/ncomms5100. [DOI] [PubMed] [Google Scholar]
- 27.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, et al. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajo K, Kubo Y. KCNQ1 channel modulation by KCNE proteins via the voltage-sensing domain. J Physiol. 2015;593:2617–2625. doi: 10.1113/jphysiol.2014.287672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno C, Oliveras A, de la Cruz A, Bartolucci C, Munoz C, Salar E, et al. A new KCNQ1 mutation at the S5 segment that impairs its association with KCNE1 is responsible for short QT syndrome. Cardiovasc Res. 2015;107:613–623. doi: 10.1093/cvr/cvv196. [DOI] [PubMed] [Google Scholar]
- 30.Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, et al. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403:196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- 31.Grahammer F, Warth R, Barhanin J, Bleich M, Hug MJ. The small conductance K+ channel, KCNQ1: expression, function, and subunit composition in murine trachea. J Biol Chem. 2001;276:42268–42275. doi: 10.1074/jbc.M105014200. [DOI] [PubMed] [Google Scholar]
- 32.Preston P, Wartosch L, Gunzel D, Fromm M, Kongsuphol P, Ousingsawat J, et al. Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl− transport. J Biol Chem. 2010;285:7165–7175. doi: 10.1074/jbc.M109.047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barro-Soria R, Perez ME, Larsson H. KCNE3 acts by promoting voltage sensor activation in KCNQ1. Proc Natl Acad Sci USA. 2015;112:E7286–7292. doi: 10.1073/pnas.1516238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loussouarn G, Park KH, Bellocq C, Baro I, Charpentier F, Escande D. Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. EMBO J. 2003;22:5412–5421. doi: 10.1093/emboj/cdg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaydman MA, Silva JR, Delaloye K, Li Y, Liang H, Larsson HP, et al. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci USA. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barro-Soria R, Ramentol R, Liin SI, Perez ME, Kass RS, Larsson H. KCNE1 and KCNE3 modulate KCNQ1 channels by affecting different gating transitions. Proc Natl Acad Sci USA. 2017;114:E7367–E7376. doi: 10.1073/pnas.1710335114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J, MacKinnon R. Cryo-EM Structure of a KCNQ1/CaM Complex Reveals Insights into Congenital Long QT Syndrome. Cell. 2017;169:1042–1050. e9. doi: 10.1016/j.cell.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dempski RE, Friedrich T, Bamberg E. The beta subunit of the Na+/K+-ATPase follows the conformational state of the holoenzyme. J Gen Physiol. 2005;125:505–520. doi: 10.1085/jgp.200409186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorraitz E, Hirayama BA, Paz A, Wright EM, Loo DDF. Active site voltage clamp fluorometry of the sodium glucose cotransporter hSGLT1. Proc Natl Acad Sci USA. 2017;114:E9980–E9988. doi: 10.1073/pnas.1713899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakajo K. Analysis of Voltage Sensor Movement in KCNQ1 - KCNE1 Channels by Voltage Clamp Fluorometry. Seibutsu Butsuri. 2018;58:144–148. [Google Scholar]