Abstract
Poly-(ADP-ribose) polymerase (PARP) inhibitors (PARPi) have shown remarkable therapeutic efficacy against BRCA1/2 mutant cancers through a synthetic lethal interaction. PARPi exert their therapeutic effects mainly through the blockade of single-stranded DNA damage repair, which leads to the accumulation of toxic DNA double-strand breaks specifically in cancer cells with DNA repair deficiency (BCRAness), including those harboring BRCA1/2 mutations. Here we show that PARPi-mediated modulation of the immune response contributes to their therapeutic effects independently of BRCA1/2 mutations. PARPi promoted accumulation of cytosolic DNA fragments due to unresolved DNA lesions, which in turn activated the DNA sensing cGAS-STING pathway and stimulated production of type I interferons to induce antitumor immunity independent of BRCAness. These effects of PARPi were further enhanced by immune checkpoint blockade. Overall, these results provide a mechanistic rationale for using PARPi as immunomodulatory agents to harness the therapeutic efficacy of immune checkpoint blockade.
Keywords: PARP1 inhibitors, cytosolic DNA, STING, immune checkpoint blockade
Introduction
Poly (ADP-ribose) polymerase (PARP) inhibitors are approved for the treatment of patients with ovarian and breast cancers harboring BRCA1 or BRCA2 (BRCA1/2) mutations (1,2). The rationale supporting the development of single agent PARP inhibitors (PARPis) in BRCA1/2 mutant cancers was based on the concept of synthetic lethality, which predicted antitumor efficacy in tumors with defects in homologous recombination (HR) repair, also known as BRCAness. PARP1 is the most abundant and ubiquitously expressed member of the PARP family and contributes the majority of PARP enzymatic activity and is thus the major target of PARPis (3). In the presence of DNA damage, PARP1 rapidly binds to DNA strand breaks and is essential for the repair of DNA single-strand breaks (SSBs) through base excision repair. In normal cells, recombinogenic DNA substrates generated by PARPis can be functionally resolved by the error-free HR repair pathway. In contrast, in cancer cells with defective HR repair, such as those deficient in BRCA1 or BRCA2, the DNA substrates generated by PARPis cannot be resolved, and therefore the cells are hypersensitive to PARPis (4,5).
Clinical studies have now also shown patient benefit with PARPis in those with BRCA1/2 wildtype tumors (6,7). A recent Phase III clinical trial confirmed that patients with platinum-sensitive, recurrent ovarian cancer receiving PARPi treatment as maintenance therapy had significantly longer progression-free survival than those on placebo, regardless of BRCA1/2 mutation status or HR repair status (7). These clinical observations raise the key question of whether PARPis can exert antitumor effects through mechanisms other than those leading to unresolved genomic lesions in tumors with DNA repair deficiency.
In this study, we show that PARPi treatment induces IFN-mediated antitumor immune responses. PARPis generate cytosolic double-strand DNA (dsDNA), which activate STING signaling and its associated-transcription programs. These critical changes amplify STING signaling and promote tumor infiltrating lymphocytes (TILs) and antitumor immunity, which can be further enhanced through immune checkpoint blockade.
Materials and Methods
Cell culture.
Cell lines were validated by short tandem repeat (STR) DNA fingerprinting using the AmpF STR identifier kit according to the manufacturer’s instructions (Applied Biosystems, catalogue no. 4322288). The STR profiles were compared to known American Type Culture Collection fingerprints; to the Cell Line Integrated Molecular Authentication database, version 0.1.200808 (Nucleic Acids Research 2009; 37:D925-D932); and to the MD Anderson fingerprint database. The STR profiles matched known DNA fingerprints or were unique. The colorectal and ovarian cancer cell lines were kindly provided by Dr. Gordon B. Mills’ laboratory at The University of Texas MD Anderson Cancer Center. Cell line authentication was performed in the MD Anderson Characterized Cell Line Core in 2012 and 2013. All media were supplemented with 10% FBS with glutamine, penicillin, and streptomycin. The ID8 mouse ovarian surface epithelial cells were kindly provided by Dr. Vahid Afshar-Kharghan’s laboratory at MD Anderson. The ID8 cells were maintained in DMEM (high-glucose, Cellgro) supplemented with 4% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, 5 μg/ml transferrin, and 5 ng/ml sodium selenite. Cells were incubated at 37°C in a humidified incubator with 5% CO2. Mycoplasma testing of these cell lines has confirmed negative results.
Antibodies and reagents.
Anti-γH2AX (JBW301) antibodies were purchase from Millipore Sigma. Anti-β-Actin (A2228), anti-α-Tubulin (T6074) and anti-γ-Tubulin (SAB4503045) antibodies were purchase from Sigma-Aldrich. Anti-STING (D2P2F, #13647), anti-cGAS (D1D3G, #15102), anti-IRF3 (D6I4C, #11904), anti-phospho-IRF3 (Ser396, D6O1M, #29047), anti-TBK1 (D1B4, #3504), anti-phospho-TBK1 (Ser172, D52C2, #5483), anti-CtIP (D76F7, #9201), anti-MRE11 (#4895) and anti-PD-L1 (13684 and 64988) antibodies were purchased from Cell Signaling Technology. Anti-CD8 (sc-7970), anti-BLM (B-4, sc-365753), anti-EXO1 (SPM394, sc-56387) and other antibodies were purchased from Santa Cruz Biotechnology. BMN673 (S7048, Talazoparib) was purchased from Selleck Chemicals. VivoGlo Luciferin was purchased from Promega. Isotype control IgG and anti-PD-L1 (BE0101, clone 10F.9G2) antibodies were purchased from Bio X Cell. The ELISA kits of CCL5 and CXCL10 were purchase from Thermofisher. The multiplexed immunofluorescence IHC kit was purchased from PerkinElmer.
RNA interference.
Knockdown was achieved by RNA interference using a lentiviral vector–based MISSION shRNA (Sigma-Aldrich). The shRNA sequences were as follows: mouse Sting (NM_028261), TRCN0000346319 (#1), AGAGGTCACCGCTCCAAATAT; TRCN0000346266 (#2), CAACATTCGATTCCGAGATAT. SMART pool ON-Target plus siRNA for CtIP (L-011376–00), BLM (L-007287–00), EXO1 (L-007287–00), STING (L-024333–02), IRF3 (L-024333–02), TBK1 (L-024333–02) and cGAS (L-015607–02) were purchased from GE Dharmacon. Specificity and efficacy of knockdown was evaluated by Western blotting.
Immunoblotting and Immunofluorescence.
Cells were washed in PBS, and cellular proteins were extracted in 8 mol/L urea lysis buffer plus protease and phosphatase inhibitors (GenDEPOT) for 30 min at 4°C. Lysates were cleared by centrifugation, and proteins were separated by gel electrophoresis. Membranes were blocked in PBS 0.1% Tween 20 (PBS-T)/5% (w/v) milk for 1 hr at room temperature. Membranes were then incubated with primary antibodies diluted in PBS-T/5% (w/v) milk at 4°C overnight. Subsequently, membranes were washed with PBS-T and incubated with horseradish peroxidase secondary antibody (1:2,000; Jackson ImmunoResearch) diluted in PBS-T/5% skim milk. Membranes were washed in PBS-T, and bound antibody was detected by enhanced chemiluminescence (GE Healthcare). For detection of subcellular localization of IRF3, phospho-IRF3, TBK1 and phospho-TBK1, immunofluorescent staining was performed essentially as described previously (8). After treatment, cells were first fixed in ice-cold methanol for 10 min at −20°C, then blocked with 10% goat serum for 30 min at room temperature. Primary antibodies (IRF3, 1:200; Phospho-IRF3, 1:200; TBK1, 1:200; phospho-TBK1, 1:100, Cell Signaling Technology) were incubated at 4°C overnight, and Alexa 488– or Alexa 594–conjugated secondary antibodies (1:500,Thermo Fisher Scientific) were incubated for 1 hr at room temperature. Slides were mounted in ProLong anti-fade mounting medium containing DAPI (Thermo Fisher Scientific) and analyzed under a fluorescence microscope. Nuclear signals of staining were further examined by ImageJ (1.51j8), and the positivity was determined as at least 5 times greater in nuclear signal compared with the average in control. At least 50 cells per sample were analyzed, and the percentage of cells with positive staining was determined.
PicoGreen staining.
PicoGreen staining was performed using Quant-iT Pico-Green dsDNA reagent and kits from Thermo Fisher Scientific. For confocal microscopy, PicoGreen was diluted into cell culture medium at the concentration of 3μl/ml, and the cells were incubated in the presence of PicoGreen at 37 °C for 1 h. The cells were washed and fixed for confocal microscopy with DAPI counterstaining.
Multiplexed IHC staining.
Tumor tissue retrieved from ID8 i.p. injection or CT26 subcutaneous injection were subjected to fixation and paraffin embedding. The sections cut from paraffin blocks were baked at 60 °C for 1 hour and deparaffinized and rehydrated with serial passage through changes of xylene and graded alcohol and washed in water. Multiplexed immunofluorescence was performed following the manufacturer’s instruction (PerkinElmer). The following antibodies were used for IHC: anti-mouse PD-L1 (D5V3B, Cell Signaling Technology, 1:100), anti-CD8 (H160, Santa Cruz, 1:200), anti-STING (D2P2F, Cell Signaling Technology, 1:100) and anti-phospho-IRF3 (D6O1M, Cell Signaling Technology, 1:100). Stained slides were counterstained with DAPI and coverslipped for review. Positivity was defined as≥5% of staining or the percentage of positive cells per slide was calculated.
Quantitative PCR (Q-PCR).
Total RNA (1–2 μg) was used in a reverse transcriptase reaction with the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). The SYBR Green Real-Time PCR Master Mixes kit (Life Technologies) was used for the thermocycling reaction in an ABI-VIIA7 RealTime PCR machine (Applied Biosystems). The Q-PCR analysis was carried out in triplicate with the following primer sets: mouse Ccl5 (Forward: 5’-ATATGGCTCGGACACCACTC-3’; Reverse: 5’-TCCTTCGAGTGACAAACACG-3’), mouse Cxcl10 (Forward: 5’-CCCACGTGTTGAGATCATTG-3’; Reverse: 5’-GTGTGTGCGTGGCTTCACT-3’), mouse Gapdh (Forward: 5’- ACCCAGAAGACTGTGGATGG-3’; Reverse: 5’- ACACATTGGGGGTAGGAAC-3’), human CCL5 (Forward: 5’-TGCCCACATCAAGGAGTATTT-3’; Reverse: 5’- CTTTCGGGTGACAAAGACG-3’), human CXCL10 (Forward: 5’- GGCCATCAAGAATTTACTGAAAGCA-3’; Reverse: 5’- TCTGTGTGGTCCATCCTTGGAA-3’), and human β-Actin (Forward: 5’- GAGCACAGAGCCTCGCCTTT-3’; Reverse: 5’-TCATCATCCATGGTGAGCTG-3’)
ELISA.
The cell culture supernatant or ascites from ID8 model were collected and processed according to the manufacturer’s instructions. The CXCL10 and CCL5 levels were determined using ELISA kits from R&D/Thermo Fisher Scientific following the standard procedures.
In vivo mouse models.
All studies were supervised and approved by the MD Anderson Institutional Animal Care and Use Committee (IACUC). Female mice were used as models to study ovarian cancer. When used in a power calculation, our sample size predetermination experiments indicated that 5 mice per group could identify the expected effects with 90% power.
Ovarian Cancer Syngeneic Model:
Luciferase labeled ID8 ovarian cancer cells (5 × 106) were injected into the peritoneal cavity of C57BL/6 mice per group (6–8 weeks old, CRL/NCI). STING knockout mice (Tmem173 gt/J) were purchased from The Jackson laboratory (Cat#: 017537). The mice were allowed to recover and were monitored closely for the next 24 hrs. Tumor progression was monitored once per week by Xenogen IVIS Spectrum In Vivo Bioluminescence Imaging System. Tumor volume was determined based on total flux (photons per second). Tumor-bearing mice were treated intraperitoneally (i.p.) with isotype control IgG or anti-PD-L1 antibody (200 μg/mouse, B7-H1, clone 10F.9G2, Bio X Cell) every three days. BMN673 was administered by daily oral gavage with a dose of 0.33 mg/kg. Mice reaching an endpoint requiring euthanasia by IACUC guidelines or weighing more than 35 grams as a result of tumor growth and/or ascites were euthanized.
Colorectal Cancer Syngeneic Model:
Murine CT26 colorectal cancer cells (2 × 105) were subcutaneously injected into the left flank of BALB/C mice (6–8 weeks old, CRL/NCI) as previously described. Mice were allowed to recover and monitored closely for the next 24 hrs. Tumor size was measured every three days and tumor volume was determined based on the calculation (width × width × length)/2. Tumor bearing mice were treated (i.p.) with isotype control IgG or anti-PD-L1 antibody (200 μg/mouse, B7-H1, clone 10F.9G2, Bio X Cell) every three days. BMN673 was administered by daily oral gavage with a dose of 0.33 mg/kg. Mice reaching an endpoint requiring euthanasia by IACUC guidelines or exceeding tumor burden limits were euthanized.
Colorectal Cancer Nude Mouse Model:
Nude mouse experiments were conducted as described previously. Briefly, murine colorectal cancer cells CT26 (2 × 105) were subcutaneously injected into the left flank of athymic nude mice (6–8 weeks old, CRL/NCI). Mice were allowed to recover and monitored closely for the next 24 hrs. Tumor size was measured every three days and the tumor volume was determined based on the calculation (width × width × length)/2. Tumor bearing mice were treated (i.p.) with isotype control IgG or anti-PD-L1 antibody (200 μg/mouse, B7-H1, clone 10F.9G2, Bio X Cell) every three days. BMN673 was administered by daily oral gavage with a dose of 0.33 mg/kg. Mice reaching an endpoint requiring euthanasia by IACUC guidelines or exceeding tumor burden limits were euthanized.
Ovarian Cancer Nude Mouse Model:
Luciferase labeled ID8 cells (5 × 106) were injected into peritoneal cavity of athymic nude mice (6–8 weeks old, CRL/NCI). Mice were allowed to recover and monitored closely for the next 24 hrs. Tumor progression was monitored once a week by Xenogen IVIS Spectrum in vivo bioluminescence imaging system. Tumor volume was determined based on total flux (photons per second). Tumor bearing mice were treated (i.p.) with isotype control IgG or anti-PD-L1 antibody (200 μg/mouse, B7-H1, clone 10F.9G2, Bio X Cell) every three days. BMN673 were administered by daily oral gavage with a dose of 0.33 mg/kg. Mice reaching an endpoint requiring euthanasia by IACUC guidelines or weighing more than 35 grams as a result of tumor growth and/or ascites were euthanized.
Statistics.
All statistical analyses were done in GraphPad Prism 7 software. Overall survival of various treatment groups was analyzed using the Cox regression model. Otherwise, unpaired t-tests were used to generate two-tailed P values.
Results
PARPi induces an accumulation of cytosolic DNA and activates STING signaling pathway
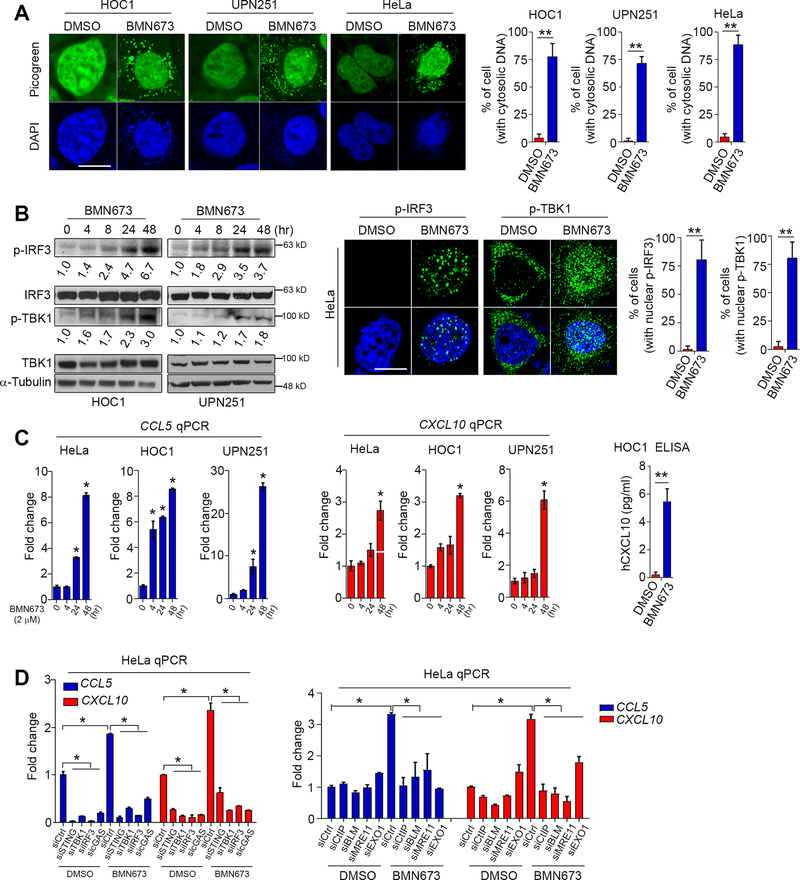
PARPi treatment markedly induced DNA double-strand breaks (DSBs) as detected by increased γ-H2AX levels, and thus caused cell cycle arrest in S phase (Supplementary Fig. S1A and S1B). The cytosolic DNA sensor cGAS is the most potent activator of the STING signaling pathway (9). After the recognition of cytosolic DNA, cGAS activates STING via generation of 2’−5’ cyclic GMP-AMP (cGAMP). STING, in turn, induces phosphorylation and nuclear translocation of IFN transcriptional regulatory factors TANK-binding kinase 1 (TBK1) and IFN regulatory factor 3 (IRF3) (10,11). We thus examined whether PARPi induces accumulation of cytosolic DNA that could activate the cGAS-STING-TBK1-IRF3 axis in ovarian cancer cell lines HOC1 (BRCA1/2 WT), UPN251 (BRCA1 deleterious and restoration mutations, functional WT) (12) and HeLa (BRCA1/2 WT).
As shown in Fig. 1A, BMN673 caused a significant accumulation of cytosolic dsDNA in multiple cell lines. Moreover, phosphorylation of IRF3 and TBK1, two key components along the STING pathway, was markedly elevated by BMN673 treatment in a time-dependent manner (Fig. 1B). PARPi also remarkably induced the translocation of phospho-IRF3, phospho-TBK1 (Fig. 1B), as well as total IRF3 and total TBK1 from the cytoplasm to the nucleus (Supplementary Fig. S2A), which indicated functional activation of STING signaling. We then examined mRNA expression of CCL5 and CXCL10, two major target genes downstream of STING activation that are involved in T-cell chemotaxis (13). We found a time-dependent increase in CCL5 and CXCL10 mRNA levels after PARPi treatment (Fig. 1C). Consistent with these changes in mRNA levels, PARPi substantially increased the production of CXCL10 as detected by ELISA (Fig. 1C). We further found that upregulation of CCL5 and CXCL10 was significantly reduced in response to PARPi treatment in cells with STING, TBK1, IRF3 or cGAS knockdown (Fig. 1D, Supplementary Fig. S2B-2D and Supplementary Fig. S3A). Together, these results demonstrate that PARPi induces accumulation of cytosolic dsDNA and activation of cGAS-STING-TBK1-IRF3 signaling to promote chemokine expression.
Fig. 1. PARPi induces accumulation of cytosolic DNA and activates the STING signaling pathway.
A, Representative images and quantitative analysis of PicoGreen staining in cells treated with DMSO or BMN673 (2 μM) for 48 hrs. DAPI (blue) was used to visualize the nuclei. Data represent mean ± s.e.m. of three independent experiments. B, Western blots (Left) and immunofluorescent images (Right) of phosphorylated IRF3 (p-IRF3) and phosphorylated TBK1 (p-TBK1) in cells treated with BMN673 (2 μM). The numbers represent the folds change of p-IRF3 and p-TBK1 from three independent experiments. For immunofluorescence staining, HeLa cells were treated with DMSO or BMN673 (2 μM) for 48 hrs. Data represent mean ± s.e.m. of three independent experiments. C, qPCR and ELISA evaluation of CCL5 and CXCL10 expression in cells under DMSO or BMN673 treatment. For ELISA analysis, HOC1 cells were treated with DMSO or BMN673 treatment for 48 hrs. Data represent mean ± s.e.m. of three independent experiments. D, qPCR evaluation of CCL5 and CXCL10 levels in HeLa cells with depletion of STING (siSTING), TBK1 (siTBK1), IRF3 (siIRF3), cGAS (sicGAS), CtIP (siCtIP), BLM (siBLM), MRE11 (siMRE11), or EXO1 (siEXO1). Cells were treated with DMSO or BMN673 (2 μM) for 48 hrs. Data represent mean ± s.e.m. of three independent experiments. Scale bar, 10 μm, *, p<0.05; **, p<0.01.
Several key factors containing and/or regulating nuclease activity, including MRE11, CtIP, BLM and EXO1, are recruited to DSBs, which can produce DNA fragments during HR repair and maintenance of replication fork stability (14). We reasoned that trapping of PARP1 by PARPi forms a barrier against DSB end resection and HR repair, which may lead to generation of dsDNA through degradation of unrepaired reversed replication forks. Indeed, we demonstrated that knockdown of these resection factors by siRNAs markedly reduced mRNA expression of CCL5 and CXCL10 (Fig. 1D and Supplementary Fig. S3B), suggesting a key role for the requirement of these factors in generating PARPi-induced immune responses.
PARPi activates STING signaling and immune checkpoint in vivo
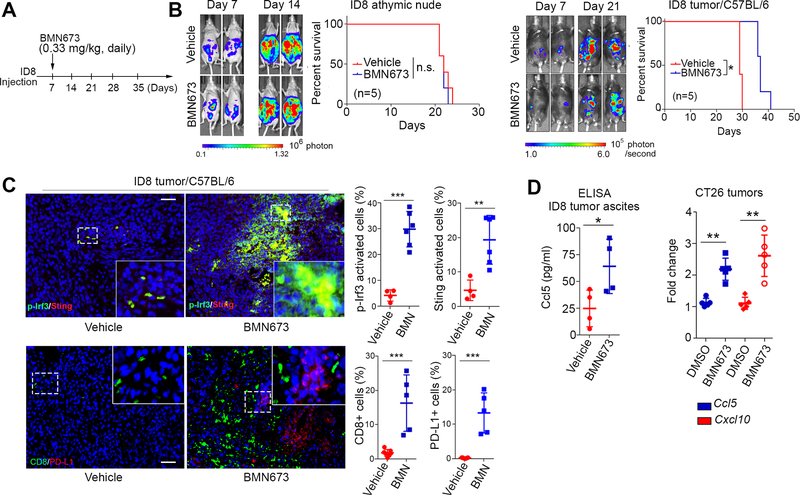
We next investigated the in vivo effects of PARPi-induced immune responses, using syngeneic immunocompetent mouse models of ovarian cancer (ID8) and colon cancer (CT26). CT26 and ID8 cells have no known mutations in genes involved in the HR repair pathway, including BRCA1/2 mutations. As expected, PARPi treatment exhibited no therapeutic effects in immune-deficient mice bearing ID8 tumors (Fig. 2A and 2B). In contrast, PARPi can reduce tumor growth and prolong survival in the ID8 syngeneic model (Fig. 2B). These data indicated that an intact T cell-mediated immune response is required for PARPi efficacy in the ID8 model.
Fig. 2. PARPi activates STING signaling and immune checkpoint in vivo.
A, Schematic of PARPi treatment in nude and syngeneic mice bearing intraperitoneal (i.p.) ID8 tumors. Mice were treated with BMN673 (0.33 mg/kg) daily by oral administration 7 days after ID8 inoculation until euthanization (n=5). B, Representative bioluminescence images of i.p. tumors in immune-deficient (Left) and immune-proficient (Right) mice on day 7 and day 21 of ID8 cell inoculation and survival curves. C, Representative images and quantitative analysis of phosphorylated Irf3 (p-Irf3), Sting, CD8 and PD-L1 in ID8 tumors 30 days after inoculation. DAPI (blue) was used to visualize the nuclei. D, ELISA evaluation of Ccl5 levels in ascites from C57BL/6 mice when euthanization was performed (Left) and qPCR evaluation of Ccl5 and Cxcl10 levels in CT26 tumors from BALB/C mice 22 days after tumor cell inoculation (Right). Dashed square, area for magnification. Scale bar, 50 μm, *, p<0.05; **, p<0.01; ***, p<0.001; n.s. not significant.
It has been reported that expression levels of CCL5 and CXCL10 positively correlate with infiltrating CD8+ cytotoxic lymphocytes in various cancers (15). We thus conducted immunohistochemistry (IHC) analysis of STING activation and immune response. In syngeneic ID8 and CT26 models, PARPi significantly upregulated the levels of Irf3 phosphorylation as well as Sting expression, indicating robust activation of the Sting signaling pathway in vivo (Fig. 2C and Supplementary Fig. S4A). Consistent with this finding, remarkably higher percentages of CD8+ T cells and PD-L1+ cells were found in PARPi-treated tumors (Fig. 2C and Supplementary Fig. S4B). Furthermore, as shown in Fig. 2D, PARPi treatment induced expression of Ccl5 and Cxcl10, which was consistent with in vitro studies (Fig. 1). Together, these data showed that PARPi treatment induces an immunogenic response through the activation of the STING pathway and enhancement of type I IFN response and TILs in vivo. However, activation of the PD-1/PD-L1 immune checkpoint pathway may counterbalance the impact of active TILs and block elimination of tumor cells despite the immunogenic microenvironment-induced by PARPi. These results raised the possibility that the combination of immune checkpoint blockade and PARPi would synergistically limit tumor growth and prolong survival.
Immune checkpoint blockade targeting PD-1/PD-L1 pathway potentiates therapeutic efficacy of PARPi in syngeneic mouse models.
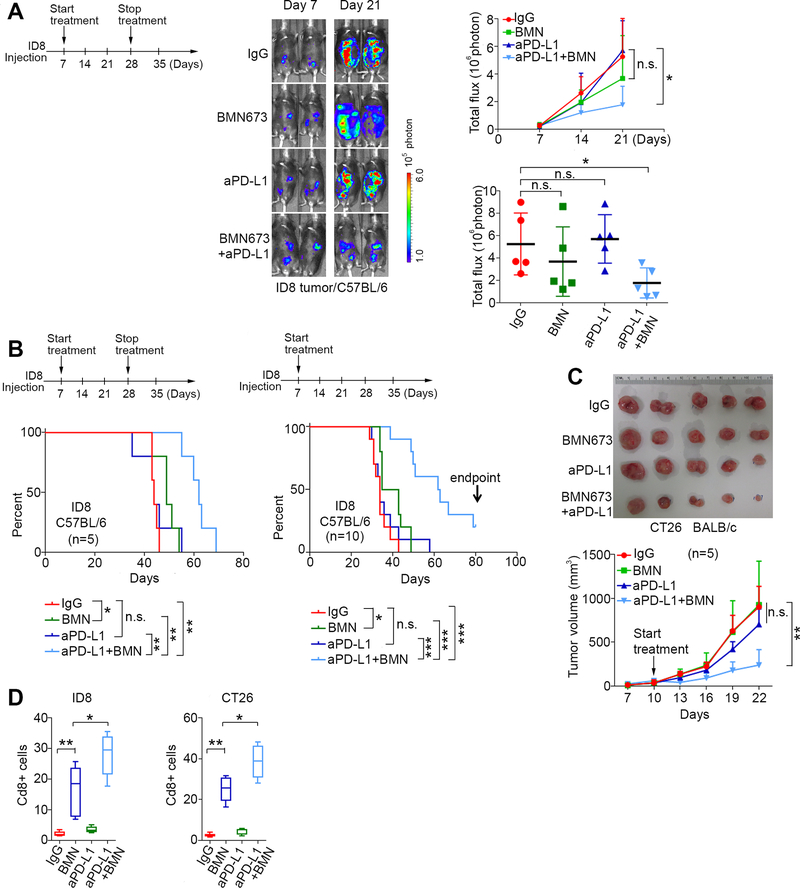
To test this possibility, we first treated mice with ID8 3 weeks and then stopped treatment. Consistent with relatively poor immunogenicity and low TILs in ID8 tumors (16), ID8 tumors did not significantly respond to anti-PD-L1 therapy or BMN673 alone after 3 weeks of treatment (Fig. 3A). Only the combination significantly reduced tumor growth while no significant changes in mouse weight were observed (Fig. 3A and Supplementary Fig. S4C). In the third week of treatment, mice developed ascites which interfered with further luciferase measurements, but the study was continued to measure survival. As shown in Fig. 3B, only the combination treatment significantly prolonged survival. Continuous treatment with the combination of BMN673 and anti-PD-L1 produced a remarkably better outcome than three-week treatment (Fig. 3B), suggesting a potential curative effect from this continuous regimen.
Fig. 3. Immune checkpoint blockade targeting PD-1/PD-L1 pathway potentiates therapeutic efficacy of PARPi in syngenic mouse models.
A, Combination treatment in C57BL6 mice bearing ID8 tumors. Schematic of treatment (Left). i.p. injections of isotype control IgG (200 μg/mouse) and anti-PD-L1 antibody (aPD-L1, 200 μg/mouse) started at day 7 and stopped at day 28 after ID8 cell inoculation. BMN673 (0.33 mg/kg) was orally administered daily. Representative bioluminescence images of ID8 tumors after 7 and 21 days of inoculation (Middle). Statistical analysis of bioluminescence changes over time (mean ± s.e.m) or at the endpoint (Day 21, each dot represents one mouse) (Right) (n=5). B, Survival curves of mice with temporary (Left, started on day 7 after ID8 inoculation and stopped on day 28) (n=5) or continuous treatment (Right, started on day 7 after inoculation and continued until the mice were euthanized) (n=10). C, Representative images and tumor volume measurements of CT26 tumors in BALB/c mice with continuous treatment (n=5). D, Percentage of Cd8+ cells in ID8 or CT26 tumors after treatments. *, p<0.05; **, p<0.01; ***, p<0.001; n.s. not significant.
We next used CT26 mouse colon tumor cells as an independent syngeneic model to validate the efficacy of combination therapy. Because CT26 tumors grew aggressively, mice were treated for 12 days before the tumor size reached the maximum volume allowed by the animal protocol. The combination therapy significantly reduced CT26 tumor burden compared to IgG control (Fig. 3C). Although BMN673 increased the percentage of tumor infiltrating Cd8+ cells (Supplementary Fig. S4B), these tumors were not sensitive to BMN673 alone. The combination of BMN673 and anti-PD-L1 treatment results in the most significant increase of Cd8+ cells in both ID8 and CT26 models (Fig. 3D).
The therapeutic effects of combining PARPi and anti-PD-L1 depend on an intact immune system.
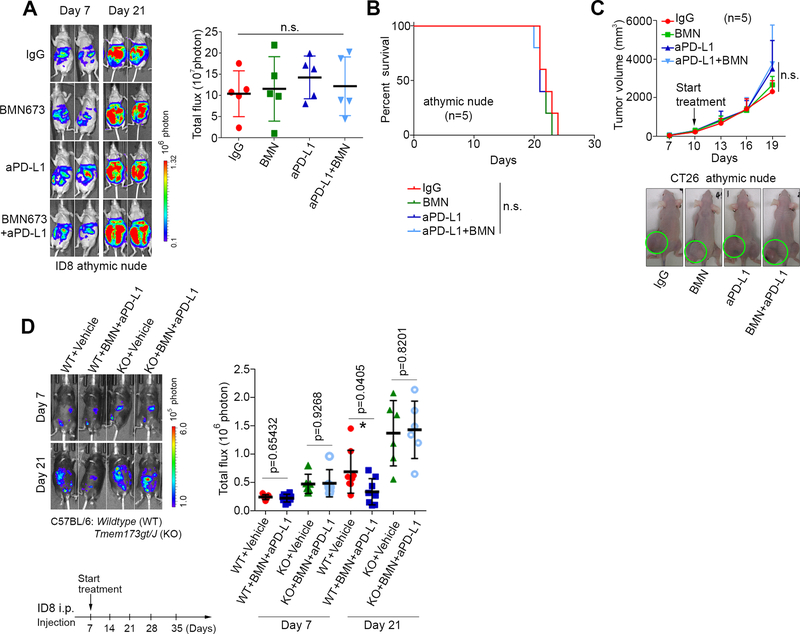
Interestingly, none of the treatments examined - BMN673 monotherapy, anti-PD-L1 monotherapy, and the combination of BMN673 and anti-PD-L1 - inhibited tumor growth or improved survival in immune compromised nude mouse models bearing ID-8 or CT26 tumors (Fig. 4A-4C). These results strongly supported the notion that an intact immune system is a prerequisite to achieve the benefits of combination therapy. More importantly, in a STING-knockout mouse model, we found that lack of STING abolished anti-tumor effects of PARPi and anti-PD-L1 treatment on ID-8 tumors, demonstrating that STING signaling is genetically required for therapeutic effects of PARPi and anti-PD-L1 treatment (Fig. 4D).
Fig. 4. An intact immune system is required for the therapeutic benefits of combining PARPi with anti-PD-L1.
A, Representative images and statistical analysis of endpoint bioluminescence in athymic nude mice bearing ID8 i.p. tumors. Data represent mean ± s.e.m, (n=5). i.p. injection of isotype control IgG (200 μg/mouse) and anti-PD-L1 (aPD-L1, 200 μg/mouse) were stated at day 7 and continued until mice were euthanized. BMN673 (0.33 mg/kg) was orally administered daily. B, Survival curves of athymic nude mice with ID8 i.p. tumors. Treatment was started on day 7 and continued until mice were euthanized. C, Representative images and tumor volume measurements of CT26 tumors in athymic nude mice with the indicated treatments (n=5). n.s., not significant. D, Representative images and statistical analysis of bioluminescence in Wildtype (WT) or STING KO (KO, Tmem173 gt/J) mice bearing ID8 i.p. tumors. i.p. injection of Vehicle (isotype control IgG, 200 μg/mouse) and anti-PD-L1 (aPD-L1, 200 μg/mouse) were stated at day 7 and continued. BMN673 (0.33 mg/kg) was orally administered daily.
Discussion
Here we show PARPi leads to an accumulation of cytosolic dsDNA and thereby activates the cGAS-STING-TBK1-IRF3 innate immune pathway, which induces type I IFN and its related immune responses (Supplementary Fig. S4D). Furthermore, PARPi treatment enhances tumor susceptibility to immune checkpoint blockade. Importantly, these responses were observed regardless of the BRCA1/2 mutation status of the cell lines assessed both in vitro and in vivo.
Recent studies have shown that micronuclei resulting from mis-segregation of DNA during cell division can be recognized by the cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS) and activate the stimulator of interferon (IFN) genes (STING) innate immune pathway (17). Our data uncovered that DNA damage induced by PARPi generates cytosolic DNA, primarily dsDNA. PARPis induces stalled or collapsed replication forks. dsDNA generated by PARPi may be related to DNA replication fork degradation/reversion or the restart of replication. Our study thus proposes a novel molecular mechanism underlying PARPi therapeutic effects, which is independent of its conventional cytotoxic effects resulting from unresolved DNA damage in DNA repair-deficient cancer cells.
The antitumor activity of PARPi has been observed in patients regardless of BRCA1/2 mutation status or the presence of HR defects (7). Our results also support this notion since the immunogenic responses induced by PARPi is not dependent on DNA repair deficiency. However, treatment with a single agent of PARPi is not sufficient to exert durable therapeutic effects. This phenomenon may be explained by the overall impact of PARPi on tumor microenvironment. CCL5 and CXCL10 are chemokines that associate with recruitment of TILs and their production is relevant to trigger proper anti-tumor immune responses (18,19). However, an increasing number of studies have demonstrated that CCL5 may favor tumor growth through myeloid cell recruitment (20,21). Of note, myeloid cells are essential to activate immune checkpoint PD-1/PD-L1 axis thereby are required for the establishment of an immunosuppressive tumor microenvironment in multiple cancer types (22–24). Significant increased number of PD-L1+ cells in ID8 and CT26 tumors indicates that PARPi may recruit myeloid cells into tumor sites therefore counterbalance the therapeutic efficacy. It is possible that the benefits of combining PARPi with anti-PD-L1 may be further strengthened by inhibiting the immunosuppressive myeloid cells.
Supplementary Material
Acknowledgments
Grant support: This research was supported by NCI Cancer Center Support Grant CA016672 to The University of Texas MD Anderson Cancer Center, Department of Defense grant OC140431, NIH R01 grant CA181663 to G. Peng. Cancer Prevention and Research Institute of Texas grant RP160242 to G. Peng and X. Shen.
Footnotes
Conflict of interests: G. B. Mills has sponsored research support from Pfizer, Abbvie, Critical Outcomes Technology, Immunomet. Karus Therapeutics, Nanostring, Millennium, Tesaro, and AstraZeneca; has ownership interests in Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindle Top Ventures and Tarveda; and is a consultant/advisory board member of AstraZeneca, Blend, Critical Outcome Technologies, ImmunoMet, Ionis Nuevolution, Signalchem Lifesciences, Symphogen, and Tarveda. G. Peng received sponsored research funding from Pfizer. T.A. Yap has sponsored research support from AstraZeneca, Bayer, Pfizer, Tesaro, Jounce, Eli Lilly, Seattle Genetics, Kyowa, Constellation, and Vertex Pharmaceuticals; and is a consultant/advisory board member of Aduro, Almac, AstraZeneca, Atrin, Bayer, Bristol-Myers Squibb, Calithera, Clovis, Cybrexa, EMD Serono, Ignyta, Jansen, Merck, Pfizer, Roche, Seattle Genetics, and Vertex Pharmaceuticals; and is a member of speaker bureau of AstraZeneca, Merck, Pfizer, and Tesaro. No potential conflicts of interest were disclosed by other authors.
References
- 1.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123–34 [DOI] [PubMed] [Google Scholar]
- 2.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 2012;366:1382–92 [DOI] [PubMed] [Google Scholar]
- 3.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017;18:610–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–7 [DOI] [PubMed] [Google Scholar]
- 5.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21 [DOI] [PubMed] [Google Scholar]
- 6.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 2011;12:852–61 [DOI] [PubMed] [Google Scholar]
- 7.Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. The New England journal of medicine 2016;375:2154–64 [DOI] [PubMed] [Google Scholar]
- 8.Pourcelot M, Zemirli N, Silva Da Costa L, Loyant R, Garcin D, Vitour D, et al. The Golgi apparatus acts as a platform for TBK1 activation after viral RNA sensing. BMC Biol 2016;14:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 2013;498:380–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corrales L, McWhirter SM, Dubensky TW Jr., Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest 2016;126:2404–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016;19:150–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stordal B, Timms K, Farrelly A, Gallagher D, Busschots S, Renaud M, et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol Oncol 2013;7:567–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longhese MP, Bonetti D, Manfrini N, Clerici M. Mechanisms and regulation of DNA end resection. EMBO J 2010;29:2864–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muthuswamy R, Berk E, Junecko BF, Zeh HJ, Zureikat AH, Normolle D, et al. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res 2012;72:3735–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, Balint K, et al. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nature medicine 2008;14:28–36 [DOI] [PubMed] [Google Scholar]
- 17.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548:466–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell 2012;21:309–22 [DOI] [PubMed] [Google Scholar]
- 19.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev 2018;63:40–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol 2012;189:5602–11 [DOI] [PubMed] [Google Scholar]
- 21.Dutta P, Nahrendorf M. Regulation and consequences of monocytosis. Immunol Rev 2014;262:167–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Velez-Delgado A, Mathew E, Li D, Mendez FM, Flannagan K, et al. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut 2017;66:124–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonios JP, Soto H, Everson RG, Moughon D, Orpilla JR, Shin NP, et al. Immunosuppressive tumor-infiltrating myeloid cells mediate adaptive immune resistance via a PD-1/PD-L1 mechanism in glioblastoma. Neuro Oncol 2017;19:796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott LA, Doherty GA, Sheahan K, Ryan EJ. Human Tumor-Infiltrating Myeloid Cells: Phenotypic and Functional Diversity. Front Immunol 2017;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.