Abstract
Aims
Antenatal hypoxia negatively impacts foetal heart development, and increases the risk of heart disease later in life. The molecular mechanisms remain largely elusive. Here, we conducted a genome-wide analysis to study the impact of antenatal hypoxia on DNA methylome and transcriptome profiling in foetal and adult offspring hearts.
Methods and results
Pregnant rats were treated with normoxia or hypoxia (10.5% O2) from Day 15 to Day 21 of gestation. Hearts were isolated from near-term foetuses and 5-month-old male and female offsprings, and DNA methylome and RNA-seq were performed. Methylome data shows a sharp dip in CpG methylation centred at the transcription start site (TSS). CpG islands (CGIs) and CpG island shores (CGSs) within 10 kb upstream of the TSS are hypomethylated, compared with CGIs and CGSs within gene bodies. Combining transcriptome, data indicate an inverse relation between gene expression and CpG methylation around the TSS. Of interest, antenatal hypoxia induces opposite changes in methylation patterns in foetal and adult hearts, with hypermethylation in the foetus and hypomethylation in the adult. Also, there is significant sex dimorphism of changes in gene expression patterns in the adult offspring heart. Notably, pathway analysis indicates that enrichment of inflammation-related pathways are significantly greater in the adult male heart than those in the female heart.
Conclusion
Our study provides an initial framework and new insights into foetal hypoxia-mediated epigenetic programming of pro-inflammatory phenotype in the heart development, linking antenatal stress, and developmental programming of heart vulnerability to disease later in life.
Keywords: Antenatal hypoxia , Heart , Methylome , Transcriptome , Inflammation
Graphical Abstract
Graphical Abstract.
1. Introduction
Heart disease is the leading cause of morbidity and mortality for the adult.1 Growing evidence indicates that many cardiovascular disorders have their roots in the foetal and early childhood development under intrauterine stress.2 The concept of ‘Developmental Origins of Adult Disease’ has been well accepted and confirmed by numerous studies over the past two decades.3,4 During the foetal development, the organism has the ability to change its gene expression pattern to provide phenotypic alteration to cope with the changing environment, known as developmental plasticity.5 Of importance, gestational hypoxia is the common and critical factor of intrauterine stress impacting developmental plasticity, and foetal hypoxia during pregnancy may occur in many complications, such as preeclampsia, placental insufficiency, cord blood compression, and gestation at high altitude.6,7 Extensive studies in experimental animals and some in humans indicate that antenatal hypoxia results in irreversible developmental programming of phenotypic changes that predispose offspring to various dysfunctions and diseases, including cardiac dysfunction and ischaemic heart disease.8
Although the molecular mechanisms underlying antenatal hypoxia-induced developmental programming remain largely elusive, emerging evidence suggests that DNA methylation, a dominant mechanism of the epigenetic regulation, plays a crucial role in this process. DNA methylation mostly occurs at the cytosine of dinucleotide sequence CpG, and methylation in the promoter regions is generally associated with transcription repression of the associated genes. Our previous studies showed that hypermethylation of single genes, e.g. protein kinase C epsilon (PKCε) and glucocorticoid receptor (GR) in the developing heart were involved in antenatal hypoxia-induced developmental plasticity of ischaemic-sensitive phenotype in the heart.9,10 However, the impact of foetal hypoxia on the alteration of global methylation pattern and transcriptomic changes in the heart development are unknown.
Using next-generation sequencing technologies coupled with DNA-seq and RNA-seq, we tested the hypothesis that antenatal hypoxia causes a global epigenomic alteration of the transcriptome in developmental programming of hypoxic/ischaemic-sensitive phenotype in the heart. We showed that antenatal hypoxia not only induced a global DNA methylome and transcriptome changes in the foetal heart, but also had a delayed and lasting effect on the heart in adult offspring. Interestingly, antenatal hypoxia induced opposite changes in the global methylation patterns in foetal and adult hearts, with hypermethylation in the foetus and hypomethylation in the adult. In addition, there are significant differences in the antenatal hypoxia responding genes and pathways between foetal and adult hearts, as well as between adult male and female hearts. Of importance, pathways related to inflammation were significantly enriched in the hearts of all groups, especially in the adult male heart. Overall, the present study provides new insights into the epigenetic programming of transcriptome in the heart development in response to the foetal stress of hypoxia, and sheds light on mechanistic understanding of developmental programming of adult heart disease.
2. Methods
2.1 Experimental animals
Pregnant Sprague-Dawley (SD) rats (Charles River Laboratories, Portage, MI, USA) were randomly divided into two groups: (i) the normoxic control and (ii) hypoxic exposure (10.5% O2) from Day 15 to Day 21 of gestation, as described previously.10 Near-term (21-day) foetuses and 5-month-old offspring were sacrificed by decapitation under isoflurane anaesthesia (5% inhalant in room air). The left ventricle of the adult heart and the whole foetal heart were used in the analyses. All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of Loma Linda University and followed the guidelines in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2 DNA and RNA extractions
For genomic DNA (gDNA) extraction, heart tissues were minced and digested with 20 µL proteinase K (20 mg/mL) in digestion buffer (10 mM Tris-HCl, pH 8.0 containing 0.5 M EDTA, 1% SDS, 10 m NaCl, and 5 mM CaCl2) for 3 h at 55°C. Next the tissue lysate was twice phenol/chloroform/isoamyl: (25/24/1) alcohol extracted and the aqueous layer was treated with RNAse A+T1 for 1 h at 37°C. The lysate was again phenol/chloroform/isoamyl alcohol extracted and the gDNA was precipitated with equal volume of isopropanol in presence of 0.3 M sodium acetate at −20°C overnight. The gDNA was finally centrifuged, washed with 70% ethanol, air dried and reconstituted in 10 mM Tris-HCl, pH 8.0. And then, gDNA was denatured with 2 N NaOH at 42°C for 15 min, treated with sodium bisulfite at 55°C for 16 h, and purified by EZ DNA Methylation-Gold KitTM (Zymo Research, Tustin, CA, USA).
Total RNA was isolated from heart tissue using TRIzol RNA Isolation reagent (Life Technologies, Carlsbad, CA, USA). Briefly, each solid tissue was homogenized in 1 mL TRIzol and the lysate was then extracted with chloroform and centrifuged at 14 000 rpm for 15 min at 4°C. Finally, total RNA was precipitated and washed thoroughly with 70% ethanol, air-dried and reconstituted in nuclease free water. RNA quality was assessed using the 2200 TapeStation (Agilent Technologies, Wilmington, DE USA). Heart samples had an average RIN number of 9.5.
2.3 Reduced representation bisulfite sequencing library construction and sequencing
High-quality gDNA was used for generation of reduced representation bisulfite sequencing (RRBS) at the Center for Genomics, Loma Linda University (Loma Linda, CA, USA) following standard protocols of the Ovation® Ultralow Methyl-seq Library Systems (NuGEN Technologies, Inc., San Carlos, USA). Briefly, 100 ng gDNA was restriction digested at 37°C for 1 h using the methyl-insensitive restriction enzyme Mspl, which cuts the DNA at CCGG sites. The fragments were directly subjected to end blunting and phosphorylation. A single nucleotide (A) was then added to the 3' ends of the fragments in preparation for ligation to a methylated adapter with a single-base T overhang. The ligation products were final repaired in a thermal cycler under the program (60°C—10 min, 70°C—10 min, hold at 4°C). The product of the final repair reaction can be input directly into the bisulfite conversion kit (QIAGEN EpiTect Fast DNA Bisulfite Kit, Cat. #59824) according to Qiagen’s protocol. Eluted the purified, bisulfite-converted DNA in 23 μL of EB and performed PCR-amplification to enrich for fragments with adapters on both ends following by Agencourt RNAClean XP Beads purification. The final libraries were quantified using Qubit 3.0 (Life Technologies, Carlsbad, CA, USA) and the average size was determined on an Agilent TapeStation 2000 (Agilent Technologies, Wilmington, DE, USA). The final library was diluted to 4 nM and further quantitated to ensure high accuracy quantification for consistent pooling of barcoded libraries and maximization of the number of clusters in the Illumina flow cell. RRBS libraries were sequenced using the Illumina Nextseq 550 (Illumina, San Diego, CA, USA) in a single-ended, 75 bp, with approximately 25 M reads/each at the Center for Genomics, Loma Linda University.
2.4 RNA-seq library construction and sequencing
RNA-seq libraries were constructed using the Ovation Universal RNA-seq System (NuGEN, San Carlos, USA). All total RNA samples (except non-template control) were spiked with 1:500 ERCC RNA spike-in control mix (Life Technologies, Cat #4456740). Before cDNA generation, samples were treated with a second round of DNase for more thorough removal of gDNA following NuGEN’s integrated DNase treatment protocol. Double-stranded cDNA was generated from approximately 100 ng of total RNA per sample. cDNA was sheared using the Covaris S220 sonication system. Each sample was sheared according to the manufacturer’s settings of 130 μL sample with a target (peak) of 200 bp. The settings were as follows: 10% duty factor and 200 cycles/burst at 7°C for 180 s. End repair followed to generate blunt ends for adaptor ligation. Unique barcodes were used for each sample for multiplexing. Targeted rRNA-depletion was performed before final library construction. cDNA libraries were amplified using 15 cycles (Artik thermal cycler from Thermo Scientific) and purified using RNAClean XP beads (Agencourt, Brea, USA). The size distribution of the libraries was checked using the 2200 TapeStation. The peak size for all samples was around 300 bp (including a 122 bp adaptor). All libraries were quantified using the Qubit 3.0 Fluorometer (Life Technologies) and stored at −20°C in non-sticky Eppendorf tubes (Life Technologies, Carlsbad, USA). RNA-seq libraries were sequenced on Illumina NextSeq 550 with 75 bpx2, PE, approximately 25 M reads/each, at the Center for Genomics, Loma Linda University.
2.5 RRBS data analysis
For the RRBS data, we used a pipeline that integrates the read quality assessment (FastQC), trimming process (TrimGalore, NuGEN diversity trimming and N6 de-duplicate scripts), alignment (Bismark),11 and differential methylation analysis using MethylKit,12 and DMAP.13 This pipeline facilitates a rapid transition from sequencing reads to a fully annotated CpG methylation report for biological interpretation. Briefly, the RRBS raw fastq data were first trimmed using Trim Galore (V0.3.7). The rat genome NCBI Rnor6.0, downloaded from iGenome (ftp://igenome:G3nom3s4u@ussdftp.illumina.com/Rattus_norvegicus/NCBI/Rnor_6.0/Rattus_norvegicus_NCBI_Rnor_6.0.tar.gz), was used as a reference genome. Reads were aligned to the rat reference genome with Bismark V0.16.334 by default parameter settings. The methylation call files including the location of each CpG sites and the methylation percentage were generated by the bismark_methylation_extractor function. The aligned sam files were further processed through DMAP to generate CpG regions profiling for foetal and adult male rats, respectively. CpG regions with coverage by a minimum 20 reads in all samples were used for follow-up analysis. Then the differential methylation region (DMR) analysis was performed by methylKit. The DMRs were selected by false discovery rate (FDR) <0.05 and methylation percentage change between control and test groups are >10%.
2.6 RNA-seq data analysis
For mRNA-seq data, we adopted the pipelines used in our recent publications for mRNA-seq data visualization, which integrated the QC (FastQC, ShortRead), trimming process (trimmomatic), alignment (Tophat2), reads quantification (cufflinks), and differentially expressed gene (DEG) analysis (cuffdiff) for mRNA-seq data analyses. Briefly, the RNA-seq raw fastq data were first trimmed using Trimmomatic. The trimmed reads were aligned to the rat reference genome (NCBI Rnor6.0) with TopHat V2.1.114 with default parameter settings. The aligned bam files were then processed using Cufflinks V2.2.115 for gene quantification. Reads that were unable to align to the rat genome were converted to fastq format using SamToFastq function in Picard V1.114 for ERCC mapping and calculation. Reads were then mapped to ERCC transcripts and quantified using TopHat V2.1.1 and Cufflinks V2.1.1 with default parameter settings. Genes with FPKM ≥1 in all samples were used for DEG analysis. The DEGs were identified by Cuffdiff with FDR <0.3, and fold change (FC) >1.2.
2.7 Real-time RT-PCR
Total RNA was isolated from heart tissues with TRIzol reagent (Life Technologies) and subjected to reverse transcription using Superscript III First-Strand Synthesis System (Life Technologies, Carlsbad, USA). The relative mRNA abundance of Hand1, HK2, Lcn2, and Ddit4 was determined by real time PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), as described previously.9 β-actin was used as an internal control. Primers used are listed in Supplementary material online, Table S1. Real-time PCR was performed in a final volume of 20 μL and the following protocol was used 95°C for 5 min, followed by 40 cycles of 95°C for 20 s, annealing for 20 s at appropriate temperature depending on the primer sequence, 72°C for 20 s. PCR was performed in triplicate in a CFX connect real time PCR machine (Bio-Rad, Hercules, USA), and threshold cycle numbers (Ct) were averaged for each sample. The relative expression of each gene of interest was calculated by 2−ΔΔCT method and expressed as fold of normoxic control.
2.8 Methylated DNA immunoprecipitation
Methylated DNA immunoprecipitation (MeDIP) assays were performed with the MeDIP kit (Active Motif, Carlsbad, USA) following the manufacturer's instructions, as described previously.9 Briefly, gDNA was extracted from heart tissues. Each sample of gDNA (20 µg) was sonicated in Bioruptor Pico (Diagenode, New Jersey, USA.) to generate DNA fragments ranging in size from 200 to 600 base pairs, which was confirmed by 2% agarose gel electrophoresis. Fragmented DNA (1 μg) of each sample was subjected to MeDIP assay. An additional quantity of fragmented DNA equivalent to 10% of the DNA being used in the IP reaction served as input DNA. The double strand DNA fragments were denatured at 95°C to produce single strand DNA, and a 5-methylcytosine (5-mC) antibody was then used to precipitate DNA containing 5-mC. The 5-mC enriched DNA and input DNA were purified with phenol/chloroform extraction under the same condition, and subjected to quantitative real-time PCR analysis. Sequences of primers flanking the promoter regions of Hand1, HK2, Lcn2, and Ddit4 genes are listed in Supplementary material online, Table S2. β-actin promoter specific primers were used as internal control. PCR was performed in triplicate in a CFX connect real time PCR machine, and the relative 5-mC abundance in each promoter of interest was calculated by 2−ΔΔCT method, and expressed as percent of normoxic control.
2.9 Statistical analysis
Circos16 was used to generate the graphs integrating DEGs at the transcriptomic level vs. DMRs at the methylome level. For DEGs analysis, FDR <0.3 and FC >1.2 were used as a threshold to select DEGs. For DMR analysis, FDR <0.05 and methylation change ≥10% were used as a threshold to declare statistical significance. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) to identify canonical pathways. Principle component analysis (PCA) was performed using Partek Genomics Suite software. For non-bioinformatic data analysis, data were presented as mean ± SEM, and were analysed using Student’s t-test or one-way ANOVA followed by Turkey’s post hoc test. P-value <0.05 was considered statistically significant. Experimental number (n) represents hearts of animals from different dams.
3. Results
3.1 Study design and the overall genomic data summary
To study the rat heart epigenome and transcriptome changes during the heart development in response to hypoxic stress, we constructed 18 RRBS methylation libraries derived from 18 SD rat hearts and 26 RNA-seq libraries derived from 26 SD rat hearts in two development stages (foetus and adult). Pregnant rats were exposed to either normoxia (control group) or hypoxia (10.5% O2) for 6 days during gestational Days 15–21, as reported previously.9,10Supplementary material online, Figure S1 shows the study design in details. The foetal hearts were collected at the Day 21 of gestation, and the adult offspring hearts were collected at 5 months old after birth. DNA and RNA were extracted from the hearts, and both RRBS DNA-seq and RNA-seq libraries were constructed. To monitor the quality of RNA-seq, we also added External RNA Control Consortium (ERCC) spike-in controls in an amount equivalent to about 1% of the mRNA in each sample before RNA-seq library construction. We generated approximately 449.1 million reads of 75 bp single-end RRBS DNA methylome data (Supplementary material online, Table S3) and 654.6 million reads (75 bp × 2) of pair-end RNA-seq transcriptomic data (Supplementary material online, Table S4), corresponding to an average of 25.0 million sequence reads per RRBS sample, and 25.2 million sequence reads per RNA-seq sample.
3.2 Overall genomic data mapping and QC
3.2.1 RRBS DNA-seq methylome data
We aligned the RRBS DNA-seq methylome data, 75 bp single reads, to the NCBI rat Rnor 6.0 genome. On average, 68.8% of the reads were aligned to the genome with an average of 17.2 million aligned reads per sample (Supplementary material online, Table S3). The aligned reads were further annotated, resulting in an average of 1 041 827 CpG sites (CpGs) with at least 10 reads covered, and an average of 221 210 CpG regions with at least 20 reads covered. The overlapping CpGs among six samples in the foetal group and 12 samples in the adult male group are 590 422 and 454 882, respectively, resulting in 43.4% and 56.3% CpGs reduction. Meanwhile, the overlapping CpG regions in the foetal samples and in the adult samples are 196 513 and 176 402, respectively, resulting in 11.2% and 20.3% reduction. Therefore, we mainly focused on the DMRs analysis instead of the differential methylation CpG sites (DMCs) analysis because of the lower reduction rates in CpG regions.
3.2.2. RNA-seq transcriptome data
We aligned the fastq reads to the NCBI rat Rnor 6.0 genome, NCBI Rnor 6.0 gene model, and ERCC transcripts. Overall, in average, 92.9% of the reads were aligned to the genome, 29.9% to the exons, 16.6% to the intron, and 0.32% to the ERCC transcripts (Supplementary material onlineTables S4 and S5). Scatterplots of ERCC log2 [fragments per kilobase of transcript per million mapped reads (FPKM)] vs. log2 (spike-in concentration) showed an overall linear relationship between observed transcripts and the true concentration of the ERCC spike-in controls (Supplementary material online, Figure S2). Overall, 13 116 (43.7%) of the 30 044 NCBI annotated genes were defined as expressed (FPKM ≥1) for all the 26 samples. More expressed genes were identified in the foetuses compared with the adult male and female samples.
3.3 Antenatal hypoxia exposure induced differential epigenomic reprogramming
3.3.1 Genome-wide CpG methylation in relation to genomic features
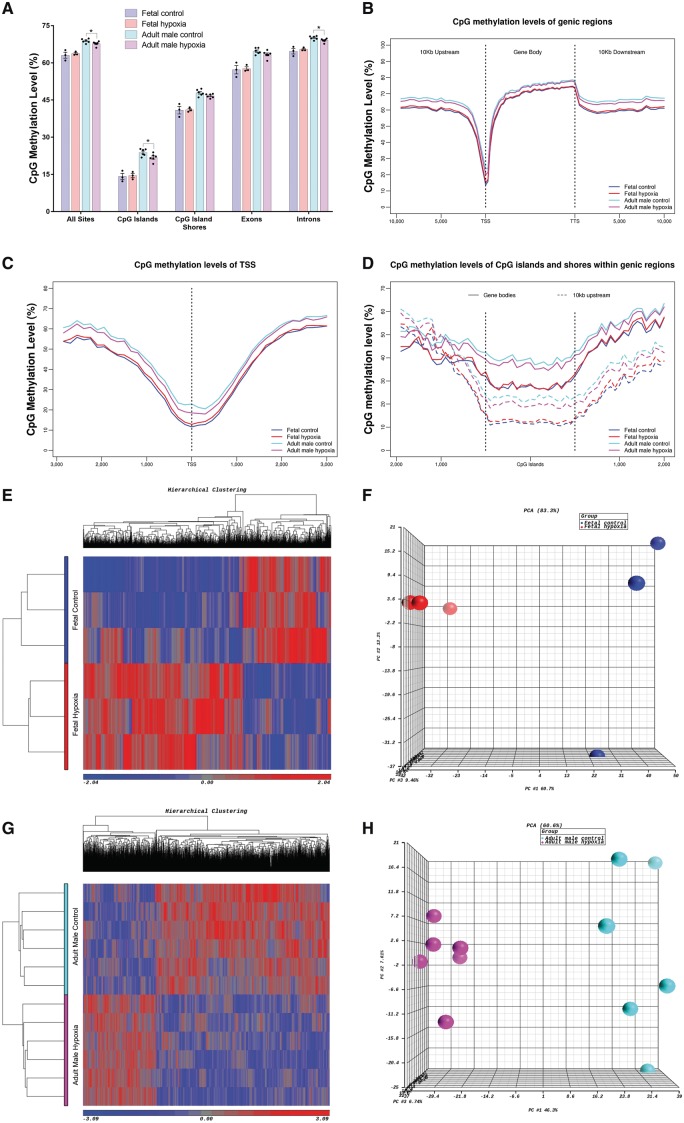
Overall, 1 041 827 CpGs, ranging from 725 525 to 1 242 160 (Supplementary material online, Table S3), were used to study the genome-wide CpG methylation patterns in terms of the relationship to genomic features. Profiling CpG methylation patterns (Figure 1A–D) revealed that the adult rat hearts are hypermethylated compared with foetal rat heart in both hypoxia and control groups. There were no significant differences on the CpG methylation level in foetal hearts between hypoxia and controls groups within all five genomic features (Figure 1A). However, we observed slightly significant differences on the CpG methylation level in adult rat hearts between hypoxia and controls within following regions: all sites (P = 0.048), CpG islands (CGIs; P = 0.041), and introns (P = 0.033) (Figure 1A). Consistently, significantly lower CpG methylation levels were observed within CGIs compared with CpG island shores (CGS) in both foetal and adult groups, under both control and hypoxia conditions (P < 0.001, Figure 1A). Also, all samples displayed a similar dip in methylation centred at the transcription start site (TSS) (Figure 1B) and then gradually increased towards the 3’ end of gene bodies (Figure 1B and C). CpG methylation was more abundant in the adult heart than foetal heart, particularly within the TSS region (Figure 1A and C). In addition, CGIs and CGS in 10 kb upstream of the TSS were hypomethylated compared with CGIs and CGS within gene bodies (Figure 1D). Meanwhile, significantly higher levels of methylation were observed in introns compared to exons across all four groups (P < 1 × 10−5). However, to our surprise, the methylation patterns between the hypoxia and control groups showed opposite trends in foetal and adult rats (Figure 1A–D). Compared to the control hearts, the hypoxia-exposed foetal hearts were more hypermethylated. In contrast, the hearts of adult offspring that exposed to antenatal hypoxia were more hypomethylated compared to the control group.
Figure 1.
Genome-wide CpG sites methylation in relation to genomic features and hypoxia-induced DMRs. (A) Average CpG methylation levels for various genomic features. Data are expressed as mean ± SEM. Statistical significance was calculated by one-way ANOVA. *P < 0.05. CpG methylation levels in relation to (B) genic regions, (C) TSS, and (D) CGI and CGS within gene bodies and up to 10 kb upstream of TSS. Heatmap and PCA plot of DMRs between the control and hypoxia groups in (E, F) foetal rats and (G, H) adult male rats. n = 3 in foetal control/hypoxia group; n = 6 in adult control/hypoxia group.
3.3.2 Hypoxia-induced methylation profiling change
Next, we used logistic regression analysis (methylKit Bioconductor package) to identify the DMRs between the hypoxia and control groups in both foetal and adult male hearts, respectively. The DMRs are defined based on FDR <0.05 and the methylation differences between two groups >10%. In foetal hearts, we identified 2828 DMRs including 1824 hypermethylated DMRs and 1004 hypomethylated DMRs. In adult male hearts, we identified 2193 DMRs including 647 hypermethylated DMRs and 1546 hypomethylated DMRs. Our differential methylation analysis showed that more hypermethylated but less hypomethylated DMRs were identified in foetal rat hearts compared with the DMRs in adult male rat hearts, which is consistent with the findings based on the above genome-wide CpG methylation pattern analysis. PCA and hierarchical clustering also showed a clear separation between the hypoxia and control groups based on the DMRs in foetal and adult male rat hearts (Figure 1E–H). Furthermore, we found 218 DMRs in foetal hearts, and 217 DMRs in adult male hearts are partially or fully overlapped, representing a total of 173 genes. However, only 101 DMRs (representing 72 genes) in foetal rat hearts and 102 DMRs in the adult male rat hearts displayed a similar methylation trend or pattern. The rest of DMRs (representing 101 genes) showed the opposite methylation trend or pattern between the foetal and adult hearts.
3.4 Antenatal hypoxia exposure induced differential gene expression
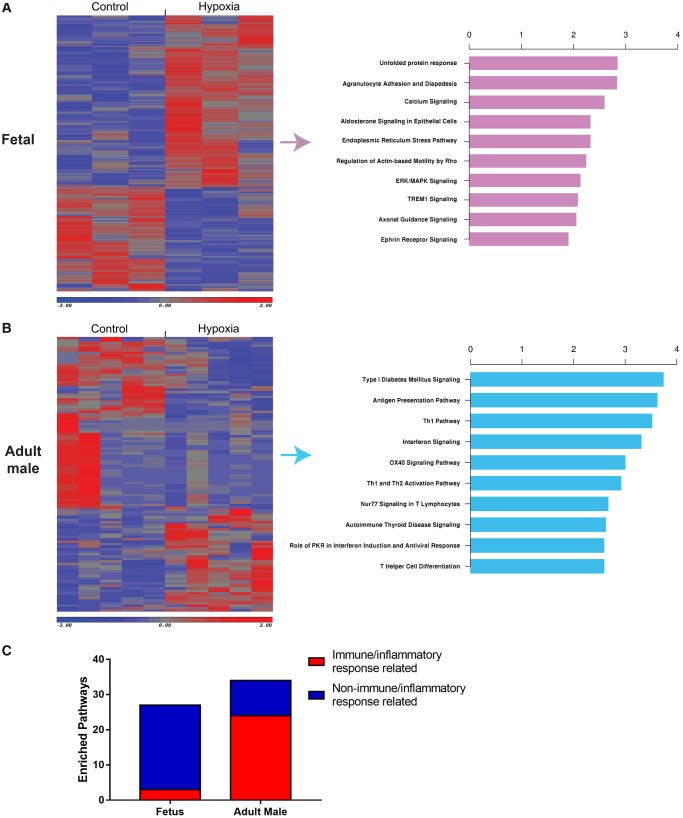
We used cuffdiff pipeline to identify DEGs, defined as FDR <0.3 and FC >1.2, between hypoxia and controls in foetal, and adult male rat hearts. Totally, 323 DEGs were identified in foetal hearts due to the hypoxia exposure. However, only 112 DEGs were identified in adult male rats (Figure 2A, Supplementary material online, Table S6). More up-regulated DEGs were identified in foetal (62.2%, 201 out of 323), comparing to adult male rats (37.5%, 42 out of 112). Based on the DEGs found in foetal and adult rat hearts, we performed pathway analysis (IPA, QIAGEN). Figure 2A and B showed the top10 enriched pathways from DEGs of foetal and adult male heart, separately. In foetal heart exposed to antenatal hypoxia, developmental/stress response related pathways were enriched. However, immune/inflammatory response related pathways (24 out of 34) were highly enriched in adult male offspring heart, compared with foetal heart (3 out of 27) (Figure 2C). Supplementary material online, Data S1 contains a list of all pathways that were significantly enriched (P < 0.05).
Figure 2.
Hypoxia-induced DEGs. (A, B) Hypoxia-induced DEGs in foetal, adult male rats, and top 10 canonical pathways. (C) Number of immune/inflammatory response related pathways over all enriched pathways in foetal and adult male heart exposed to antenatal hypoxia. n = 3 in foetal control/hypoxia group; n = 5 in adult control/hypoxia group.
3.5 Integrated analysis of hypoxia-induced RRBS/DNAm, mRNA-seq gene expression
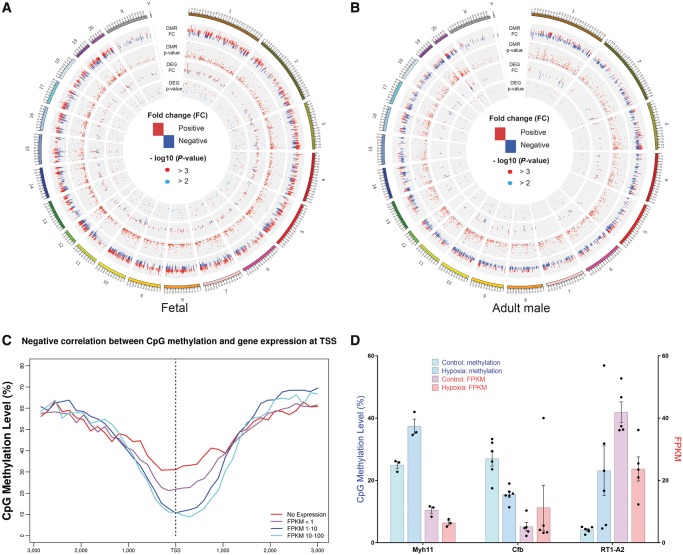
We use Circos plotting to illustrate the global relationship between DNA methylome (DMRs) and transcriptome (DEGs) in response to hypoxia in foetal (Figure 3A) and adult male (Figure 3B) hearts across all chromosomes. Overall, there are more hypermethylated DMRs across chromosomes in foetal hearts, compared with adult offspring heart. Interestingly, there are many hypomethylated DMRs in the starting region of chromosome 13 in foetal rats but no DMRs in that region in adult male rats (Supplementary material online, Table S7). In addition, there is a negative correlation between the TSS CpG methylation and the gene expression in the pooled data from both foetal and adult hearts (Figure 3C). We further investigated the correlation between methylation patterns of DMRs and regulation directions of DEGs for the 84 overlapping DMRs and DEGs. Supplementary material online, Data S2 shows the detailed DNA methylation status and the corresponding gene expression for the 84 overlapping DMRs and DEGs. Among these overlapping genes, there are three genes (Myh11, Cfb, and RT1-A2) where the DMRs are located in promoter regions and 11 genes (Myh11, Atp1a3, Grip2, Pygl, Cntnap1, Myl4, Ephb3, Ftcd, Nfkbia, Cfb, and RT1-A2) where DMRs are located in exon regions. Three example genes (Myh11, Cfb, and RT1-A2) with DMRs located in promoter regions showed a strong negative correlation between gene expression (FPKM) and methylation rate (beta value) of CpG regions under control and hypoxic conditions (Figure 3D). In addition, 9 out of 11 genes with DMRs located in exon regions also showed a significant negative correlation of methylation and gene expression. However, only 36 out of 73 genes displayed a negative correlation when the DMRs are not located in promoter or exon regions.
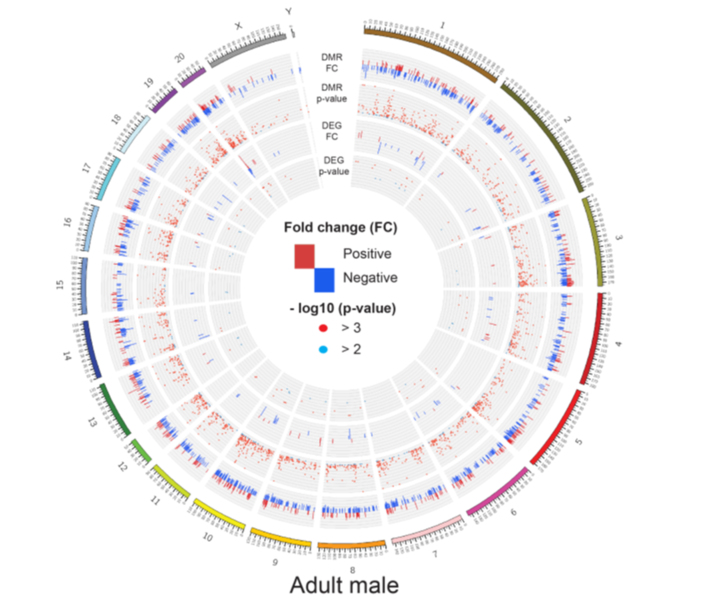
Figure 3.
Integration of DMRs and DEGs. (A, B) Circos plot of DMRs and DEGs in foetal and adult male rats. Positive fold-changes in DMRs represent hypermethylation in hypoxia group while negative fold-changes in DMRs represent hypermethylation in control group. Positive fold-changes in DEGs represent up-regulation in hypoxia group while negative fold-changes in DEGs represent up-regulation in control group. Red dots represent P-value <0.001 while blue dots represent 0.001 ≤ P-value <0.01 (n = 3–6). (C) Negative correlation between CpG methylation and gene expression at TSS. (D) Three example genes (Myh11, Cfb, and RT1-A2) with DMRs located in promoter regions show a negative correlation between gene expression (FPKM) and methylation rate (beta value) of CpG regions under control and hypoxia environment. Data are expressed as mean ± SEM. n = 3–6.
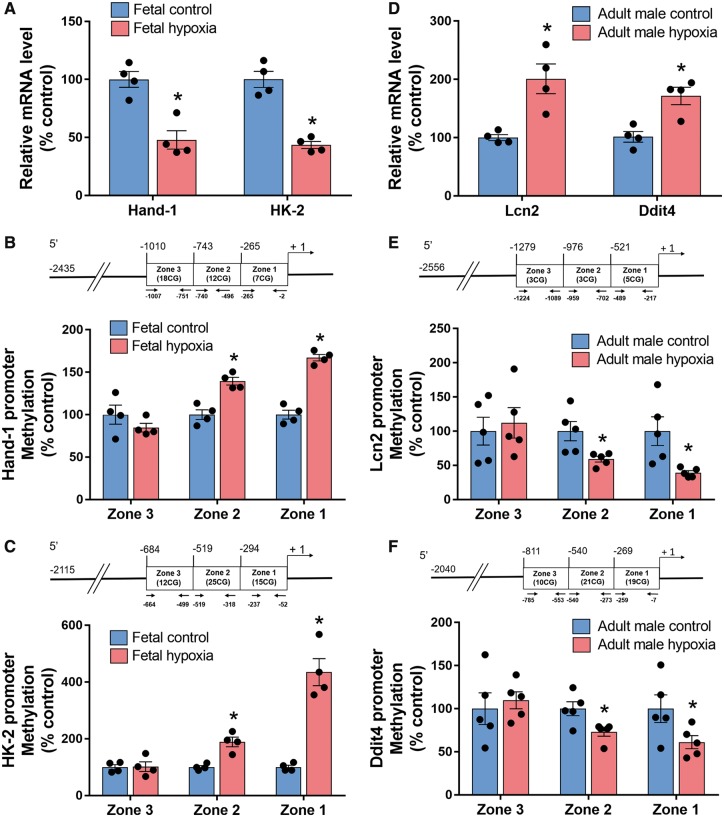
We validated expressional changes in two of the top down-regulated genes in the foetal heart and two of the top up-regulated genes in the adult heart with qPCR. Consistent with the RNAseq data, we found that hypoxia significantly decreased mRNA abundance of Hand1 that encodes heart- and neural crest derivatives-expressed protein 1 (HAND1), and Hk2 that encodes hexokinase 2, in the foetal heart (Figure 4A). In addition, we demonstrated that promoter methylation of both Hand1 and Hk2 genes were increased by hypoxia (Figure 4B and C). In the adult heart, we demonstrated that antenatal hypoxia resulted in a decrease in promoter methylation and an increase in gene expression of Lcn2 that encodes lipocalin-2 (Lcn2), also known as neutrophil gelatinase-associated lipocalin (NGAL), and Ddit4 that encodes DNA-damage-inducible transcript 4 (Figure 4D and F).
Figure 4.
Validation of gene expression and DNA methylation by real-time PCR and MeDIP. (A) mRNA abundance of Hand-1 and HK-2 genes between hypoxia and control groups in foetal heart. (B and C) DNA methylation in the promoter region of Hand-1 (B) and HK-2 (C) genes was detected by MeDIP in the foetal heart. (D) mRNA abundance of Lcn2 and Ddit4 genes between hypoxia and control groups in adult male heart. (E, F) DNA methylation in the promoter region of Lcn2 (E) and Ddit4 (F) genes was detected by MeDIP in the adult male heart. Data are expressed as mean ± SEM. Statistical significance was calculated by Student’s t-test. *P < 0.05 vs. control group. n = 4–5.
3.6 Developmental stage-dictated differential genes and pathways participating in hypoxia-induced epigenomic and transcriptomic responses
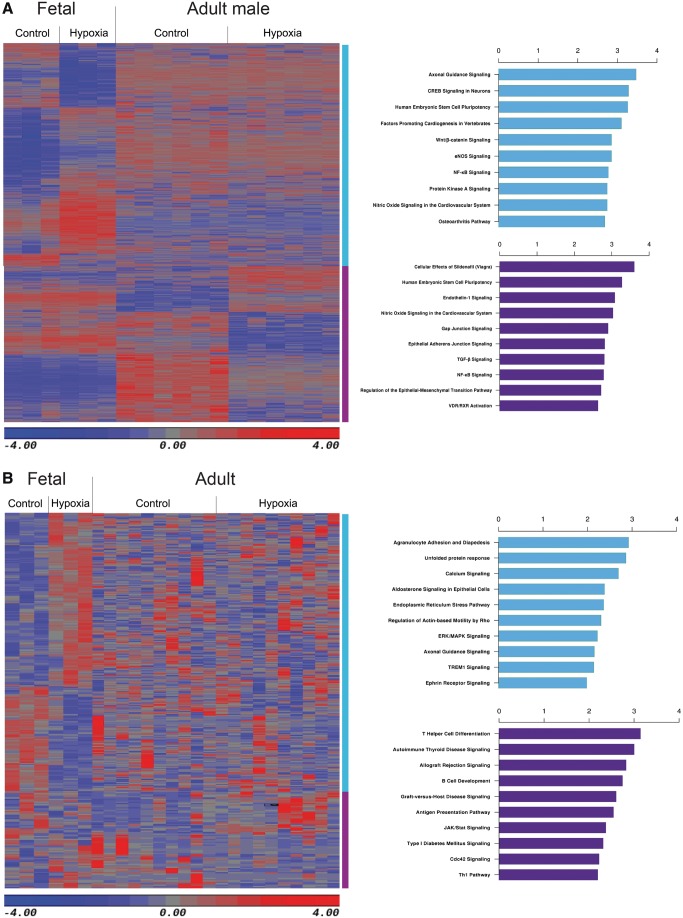
Interestingly, we found that there are less than 10% of overlapping DMRs between foetal and adult male rat hearts. Except for the overlapping DMRs, we generated new heat map combining all the DNA regions from the DMRs of both foetal and adult male heart. Pathway analysis was performed on a total of 2610 DMRs derived from the foetal hearts and 2076 DMRs derived from the adult male rat hearts, respectively. Figure 5A shows the heatmap of exclusive DMRs and the 10 most enriched pathways in foetal and adult male rat hearts, respectively. We also performed a similar analysis on the exclusive 315 DEGs derived from foetal hearts, and 71 DEGs derived from adult rat hearts. Figure 5B shows the heatmap of exclusive DEGs and the 10 most enriched pathways, derived from foetal hearts and adult rat hearts, respectively.
Figure 5.
Development-dependent DMRs and DEGs. (A) Heatmap and top 10 canonical pathways of the exclusive DMRs between hypoxia and control group in foetal and adult male rats. (B) Heatmap and top 10 canonical pathways of the exclusive DEGs between hypoxia and control group in foetal and adult rats. Blue and purple bars represent exclusive DMRs or DEGs in foetal and adult rats respectively. n = 3–6.
3.7 Sex-dependent hypoxia exposure caused differential gene expression
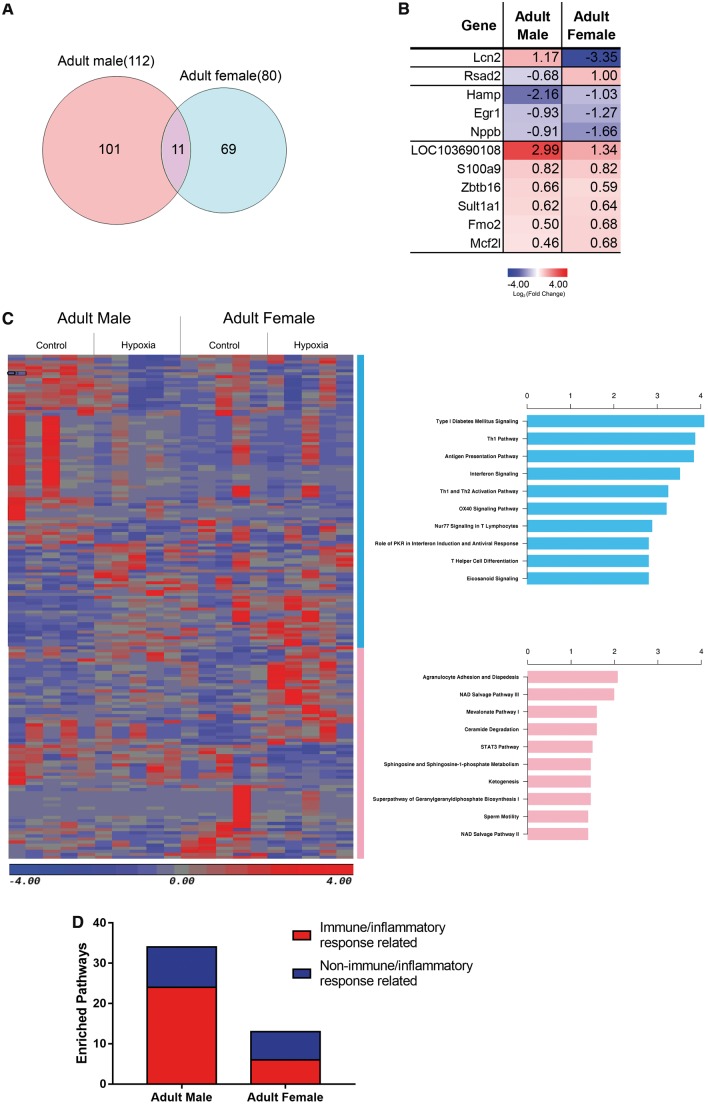
Moreover, we identified 112 DEGs in adult male offspring and 80 DEGs in female offspring rat heart after the antenatal hypoxia exposure. Venn diagram noticed only 11 overlapping DEGs between the adult male and female (Figure 6A and B). Among the 11 genes, the majority of them displayed a concordant regulation direction (9 out of 11, 81.8%). Notably, Lcn2 was up-regulated in male and dramatically down-regulated in female offspring heart (Figure 6B). Pathway analysis was also performed based on the exclusive DEGs of the male and female heart, and the most enriched pathways were listed, respectively (Figure 6C). To our surprise, and immune/inflammation response related pathways were highly enriched in offspring heart of adult male, compared with adult female (Figure 6D).
Figure 6.
Sex-specific DEGs. (A) Venn diagram of the hypoxia-induced DEGs in adult male and female heart. (B) Average gene expression of antenatal hypoxia exposed group compared with control group in male and female rat heart over the 11 overlapped genes. (C) Heatmap and top 10 canonical pathways of the exclusive DEGs between hypoxia and control group in adult male and female rats. (D) Number of immune/inflammatory response related pathways over all enriched pathways in adult male and female heart exposed to antenatal hypoxia. n = 5 for each group.
4. Discussion
Hypoxia is the most important and clinically relevant stress to the foetal development during gestation. Foetal hypoxia has profound adverse effects on developmental plasticity. Both animal and human studies indicate that antenatal hypoxia results in developmental programming of phenotypic changes that predispose offspring to various physiological dysfunctions and diseases, including cardiac dysfunction and ischaemic heart disease, hypertension, metabolic disease, and other conditions.7
In the present study, we for the first time reveal that antenatal hypoxic stress alters the pattern of global DNA methylation, not only in the foetal heart but also in the adult offspring heart, demonstrating epigenetic programming of heart development in response to foetal stress. In general, both foetal and adult hearts showed similar global DNA methylation features with hypomethylation in the CGIs and a sharp dip in the CpG methylation centred at the TSS. This was consistent with the previous report that indicates low DNA methylation exists within TSS region in newborn and adult cardiomyocytes.17 Notably, our data also demonstrate that antenatal hypoxia differentially affects DNA methylation in foetal and adult offspring hearts exposed to antenatal hypoxia. Compared to the normoxia control, antenatal hypoxia exposure induced a slight DNA hypermethylation in the foetal heart, but a global hypomethylation in the adult heart. Moreover, these hypermethylated DMRs in foetal heart were not the same DMRs found hypomethylated in adult heart, which suggests that antenatal hypoxia epigenetically regulate a distinct set of genes in the foetal and adult heart. Growing evidence indicates that changes in DNA methylation profile strongly associated with heart diseases.18–20 Interestingly, similar methylation pattern, global promoter hypermethylation, has been reported in patients with congenital heart disease (e.g. tetralogy of Fallot).19 Meanwhile, DNA hypomethylation at gene promoters is observed in the heart of heart failure patient,18,20 which shows the consistent global epigenomic pattern in the adult rat heart exposed to antenatal hypoxia in the present study. Thus, our methylome data may partly explain why antenatal hypoxic stress increases the risk of cardiovascular disease in foetus and adult offspring. The reversal of the global DNA methylation pattern could be a promising strategy to reduce such risk. Indeed, our recent study indicates that treatment of newborn rat pups with DNA methylation inhibitor, 5-aza-2′-deoxycytidine, prevented the antenatal hypoxia-induced increase of vulnerability to heart ischaemia and reperfusion injury.9
In the present study, the finding of 60–70% global CpG methylation genome-wide in the heart is consistent with the notion that 60–80% CpGs in mammalian genomes are generally methylated.21 The levels of DNA methylation were further evaluated in different genomic contexts, e.g. TSS, gene bodies. Of importance, we found that CpG methylation levels were the lowest at CGIs and gradually increased in CGS, exons and introns. CGIs are prevalent at TSSs and over 70% CGIs exist around the TSS. Indeed, our analysis of DNA methylation levels of genic regions demonstrated a sharp dip of CpG methylation at the TSS. This is consistent with previous findings that CGIs found at gene promoters are generally hypomethylated.21 Hypomethylation of TSS regions is of critical importance and provides a mechanism of rigorous and robust methylation regulation of gene expression. In the present study, genome-wide analysis of gene transcription and DNA methylation demonstrated a negative correlation between CpG methylation within the TSS region and gene expression in the heart, supporting the regulatory principle on the global perspective that CGIs methylation at the TSS is negatively related to gene transcription.22 Of interest, we found that genome-wide CpG methylation was more abundant in the adult heart than foetal heart, particularly within the TSS region. This finding is consistent with previous studies in mice showing gain of mCpG in the heart during postnatal development.17,23 The state of hypomethylation around the TSS depends on active and continuous exclusion of DNA methyltransferases. It has been established that active transcription factor binding strongly influences the unmethylated state of promoter CGIs, which can progressively accrue heritable methylation if they are depleted of transcription factor binding.21 The finding of increased methylation within the TSS region in the adult heart suggests a developmental repression of gene transcription (e.g. foetal genes) from the foetal heart to the terminally differentiated heart in the adult. In response to hypoxia, the foetal heart underwent hypermethylation. These findings are consistent with our previous studies measuring global DNA methylation using 5-mC DNA ELISA, which showed a development-dependent increase of global DNA methylation in the heart, as well as hypoxia-mediated increase of global methylation in the foetal heart.9 In addition, our previous studies of single gene approach also demonstrated that foetal hypoxia increased promoter methylation and decreased gene expression of PKCε and GR in the heart.9,10,24 To further validate the negative correlation of DNA methylation and gene expression, we performed qPCR to measure expressional changes in two of the top down-regulated genes found in the RNAseq data in the foetal heart. Consistent with the RNAseq data, we found that foetal hypoxia significantly decreased mRNA abundance of Hand1 that encodes heart- and neural crest derivatives-expressed protein 1 (HAND1), and Hk2 that encodes hexokinase 2, in the heart. Of importance, we demonstrated that promoter methylation of both Hand1 and Hk2 genes were increased by hypoxia. Both HAND1 and hexokinase 2 are important regulatory proteins in the heart. HAND1 controls the balance between proliferation and differentiation in the developing heart.25 Hexokinase 2 plays a crucial role to maintain mitochondrial membrane potential and functions as a molecular switch from glycolysis to autophagy to ensure cellular energy homeostasis under stress.26,27 In contrast to the foetal heart, the adult offspring heart displaced a decrease in global DNA methylation around the TSS, resulting from antenatal hypoxia. This is intriguing and suggests that antenatal stress of hypoxia impacts epigenetic programming in postnatal development and retards the development-mediated increase in DNA methylation in the heart with possible retaining of certain ‘foetal gene expression programme’ in the adult heart. We validated two of the top up-regulated genes found in RNAseq in the adult heart with qPCR and measuring promoter methylation. We demonstrated that antenatal hypoxia resulted in a decrease in promoter methylation and an increase in gene expression of Lcn2 that encodes Lcn2, also known as NGAL, and Ddit4 that encodes DNA-damage-inducible transcript 4.
The hypoxia-induced methylation/expression changes in the heart are likely to have a significant impact on heart development and disease vulnerability. Indeed, we demonstrated previously in the same animal model that foetal hypoxia caused a premature exit of the cell cycle of cardiomyocytes, decreased cardiomyocytes proliferation, and resulted in fewer but larger myocytes in the adult heart.28–32 In the adult heart, it has been demonstrated that antenatal hypoxia leads to pathological cardiac remodelling with left ventricular hypertrophy and an increase in heart vulnerability to ischaemia and reperfusion injury.9,10,24,33–36 In addition, foetal hypoxia had a negative impact on mitochondrial function by altering mitochondria associated proteins levels and reducing cytochrome c oxidase activity in the foetal heart, which persisted into the adult heart.37–39 This caused a mismatch between myocardial glycolysis and glucose oxidation rates, as well as an increase in myocardial production of acetyl-CoA during reperfusion after ischaemia, and this mismatch between energy production and cardiac performance contributed to the increased heart vulnerability to ischaemia and reperfusion injury in the adult heart resulting from antenatal hypoxia.36,40
The finding that about 21 Mb segment of chromosome 13 undergoing DNA hypomethylation in the foetal heart, but not in the adult heart is intriguing. This part of chromosome 13 plays an important role in blood pressure regulation and cardiomyocyte survival.41,42 Introgression of chromosome 13 from Brown Norway rats to Dahl salt-sensitive rats reduced detrimental hypertension caused by high salt diet feeding.42 Further study showed that specific genomic region located in the p-end of chromosome 13 (around 10 Mb) related to the blood pressure regulation.41 We found that this region is overlapped with the 21 Mb segment that only been demethylated in foetal heart. Of interest, PH domain leucine-rich repeat protein phosphatase-1(PHLPP-1), a gene located within this region of chromosome 13, is closely associated with the survival of cardiomyocytes. Knockdown of PHLPP-1 increased the phosphorylation of Akt in mitochondria, thereby improved the function of mitochondria in neonatal cardiomyocytes.43 In addition, PHLPP-1 knockout heart reduced the infarction caused by the ischaemia/reperfusion.43 Furthermore, PHLPP-1 deletion attenuated the pathological hypertrophy of heart.44 The hypomethylation of PHLPP-1 in chromosome 13 in the foetal heart may suggest that the up-regulation of PHLPP-1 expression after antenatal hypoxia and increase the heart sensitivity to ischaemia/reperfusion injury later in life. The different methylation and expression patterns between foetal and adult hearts imply that the antenatal hypoxia-induced programming of certain genes is settled in the early developmental stage.
Furthermore, we found that antenatal hypoxia caused a distinct gene expression pattern in foetal and adult offspring hearts. During normal heart development, dramatic metabolic and functional changes occur in cardiomyocytes before birth, adapting to the environmental cues.45 Pathway analyses of DEGs indicate that multiple oxygen stress-adaptive pathways are motivated in the foetal heart suffered hypoxia, such as endoplasmic reticulum (ER) stress pathway. The ER stress itself is strongly correlated with the onset of heart disease. For example, ER stress leads to degeneration and cell death of cardiomyocytes, contributing to the development of ischaemic heart disease.46,47 On the other hand, ER stress and coping response (e.g. unfolded protein response) could be an adapt strategy of the foetal heart to mitigate or eliminate the hypoxia stress.48 Thus, it is possible that the foetal heart initiates an adaptive mechanism to balance the detrimental impact of antenatal hypoxia. However, these adaptive processes may not compensate the damage and disrupt the regular gene programming, leading to the dysfunction of heart after birth. Of importance, a striking finding in the offspring heart is the significant enrichment of immune response/pro-inflammatory related signal pathways (e.g. interferon signalling, antigen presentation pathway). Inflammatory biomarkers, like C-reactive protein and interleukin-6, have been confirmed as risk factors for cardiovascular disease by abundant prospective cohort studies.49 Furthermore, inflammation interplays with ER stress and both are underlying drivers for cardiovascular diseases.46,50 ER stress activates several pro-inflammation cytokine pathways by regulators such as Jun amino-terminal kinases, nuclear transcription factor-κB.51–53 It is possible that activity of pro-inflammatory genes found in the adult offspring heart is the consequence of the maladaptive coping response to oxygen stress in the foetal heart. Notably, our data indicate that the antenatal hypoxia has a long-term effect on developmental plasticity, causing offspring hearts in a pre-inflammatory state and increasing the vulnerability of adult heart disease.
Moreover, an apparent sex dichotomy of gene expression in adult offspring heart is observed in our study. Several immune response/inflammation-related gene pathways (e.g. antigen presentation, Th1, and Th2 activation pathways) were enriched in the male heart; in contrast, multiple gene pathways related to energy restoration/protection were mobilized in the female heart, such as NAD salvage pathways. The previous study showed that the NAD salvage pathway played an essential role in post-hypoxic bioenergetic recovery and cardiomyocyte protection in ischaemic heart disease.54 Among the 11 overlapped genes between adult male and female offspring hearts exposed to antenatal hypoxia, it is worth to note that Lcn2 was found up-regulated in male and down-regulated in female. Interestingly, the Lcn2 is a critical regulator of innate immune response55 and a strong predictor of cardiovascular disease.56,57 Lcn2 expression in cardiomyocytes significantly increased in cardiac infarction and serum Lcn2 levels correlated with the clinical deterioration of heart failure patients.56 The up-regulation of Lcn2 induced cardiomyocyte apoptosis mediated via the mitochondrial pathway and intracellular iron overload.58 A recent study also indicated that Lcn2 contributed to cardiac pathologic hypertrophy.59 The differential expression of Lcn2 between male and female hearts provides a mechanistic understanding of why female offspring is more resistant to cardiac ischaemia/reperfusion injury resulting from foetal stress.10,59,60 In line with our finding, a prospective clinical study confirmed that the baseline serum Lcn2 level is significantly higher in the male subjects who developed cardiovascular disease than those did not.61 However, this difference was not observed in the female subjects.61 Our finding suggests that male and female hearts initiate distinct molecular mechanisms after antenatal hypoxia. Male hearts undergo an extended pro-inflammatory status, and female hearts launch a long-lasting adaptive mechanism to balance hypoxic impact, and these differences on gene expression may explain the gender difference in the occurrence and severity of cardiovascular disease observed in our previous and other studies.10,60,62
Overall, the present study provides novel insights into the alteration of global DNA methylation and transcriptional pattern of foetal and adult offspring heart that exposed to antenatal hypoxia. The hypoxia stress not only induces a global epigenomic and transcriptomic reprogram in the foetal heart, more importantly, also has a long-lasting effect on the adult offspring heart, and this effect is developmental stage and gender dependent. Moreover, our pathway analyses indicate that pathways related to inflammation are significantly enriched in the hearts after antenatal hypoxic exposure, especially in the adult male offspring. Thus, our study provides an initial framework and new insights into foetal hypoxia-mediated epigenetic programming of pro-inflammatory phenotype in the heart development, linking antenatal stress, and developmental programming of heart vulnerability to disease later in life. Whereas it is arguable that the present study is a descriptive study, to demonstrate a true cause-and-effect of foetal hypoxia-induced methylation and expression changes in adult hearts directly impacting disease vulnerability is extremely challenging. With the recent development of CRISPR/Cas9 genome editing, it may become possible to knock-out and knock-in genes in order to start to test the causal effect of foetal hypoxia-induced gene expression changes impacting disease vulnerability in the adult heart. Although this is clearly beyond the scope of the present study, an unbiased assessment of methylation and transcriptional activity across the genome and the findings of foetal hypoxia-induced changes in gene expression in the heart at different developmental stage and sex in the present study provide a basis and a very useful reference set of genes for future investigations.
Supplementary Material
Acknowledgements
The authors would like to thank Mandy Liu, Yonghong Zhang, and Seta Stanbouly for their technical assistance.
Conflict of interest: none declared.
Funding
This work was partially supported by the National Institutes of Health (NIH) [HL118861 to L.Z.; S10OD019960 to C.W.], Ardmore Institute of Health [2150141 to C.W.], and Charles A. Sims' gift.
Time for primary review: 10 days
References
- 1.Writing Group Members, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB; American Heart Association Statistics Committee; Stroke Statistics Subcommittee. Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation 2016;133:e38–e360. [DOI] [PubMed] [Google Scholar]
- 2. McMillen IC, Robinson JS.. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 2005;85:571–633. [DOI] [PubMed] [Google Scholar]
- 3. Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS.. Fetal nutrition and cardiovascular disease in adult life. Lancet 1993;341:938–941. [DOI] [PubMed] [Google Scholar]
- 4. Barker DJ, Osmond C.. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1986;1:1077–1081. [DOI] [PubMed] [Google Scholar]
- 5. Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE.. Developmental plasticity and human health. Nature 2004;430:419–421. [DOI] [PubMed] [Google Scholar]
- 6. Fajersztajn L, Veras MM.. Hypoxia: from Placental Development to Fetal Programming. Birth Defects Res 2017;109:1377–1385. [DOI] [PubMed] [Google Scholar]
- 7. Ducsay CA, Goyal R, Pearce WJ, Wilson S, Hu XQ, Zhang L.. Gestational hypoxia and developmental plasticity. Physiol Rev 2018;98:1241–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hauton D, Al-Shammari A, Gaffney EA, Egginton S.. Maternal hypoxia decreases capillary supply and increases metabolic inefficiency leading to divergence in myocardial oxygen supply and demand. PLoS One 2015;10:e0127424.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiong F, Lin T, Song M, Ma Q, Martinez SR, Lv J, MataGreenwood E, Xiao D, Xu Z, Zhang L.. Antenatal hypoxia induces epigenetic repression of glucocorticoid receptor and promotes ischemic-sensitive phenotype in the developing heart. J Mol Cell Cardiol 2016;91:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patterson AJ, Chen M, Xue Q, Xiao D, Zhang L.. Chronic prenatal hypoxia induces epigenetic programming of PKC{epsilon} gene repression in rat hearts. Circ Res 2010;107:365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 2012;13:R87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stockwell PA, Chatterjee A, Rodger EJ, Morison IM.. DMAP: differential methylation analysis package for RRBS and WGBS data. Bioinformatics 2014;30:1814–1822. [DOI] [PubMed] [Google Scholar]
- 14. Trapnell C, Pachter L, Salzberg SL.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilsbach R, Preissl S, Gruning BA, Schnick T, Burger L, Benes V, Wurch A, Bonisch U, Gunther S, Backofen R, Fleischmann BK, Schubeler D, Hein L.. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun 2014;5:5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meder B, Haas J, Sedaghat-Hamedani F, Kayvanpour E, Frese K, Lai A, Nietsch R, Scheiner C, Mester S, Bordalo DM, Amr A, Dietrich C, Pils D, Siede D, Hund H, Bauer A, Holzer DB, Ruhparwar A, Mueller-Hennessen M, Weichenhan D, Plass C, Weis T, Backs J, Wuerstle M, Keller A, Katus HA, Posch AE.. Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation 2017;136:1528–1544. [DOI] [PubMed] [Google Scholar]
- 19. Grunert M, Dorn C, Cui H, Dunkel I, Schulz K, Schoenhals S, Sun W, Berger F, Chen W, Sperling SR.. Comparative DNA methylation and gene expression analysis identifies novel genes for structural congenital heart diseases. Cardiovasc Res 2016;112:464–477. [DOI] [PubMed] [Google Scholar]
- 20. Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett MR, Foo RS.. Distinct epigenomic features in end-stage failing human hearts. Circulation 2011;124:2411–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith ZD, Meissner A.. DNA methylation: roles in mammalian development. Nat Rev Genet 2013;14:204–220. [DOI] [PubMed] [Google Scholar]
- 22. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–492. [DOI] [PubMed] [Google Scholar]
- 23. He Y, Hariharan M, Gorkin DU, Dickel DE, Luo C, Castanon RG, Nery JR, Lee AY, Williams BA, Trout D, Amrhein H, Fang R, Chen H, Li B, Visel A, Pennacchio L, Ren B, Ecker J, Spatiotemporal DNA methylome dynamics of the developing mammalian fetus. bioRxiv 2017; https://www.biorxiv.org/content/early/2017/07/21/166744.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patterson AJ, Xiao DL, Xiong FX, Dixon B, Zhang LB.. Hypoxia-derived oxidative stress mediates epigenetic repression of PKC epsilon gene in foetal rat hearts. Cardiovasc Res 2012;93:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Risebro CA, Smart N, Dupays L, Breckenridge R, Mohun TJ, Riley PR.. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 2006;133:4595–4606. [DOI] [PubMed] [Google Scholar]
- 26. Tan VP, Miyamoto S.. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy 2015;11:963–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pasdois P, Parker JE, Griffiths EJ, Halestrap AP.. Hexokinase II and reperfusion injury: TAT-HK2 peptide impairs vascular function in Langendorff-perfused rat hearts. Circ Res 2013;112:e3–e7. [DOI] [PubMed] [Google Scholar]
- 28. Bae S, Xiao YH, Li G, Casiano C, Zhang L.. Effect of maternal chronic hypoxic exposure during gestation on apoptosis in fetal rat heart. Am J Physiol Heart Circ Physiol 2003;285:H983–H990. [DOI] [PubMed] [Google Scholar]
- 29. Tong W, Xue Q, Li Y, Zhang L.. Maternal hypoxia alters matrix metalloproteinase expression patterns and causes cardiac remodeling in fetal and neonatal rats. Am J Physiol Heart Circ Physiol 2011;301:H2113–H2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tong W, Xiong F, Li Y, Zhang L.. Hypoxia inhibits cardiomyocyte proliferation in fetal rat hearts via upregulating TIMP-4. Am J Physiol Regul Integr Comp Physiol 2013;304:R613–R620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li G, Bae S, Zhang L.. Effect of prenatal hypoxia on heat stress-mediated cardioprotection in adult rat heart. Am J Physiol Heart Circ Physiol 2004;286:H1712–H1719. [DOI] [PubMed] [Google Scholar]
- 32. Zhang L. Prenatal hypoxia and cardiac programming. J Soc Gynecol Investig 2005;12:2–13. [DOI] [PubMed] [Google Scholar]
- 33. Li G, Xiao YH, Estrella JL, Ducsay CA, Gilbert RD, Zhang L.. Effect of prenatally chronic hypoxia on heart susceptibility to ischemia/reperfusion injury in adult rat. J Soc Gynecol Investig 2003;10:265–274. [DOI] [PubMed] [Google Scholar]
- 34. Xue Q, Dasgupta C, Chen M, Zhang L.. Fetal hypoxia causes programming of AT2R and cardiac vulnerability to ischemic injury in rat offspring. Cardiovasc Res 2011;89:300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Y, Williams SJ, O’Brien D, Davidge ST.. Hypoxia or nutrient restriction during pregnancy in rats leads to progressive cardiac remodeling and impairs postischemic recovery in adult male offspring. FASEB J 2006;20:1251–1253. [DOI] [PubMed] [Google Scholar]
- 36. Rueda-Clausen CF, Morton JS, Lopaschuk GD, Davidge ST.. Long-term effects of intrauterine growth restriction on cardiac metabolism and susceptibility to ischaemia/reperfusion. Cardiovasc Res 2011;90:285–294. [DOI] [PubMed] [Google Scholar]
- 37. Xiao D-L, Ducsay CA, Zhang L.. Chronic hypoxia and developmental regulation of cytochrome c expression in rats. J Soc Gynecol Investig 2000;7:279–283. [PubMed] [Google Scholar]
- 38. Al Hasan YM, Pinkas GA, Thompson LP.. Prenatal hypoxia reduces mitochondrial protein levels and cytochrome c oxidase activity in offspring guinea pig hearts. Reprod Sci 2014;21:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al Hasan YM, Evans LC, Pinkas GA, Dabkowski ER, Stanley WC, Thompson LP.. Chronic hypoxia impairs cytochrome oxidase activity via oxidative stress in selected fetal guinea pig organs. Reprod Sci 2013;20:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shah A, Reyes LM, Morton JS, Fung D, Schneider J, Davidge ST.. Effect of resveratrol on metabolic and cardiovascular function in male and female adult offspring exposed to prenatal hypoxia and a high-fat diet. J Physiol 2016;594:1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW Jr.. Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics 2007;31:228–235. [DOI] [PubMed] [Google Scholar]
- 42. Cowley AW, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ.. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension 2001;37:456–461. [DOI] [PubMed] [Google Scholar]
- 43. Miyamoto S, Purcell NH, Smith JM, Gao T, Whittaker R, Huang K, Castillo R, Glembotski CC, Sussman MA, Newton AC, Brown JH.. PHLPP-1 negatively regulates Akt activity and survival in the heart. Circ Res 2010;107:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moc C, Taylor AE, Chesini GP, Zambrano CM, Barlow MS, Zhang X, Gustafsson AB, Purcell NH.. Physiological activation of Akt by PHLPP1 deletion protects against pathological hypertrophy. Cardiovasc Res 2015;105:160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martinez SR, Gay MS, Zhang L.. Epigenetic mechanisms in heart development and disease. Drug Discov Today 2015;20:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Groenendyk J, Agellon LB, Michalak M.. Coping with endoplasmic reticulum stress in the cardiovascular system. Annu Rev Physiol 2013;75:49–67. [DOI] [PubMed] [Google Scholar]
- 47. Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE.. Activation of endoplasmic reticulum stress response during the development of ischemic heart disease. Am J Physiol Heart Circ Physiol 2006;291:H1411–H1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC.. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res 2006;99:275.. [DOI] [PubMed] [Google Scholar]
- 49. Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH.. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med 1997;336:973–979. [DOI] [PubMed] [Google Scholar]
- 50. Zhang C, Syed TW, Liu R, Yu J.. Role of endoplasmic reticulum stress, autophagy, and inflammation in cardiovascular disease. Front Cardiovasc Med 2017;4 https://www.frontiersin.org/articles/10.3389/fcvm.2017.00029/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH.. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1 alpha-mediated NF-kappa B activation and down-regulation of TRAF2 expression. Mol Cell Biol 2006;26:3071–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaneko M, Niinuma Y, Nomura Y.. Activation signal of nuclear factor-kappa B in response to endoplasmic reticulum stress is transduced via IRE1 and tumor necrosis factor receptor-associated factor 2. Biol Pharm Bull 2003;26:931–935. [DOI] [PubMed] [Google Scholar]
- 53. Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D.. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000;287:664–666. [DOI] [PubMed] [Google Scholar]
- 54. Gero D, Szabo C.. Salvage of nicotinamide adenine dinucleotide plays a critical role in the bioenergetic recovery of post-hypoxic cardiomyocytes. Br J Pharmacol 2015;172:4817–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A.. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004;432:917–921. [DOI] [PubMed] [Google Scholar]
- 56. Yndestad A, Landro L, Ueland T, Dahl CP, Flo TH, Vinge LE, Espevik T, Froland SS, Husberg C, Christensen G, Dickstein K, Kjekshus J, Oie E, Gullestad L, Aukrust P.. Increased systemic and myocardial expression of neutrophil gelatinase-associated lipocalin in clinical and experimental heart failure. Eur Heart J 2009;30:1229–1236. [DOI] [PubMed] [Google Scholar]
- 57. Hemdahl A-L, Gabrielsen A, Zhu C, Eriksson P, Hedin U, Kastrup J, Thorén P, Hansson GK.. Expression of neutrophil gelatinase-associated lipocalin in atherosclerosis and myocardial infarction. Arterioscler Thromb Vasc Biol 2006;26:136–142. [DOI] [PubMed] [Google Scholar]
- 58. Xu G, Ahn J, Chang S, Eguchi M, Ogier A, Han S, Park Y, Shim C, Jang Y, Yang B, Xu A, Wang Y, Sweeney G.. Lipocalin-2 induces cardiomyocyte apoptosis by increasing intracellular iron accumulation. J Biol Chem 2012;287:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marques FZ, Prestes PR, Byars SG, Ritchie SC, Wurtz P, Patel SK, Booth SA, Rana I, Minoda Y, Berzins SP, Curl CL, Bell JR, Wai B, Srivastava PM, Kangas AJ, Soininen P, Ruohonen S, Kahonen M, Lehtimaki T, Raitoharju E, Havulinna A, Perola M, Raitakari O, Salomaa V, Ala-Korpela M, Kettunen J, McGlynn M, Kelly J, Wlodek ME, Lewandowski PA, Delbridge LM, Burrell LM, Inouye M, Harrap SB, Charchar FJ.. Experimental and human evidence for lipocalin-2 (Neutrophil Gelatinase-Associated Lipocalin [NGAL]) in the development of cardiac hypertrophy and heart failure. J Am Heart Assoc 2017;6 https://www.ahajournals.org/doi/full/10.1161/JAHA.117.005971?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xue Q, Zhang L.. Prenatal hypoxia causes a sex-dependent increase in heart susceptibility to ischemia and reperfusion injury in adult male offspring: role of protein kinase C epsilon. J Pharmacol Exp Ther 2009;330:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu GY, Li HT, Fang QC, Jiang S, Zhang L, Zhang J, Hou XH, Lu JX, Bao YQ, Xu AM, Jia WP.. Elevated circulating lipocalin-2 levels independently predict incident cardiovascular events in men in a population-based cohort. Arterioscler Thromb Vasc Biol 2014;34:2457–2464. [DOI] [PubMed] [Google Scholar]
- 62. Elahi MM, Matata BM.. Gender differences in the expression of genes involved during cardiac development in offspring from dams on high fat diet. Mol Biol Rep 2014;41:7209–7216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.