Abstract
Emerging data indicate that interferon‐induced transmembrane protein 1 (IFITM1) plays an important role in many cancers. However, it remains unclear whether IFITM1 is functionally indispensable in nonsmall cell lung cancer (NSCLC). Here, using NSCLC cell lines and patient‐derived samples, we show that IFITM1 is essentially required for the progression of NSCLC in vitro and in vivo. Specifically, IFITM1 depletion resulted in a significant reduction in sphere formation, migration, and invasion of NSCLC cells in vitro; these events were inversely correlated with the ectopic expression of IFITM1. In addition, tumor development was significantly impaired in the absence of IFITM1 in vivo. Mechanistically, epidermal growth factor receptor/sex‐determining region Y‐box 2 (EGFR/SOX2) signaling axis was compromised in the absence of IFITM1, and the ectopic expression of SOX2 partially rescued the defects caused by IFITM1 depletion. More importantly, using 226 patient‐derived samples, we demonstrate that a high level of IFITM1 expression is associated with a poor overall survival (OS) rate in adenocarcinoma but not in squamous cell carcinoma. Collectively, these data suggest that IFITM1 is a poor prognostic marker of adenocarcinoma and an attractive target to develop novel therapeutics for NSCLC.
Keywords: NSCLC, IFITM1, adenocarcinoma, CSC, EMT, EGFR, SOX2
Short abstract
What's new?
Interferon response genes play key roles in pathogen defense but emerging evidence also link them with cancer. The authors report that interferon‐induced transmembrane protein 1 (IFITM1) critically regulates epidermal growth factor receptor‐mediated signaling in nonsmall lung cancer models and is associated with a poor prognosis of patients with adenocarcinoma. This expands the function of this innate defense factor and might lead to improved clinical management of individuals afflicted with lung cancer.
Introduction
Despite marked advancement in cancer therapy, lung cancer is the disease that is the most commonly diagnosed (1.82 million) and the leading cause of death in cancer patients (1.6 million deaths) in 2012.1 Lung cancer is classified into two categories, small cell lung cancer (SCLC) and nonsmall cell lung cancer (NSCLC); NSCLC accounts for approximately 85% of all lung cancer cases and the 5‐year survival rate of NSCLC patients is as low as about 15%.2 This highlights a need to understand the cellular and molecular events leading to the progression of this disease. Cancer relapse and metastasis are suggested to be the most important factors contributing to the progression and poor prognosis of cancer.3 Epithelial–mesenchymal transition (EMT) is a key event in tumor metastasis.4 EMT is a reversible process, during which epithelial cells exhibit some properties of mesenchymal cells (including proteolysis and motility) and lose many of their epithelial characteristics (such as cell–cell adhesion and cell polarity).5 Other events, including invasion, chemo‐resistance and the acquisition of stem cell‐like characteristics, are also known to be related to EMT.4 In recent studies, cancer stem cells (CSCs) or tumor initiating cells (TICs), a subpopulation of cells within a tumor with dysregulated self‐renewal properties of normal stem cells, are believed to be responsible for tumor initiation and progression as well as chemotherapy resistance.6 These events could explain rapid tumor relapse and a poor prognosis in cancer patients.7, 8 Several signaling pathways, such as Sonic Hedgehog,9 Wnt/β‐catenin,10 EGFR11 and SOX2,12 have been described to regulate the behavior of CSCs and EMT, and contribute to the development of several cancers, such as lung cancer.9
EGFR is a receptor tyrosine kinase (RTK) that is stimulated by its ligands such as EGF, amphiregulin, betacellulin, epiregulin, neuregulin, heparin‐binding EGF, and transforming growth factor alpha (TGF‐α).13 The receptors then dimerize and autophosphorylate, leading to the initiation of various downstream signaling pathways, mainly mitogen‐activated protein kinase (MAPK) and phosphoinositide 3‐kinase (PI3K)‐activated AKT pathways.14 EGFR is found to be overexpressed in various cancers, including head and neck cancer.15 In cancer cells, it has been demonstrated that EGFR can be aberrantly activated by several mechanisms, including EGFR gene amplifications, activating mutations in the extracellular or tyrosine kinase domains, and autocrine/paracrine signaling mechanisms.16, 17 High expression of EGFR is correlated with metastatic disease and a poor prognosis of many cancers.18, 19 Specifically, in lung cancer patients, EGFR overexpression is associated with reduced survival, high risk of lymph node metastasis, and poor chemo‐sensitivity.20, 21 Thus, it is required to better characterize essential molecules implicated in EGFR signaling.
Interferon‐induced transmembrane protein 1 (IFITM1), a member of the interferon (IFN)‐inducible transmembrane protein family, was initially identified as Leu 13.22 It is also known as 9–27 or CD225, and is a critical component of a membrane complex that is implicated in many essential functions such as proliferation, homotypic adhesion in lymphocytes, and metastasis.23, 24, 25 Originally, IFITM1 was identified for its role in host‐pathogen interaction after the induction by IFN‐α and IFN‐γ in response to pathogens.24 However, it has been recently shown that IFITM1 is highly expressed and functionally important in several cancers, including cervical, esophageal, ovarian, brain, and colon cancer.26, 27 Specifically, the importance of IFITM1 was reported in glioma cells and head and neck squamous cell carcinoma (HNSCC), where IFITM1‐depleted cancer cells displayed low levels of proliferation and invasion.27 Consistent with other studies, we reported that IFITM1 was associated with a poor prognosis of the colorectal cancer (CRC).28 However, it still remains unclear whether IFITM1 is essential for the maintenance of CSCs in lung cancer and may be further associated with a prognosis of NSCLC. Using lung cancer cell lines and patient‐derived samples, we addressed these questions and demonstrated that IFITM1 is essential in modulating EGFR/SOX2 signaling axis in lung cancer. Furthermore, IFITM1 is implicated in the progression of NSCLC in vivo and associated with a poor prognosis of adenocarcinoma.
Materials and Methods
Cell lines
Nonsmall Cell Lung Cancer cell lines A549, H1299 and H1650 cells were grown in RPMI‐1640 medium (Corning, USA) supplemented with 10% Fetal Bovine Serum (Corning, USA), 1% MEM essential amino acids (Corning, USA) and 1% penicillin/streptomycin (Gibco, USA) at 37 °C in a humidified atmosphere containing 5% CO2.
Lentiviral constructs and transfection
Short hairpin RNA (shRNA) constructs (870‐shIFITM1 and 642‐shIFITM1) and overexpression construct of IFITM1 were described previously.28 The open reading frame (ORF) of SOX2 (Forward, 5′‐GCCGGAATTCATGTACAACATGATGGAGACGGAG‐3′ and Reverse, 5′‐GCCGCTCGAG TCACATGTGTGAGAGGGG‐3′) were amplified and cloned into MSCV‐ires‐hCD2. Virus was produced in 293 T cells transfected with viral constructs along with psPAX2 and pMD2 or VSVSG and gag/pol constructs. Viral supernatants were collected on day 2 and 3 after transfection and used to infect target cells.
RNA extraction and realtime qPCR (RT‐qPCR)
RNA was isolated using Trizol (Invitrogen, USA) or Hybrid R (Gene All, Korea), and converted to cDNA using ReverTra Ace® qPCR Kit (TOYOBO, Japan) according to the manufacturer's instructions. To determine the level of gene expression, RT‐qPCR was performed using the qPCR Master Mix Kit (TOYOBO, Japan). Primer sequences for RT‐qPCR are shown in Supporting Information Table 1.
Western blotting analysis
Cell lysates were harvested using RIPA lysis buffer for 30 min on ice and centrifuged at 13,000 rpm for 10 min at 4 °C. Protein concentration of the supernatant was determined by Bio‐Rad Protein Assay (Bio‐Rad Laboratories, Inc., USA). An equal amount of each protein extract was resolved using 10% polyacrylamide gel and electro‐transferred onto 0.45 μm hybridization nitrocellulose filter (HATF) membrane (Millipore, USA) using Trans‐blot Turbo (Bio‐Rad Laboratories, Inc., USA). Membranes were immunoblotted with either rabbit polyclonal anti‐IFITM1 antibody (GeneTex, USA), rabbit polyclonal anti‐actin antibody (Abcam, USA), mouse monoclonal anti‐CAV1 antibody (Abnova, Taiwan), rabbit polyclonal anti‐pEGFR (Tyr1068) antibody (Cell Signaling, USA), rabbit polyclonal anti‐EGFR antibody (Cell Signaling, USA), rabbit monoclonal anti‐Sox2 antibody (Cell Signaling, USA), rabbit polyclonal anti‐p‐AKT (Ser473) antibody (Cell Signaling, USA), rabbit polyclonal anti‐Akt antibody (Cell Signaling, USA), rabbit polyclonal anti‐p‐β‐Catenin(Ser33/37/Thr41) antibody (Cell Signaling, USA), or rabbit polyclonal anti‐β‐Catenin antibody (Cell Signaling, USA) overnight at 4 °C. Membranes were incubated with either HRP‐conjugated anti‐rabbit immunoglobulin (Cell Signaling, USA) or HRP‐linked anti‐mouse immunoglobulin (Cell Signaling, USA) for 1 h at room temperature. The protein signal was detected by enhanced chemiluminescence (Thermo, USA) using the Amersham Imager 600 (GE Healthcare Life Sciences, UK).
Cell proliferation assay (MTT assay)
MTT assay was used to evaluate cell proliferation by using Cell Proliferation Kit I according to the manufacturer's instructions (Roche, Germany). Briefly, cells (5 × 103) were seeded into a 96‐well plate and incubated for an additional 72 h. Cells were incubated in 5 mg/ml of MTT solution for 4 h and then solubilized with 100 μL solubilization solution (10% SDS in 0.01 M HCl) overnight. Absorbance was read at 575 nm and 650 nm using a plate reader.
Sphere forming assay
NSCLC cell lines were cultured and serially plated on a low adherent 96‐well plate at low density (~500 cells per well) under serum‐free conditions and supplemented with 20 ng/mL of epidermal growth factor (EGF) (Life Technologies, Foster City, CA), 10 ng/ml of basic fibroblast growth factor (bFGF) (Life Technologies), and N2 supplement (Life Technologies) for 10 days according to published protocols.29 The experiment was conducted in three independent replicates for H1650 and A549 cells.
Migration and invasion assay
Cell migration and invasion were analyzed in vitro using the transwell insert system (Corning, USA) without coating or with coating by 20 μL of Matrigel (BD, USA), respectively. The culture insert was attached on bottom of a 24‐well plate, and 100 μL of serum‐free media containing 2 × 104 cells were seeded into each well of the insert. Six‐hundred μL of media containing 10% FBS was added outside the transwell culture insert. Cells were incubated at 37 °C for 16 h and 24 h in a humidified atmosphere with 5% CO2 for migration and invasion, respectively. Transwells were washed twice with PBS and cleaned using cotton swap. The cells were fixed with 1% formaldehyde for 15 min, washed twice with PBS, stained with 0.1% of crystal violet for 15 min and then observed using a microscope (Leica, Germany).
Tumorigenicity by subcutaneous injection in NSG mice
1 × 106 cells of control or IFITM‐knockdown H1650 were suspended in 1:1 ratio of PBS: Matrigel (Corning, Corning, NY) to 100 μL per injection, and injected (25 g needle) subcutaneously into the back of NSG mice (NOD.Cg‐Prkdc scid Il2rg tm1Wjl/SzJ, The Jackson Laboratory). Mice were monitored for tumor growth for up to 60 days. Tumor volume (TV) was calculated according to the formula: TV (mm3) = πW2×L/6, where W and L were the shortest and the longest diameters, respectively. On day 60 after transplantation, tumor tissues collected from sacrificed mice were processed for morphological analysis and molecular study. All animal experiments were performed according to protocols approved by Soonchunhyang University Institutional Animal Care and Use Committee.
Orthotopic lung transplantation
A small skin incision was made over the ribcage to visualize the lungs after anesthesia. 1 × 106 cells of control or IFITM‐knockdown H1650 were suspended in 1:1 ratio of PBS: Matrigel (Corning, Corning, NY) to 50 μL per injection, and injected (25 g needle) into NSG mice (NOD.Cg‐Prkdc scid Il2rg tm1Wjl/SzJ, The Jackson Laboratory) in the intercostal space between ribs 5 and 6 of the left lung. Mice were monitored for tumor growth and metastasis for up to 32 days. The lungs and liver tissues were assessed to count tumor nodules, and then were processed for H&E analysis. All animal experiments were performed according to protocols approved by Soonchunhyang University Institutional Animal Care and Use Committee.
Human nonsmall cell lung cancer (NSCLC) specimens
This retrospective study was approved by the Institutional Review Board of Ajou University School of Medicine. Inform consent was granted a waiver due to the retrospective nature of our study. All analyses were performed in accordance with ethical guidelines for clinical research. A total of 226 patients confirmed to have NSCLC after surgical resection between January 2009 and December 2013 were enrolled.
Histopathological analysis and immunohistochemistry assay
Histological subclassification was performed by two pathologists according to the 2015 World Health Organization Classification of Lung Tumors.30 Formalin‐fixed, paraffin‐embedded tissue samples were arranged in a Benchmark XT automatic immunohistochemical staining device (Ventana Medical Systems, Tucson, AZ). Samples were incubated with rabbit polyclonal anti‐IFITM1 antibody (1:100 dilution; GeneTex, USA), rabbit monoclonal anti‐Sox2 antibody (1:300 dilution; Cell Signaling, USA) and rabbit polyclonal anti‐EGFR antibody (1:300 dilution; Cell Signaling, USA). IFITM1 intensity was evaluated on a four‐point intensity scale (0, none, 1, faint; 2, moderate; 3, strong). The proportion of membranous and/or cytoplasmic expression of IFITM1 was also evaluated. A sample was considered IFITM1 positive if >30% of definitive tumor Cells exhibited moderate or strong immunohistochemical reactivity with the anti‐ IFITM1 antibody. The staining results were scored based on staining intensity: 0, none; 1, faint; 2, moderate; 3, strong. A sample was considered SOX2 and EGFR positive if >30% of definitive tumor Cells exhibited moderate or strong immunohistochemical reactivity with the anti‐ SOX2 antibody and anti‐EGFR antibody. Overall survival (OS) was defined as the time between the first day of diagnosis and the date of death regardless of the cause of death. The follow‐up of patients still alive without events was censored at the date of their last follow‐up.
Statistical analysis
The results of RT‐qPCR, migration and invasion were analyzed with Student's T‐test. Hazard ratio and 95% confidence interval of clinicopathological data were evaluated using Cox regression models. Categorical variables were compared using the chi‐squared test, and continuous variables were compared using an independent‐sample t‐test Kaplan–Meier method was used to analyze disease‐free survival rate using the log‐rank test. A p‐value of less than 0.05 was considered statistically significant in all assessments.
Results
IFITM1 expression is highly induced in a stem cell selective medium
IFITM1 is a highly expressed prognostic marker in several cancers such as cervical, esophageal, ovarian, brain, and colon cancers.26, 31, 32 We analyzed the expression of IFITM1 in several patient‐derived NSCLC cell lines using both RT‐qPCR and immunoblotting. Both A549 (with wild‐type EGFR expression) and H1650 (with activating mutation of EGFR [L858R]) cell lines displayed high levels of IFITM1 expression compared to H1299 cells (wild type for EGFR expression) (Fig. 1 a‐c). This suggests that IFITM1 is highly expressed in NSCLC cell lines; however, high expression of IFITM1 protein is not related to the mutational status of EGFR. Next, we determined whether the expression of IFITM1 was affected by culture conditions, such as conventional medium and stem cell selective medium. To test this, H1650 and A549 cells were cultured for 4 days either in conventional culture media without FGF and EGF (on regular culture plates) or stem cell selective media with FGF and EGF (on ultra‐low attachment plates). We observed that IFITM1 expression was highly upregulated when H1650 and A549 cell lines were cultured in stem cell selective medium compared to conventional culture medium (Fig. 1 d and e).
Figure 1.
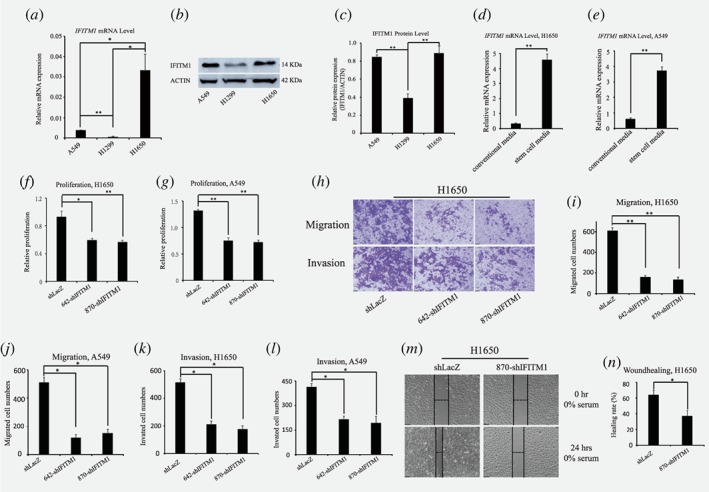
IFITM1 depletion impairs cell proliferation and motility in NSCLC cell lines. (a ‐ c) IFITM1 expression was evaluated in NSCLC cell lines including A549, H1299 and H1650 by RT‐qPCR and western blots. (a) RT‐qPCR was performed to evaluate the mRNA expression of IFITM1 from cells indicated. (b and c) Immunoblot with anti‐IFITM1 antibody was conducted to analyze IFITM1 protein level, and actin was used as the loading control. (b) A representative image of immunoblots was shown. (c) Relative protein expression of IFITM1 was quantified using Image J. (d and e) H1650 and A549 were cultured in either conventional media or stem cell selective media. RNA was isolated and RT‐qPCR was performed to evaluate the mRNA expression of IFITM1. (f and g) NSCLC cell lines were infected with either control (shLacZ) virus or IFITM1 knockdown virus (642‐shIFITM1 and 870‐shIFITM1). Control (shLacZ) or IFITM1‐depleted (shIFITM1) H1650 (f) and A549 (g) were incubated for 72 h to determine the proliferation rate by MTT assay. (h‐l) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) NSCLC cell lines were seeded in a matrigel‐uncoated and coated transwell, followed by incubation for 16 h and 24 h for assessing migration and invasion, respectively. Cells that had migrated to the lower surface of the transwell were stained and quantified. Imaging was done using an inverted microscope (magnification: 100x) and representative images are shown (h). Data shown (i and j, migration; k and l, invasion) are from three independent experiments. (m and n) Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were seeded and subjected to in vitro scratch assays with images captured at 0 and 24 h after incubation. The lines define the areas lacking cells. (m) Representative images are shown. Data are shown as mean ± SEM of three independent experiments (*p < 0.05, **p < 0.001). [Color figure can be viewed at wileyonlinelibrary.com]
Depletion of IFITM1 impairs the proliferation and sphere‐forming ability of NSCLC cells
To determine the functional importance of IFITM1 in NSCLC, we used shRNAs to inhibit the expression of IFITM1 in cancer cell lines. NSCLC cell lines, H1650 and A549, were transduced with either nonsense control (shLacZ) or two independent IFITM1 shRNAs (642‐shIFITM1 and 870‐shIFITM1) and either sorted using flow cytometry or selected with puromycin. Both RNA and protein were extracted from transduced H1650 and A549, and the knockdown efficiency of IFITM1 was determined by RT‐qPCR and immunoblotting. Both mRNA and protein levels of IFITM1 significantly decreased in IFITM1 shRNA‐transduced cells compared to nonsense control (Supporting Information Fig. 1a‐f). As IFITM2 and IFITM3 bear a high homology with IFITM1,33 we determined whether IFITM1 depletion also affected the expression of other IFITM family members. RT‐qPCR results demonstrated that the expression levels of both IFITM2 and IFITM3 did not change by the depletion of IFITM1 (Supporting Information Fig. 1g and h). These data confirmed that IFITM1 was successfully depleted at both RNA and protein levels without affecting the expression of other IFITM family members such as IFITM2 and IFITM3. To test whether IFITM1 was involved in the growth of NSCLC cell lines, we determined the proliferation ability of IFITM1‐depleted cells using MTT assay. Control or IFITM1 shRNA‐transduced NSCLC cell lines were incubated for 72 h to determine proliferation in vitro. Relative to control cells, the proliferation rate of IFITM1‐depleted H1650 and A549 cell lines decreased by approximately 36% and 43%, respectively, suggesting that cell proliferation was modestly impaired in the absence of IFITM1 in NSCLC cell lines (Fig. 1 f and g). As sphere forming cells were shown to induce extensive cancer proliferation and also display cancer stem cell‐like characteristics, we asked whether IFITM1 contributed to the sphere forming ability of NSCLC cell lines. Control or IFITM1 shRNA‐transduced cell lines were serially plated up to tertiary plating on ultra‐low attachment plate for 10 days, and the number of spheres formed were counted. In the absence of IFITM1, the sphere forming ability of NSCLC cell lines such as H1650 and A549 was gradually decreased starting from secondary plating, reaching a significant difference on tertiary plating (Supporting Information Fig. 2 a‐c). Considering that the sphere forming ability is a unique property of cancer stem cells, the above data suggest that IFITM1 plays a critical role in the maintenance of cancer stem cells.
IFITM1 is required for the mobility of NSCLC cells
As IFITM1 promoted cell invasion in several cancers such as glioma cell lines, head and neck squamous cell carcinoma (HNSCC), and colon cancer,27, 28, 34 we investigated whether IFITM1 was indispensable for the mobilization of NSCLC cell lines. NSCLC cell lines (H1650 and A549) were transduced with either shLacZ or IFITM1 shRNAs (642‐shIFITM1 and 870‐shIFITM1), and their migration was examined by seeding them on an insert of the transwell system, followed by an incubation for 16 h. Thereafter, the cells on the inserts of the transwell system were stained and counted to evaluate their migratory ability. Relative to control‐transduced cells, IFITM1 shRNA‐transduced H1650 and A549 cells migrated 24% and 26%, respectively (Fig. 1 h‐j). Using a matrigel‐coated insert of the transwell system, we examined whether IFITM1 was required for the invasion of cancer cells. As shown in Fig. 1 h, k, and l, IFITM1 shRNA‐transduced H1650 and A549 cells exhibited 37.5% and 49.5% of cell invasion ability, respectively, relative to control‐transduced cells. Consistent with the above results, scratch‐induced wound healing assay showed that wound healing was significantly delayed in the absence of IFITM1 (Fig. 1 m and n). These results indicate that IFITM1 is required for the mobility of NSCLC cells.
Ectopic expression of IFITM1 promotes sphere‐forming, proliferation and migration ability of NSCLC cells
Above mentioned shRNA depletion experiments showed that IFITM1 was required for essential functions such as proliferation, and migration in NSCLC cell lines. Thus, we investigated whether IFITM1 was sufficient to drive these functions by gain of function of IFITM1 in H1299 and H1650. Here, we ectopically expressed IFITM1 using a lentiviral system and determined the expression of IFITM1 using RT‐qPCR and immunoblotting. As shown in Figure 2 a‐f, IFITM1 was highly upregulated at both mRNA and protein levels upon IFITM1 overexpression. In line with the results obtained from shRNA‐mediated knockdown experiments (Fig. 1), ectopic expression of IFITM1 increased the sphere‐forming, proliferation, and migration ability of H1299 and H1650 cells in vitro (Fig. 2 g‐o). Collectively, these results suggest that IFITM1 overexpression is sufficient to promote the progression of NSCLC cells.
Figure 2.
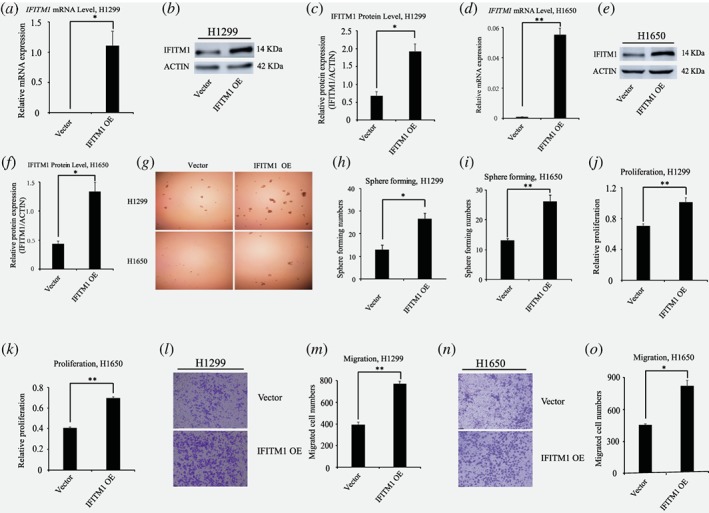
Ectopic expression of IFITM1 promotes the sphere‐forming, proliferation and migration of NSCLC cell lines in vitro. (a ‐ f) Both H1299 and H1650 cancer cell lines were infected with either control (vector) or IFITM1 over‐expression construct (IFITM1 OE). Both RNA and protein were isolated to examine the overexpression of IFITM1 by RT‐qPCR (a and d) and immunoblots (b, c, e and f). A representative image was shown (b and e) and the image intensity was analyzed by Image J and quantified (c, H1299; f, H1650). (g‐i) H1299 and H1650 cells infected with control (vector) or IFITM1‐overexpression construct (IFITM1 OE) were cultured on ultra‐low attachment plates to determine the sphere‐forming ability. Representative images are shown (g). The data are shown as mean ± SEM of three independent (h, H1299; i, H1650). (j and k) NSCLC cell lines including H1299 (j) and H1650 (k) infected with control (vector) or IFITM1‐overexpression construct (IFITM1 OE) were incubated for 96 h to determine the proliferation by MTT assay. Data shown are from three independent experiments. (l‐o) H1299 and H1650 cell lines infected with control (vector) or IFITM1‐expression construct (IFITM1 OE) were seeded in a transwell for assessing migration. Cells were incubated for 16 h, stained, and quantified. (l and n) Representative images are shown. The data are shown as mean ± SEM of three independent (m, H1299; o, H1650) (*p < 0.05, **p < 0.001). [Color figure can be viewed at wileyonlinelibrary.com]
IFITM1 is essential for the maintenance of EMT signature of NSCLC cell lines in vitro
We have previously shown that EMT signaling is defective in the absence of IFITM1 in CRC.28 Thus, we questioned whether IFITM1 depletion gave rise to a defective EMT signature in NSCLC. NSCLC cell lines, H1650 and A549, were treated with TGF‐β to induce EMT. Both RNA and protein were isolated and analyzed to determine whether EMT‐related genes were differentially regulated in response to TGF‐β treatment. In the absence of IFITM1, several EMT markers such as E‐cadherin (CDH1), N‐cadherin (CDH2), SNAIL1, SNAIL2, Fibronectin (FN1), matrix metalloproteinase (MMP2), MMP9, etc., were either upregulated or downregulated in H1650 and A549 cells (Fig. 3 a‐c; Supporting Information Fig. S3a‐c). Specifically, CDH1 expression was upregulated in the absence of IFITM1, whereas the expression of CDH2 and SNAIL was downregulated at both the RNA and protein levels. Additionally, using a gelatin digestion assay, we showed that the enzymatic activity of MMP2 was inhibited in the absence of IFITM1 (Fig. 3 c).
Figure 3.
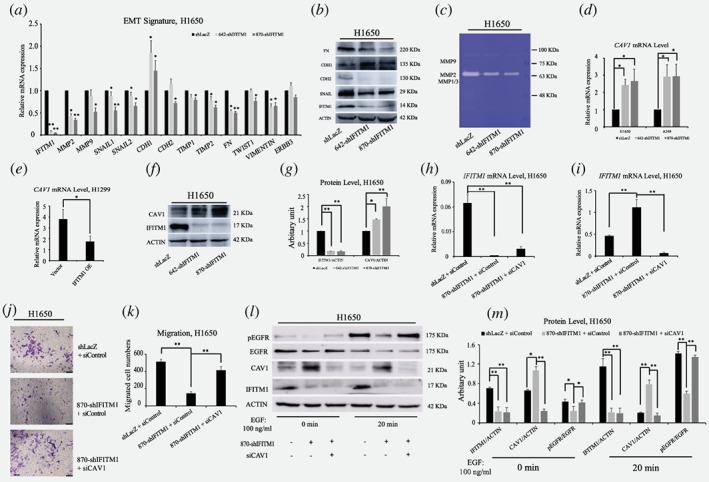
IFITM1 is essential for the maintenance of EMT signature of NSCLC cell lines in vitro. (a) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) H1650 cells were treated with 5 ng/ml of TGF‐β, and the RNA levels of EMT genes were determined by RT‐qPCR. (b) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) H1650 cell lysates were isolated and the protein level of EMT genes were probed with antibodies indicated. ACTIN was used as a loading control. (c) Supernatants obtained from control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) H1650 cells were analyzed for enzymatic activity of MMP1/3, MMP2 and MMP9. (d‐g) NSCLC cell lines were transduced with either knocking down IFITM1 or overexpression of IFITM1, and the expression level of CAV1 was determined by RT‐qPCR (d and e) and immunoblot (f and g). ACTIN was used as a loading control. Representative images were shown (f) and quantification of protein levels was performed using Image J software from three separate experiments (g). (h‐m ) Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were transiently transfected with either negative control (siNeg) or siCaveolin‐1 (siCAV1), and the expression of IFITM1 and CAV1 was determined by RT‐qPCR (h and i) and immunoblot (l and m). The transduced cells were either tested to determine migratory ability (j and k), or treated with 100 ng/ml of EGF to determine pEGFR level (l and m). Representative images were shown (l) and quantification of protein levels was performed using Image J software (m). The data are shown as mean ± SEM of three independent. (*p < 0.05, **p < 0.001) [Color figure can be viewed at wileyonlinelibrary.com]
EGFR/SOX2 signaling axis in NSCLC cells is dependent on IFITM1 in vitro
It has previously been shown that IFITM1 interacts with caveolin 1 (CAV1)28, 35 and that CAV1 colocalizes with EGFR in a phosphorylation‐dependent manner.36, 37, 38 However, a direct relationship between IFITM1 and EGFR still remains unknown. To reveal the mechanisms by which IFITM1 deficiency led to the impaired ability of EMT signaling and cancer stem cell‐like properties in NSCLC cell lines, we tested whether IFITM1 was related to CAV1 and EGFR signaling. The expression level of CAV1 was determined by RT‐qPCR and immunoblotting in IFITM1‐depleted H1650 and A549 cells. We found that CAV1 expression was significantly upregulated in the absence of IFITM1 (Fig. 3 d, f, and g; Supporting Information Fig. S3 d and e). Consistently, ectopic expression of IFITM1 in H1299 cells led to a downregulation of CAV1 (Fig. 3 e). Because CAV‐1 was known to be a negative regulator of EGFR,38, 39 we hypothesized that the activity of EGFR would be compromised in the absence of IFITM1 due to the high level of CAV1. To test this hypothesis, control or IFITM1‐knocked down cells were transiently transfected with either control (siControl) or siCaveolin‐1 (siCAV1). Then, we determined whether the impaired migratory ability of IFITM1‐depleted cells was rescued by siCAV1. As shown in Figure 3 h‐k, the reduced migratory ability of H1650 in the absence of IFITM1 was partially rescued by the simultaneous suppression of CAV‐1 and IFITM1. Further, pEGFR level was also decreased in the absence of IFITM1 partly because of a higher level of CAV1 (Fig. 3 l and m). Importantly, the reduced EGFR phosphorylation in the absence of IFITM1 was partially rescued by co‐knockdown of IFITM1 and CAV1 (Fig. 3 l and m). These results suggest that the regulation of EGFR signaling by IFITM1 is mediated partly by Caveolin‐1. To further determine the influences of IFITM1 depletion on EGFR/AKT signaling axis, we examined several downstream targets, including pAKT, pSTAT3, SOX2, MYC, and EGR1, which were associated with EMT, CSC, and cancer progression.40, 41, 42, 43 H1650 cells were stimulated with EGF for 10 to 40 min, and the phosphorylation of downstream signaling, such as pEGFR, pAKT, and pSTAT3, was studied by western blot analysis. In control cells, EGFR phosphorylation was found to be the highest after 20 min treatment with EGF, whereas IFITM1‐depleted cells displayed very low level of EGFR phosphorylation under the same conditions (Fig. 4 a). The levels of pAKT and pSTAT3 were consistently much lower in the absence of IFITM1 (Fig. 4a). In addition, the RNA levels of SOX2, MYC, and EGR1 significantly decreased after knocking down IFITM1 (Fig. 4 b, d, and f). As expected, IFITM1 overexpression was associated with increased RNA levels of these genes (Fig. 4 c, e, and g). Consistently with the results of RNA expressions, the levels of SOX2 and pAKT decreased after IFITM1 knockdown, whereas there was an increase in the levels of these proteins in response to IFITM1 overexpression (Fig. 4 h‐m). Additionally, we observed that the level of phospho‐β‐catenin was also affected in an IFITM1‐dependent manner (Fig. 4 h‐m). Furthermore, several surface markers such as CD44 and CD133 marking cancer stem cell population were analyzed using flow cytometry.29, 44, 45, 46 IFITM1‐depletion decreased the expression of CD133 and CD44 in A549 cells (Supporting Information Fig. S4). Collectively, these in vitro data suggest that IFITM1 is essential for the maintenance of EMT and CSC‐like properties in NSCLC cells via modulation of the EGFR/SOX2 signaling axis.
Figure 4.
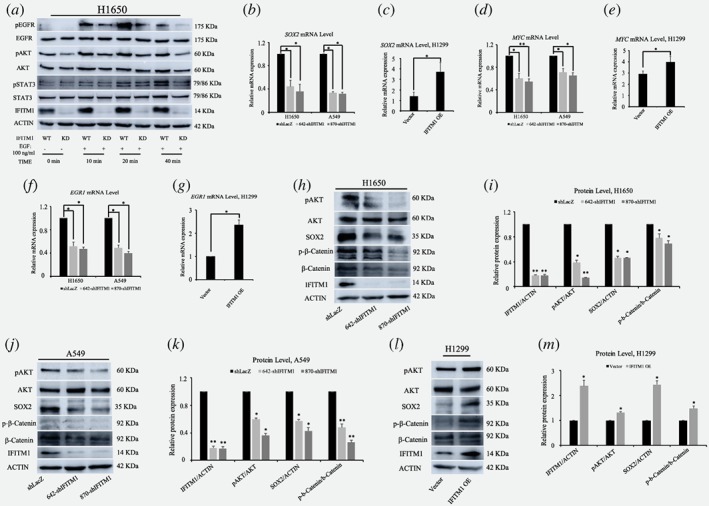
Impaired EGFR/SOX2 signaling axis in the absence of IFITM1. (a) Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were treated with 100 ng/ml of EGF, and cell lysates were isolated to probe with antibodies indicated. ACTIN and unphosphorylated proteins were used as the loading control. (b ‐ g) RNA was isolated from NSCLC cell lines either after knocking down IFITM1 (b, d and f) or overexpression of IFITM1 (c, e and g), and the expression level of SOX2 (b, c), MYC (d, e), and EGR1 (f, g) were determined by RT‐qPCR. (h‐m) Cell lysates were isolated from NSCLC cell lines either after knocking down IFITM1 (h‐k) or overexpression of IFITM1 (l, m), and the protein level was determined by immunoblots. ACTIN and unphosphorylated proteins were used as the loading control. Representative images were shown (h, H1650; j, A549; l, H1299) and quantification of protein levels was performed using Image J software from three separate experiments (i, H1650; k, A549; m, H1299). (*p < 0.05, **p < 0.001). [Color figure can be viewed at wileyonlinelibrary.com]
Defects resulting from the depletion of IFITM1 are partially rescued by ectopic expression of SOX2 in vitro
Since SOX2 was found to be a downstream signaling of IFITM1 in NSCLC cell lines, we investigated whether the functional defects caused by IFITM1 deficiency could be rescued by ectopic expression of SOX2. H1650 cells were transduced with either shLacZ+Vector, shIFITM1 + Vector, or shIFITM1 + SOX2 overexpression (OE). RNA was isolated from transduced cells, and the expression levels of IFITM1 and SOX2 were determined by RT‐qPCR. While the reduced RNA level of SOX2 in IFITM1‐depleted cells increased by overexpression of SOX2, the level of IFITM1 expression was not affected by SOX2 overexpression (Fig. 5 a, b, g, and h). To test the functional effects of SOX2 overexpression on cell migration, the transduced cells were seeded on a transwell, and the mobility of cells was determined. IFITM1‐depleted cells (870‐shIFITM1 + Vector) displayed 24.6% of migration compared to the control cells (shLacZ+Vector). Importantly, overexpression of SOX2 in IFITM1‐depleted cells (870‐shIFITM1 + SOX2 OE) showed 75.8% of migration relative to cells transduced with control vector (shLacZ+Vector); this result suggested that the defects caused by IFITM1 deficiency were partially rescued by the ectopic expression of SOX2 (Fig. 5 c and d). At the molecular level, we found that MYC, EGR1, and phospho‐β‐catenin were partially upregulated by SOX2 overexpression (Fig. 5 e‐h). These data suggest that SOX2 partially complements the function of IFITM1 as a downstream signaling of IFITM1.
Figure 5.
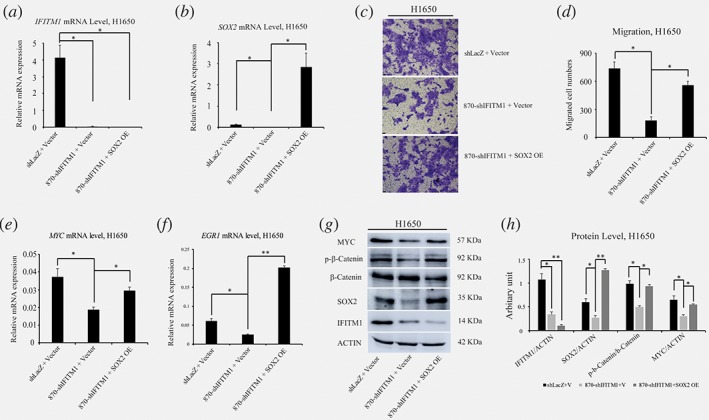
The defects caused by IFITM1 depletion is partly rescued by SOX2. (a and b) H1650 cells were transduced with either shLacZ+Vector, 870‐shIFITM1 + Vector, or 870‐shIFITM1 + SOX2 OE, and RNA was isolated to determine the level of IFITM1 (a) and SOX2 (b). (c and d) H1650 cells transduced with control (shLacZ+Vector), IFITM1‐depleted (shIFITM1 + Vector) or IFITM1‐depleted plus SOX2‐overexpression (shIFITM1 + SOX2‐OE) were seeded in a transwell for assessing migration. Cells were incubated for 16 h, stained, and quantified. Representative images are shown (c) and data shown are from more than three independent experiments (d). (e‐h) RNA and cell lysates were obtained from each cells indicated and analyzed by RT‐qPCR (e and f) and immunoblots (g and h). Representative images are shown (g) and quantification of protein levels was performed using Image J software from three independent experiments (h). The data are shown as mean ± SEM (*p < 0.05, **p < 0.001). [Color figure can be viewed at wileyonlinelibrary.com]
IFITM1 is required for the progression of NSCLC cells in vivo
Because our in vitro data highlighted the essential functions of IFITM1 in NSCLC cells, we investigated whether this phenotype was also observed in vivo. Therefore, control or IFITM1 shRNA‐transduced H1650 cells were subcutaneously injected into immunocompromised Nod Scid Gamma (NSG) mice, and tumor size was monitored every 3 days. Tumors were isolated and analyzed 60 days after transplantation. Starting from 24 days after transplantation, control mice began to show differential tumor growth compared to mice with IFITM1 shRNA‐transduced cancer cells (Fig. 6 a). At 60 days after injection, control mice developed large size tumors, whereas the mice injected with shIFITM1‐transduced cancer cells displayed much smaller size tumors (control, 864.5 ± 34.2 mm3 vs. shIFITM1, 431.8 ± 15.7 mm3; p < 0.05; Fig. 6 a and b). When tumors were weighted after dissection, the weight of IFITM1 shRNA‐transduced tumor was 36.8% of that of control (Fig. 6 b and c). To check the levels of IFITM1 expression in tumors, both RNA and protein were isolated and analyzed by RT‐qPCR and immunoblotting. IFITM1 expression was significantly lower in IFITM1 shRNA‐transduced tumors at both RNA and protein levels compared to their expression levels in tumors from the control mice (Fig. 6 d‐f). For the in vivo validation of our in vitro IFITM1 depletion effects, levels of pEGFR, pAKT, and SOX2 proteins in IFITM1 shRNA‐transduced tumors were determined by immunoblotting. In agreement with our in vitro data, the levels of pEGFR, pAKT and SOX2 significantly decreased in the absence of IFITM1 (Fig. 6 e and f). Consistently, when control or IFITM1 shRNA‐transduced H1650 cells were orthotopically injected into immunocompromised NSG mice, IFITM1‐depleted cells displayed a much lower number of tumor nodules in the lungs as well as the liver compared to the control (lung, 7.7 vs. 2.5; liver, 29 vs. 3.5; Fig. 6 g and h; Supporting Information Fig. S5). These data indicate that IFITM1 is required for the tumor progression of NSCLC cells in vitro and in vivo.
Figure 6.
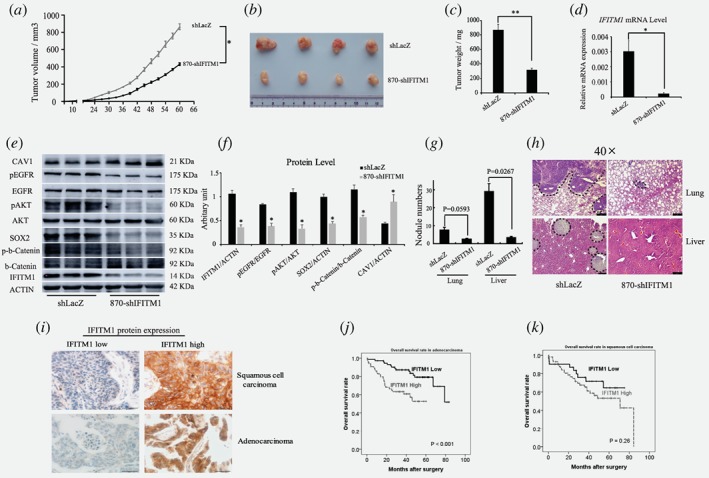
IFITM1 is required for the progression of lung cancer in vivo, and is associated with a poor prognosis of adenocarcinoma patients. (a‐c) Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were subcutaneously injected into NSG mice, and then mice were monitored for tumor growth. (a) Tumor volume was measured every 3 days up to 60 days and the tumor growth curve displayed over time. (b) A representative image of tumors was shown. (c) Tumors isolated from each mouse were weighed. (d) RNA was isolated from each tumor and IFITM1 expression was analyzed by RT‐qPCR. (e, f) Cell lysates obtained from each tumor were analyzed by immunoblots to determine the expression of key signaling molecules indicated. A representative image was shown (e) and the image intensity was analyzed by Image J and quantified (f). The data are shown as mean ± SEM (n = 6, *p < 0.05, **p < 0.001). (g and h) Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were orthotopically injected into the lungs of NSG mice, and then mice were monitored for tumor metastasis. (g) Quantification of the number of nodules in the lungs and liver (shLacZ, n = 3; 870‐shIFITM1, n = 2). (h) Representative images of HE staining. The dotted lines indicated the area of tumor nodules. (i‐k) A total of 226 NSCLC patients‐derived cancer samples were stained with IFITM1 antibody and evaluated based on staining frequency. (i) Representative images of immunohistochemistry are shown. Survival rates of adenocarcinoma (j) and squamous cell carcinoma (k) were determined based on the expression of IFITM1 expression using Kaplan–Meier curve (j, p < 0.001; k, p = 0.26). [Color figure can be viewed at wileyonlinelibrary.com]
IFITM1 is associated with a poor prognosis in NSCLC patients
To establish the biological relevance of our results to the prognosis in NSCLC patients, we analyzed 226 NSCLC patient‐derived samples and measured the expression of IFITM1 protein. Then, the association between IFITM1 expression and patient's survival rate was determined. The age range of patients included in our study was 35 to 86 years (median, 64 years), and demographic data of these patients were provided in Supporting Information Table 2. our study comprised 138 (61.1%) adenocarcinoma and 88 (38.9%) squamous cell carcinoma patients, and their median follow‐up time was 39.9 months (range, 1–86 months). When the correlation between IFITM1 expression and the 5‐year overall survival (OS) rate was analyzed, we found that IFITM1 expression was closely associated with the OS rate of patients with adenocarcinoma but had no correlation with the OS rates of squamous cell carcinoma patients (Fig. 6 i‐k). Specifically, IFITM1‐positive patients with adenocarcinoma had a significantly lower 5‐year overall survival (OS) rate than that of IFITM1‐negative patients (52% vs. 79%, p < 0.001; Fig. 6 j). However, in case of squamous cell carcinoma, IFITM1 expression did not affect the 5‐year OS rates (53% vs. 64%, p = 0.26; Fig. 6 k). IFITM1 positivity was not correlated with other clinicopathological variables, including histologic subtype, pathologic stage, lympho‐vascular invasion, and smoking history (Supporting Information Table 3). In multivariate analysis, IFITM1 expression was an independent prognostic marker for OS in adenocarcinoma patients (hazard ratio = 3.322, p = 0.001; Supporting Information Table 4). These results suggest that IFITM1 is closely associated with the poor prognosis of adenocarcinoma and, thus, serves as an attractive therapeutic target to block the progression of the disease.
Discussion
Several groups including ours previously showed that IFITM1 was associated with the progression of several cancers such as glioma, head and neck cancer, and CRC.27 Additionally, it has been shown that IFITM1 is related to Wnt signaling and cisplatin resistance.47 However, it remains unknown whether IFITM1 is correlated with a prognosis and is functionally indispensable in NSCLC. In our study, we used several NSCLC cell lines and patient‐derived samples to address these questions. In agreement with data obtained from the other cancer studies mentioned above, our gain‐ and loss‐of‐function experiments demonstrated that IFITM1 was required for proliferation, sphere formation, migration, invasion, and tumorigenesis via EGFR/SOX2 signaling axis in vitro and in vivo. More importantly, we showed that IFITM1 was closely associated with a poor prognosis of adenocarcinoma, but not squamous cell carcinoma; this was evidenced by a significantly low OS rate of IFITM1‐positive adenocarcinoma patients versus IFITM1‐negative patients. However, it remains not clear how IFITM1 is induced in the lung cancer. Kim et al. reported that CD147 stimulation, which was implicated in tumor invasion, metastasis, and angiogenesis,48 could induce the expression of IFITM1 using a leukemic cell line.49 KRAS mutation was associated with higher expression of IFITM1 in rectal cancer.50 Analysis of the cancer genome atlas (TCGA) database identifies that three cases of IFITM1 mutations and one case of IFITM1 mutation are found in adenocarcinoma and squamous cell carcinoma, respectively. Here, we showed that IFITM1 was highly upregulated in stem cell selective medium compared to conventional culture medium. Because EGF was a component of stem cell selective medium, we determined whether EGF could induce IFITM1. However, we did not observe any differential expression of IFITM1 in the presence or absence of EGF (Supporting Information Fig. S6). Thus, we expect that unknown factors, which needs to be explored further, may be enriched in stem cell selective medium and induce IFITM1 expression.
SOX2 is a member of the SOX (sex‐determining region Y [SRY]‐box protein) family and is characterized by the presence of a conserved high mobility group (HMG) DNA‐binding domain of the SRY protein.51 It is known to be upregulated in various human cancers,52 including lung cancer.53 Notably, SOX2 has recently been used as a CSC marker in several cancers, including CRC54 and lung53 cancers. Here, we also showed that EGFR‐SOX2 signaling was compromised in the absence of IFITM1, resulting in an impairment of EMT and cancer stem cell‐like properties such as mobility, sphere formation, and expression of cancer stem cell markers including CD133 and CD44. Previously, we showed that IFITM1 was essential for epithelial to mesenchymal transition (EMT), an important process implicated in various aspects of tumor progression, such as invasion, metastasis, and drug resistance,28, 55 in colorectal cancer (CRC) by the regulation of CAV1. In NSCLC cell lines (A549 and H1650) used in our study, EMT signature and CAV1 were consistently dysregulated, suggesting that IFITM1 broadly affected the EMT signature in both CRC and NSCLC cells. We also found that cancer stem cell‐like properties, mediated by EGFR signaling pathway, were critically impaired in the absence of IFITM1. In order to investigate the molecular mechanisms by which EGFR signaling pathway was dysregulated in an IFITM1‐dependent manner, we focused on the downstream targets of EGFR, such as pSTAT3, SOX2, MYC and EGR1. While MYC overexpression or dysregulation is related to the numerous oncogenic cascades, it has been reported to function as a tumor suppressor as well.56, 57 In addition, the reciprocal relationship between MYC and SOX2 has been implicated in the maintenance of stem cells and cancer development.41 They occupy more than 85% of the same gene promoters, and their interaction is related to the promotion of self‐renewal, growth regulation, and cell cycle progression.57 EGR1, a downstream target of EGFR signaling,58 has been reported to interact with MYC and its interaction leads to the induction of apoptosis.43, 56 Our observations showed that IFITM1 depletion reduced the levels of pEGFR, SOX2, MYC, and EGR1; these effects were partly rescued by ectopic expression of SOX2. It has been reported that CAV1 interacts with EGFR, and IFITM1 colocalizes with CAV1.28, 35, 37 In our study, we found that CAV1 was upregulated, and the pEGFR level was down regulated in the absence of IFITM1, which was rescued by simultaneous knockdown of CAV1 and IFITM1. Thus, we suggest that IFITM1 is essential for the EGFR‐SOX2 signaling cascade and regulates the signal transduction possibly through direct or indirect interaction with EGFR in a CAV1‐dependent manner.
We conducted IHC analyses of IFITM1, SOX2, and EGFR to see the correlation in patient‐derived samples. The positivity of IFITM1 expression was 65% and 45% in squamous cell carcinoma and adenocarcinoma, respectively (Supporting Information Table 5). As reported,59, 60 the positivity of SOX2 expression was also much higher in squamous cell carcinoma than that in adenocarcinoma (Supporting Information Tables 6 and 7). SOX2 expression tended to show more positivity in the IFITM1‐positive group (30.9% versus 21.4%), although it was not statistically different. EGFR expression was not different between IFITM1‐positive and negative group (Supporting Information Table 8). As we showed that IFITM1 was associated with OS in adenocarcinoma, but not in squamous cell carcinoma (Fig. 6 i‐k), it was a little bit paradoxical that the positivity of IFITM1 and SOX2 expression was higher in squamous cell carcinoma than that in adenocarcinoma. There are possibly several explanations to this result; (1) the sample number of squamous cell carcinoma was too small to result in a statistically conclusive analysis (n = 88); (2) there might be a discrepancy between established human cell lines and patient‐derived samples that we used in our study; (3) there might be a multistep complex progression of squamous cell carcinoma, because OS of squamous cell carcinoma was considerably different between IFITM1‐positive and ‐negative in 20 to 80 months after surgery, but not in the initial period of OS tracking (Fig. 6 k); (4) there may be unknown factors other than IFITM1, complicating the analysis. Thus, the significance and association of IFITM1 and SOX2 in NSCLC are more complex, and further studies are required to get insights to better understand these molecules.
To the best of our knowledge, our study is the first to show that IFITM1 plays an essential role in maintaining EMT and cancer stem cell‐like properties of NSCLC via EGFR/SOX2 signaling axis. Therefore, IFITM1 may represent an attractive target to achieve better therapeutic outcomes in NSCLC, and hence it is strongly suggested to develop chemotherapeutic agents and blocking antibodies targeting IFITM1.
Supporting information
Supplementary Figure 1. IFITM1 depletion does not affect the expression level of IFITM2 and IFITM3.
NSCLC cell lines were infected with either control (shLacZ) virus or IFITM1 knockdown virus (642‐shIFITM1 and 870‐shIFITM1) and selected with puromycin or sorted by flow cytometry. Both RNA and proteins were isolated to determine the expression of IFITM1. RT‐qPCR was performed to determine IFITM1, 2, 3 mRNA expression in H1650 (a, g) and A549 (d, h) cells. Immunoblots with anti‐IFITM1 antibody were conducted to analyze IFITM1 protein level in H1650 (b, c) and A549 (e, f), and Quantification of protein levels was performed using Image J software from three separate experiments. (*p < 0.05, **p < 0.001).
Supplementary Figure 2. IFITM1 depletion impairs sphere forming ability of NSCLC cells
Control (shLacZ) or IFITM1‐depleted (shIFITM1) H1650 (a) and A549 (b) were serially plated to determine sphere forming ability. Each plate was incubated in stem cell selective media for 10 days. Then spheres were washed with PBS, dissociated to single cells by trypsin and plated again. The data are shown as mean ± SEM of three independent experiments. (c) Representative images of tertiary plating are presented. (*p < 0.05, **p < 0.001).
Supplementary Figure 3. IFITM1 is essential for the maintenance of EMT signature of NSCLC cell lines in vitro.
(a) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) A549 cells were treated with 5 ng/ml TGF‐β, and the RNA level of EMT genes were determined by RT‐qPCR. (b and c) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) H1650 cell lysates were isolated and the protein level of EMT genes were probed with antibodies indicated. ACTIN was used as a loading control. Quantification of protein levels was performed using Image J software from three separate experiments (b, mesenchymal signature; c, epithelial signature). (d, e) The cell lysates were isolated from control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) A549 cells and probed with antibodies indicated. ACTIN was used as a loading control. Representative images were shown (d) and quantification of protein levels was performed using Image J software from three separate experiments (e). (*p < 0.05, **p < 0.001).
Supplementary Figure 4. IFITM1 depletion modestly impairs the expression of cancer stem cell makers of A549 cell line
A549 cell line was transduced with either control (shLacZ) or IFITM1 (642‐shIFITM1 and 870‐shIFITM1) knockdown virus and selected with puromycin or sorted by flowcytometry. Cells were cultured on ultra low attachment plates with stem cell selective medium for 3 days. Then, cell surface maker CD133 (a, b) and CD44 (c, d) were analyzed with flow cytometry. (b, d) Representative images of histogram of CD133 and CD44. Data shown are from more than three experiments (a, c). (*p < 0.05, **p < 0.001).
Supplementary Figure 5. Macroscopic view of tumor nodules on the lungs and liver
Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were orthotopically injected into the lungs of NSG mice, and then mice were monitored for tumor metastasis. Macroscopic view of tumor nodules on the lungs and liver.
Supplementary Figure 6. EGF does not induce the expression of IFITM1.
NSCLC cells were cultured with or without 100 ng/ml of EGF in conventional media for 1 and 3 days. Then, RNA was isolated and RT‐qPCR was performed to evaluate the mRNA expression of IFITM1 in H1650 (a and b) and A549 (c and d).
Supplementary Table 1. Primer sequences for RT‐qPCR
Supplementary Table 2. Demographic and clinical characteristics
Supplementary Table 3. Correlation of IFITM1 expression with pathologic characteristics in patients with adenocarcinoma
Supplementary Table 4. Multivariate analyses of overall survival in patients with adenocarcinoma
Supplementary Table 5. IFITM1 expression in squamous cell carcinoma and adenocarcinoma
Supplementary Table 6. Correlation of IFITM1 expression with SOX2, EGFR expression in squamous cell carcinoma
Supplementary Table 7. Correlation of IFITM1 expression with SOX2, EGFR expression in patients with adenocarcinoma
Supplementary Table 8. Correlation of IFITM1 expression with SOX2, EGFR expression in all patients.
Acknowledgements
This work was supported by the new faculty research fund of Ajou University School of Medicine awarded to Y.W.G., the grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHID), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI15C1647) and Global Research Development Center (NRF‐2016K1A4A3914725) awarded to H.Y.K.
Conflict of interest: The authors declare no conflict of interest.
Contributor Information
Dongjun Jeong, Email: juny1024@sch.ac.kr.
Hyog Young Kwon, Email: hykwon@sch.ac.kr.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Center MM, Ward E, et al. Cancer occurrence. Methods Mol Biol 2009;471:3–29. [DOI] [PubMed] [Google Scholar]
- 3. Swiderska M, Choromanska B, Dabrowska E, et al. The diagnostics of colorectal cancer. Contemp Oncol (Pozn) 2014;18:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tam WL, Weinberg RA. The epigenetics of epithelial‐mesenchymal plasticity in cancer. Nat Med 2013;19:1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thiery JP, Acloque H, Huang RY, et al. Epithelial‐mesenchymal transitions in development and disease. Cell 2009;139:871–90. [DOI] [PubMed] [Google Scholar]
- 6. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265–73. [DOI] [PubMed] [Google Scholar]
- 7. Alamgeer M, Peacock CD, Matsui W, et al. Cancer stem cells in lung cancer: evidence and controversies. Respirology 2013;18:757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houthuijzen JM, Daenen LG, Roodhart JM, et al. The role of mesenchymal stem cells in anti‐cancer drug resistance and tumour progression. Br J Cancer 2012;106:1901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leprieur EG, Tolani B, Li H, et al. Membrane‐bound full‐length sonic hedgehog identifies cancer stem cells in human non‐small cell lung cancer. Oncotarget 2017;8:103744–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gong L, Song J, Lin X, et al. Serine‐arginine protein kinase 1 promotes a cancer stem cell‐like phenotype through activation of Wnt/beta‐catenin signalling in NSCLC. J Pathol 2016;240:184–96. [DOI] [PubMed] [Google Scholar]
- 11. Abhold EL, Kiang A, Rahimy E, et al. EGFR kinase promotes acquisition of stem cell‐like properties: a potential therapeutic target in head and neck squamous cell carcinoma stem cells. PLoS One 2012;7:e32459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rybak AP, Tang D. SOX2 plays a critical role in EGFR‐mediated self‐renewal of human prostate cancer stem‐like cells. Cell Signal 2013;25:2734–42. [DOI] [PubMed] [Google Scholar]
- 13. Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cell 2014;3:304–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prasad SB, Yadav SS, Das M, et al. PI3K/AKT pathway‐mediated regulation of p27(Kip1) is associated with cell cycle arrest and apoptosis in cervical cancer. Cell Oncol 2015;38:215–25. [DOI] [PubMed] [Google Scholar]
- 15. Sweeny L, Dean NR, Magnuson JS, et al. EGFR expression in advanced head and neck cutaneous squamous cell carcinoma. Head Neck 2012;34:681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paugh BS, Zhu X, Qu C, et al. Novel oncogenic PDGFRA mutations in pediatric high‐grade gliomas. Cancer Res 2013;73:6219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non‐small‐cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng Y, Kurland BF, Wang J, et al. High epidermal growth factor receptor expression in metastatic colorectal cancer lymph nodes may be more prognostic of poor survival than in primary tumor. Am J Clin Oncol 2009;32:245–52. [DOI] [PubMed] [Google Scholar]
- 19. Laimer K, Spizzo G, Gastl G, et al. High EGFR expression predicts poor prognosis in patients with squamous cell carcinoma of the oral cavity and oropharynx: a TMA‐based immunohistochemical analysis. Oral Oncol 2007;43:193–8. [DOI] [PubMed] [Google Scholar]
- 20. Peled N, Yoshida K, Wynes MW, et al. Predictive and prognostic markers for epidermal growth factor receptor inhibitor therapy in non‐small cell lung cancer. Ther Adv Med Oncology 2009;1:137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scagliotti GV, Selvaggi G, Novello S, et al. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res 2004;10:4227s–32s. [DOI] [PubMed] [Google Scholar]
- 22. Takahashi S, Doss C, Levy S, et al. TAPA‐1, the target of an antiproliferative antibody, is associated on the cell surface with the Leu‐13 antigen. J Immunol 1990;145:2207–13. [PubMed] [Google Scholar]
- 23. Bradbury LE, Kansas GS, Levy S, et al. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody‐1 and Leu‐13 molecules. J Immunol 1992;149:2841–50. [PubMed] [Google Scholar]
- 24. Lewin AR, Reid LE, McMahon M, et al. Molecular analysis of a human interferon‐inducible gene family. Eur J Biochem 1991;199:417–23. [DOI] [PubMed] [Google Scholar]
- 25. Sato S, Miller AS, Howard MC, et al. Regulation of B lymphocyte development and activation by the CD19/CD21/CD81/Leu 13 complex requires the cytoplasmic domain of CD19. J Immunol 1997;159:3278–87. [PubMed] [Google Scholar]
- 26. Chattopadhyay I, Phukan R, Singh A, et al. Molecular profiling to identify molecular mechanism in esophageal cancer with familial clustering. Oncol Rep 2009;21:1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatano H, Kudo Y, Ogawa I, et al. IFN‐induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res 2008;14:6097–105. [DOI] [PubMed] [Google Scholar]
- 28. Sari IN, Yang YG, Phi LT, et al. Interferon‐induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget 2016;7:86039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leung EL, Fiscus RR, Tung JW, et al. Non‐small cell lung cancer cells expressing CD44 are enriched for stem cell‐like properties. PLoS One 2010;5:e14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243–60. [DOI] [PubMed] [Google Scholar]
- 31. Gyorffy B, Dietel M, Fekete T, et al. A snapshot of microarray‐generated gene expression signatures associated with ovarian carcinoma. Int J Gynecol Cancer 2008;18:1215–33. [DOI] [PubMed] [Google Scholar]
- 32. Seyfried NT, Huysentruyt LC, Atwood JA 3rd, et al. Up‐regulation of NG2 proteoglycan and interferon‐induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer Lett 2008;263:243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey CC, Zhong G, Huang IC, et al. IFITM‐family proteins: the Cell's first line of antiviral defense. Annu Rev Virol 2014;1:261–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu F, Ng SS, Chow BK, et al. Knockdown of interferon‐induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neurooncol 2011;103:187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu F, Xie D, Ng SS, et al. IFITM1 promotes the metastasis of human colorectal cancer via CAV‐1. Cancer Lett 2015;368:135–43. [DOI] [PubMed] [Google Scholar]
- 36. Senetta R, Miracco C, Lanzafame S, et al. Epidermal growth factor receptor and caveolin‐1 coexpression identifies adult supratentorial ependymomas with rapid unfavorable outcomes. Neuro Oncol 2011;13:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jankovic J, Tatic S, Bozic V, et al. Inverse expression of caveolin‐1 and EGFR in thyroid cancer patients. Hum Pathol 2017;61:164–72. [DOI] [PubMed] [Google Scholar]
- 38. Cui Y, Zhu T, Song X, et al. Downregulation of caveolin‐1 increased EGFR‐TKIs sensitivity in lung adenocarcinoma cell line with EGFR mutation. Biochem Biophys Res Commun 2018;495:733–9. [DOI] [PubMed] [Google Scholar]
- 39. Lu Z, Ghosh S, Wang Z, et al. Downregulation of caveolin‐1 function by EGF leads to the loss of E‐cadherin, increased transcriptional activity of beta‐catenin, and enhanced tumor cell invasion. Cancer Cell 2003;4:499–515. [DOI] [PubMed] [Google Scholar]
- 40. Shafarenko M, Liebermann DA, Hoffman B. Egr‐1 abrogates the block imparted by c‐Myc on terminal M1 myeloid differentiation. Blood 2005;106:871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwan KY, Shen J, Corey DP. C‐MYC transcriptionally amplifies SOX2 target genes to regulate self‐renewal in multipotent otic progenitor cells. Stem Cell Rep 2015;4:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park SB, Seo KW, So AY, et al. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf‐1 and c‐MYC. Cell Death Differ 2012;19:534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boone DN, Qi Y, Li Z, et al. Egr1 mediates p53‐independent c‐Myc‐induced apoptosis via a noncanonical ARF‐dependent transcriptional mechanism. Proc Natl Acad Sci U S A 2011;108:632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu D, Li WM, Mo XM, et al. Multiparametric flow cytometry analyzes the expressions of immunophenotype CD133, CD34, CD44 in lung cancer naive cell. Sichuan Da Xue Xue Bao Yi Xue Ban 2008;39:827–31. [PubMed] [Google Scholar]
- 45. Chan KM, Wu TH, Cheng CH, et al. Prognostic significance of the number of tumors and aggressive surgical approach in colorectal cancer hepatic metastasis. World J Surg Oncol 2014;12:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okudela K, Woo T, Mitsui H, et al. Expression of the potential cancer stem cell markers, CD133, CD44, ALDH1, and beta‐catenin, in primary lung adenocarcinoma‐‐their prognostic significance. Pathol Int 2012;62:792–801. [DOI] [PubMed] [Google Scholar]
- 47. Andreu P, Colnot S, Godard C, et al. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res 2006;66:1949–55. [DOI] [PubMed] [Google Scholar]
- 48. Kim JY, Kim WJ, Kim H, et al. The stimulation of CD147 induces MMP‐9 expression through ERK and NF‐kappaB in macrophages: implication for atherosclerosis. Immune Netw 2009;9:90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim JY, Kim H, Suk K, et al. Activation of CD147 with cyclophilin a induces the expression of IFITM1 through ERK and PI3K in THP‐1 cells. Mediators Inflamm 2010;2010:821940–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gaedcke J, Grade M, Jung K, et al. Mutated KRAS results in overexpression of DUSP4, a MAP‐kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer 2010;49:1024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de la Rocha AM, Sampron N, Alonso MM, et al. Role of SOX family of transcription factors in central nervous system tumors. Am J Cancer Res 2014;4:312–24. [PMC free article] [PubMed] [Google Scholar]
- 52. Boumahdi S, Driessens G, Lapouge G, et al. SOX2 controls tumour initiation and cancer stem‐cell functions in squamous‐cell carcinoma. Nature 2014;511:246–50. [DOI] [PubMed] [Google Scholar]
- 53. Drilon A, Rekhtman N, Ladanyi M, et al. Squamous‐cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol 2012;13:e418–26. [DOI] [PubMed] [Google Scholar]
- 54. Lundberg IV, Edin S, Eklof V, et al. SOX2 expression is associated with a cancer stem cell state and down‐regulation of CDX2 in colorectal cancer. BMC Cancer 2016;16:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial‐mesenchymal transition to suppression of anoikis. J Cell Sci 2013;126:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wirth M, Stojanovic N, Christian J, et al. MYC and EGR1 synergize to trigger tumor cell death by controlling NOXA and BIM transcription upon treatment with the proteasome inhibitor bortezomib. Nucleic Acids Res 2014;42:10433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu C, Zhang HF, Gupta N, et al. A positive feedback loop involving the Wnt/beta‐catenin/MYC/Sox2 axis defines a highly tumorigenic cell subpopulation in ALK‐positive anaplastic large cell lymphoma. J Hematol Oncol 2016;9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arora S, Wang Y, Jia Z, et al. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP‐on‐chip. Genome Biol 2008;9:R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage‐survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hussenet T, du Manoir S. SOX2 in squamous cell carcinoma: amplifying a pleiotropic oncogene along carcinogenesis. Cell Cycle 2010;9:1480–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. IFITM1 depletion does not affect the expression level of IFITM2 and IFITM3.
NSCLC cell lines were infected with either control (shLacZ) virus or IFITM1 knockdown virus (642‐shIFITM1 and 870‐shIFITM1) and selected with puromycin or sorted by flow cytometry. Both RNA and proteins were isolated to determine the expression of IFITM1. RT‐qPCR was performed to determine IFITM1, 2, 3 mRNA expression in H1650 (a, g) and A549 (d, h) cells. Immunoblots with anti‐IFITM1 antibody were conducted to analyze IFITM1 protein level in H1650 (b, c) and A549 (e, f), and Quantification of protein levels was performed using Image J software from three separate experiments. (*p < 0.05, **p < 0.001).
Supplementary Figure 2. IFITM1 depletion impairs sphere forming ability of NSCLC cells
Control (shLacZ) or IFITM1‐depleted (shIFITM1) H1650 (a) and A549 (b) were serially plated to determine sphere forming ability. Each plate was incubated in stem cell selective media for 10 days. Then spheres were washed with PBS, dissociated to single cells by trypsin and plated again. The data are shown as mean ± SEM of three independent experiments. (c) Representative images of tertiary plating are presented. (*p < 0.05, **p < 0.001).
Supplementary Figure 3. IFITM1 is essential for the maintenance of EMT signature of NSCLC cell lines in vitro.
(a) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) A549 cells were treated with 5 ng/ml TGF‐β, and the RNA level of EMT genes were determined by RT‐qPCR. (b and c) Control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) H1650 cell lysates were isolated and the protein level of EMT genes were probed with antibodies indicated. ACTIN was used as a loading control. Quantification of protein levels was performed using Image J software from three separate experiments (b, mesenchymal signature; c, epithelial signature). (d, e) The cell lysates were isolated from control (shLacZ) or IFITM1‐depleted (642‐shIFITM1 and 870‐shIFITM1) A549 cells and probed with antibodies indicated. ACTIN was used as a loading control. Representative images were shown (d) and quantification of protein levels was performed using Image J software from three separate experiments (e). (*p < 0.05, **p < 0.001).
Supplementary Figure 4. IFITM1 depletion modestly impairs the expression of cancer stem cell makers of A549 cell line
A549 cell line was transduced with either control (shLacZ) or IFITM1 (642‐shIFITM1 and 870‐shIFITM1) knockdown virus and selected with puromycin or sorted by flowcytometry. Cells were cultured on ultra low attachment plates with stem cell selective medium for 3 days. Then, cell surface maker CD133 (a, b) and CD44 (c, d) were analyzed with flow cytometry. (b, d) Representative images of histogram of CD133 and CD44. Data shown are from more than three experiments (a, c). (*p < 0.05, **p < 0.001).
Supplementary Figure 5. Macroscopic view of tumor nodules on the lungs and liver
Control (shLacZ) or IFITM1‐depleted (870‐shIFITM1) H1650 cells were orthotopically injected into the lungs of NSG mice, and then mice were monitored for tumor metastasis. Macroscopic view of tumor nodules on the lungs and liver.
Supplementary Figure 6. EGF does not induce the expression of IFITM1.
NSCLC cells were cultured with or without 100 ng/ml of EGF in conventional media for 1 and 3 days. Then, RNA was isolated and RT‐qPCR was performed to evaluate the mRNA expression of IFITM1 in H1650 (a and b) and A549 (c and d).
Supplementary Table 1. Primer sequences for RT‐qPCR
Supplementary Table 2. Demographic and clinical characteristics
Supplementary Table 3. Correlation of IFITM1 expression with pathologic characteristics in patients with adenocarcinoma
Supplementary Table 4. Multivariate analyses of overall survival in patients with adenocarcinoma
Supplementary Table 5. IFITM1 expression in squamous cell carcinoma and adenocarcinoma
Supplementary Table 6. Correlation of IFITM1 expression with SOX2, EGFR expression in squamous cell carcinoma
Supplementary Table 7. Correlation of IFITM1 expression with SOX2, EGFR expression in patients with adenocarcinoma
Supplementary Table 8. Correlation of IFITM1 expression with SOX2, EGFR expression in all patients.


