Abstract
Computational modeling of tumor growth has become an invaluable tool to simulate complex cell–cell interactions and emerging population-level dynamics. Agent-based models are commonly used to describe the behavior and interaction of individual cells in different environments. Behavioral rules can be informed and calibrated by in vitro assays, and emerging population-level dynamics may be validated with both in vitro and in vivo experiments. Here, we describe the design and implementation of a lattice-based agent-based model of cancer stem cell driven tumor growth.
Keywords: Agent-based model, Tumor growth, Cancer stem cell, Calibration, Domain, Search order, High performance, Digital cell line
1. Introduction
Agent-based modeling has a long history in quantitative oncology [1–3], including stem cell dynamics [4–9], and heterogeneity [10–12]. Such models can help predict disease progression and make invaluable recommendations for therapeutic interventions [13, 14]. Agent-based models simulate the behavior and interaction of individual cells. Behavioral rules can be dependent on environmental conditions, which include chemicals in the extracellular environment [15–17], supporting structures in the extracellular matrix [18, 19], fluid dynamics [20], physical forces [21], or presence and interactions with other cells [22, 23]. Computer simulations are usually initialized with a single cell or a cluster of individual cells, and the status and behavior of each cell are typically updated at discrete time points based on their internal rules and current environmental conditions. Such models may help identify if cancer stem cells comprise a subpopulation of specific proportion in a tumor [24], and how to deliver radiotherapy doses efficiently to eradicate cancer stem cells [25].
2. Materials
Implement all classes and functions in a concurrent version system to allow shared programming and efficient debugging.
2.1. Lattice
A finite 2D (or 3D if necessary) lattice, where each site can be occupied by a single or a population of cells (crowding).
Lattice size. Simulations are commonly initialized with either (a) a small number of cells to observe emergent population-level behavior, or (b) a populated tissue architecture. Define lattice size to accommodate anticipated final cell number. Account for possible boundary effects in case of a growing population. Set the size of a single lattice site to the size of a cell.
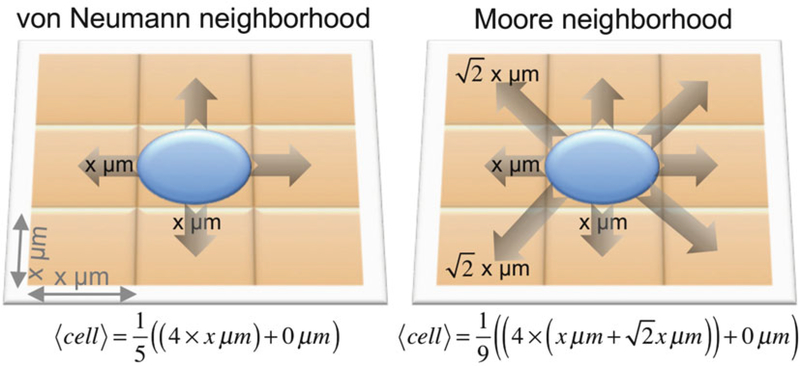
Neighborhood. Determine if cells interact with their four orthogonal neighbors (north, south, east, west; von Neumann neighborhood), or with the adjacent eight neighboring lattice points (northwest, north, northeast, east, west, southeast, south, southwest; Moore neighborhood) (Fig. 1).
Boundary conditions. Define behavior of cells on the boundary of the lattice, set to either periodic (the boundary “wraps”, so cells on the left edge interact with cells on the far right edge) or no-flux reflective (cells on the edge of the lattice only interact with interior and edge cells). Note that simulations with no-flux boundary conditions may introduce boundary effects, i.e., accumulation of cells near the boundary. For dynamically expanding arrays no boundary conditions are necessary.
Fig. 1.
Schematic of expected cell displacement, cell, in the von Neumann (left) and Moore (right) neighborhoods on a two-dimensional lattice with lattice sizes of x2 μm2
2.2. Cell
Each cell is an individual entity with the following basic attributes:
Time to next division event (tc).
Type of the cell—stem/non-stem (isStem).
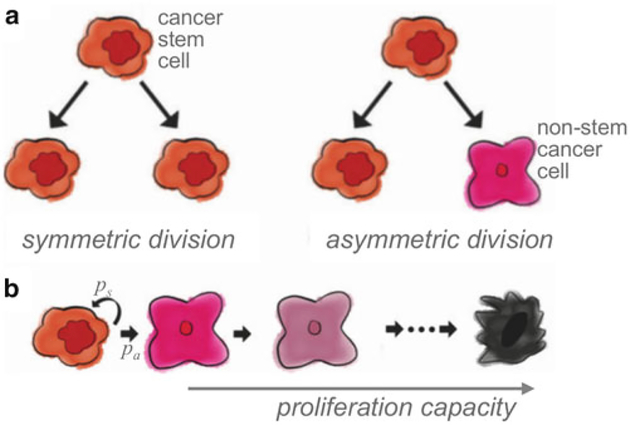
Probability of symmetric division (ps). Cancer stem cells divide either symmetrically to produce two identical cancer stem cells, or asymmetrically to produce a cancer stem cell and a non-stem cancer cell [26]. In the case of cancer stem cell define probabilities of symmetric division, 0 < ps ≤ 1, and asymmetric division pa = 1 − ps (Fig. 2).
Telomere length (p). Set the telomere length of the initial cell or cell population as a molecular clock [27–29], which is a quantification of the Hayflick limit [30].
Current cell cycle phase (if required). If information about specific cell cycle phases is required (such as for simulations of cell cycle specific chemotherapeutics) define cell cycle phases. Cell cycle length can be divided into fractions comparable to experimentally measured cell cycle distributions.
Probability of spontaneous death (α). Cancer cells (isStem = false) accumulate mutations that introduce genomic instability which may lead to premature cell death. Define a probability of spontaneous cell death, α ≥ 0, at which rate cells may die during cell division attempts. Cancer stem cells (isStem = true) have a superior DNA damage repair machinery [31–33] and, thus, the probability of spontaneous cell death can be set to zero. For more biological realism, define a cell death rate that is larger than zero, but less than the self-renewal rate to ensure net population growth.
Fig. 2.
(a) Schematic of cancer stem cell symmetric and asymmetric division. (b) Schematic representation of cancer stem cell symmetric (ps) and asymmetric division (pa) and non-stem cancer cell proliferation capacity. Adapted from [24]
Each cell is equipped with a set of basic functions:
- Procedure advance time (input arguments = time increment Δt, list of available sites in the direct neighborhood).
- Decrease time to next division (tc) by Δt. If tc < 0 set tc = 0.
- Update current cell cycle phase (if necessary).
- If tc ≤ 0 and there is available space, then perform division and generate new times to next division for both resulting cells.
-
Procedure divide (input argument = list of available sites in the direct neighborhood).
Choose a random number 0≤n≤1 from the uniform distribution. If n < α, then simulate cell death, i.e., remove cell from the simulation and instantaneously make a corresponding lattice point available. If required, instead of instantaneous cell removal, site on the lattice can be made available for new cells after specified amount of time, e.g., in order to simulate duration of apoptosis process. Otherwise:- If the cell is not-stem (isStem = false) then decrease the proliferation capacity (p) by one, as non-stem cancer cells do not upregulate telomerase and are thus not long-lived and cannot initiate, retain, and reinitiate tumors [34, 35]. Simulate cell death by removing the cell from the simulation if the proliferation capacity is exhausted, i.e., less or equal to zero. Otherwise, new cell is a clone of the mother cell, i.e., non-stem cancer cell only produces non-stem cancer cell and decremented proliferation capacity is inherited by the daughter cell. Place new cell at vacant neighboring lattice point at random.
- If the cell is cancer stem cell (isStem = true), then draw a number from uniform distribution to decide if the division is symmetric based on the probability ps. If division is symmetric, new cell is a perfect clone of the mother cell; that is a cancer stem cells do not erode telomeres and retain identical proliferation capacity after mitosis. Otherwise, non-stem cancer cell offspring of a cancer stem cell inherit the current telomere length and determined proliferation capacity, i.e., new cell is identical except for attribute isStem, which set to false. Place new cell at vacant neighboring lattice point at random.
Procedure generate time to next division. Cells divide on average every 24 h. Derive specific cell cycle times tc (hours), averages and standard deviations from proliferation rate calculations from clonogenic assays or live microscopy imaging [36].
- Procedure random migration (input argument = list of available sites in the direct neighborhood). Cells may perform a random walk, and probabilities of migrating into adjacent lattice points can be obtained from a discretized diffusion equation [37, 38]. Assuming Moore neighborhood (see Sect. 2.1 above) and a cell at position (x0,y0): at time t can move based on available lattice points in the immediate eight-cell Moore neighborhood N(x0,y0) with probabilities
With
With probability 1/(1+number of unoccupied neighboring lattice points) the cell remains temporarily stationary, and a cell that completely surrounded by other cells is not moving. Note that in this implementation, movement to all adjacent lattice points is equally weighted. This can be modified to account for increased distance to diagonal lattice points. Procedure directed migration (if required) (input arguments = list of available sites in the direct neighborhood, function F whose gradient ∇F describes the migration stimulus, such as chemoattractant or chemorepellant). Vector describes the vector connecting the current cell position (x0, y0) to the center of one of the adjacent lattice points (xi, yi). Directed migration only occurs towards available lattice sites (S) when . Weight each available site S by the factor cos(θ), where θ is the angle between the gradient ∇F and the movement direction for each possible movement). (Notice that cos(θ) = 1 when ∇F and are parallel, and cos(θ) = 0 when they are perpendicular, to give greatest weight to travel along the chemoattractant gradient direction.)
Sum each cos(θ) over the S available sites to get the averaged cosine value AS. Define the probability of mobilization G through an arbitrary chemotactic/haptotactic responsiveness function via G = H(|∇F|AS/(AS + 1)), where H has the properties:
H(0) = 0 (no directed motion when no lattice sites are available or the chemical gradient is zero),
H is a (monotonically) increasing function (directed motility increases with the magnitude chemotactic signal and alignment with the lattice), and
H tends to 1 as |∇F|AS/(AS + 1) approaches ∞(directed motility increases with the chemotactic signal, but saturates at a maximal level scaled to 1).
If a cell moves, its direction is weighted by the respective cosines for movement, so that cos(θ)G/(AS) is the probability of movement to the available site at angle θ. This method is explained in detail elsewhere [39].
3. Methods
3.1. Programming Environment
Define programming language. Agent-based models can be implemented and simulated in any programming language; most prominent languages including C++, Java, Julia, Python, and Matlab. Each of these languages offers different computational speed and coding feasibility. C++ is considered as the environment offering the best performance, but has a high programming complexity. Matlab offers a great number of built-in functions and is easy to code in, but has significantly lower performance (e.g., when using nested loops).
Define graphical output. Visualize simulation solutions of agent-based models using existing implementations of graphical programming or implement specific visualization tools.
Agent-based software packages. Utilize predeveloped agent-based software packages; most prominent include Netlogo [40, 41], CompuCell3D [36, 42], Chaste [43], or Swarm [44].
3.2. Simulation Procedure
Time step. Define the simulation time step, Δt, such that Δt is smaller than the fastest biological process that is being considered in the model. In a model of cell proliferation (~1/day) and cell migration (~1 cell width/h), set Δt ≤ 1 h.
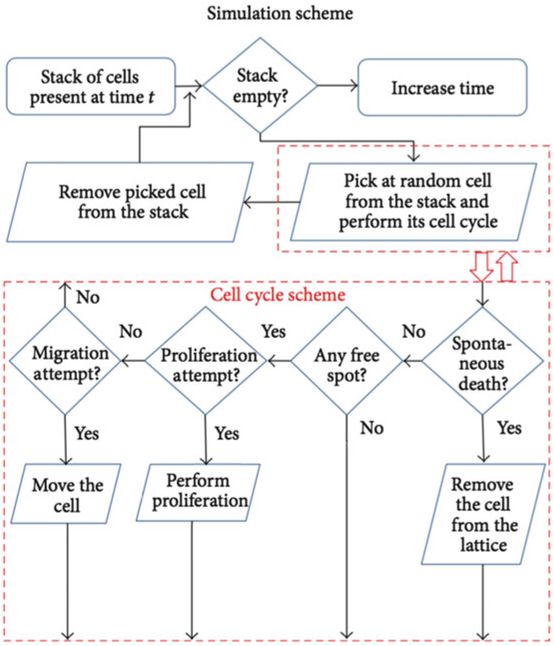
Develop a simulation flowchart to conceptualize and visualize the simulation procedure. At each defined discrete simulation time step, consider mutually exclusive processes that occur with lowest rate constant first. Let us assume that cell migration and cell proliferation are mutually exclusive (e.g., cells could migrate through most of G1, S, G2 phases). Check if maturation age is reached; if not, check if migration time is reached (Fig. 3).
Lattice update sequence. Update cells in random order to avoid lattice geometry effects. Maintain a list of “live” cell agents and select cells for update from this list at random.
Cell update sequence. Consider vacancy in cell neighborhood in random order to avoid lattice geometry effects. Maintain a list of vacant adjacent lattice sites for each cell agent and select for update from this list at random.
Fig. 3.
Sample simulation flowchart with cell cycle scheme
4. Notes
Homogenous populations. The presented model produces a heterogeneous population of stem and non-stem cancer cells. If ps = 1, the population remains (if only initialized with cancer stem cells) or will become (if initialized with both stem and non-stem cancer cells) homogenous comprised of only cancer stem cells. If ps = 0, only non-stem cancer cells are produced and the resulting population size oscillates around a dynamic equilibrium or, if the probability of cancer stem cell death is positive and non-zero, the population will inevitably die out [45].
Plasticity. The presented model considers cancer stemness a cell phenotype, whereas recent literature may suggest stemness to be a reversible trait [46, 47]. The model can be extended to simulate phenotypic plasticity through cancer stem cell differentiation (that is, the cancer stem cell phenotype is set to non-stem cancer cell) and non-stem cancer cell de-differentiation (that is, the non-stem cancer cell phenotype is set to cancer stem cell) [48].
Senescence. The presented model considers cell death after exhaustion of proliferation potential. Alternatively, cells may enter senescence [49] and, although mitotically inactive, continue to consume resources that influence the behavior of remaining cancer cells [50].
Evolution. The presented model considers fixed parameters or rate constants for each cell trait. Rate constants can change due to mutations and genetic drift [51], which allows for simulations of the evolution of cancer stem cell traits under different environmental conditions [12, 52].
Carcinogenesis. Such modeling framework can be adapted to model tissue homeostasis and mutation/selection cascades during cancer development [16, 53].
To ensure model results are reproducible, simulation inputs (parameters) and outputs should be recorded using open, standardized data formats, using biologically driven data elements that can be reused in independent models. The MultiCellDS (multicellular data standard) Project assembled a cross-disciplinary team of biologists, modelers, data scientists, and clinicians to draft a standard for digital cell lines, which record phenotype parameters such cell cycle time scales, apoptosis rates, and maximum proliferative capacity. Separate digital cell lines can represent cancer stem cells and non-stem cells. Similarly, standardized simulation outputs can record the position, cell cycle status, and other properties of simulated cells. Using standards will allow better software compatibility, facilitate cross-model validation, and streamline development of user-friendly software to start, modify, visualize, and analyze computational models. See MultiCellDS.org for further details.
Model assumptions (e.g., Fig. 3) should also be recorded with standardized data formats. The Cell Behavior Ontology (CBO) [54] provides a “dictionary” (ontology) of applicable stem cell behaviors, but it may not be able to fully capture the entire model logic in its present form. Extensions to the Systems Biology Markup Language (SBML) [55], such as SBML-Dynamic [56], are currently being developed to leverage the model structure of SBML and the ontology of CBO to annotate the mathematical structure of agent-based models, but the standards are not yet complete.
Treatment. The presented agent-based model can be extended to account for the effects of cancer therapy including radiation [57–59], chemotherapy [60], oncolytic viruses [61], or immunotherapy [62].
Analysis. Agent-based models can be rigorously analyzed for sensitivity and stability [63, 64]. Comparison of coarse-grained model behavior (e.g., growth curves) with known analytical results can quality-check calibration protocols and the computational implementation.
Dynamically expanding domains. Tradeoffs between lattice size and computing speed can be avoided using dynamically growing domains [65].
Hybrid models. The discussed model setup can be extended to a hybrid discrete-continuous framework where single cells are modeled as discrete agents, and environmental chemicals diffuse on a mapped continuum layer [66–68]. In those models cell cycle time may vary with environmental conditions (e.g., [16] and [69]).
Cell neighborhood. In more complex cases daughter cells can populate empty lattice sites within a given distance to approximate tissue deformability.
Acknowledgments
JP and HE were partially supported by the Personalized Medicine Award 09-33000-15-03 from the DeBartolo Family Personalized Medicine Institute Pilot Research Awards in Personalized Medicine (PRAPM). PM was supported by the Breast Cancer Research Foundation and the National Institutes of Health [1R01CA180149].
References
- 1.Anderson A, Chaplain MAJ, Rejniak K (2007) Single-cell-based models in biology and medicine. Springer Science & Business Media, Birkhäuser Basel, ISBN 978-3-7643-8101-1 [Google Scholar]
- 2.Enderling H, Rejniak KA (2013) Simulating cancer: computational models in oncology. Frontiers Oncol 3:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Butner JD, Kerketta R et al. (2015) Simulating cancer growth with multiscale agent-based modeling. Semin Cancer Biol 30:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.d’Inverno M, Saunders R (2004) Agent-based modelling of stem cell self-organisation in a niche In: Engineering Self-Organising Systems. Springer, Berlin, pp 52–68 [Google Scholar]
- 5.Mallet DG, de Pillis LG (2006) A cellular automata model of tumor-immune system interactions. J Theor Biol 239:334–350 [DOI] [PubMed] [Google Scholar]
- 6.Chao DL, Eck JT, Brash DE et al. (2008) Preneoplastic lesion growth driven by the death of adjacent normal stem cells. Proc Natl Acad Sci U S A 105:15034–15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enderling H, Hlatky L, Hahnfeldt P (2009) Migration rules: tumours are conglomerates of self-metastases. Br J Cancer 100:1917–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enderling H, Anderson ARA, Chaplain MAJ et al. (2009) Paradoxical dependencies of tumor dormancy and progression on basic cell kinetics. Cancer Res 69:8814–8821 [DOI] [PubMed] [Google Scholar]
- 9.Norton K-A, Popel AS (2014) An agent-based model of cancer stem cell initiated avascular tumour growth and metastasis: the effect of seeding frequency and location. J R Soc Interface 11, pii: 20140640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlee P, Anderson ARA (2008) A hybrid cellular automaton model of clonal evolution in cancer: the emergence of the glycolytic pheno-type. J Theor Biol 250:705–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sottoriva A, Verhoeff JJC, Borovski T et al. (2010) Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res 70:46–56 [DOI] [PubMed] [Google Scholar]
- 12.Poleszczuk J, Hahnfeldt P, Enderling H (2015) Evolution and phenotypic selection of cancer stem cells. PLoS Comput Biol 11: e1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson ARA, Quaranta V (2008) Integrative mathematical oncology. Nat Rev Cancer 8:227–234 [DOI] [PubMed] [Google Scholar]
- 14.Yankeelov TE, Quaranta V, Evans KJ et al. (2015) Toward a science of tumor forecasting for clinical oncology. Cancer Res 75:918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xavier JB, Foster KR (2007) Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci U S A 104:876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatenby RA, Smallbone K, Maini PK et al. (2007) Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br J Cancer 97:646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enderling H, Hlatky L, Hahnfeldt P (2012) The promoting role of a tumour-secreted chemorepellent in self-metastatic tumour progression. Math Med Biol 29:21–29 [DOI] [PubMed] [Google Scholar]
- 18.Enderling H, Alexander NR, Clark ES et al. (2008) Dependence of invadopodia function on collagen fiber spacing and cross-linking: computational modeling and experimental evidence. Biophys J 95:2203–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlüter DK, Ramis-Conde I, Chaplain MAJ (2012) Computational modeling of single-cell migration: the leading role of extracellular matrix fibers. Biophys J 103:1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rejniak KA (2007) An immersed boundary framework for modelling the growth of individual cells: an application to the early tumour development. J Theor Biol 247:186–204 [DOI] [PubMed] [Google Scholar]
- 21.Drasdo D, Höhme S (2005) A single-cell-based model of tumor growth in vitro: mono-layers and spheroids. Phys Biol 2:133–147 [DOI] [PubMed] [Google Scholar]
- 22.Ramis-Conde I, Drasdo D, Anderson ARA et al. (2008) Modeling the influence of the E-cadherin-beta-catenin pathway in cancer cell invasion: a multiscale approach. Biophys J 95:155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlüter DK, Ramis-Conde I, Chaplain MAJ (2015) Multi-scale modelling of the dynamics of cell colonies: insights into cell-adhesion forces and cancer invasion from in silico simulations. J R Soc Interface 12, piii: 20141080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enderling H (2014) Cancer stem cells: small subpopulation or evolving fraction? Integr Biol 7:14–23 [DOI] [PubMed] [Google Scholar]
- 25.Alfonso JCL, Jagiella N, Núñez L et al. (2014) Estimating dose painting effects in radiotherapy: a mathematical model. PLoS One 9: e89380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dingli D, Traulsen A, Michor F (2007) (A) symmetric stem cell replication and cancer. PLoS Comput Biol 3:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olovnikov AM (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41:181–190 [DOI] [PubMed] [Google Scholar]
- 28.Blackburn EH, Gall JG (1978) A tandemly repeated sequence at the termini of the extra-chromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120:33–53 [DOI] [PubMed] [Google Scholar]
- 29.Harley CB (1991) Telomere loss: mitotic clock or genetic time bomb? Mutat Res DNAging 256:271–282 [DOI] [PubMed] [Google Scholar]
- 30.Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37:614–636 [DOI] [PubMed] [Google Scholar]
- 31.Bao S, Wu Q, McLendon RE et al. (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760 [DOI] [PubMed] [Google Scholar]
- 32.Maugeri-Saccà M, Bartucci M, De Maria R (2012) DNA damage repair pathways in cancer stem cells. Mol Cancer Ther 11:1627–1636 [DOI] [PubMed] [Google Scholar]
- 33.Skvortsov S, Debbage P, Lukas P et al. (2015) Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin Cancer Biol 31:36–42 [DOI] [PubMed] [Google Scholar]
- 34.Allsopp RC, Morin GB, DePinho R et al. (2003) Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Hematopoiesis 102:517–520 [DOI] [PubMed] [Google Scholar]
- 35.Shay JW, Wright WE (2010) Telomeres and telomerase in normal and cancer stem cells. FEBS Lett 584:3819–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, McDonald JT, Hlatky L et al. (2013) Acute and fractionated irradiation differentially modulate glioma stem cell division kinetics. Cancer Res 73:1481–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson ARA, Chaplain MAJ, Newman EL et al. (2000) Mathematical modelling of tumour invasion and metastasis. Comput Math Methods Med 2:129–154 [Google Scholar]
- 38.Anderson ARA, Weaver AM, Cummings PT et al. (2006) Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell 127:905–915 [DOI] [PubMed] [Google Scholar]
- 39.Enderling H, Hlatky L, Hahnfeldt P (2010) Tumor morphological evolution: directed migration and gain and loss of the self-metastatic phenotype. Biol Direct 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kareva I (2015) Immune evasion through competitive inhibition: the shielding effect of cancer non-stem cells. J Theor Biol 364:40–48 [DOI] [PubMed] [Google Scholar]
- 41.Bravo R, Axelrod DE (2013) A calibrated agent-based computer model of stochastic cell dynamics in normal human colon crypts useful for in silico experiments. Theor Biol Med Model 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swat MH, Thomas GL, Belmonte JM et al. (2012) Multi-scale modeling of tissues using CompuCell3D. Methods Cell Biol 110:325–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirams GR, Arthurs CJ, Bernabeu MO et al. (2013) Chaste: an open source C++ library for computational physiology and biology. PLoS Comput Biol 9:e1002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minar N, Burkhart R, Langton C et al. (1996) The swarm simulation system: a toolkit for building multi-agent simulations Working Paper 96-06-042, Santa Fe Institute, Santa Fe [Google Scholar]
- 45.Enderling H (2013) Cancer stem cells and tumor dormancy. Adv Exp Med Biol 734:55–71 [DOI] [PubMed] [Google Scholar]
- 46.Marjanovic ND, Weinberg RA, Chaffer CL (2013) Cell plasticity and heterogeneity in cancer. Clin Chem 59:168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwitalla S (2014) Tumor cell plasticity: the challenge to catch a moving target. J Gastroenterol 49:618–627 [DOI] [PubMed] [Google Scholar]
- 48.Poleszczuk J, Enderling H (2016) Cancer stem cell plasticity as tumor growth promoter and catalyst of population collapse. Stem Cells Int. 2016:3923527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campisi J, Kim SH, Lim CS et al. (2001) Cellular senescence, cancer and aging: the telo-mere connection. Exp Gerontol 36:1619–1637 [DOI] [PubMed] [Google Scholar]
- 50.Poleszczuk J, Hahnfeldt P, Enderling H (2014) Biphasic modulation of cancer stem cell-driven solid tumour dynamics in response to reactivated replicative senescence. Cell Prolif 47:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sottoriva A, Vermeulen L, Tavaré S (2011) Modeling evolutionary dynamics of epigenetic mutations in hierarchically organized tumors. PLoS Comput Biol 7:e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott JG, Hjelmeland AB, Chinnaiyan P et al. (2014) Microenvironmental variables must influence intrinsic phenotypic parameters of cancer stem cells to affect tumourigenicity. PLoS Comput Biol 10:e1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carulli AJ, Samuelson LC, Schnell S (2014) Unraveling intestinal stem cell behavior with models of crypt dynamics. Integr Biol 6:243–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sluka JP, Shirinifard A, Swat M et al. (2014) The cell behavior ontology: describing the intrinsic biological behaviors of real and model cells seen as active agents. Bioinformatics 30:2367–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hucka M, Finney A, Sauro HM et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19:524–531 [DOI] [PubMed] [Google Scholar]
- 56.Myers C, Myers C (2011) Dynamic structures in SBML. Nat Precedings, 10.1038/npre.2011.6342.1 [DOI]
- 57.Enderling H, Park D, Hlatky L et al. (2009) The importance of spatial distribution of stemness and proliferation state in determining tumor radioresponse. Math Model Nat Phenom 4:117–133 [Google Scholar]
- 58.Powathil GG, Kohandel M, Sivaloganathan S et al. (2007) Mathematical modeling of brain tumors: effects of radiotherapy and chemotherapy. Phys Med Biol 52:3291–3306 [DOI] [PubMed] [Google Scholar]
- 59.Kempf H, Hatzikirou H, Bleicher M et al. (2013) In silico analysis of cell cycle synchronisation effects in radiotherapy of tumour spheroids. PLoS Comput Biol 9:e1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Powathil GG, Gordon KE, Hill LA et al. (2012) Modelling the effects of cell-cycle heterogeneity on the response of a solid tumour to chemotherapy: biological insights from a hybrid multiscale cellular automaton model. J Theor Biol 308:1–19 [DOI] [PubMed] [Google Scholar]
- 61.Wodarz D, Hofacre A, Lau JW et al. (2012) Complex spatial dynamics of oncolytic viruses in vitro: mathematical and experimental approaches. PLoS Comput Biol 8:e1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shengjun W, Yunbo G, Liyan S et al. (2012) Quantitative study of cytotoxic T-lymphocyte immunotherapy for nasopharyngeal carcinoma. Theor Biol Med Model 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerlee P, Anderson ARA (2007) Stability analysis of a hybrid cellular automaton model of cell colony growth. Phys Rev E Stat Nonlin Soft Matter Phys 75:051911. [DOI] [PubMed] [Google Scholar]
- 64.Gerlee P, Anderson ARA (2010) Diffusion-limited tumour growth: simulations and analysis. Math Biosci Eng 7:385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poleszczuk J, Enderling H (2014) A high-performance cellular automaton model of tumor growth with dynamically growing domains. Appl Math 5:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson ARA (2005) A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math Med Biol 22:163–186 [DOI] [PubMed] [Google Scholar]
- 67.Gerlee P, Anderson ARA (2007) An evolutionary hybrid cellular automaton model of solid tumour growth. J Theor Biol 246:583–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enderling H, Hlatky L, Hahnfeldt P (2012) Immunoediting: evidence of the multifaceted role of the immune system in self-metastatic tumor growth. Theor Biol Med Model 9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macklin P, Edgerton ME, Thompson AM et al. (2012) Patient-calibrated agent-based modelling of ductal carcinoma in situ (DCIS): from microscopic measurements to macroscopic predictions of clinical progression. J Theor Biol 301:122–140 [DOI] [PMC free article] [PubMed] [Google Scholar]