Abstract
Proximity-dependent labeling methods for detecting candidate protein-protein interactions (PPIs) or mapping the protein constituency of subcellular domains have become increasingly utilized by the scientific community. One such method, BioID, allows for the identification of not only strong interactions but also weak and transient associations between a protein of interest (POI) or targeting motif and adjacent proteins. A promiscuous biotin ligase is fused to a POI or targeting motif, expressed in living cells, and induced to biotinylate proximal proteins during a defined labeling period by biotin supplementation. This generates a history of protein-protein associations that occurred with the POI or the protein constituency within a discrete subcellular domain during the labeling period. Biotinylated proteins are subsequently isolated, identified via mass spectrometry, and investigated as candidate interactors with the POI or as constituents within a subcellular domain. The BioID method has been utilized by numerous research groups and is continually being optimized, applied to new models, and modified for use in novel applications. Here we describe a protocol by which a BioID fusion protein can be validated and utilized for BioID pull-downs.
Keywords: BioID, Biotinylation, Protein-protein interactions, Proximity labeling
1. Introduction
Defining the constituents of protein complexes and mapping protein-protein interactions (PPIs) is a powerful approach to ascertain protein function and how proteins cooperate to accomplish cellular processes. There are several methods currently available to help identify PPIs; however, based on their respective limitations each method is optimal for specific applications. Two conventional approaches for identifying PPIs are affinity complex purification and yeast two-hybrid assays. Affinity complex purification often works well for identifying strong and stable interactions between proteins, but fails to detect weak and transient associations. Yeast two-hybrid can assay PPIs from a wide variety of cellular sources, yet risks considerable potential false positives and false negatives due to an unnatural cellular environment and the frequent use of protein fragments. Not all PPIs are characterized by high-affinity binding, and newer methods of protein proximity labeling, incluing BioID, have been developed to aid the detection of weak and transient interactions in live cells. These methods covalently label interacting and proximal proteins with modifications useful for subsequent detection and isolation, usually biotin. Fundamentally, a labeling enzyme fused to a POI is expressed within or on cells and proteins proximate to the POI are labeled, isolated, and identified by mass spectrometry.
There are two classes of labeling enzymes utilized with proximity labeling methods in living cells, peroxidases and biotin ligases. The most utilized of these methods are BioID (proximity- dependent biotin identification) [1] and APEX (ascorbate peroxidase based) [2], or their variants. In this protocol we describe the BioID method and its variants. Derived from the E. coli biotin ligase BirA, the BioID ligase is a mutated form of BirA that enables promiscuous biotinylation of proximate proteins [1, 3, 4]. When fused to a POI and expressed in living cells, BioID allows for the labeling of proximate proteins over a defined labeling period to generate a history of protein associations (Fig. 1). In its natural environment, wild-type BirA specifically biotinylates acetyl CoA carboxylase by first generating a reactive biotinyl-AMP (bioAMP) and then releasing it for covalent attachment to a particular lysine on the carboxylase [4]. The mutant BirA (R118G), hereafter called BioID, generates bioAMP but with reduced affinity for the reactive bioAMP molecule, leading to the premature release and attachment of the bioAMP to proximal proteins [5]. Thus, BioID covalently attaches biotin to lysine residues which conveniently tend to be exposed on the surface of proteins. The labeling radius of BioID has been measured in living cells to be approximately 10 nm [6]; however, this presents a limitation when applied to larger POI whereby proteins that associate with regions of the POI distal to the biotin ligase may not be efficiently labeled. In an attempt to overcome this spatial constraint, the labeling radius can be increased by adding a flexible linker of a defined length between the BioID and POI [7]. Flexible linkers can also be used to prevent steric hindrance between the BioID ligase and the POI. The requirement to fuse the 35 kDa BIoID ligase to the POI can potentially impair localization, associations with other proteins, and/or function. In an attempt to reduce these effects, BioID2, a smaller promiscuous biotin ligase derived from Aquifex aeolicus, was developed and demonstrated to improve the localization of a nuclear envelope transmembrane protein, a class of proteins susceptible to mislocalization due to bulky fusions [7]. Unless specific differences are being described, the term BioID will be used interchangeably to refer to these two promiscuous biotin ligases. Likely due to the nature of the mutation which provides the ligase with its promiscuity, the labeling of proximate proteins requires biotin supplementation, typically 10–50 μM. This is well above biotin concentrations found in typical cell culture media. The requirement for biotin supplementation allows for inducibility of the biotinylation process, even with constant expression of the BioID fusion protein. The optimal duration of BioID labeling is 15–18 h, after which there is not an appreciable increase in overall protein biotinylation. This prolonged labeling time may be problematic for certain applications, leading to attempts to increase the rate of labeling kinetics [8, 9]. A toolbox of BioID ligases with a range of biotinylation rates would be beneficial to enable selection of an optimal BioID ligase for distinct applications.
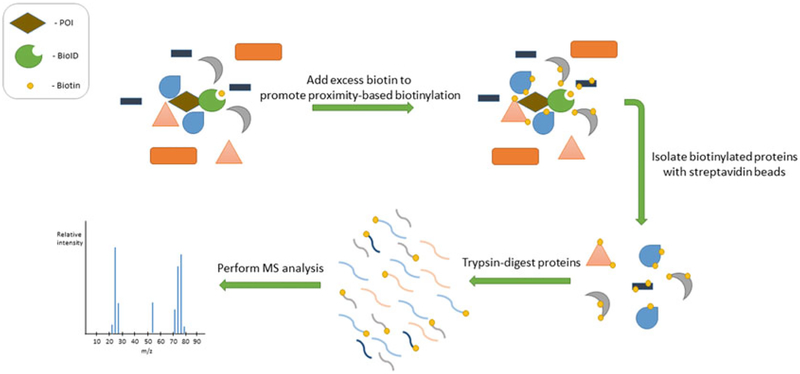
Fig. 1.
BioID method overview. Upon induction of biotinylation by the addition of biotin, BioID labels proteins based on proximity to fusion protein. Following cell lysis and protein denaturation, biotinylated proteins are isolated by biotin affinity purification and processed for identification by mass spectrometry
BioID can also be used to map the protein constituents of a specific subcellular compartment. This application has been termed Zip Code-BioID [10]. The conventional approach to identify protein constituents of discrete subcellular domains is to purify the organelle via subcellular fractionation. However, many subcellular domains are not able to be isolated, and even when they are it is often without the ability to distinguish topological location of the proteins (e.g., lumenal or the cytosolic facing proteins of the endoplasmic reticulum). Additional limitations include difficulty detecting loosely and/or transiently associated proteins or, at the other end of the spectrum, contamination by undesired proteins due to impure fractionation. A minimal targeting motif [10] or even a full-length protein can be used to target a labeling enzyme to a specific subcellular compartment or domain. An example of the latter was the use of BioID on TbMORN1, the only known protein constituent that localized to the trypanosome bilobe that could not be conventionally isolated to identify other constituents of that discrete domain [11]. What distinguishes Zip Code-BioID is the intent to assess the protein constituency of a domain and not to identify specific PPIs. A flexible linker can be utilized to expand the labeling radius to aid in labeling, if desired [7].
BioID has become widely utilized since its first reported application in 2012, and has been cited and/or applied in over 200 published scientific articles to reveal candidate PPIs in multiple subcellular compartments in mammalian cells (see ref. 12 for review); unicellular organisms such as Saccharomyces cerevisiae [13], Trypanosoma brucei [11], Toxoplasma gondii [14], Dictyostelium discoideum [15], and Plasmodium berghei [16]; and in plant [17] and mouse studies [18–20]. BioID has proven useful for studying labile proteins like kinases [21, 22], phosphatases [23], and E3 ligases [24], as well as insoluble protein networks and complexes such as the nuclear lamina [1, 25], nuclear pore complex [6], and centrosome [22, 26, 27]. BioID has recently been applied to detect RNA-protein interactions [9] and other groups have developed split-BioIDs to enable proximity labeling only when two specific proteins are associated [28, 29]. A CRISPR-mediated introduction of BioID-fusion protein has also been successfully performed which overcomes the impact of protein overexpression [30]. In an attempt to improve identification of specific biotinylation sites on candidate proteins, typically a difficult prospect in part due to nearly covalent affinity of biotinylated peptides to avidin/ streptavidin matrix, approaches have been developed to selectively isolate biotinylated peptides using antibodies [31, 32]. What follows is a protocol to enable efficient capture and identification of proteins biotinylated by BioID.
2. Materials
2.1. Validation of Fusion Protein
BioID fusion protein expression vector (see Note 1).
cDNA or PCR template for POI.
Cells of choice and appropriate medium.
6-Well tissue-culture plates.
No. 1.5 glass coverslips.
1 mM of Biotin solution (20×): Dissolve 12.2 mg of biotin in 50 mL of serum-free DMEM (or standard tissue culture medium). Vortex to dissolve biotin completely. Sterilize by passing through a 0.22 μm syringe-driven filter unit (Millex). Dispense into sterile 50 ml tube; cap tightly. Store for up to 8 weeks at 4 °C.
Fixative, paraformaldehyde (PFA) (see Note 2).
Phosphate-buffered saline (PBS).
Phosphate-buffered saline with Triton X-100 (TX-100) (PBST): 0.4% TX-100 in 1 PBS.
Antibodies specific to BioID fusion protein (e.g., anti-myc/ HA) or chicken anti-BioID (BID-CP-100, BioFront) or chicken anti-BioID2 (BID2-CP-100, BioFront).
Secondary antibodies to detect chosen primary antibody (Alexa Fluor and HRP conjugates).
Streptavidin-Alexa Fluor.
DNA-labeling reagent (e.g., Hoechst, DAPI).
SDS-PAGE sample buffer (prepare fresh daily): 50 mM Tris-Cl, pH 6.8, 12% sucrose, 2% SDS, 0.004% bromophenol blue, 20 mM dithiothreitol (DTT).
Streptavidin-HRP (AB7403, Abcam).
ABS blocking buffer: 10% (v/v) Adult bovine serum and 1% (w/v) Triton X-100 in PBS. Store for up to 4 weeks at 4 °C.
- Enhanced chemiluminescence (ECL) reagent:
- Solution 1 (wrap in tin foil to protect from light): In 15 mL conical tube, mix 1 mL of 1 M Tris-Cl, pH 8.5, 45 μL coumaric acid, 100 μL luminol, and 8.9 mL H2O.
- Solution 2: In 15 mL conical tube, mix 1 mL of 1 M Tris-Cl, pH 8.5, 6 μL 30% hydrogen peroxide (H2O2), and 9 mL H2O.
- Store solutions 1 and 2 in the dark at 4 °C for up to 1 month. Mix equal volumes of solutions 1 and 2 together and immediately add to membrane for 1 min prior to exposure to film or ChemiDoc detection.
30% H2O2.
Probe sonicator (Branson Sonifier-250 or equivalent).
2.2. Large-Scale BioID Pull-Down
Two 10 cm dishes of cells for each experimental condition (cells expressing BioID constructs or control cells) (see Notes 3 and 4).
Complete cell culture medium.
Lysis buffer (must be prepared fresh daily): 8 M Urea in 50 mM Tris-Cl, pH 7.4, 1 protease inhibitor (Halt Protease Inhibitor Cocktail, EDTA free), 1 mM DTT (dithiothreitol).
20% TX-100 in water.
50 mM Tris-Cl, pH 7.4.
Pierce Universal Nuclease.
Gelatin-conjugated Sepharose 4B.
Streptavidin Sepharose High Performance Beads.
Wash buffer (prepare fresh daily): 8 M Urea in 50 mM Tris-Cl, pH 7.4.
1 mM Biotin in 50 mM ammonium bicarbonate (NH4HCO3) (store aliquots at−20 °C).
1× SDS-PAGE sample buffer (prepare fresh daily): See Sub-heading 2.1.
DNase/RNase-free tubes, 5 mL conical and 2 mL microcentrifuge.
Tube cap opener (see Note 5).
Probe sonicator (Branson Sonifier-250 or equivalent).
Rotator.
3. Methods
3.1. Fusion Protein Validation
Use a standard PCR method to fuse the gene of interest in-frame with BioID in preferred expression vector (see Notes 6 and 7).
For each condition (e.g., control, BioID-POI) plate two wells of cells in a standard 6-well plate, one well for western blot (WB) analysis and the other well with a glass coverslip to perform immunofluorescence (IF).
Transiently transfect cells of choice with BioID-POI expression vector. Process mock-transfected or nontransfected cells in parallel. Supplement cells with biotin (50 μM final concentration) at the time of transfection, or if transfection protocol requires refreshing medium a few hours after transfection add biotin with fresh medium (see Note 8).
Process the cells on coverslips for analysis by immunofluorescence microscopy following transient transfection and biotin incubation for 15–18 h (see Note 9). Confirm expression, localization, and biotinylation (Fig. 2a).
Prepare cells for WB analysis 15–18 h following transient transfection and addition of biotin: First, rinse them with PBS to remove serum proteins, then lyse them in appropriate volume of SDS-PAGE sample buffer (e.g., 200 μL per 1×106 cells), heat them to 98 °C for 5 min to denature proteins, and finally sonicate them to shear DNA.
Perform standard WB SDS-PAGE separation and membrane transfer process.
Probe membrane with streptavidin-HRP at 1:20,000 in PBST for 1 h at room temperature (see Note 10).
Wash membrane two to three times with PBS.
Soak membrane in ABS-blocking buffer to reduce background signal for 5 min.
Rinse off ABS by washing membrane in PBS two to three times.
Add enhanced chemiluminescence (ECL) reagent to observe biotinylated proteins and capture WB image.
Quench HRP signal by agitating membrane in 30% H2O2 at room temperature for 20 min (see Note 11).
Rinse with PBS to remove residual hydrogen peroxide.
Confirm expression and migration of BioID fusion protein by probing with BioID-specific antibody (see Note 12).
For generation of BioID-expressing stable cell lines, plate desired cell type and begin process via stable transfection or viral transduction and perform drug selection if relevant (e.g., puromycin resistance).
For subcloning, screen each subclone by IF. For subclones with correct expression and localization, continue to WB analysis (see Note 13). For screening a virally transduced population, perform both IF and WB.
Freeze down multiple vials of stably expressing cells for future BioID experiments and store in liquid nitrogen.
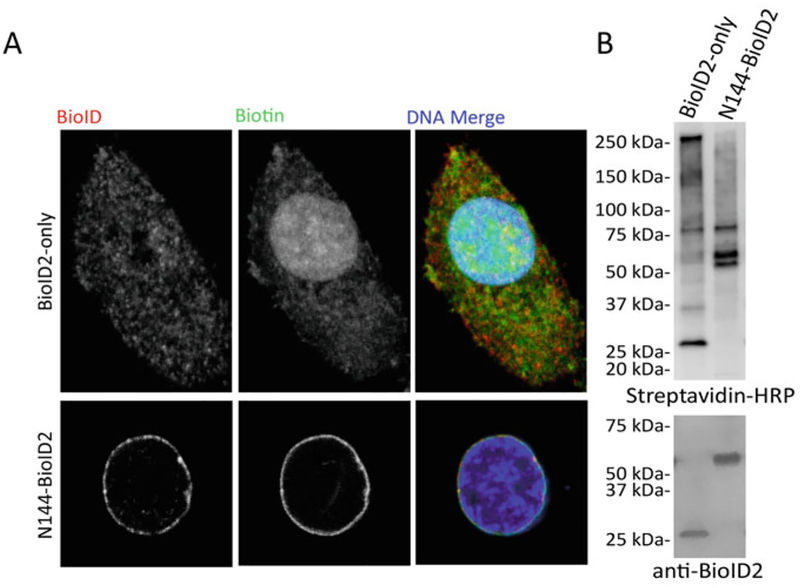
Fig. 2.
Representative analysis of BioID efficacy by fluorescence microscopy and Western blot. (a) Confocal images of A549 cells stably expressing BioID2-only or N144-BioID2 [10] constructs. Fusion proteins were detected by anti-BioID2 antibody (red) and biotinylation was detected by streptavidin-488 Alexa Fluor (green). DNA was labeled with Hoechst (blue in merged). Scale bar is 5 μm. (b) Following BioID pull-down and SDS-PAGE, isolated biotinylated proteins were visualized with streptavidin-HRP and fusion proteins were detected by anti-BioID2 antibody
3.2. BioID Pull-Down
3.2.1. Day 1 (Cell Lysis)
Begin with two 10 cm plates for each experimental condition (BioID-POI and BioID-only control) (see Notes 3 and 4).
When cells reach approximately 80% confluency, re-feed cells with fresh complete media containing 50 μM biotin (1×).
Incubate for 15–18 h.
Aspirate to remove media and rinse cells twice with 2 mL of 1×PBS to remove residual biotin.
Add 540 μL of lysis buffer to each plate and scrape gently to harvest cells.
Collect the lysed cells in a 15 mL conical tube and vortex briefly.
Add 0.1 μL Pierce Universal Nuclease to each sample.
Incubate at room temperature for 10 min.
Briefly vortex, and then place samples on ice.
Add 60 μL of 20% TX-100 (for 1% final concentration) and vortex briefly.
Sonication should occur on ice. Position the tip of the sonicator immediately above the bottom of the tube. Apply sonication for 30 s at 30% duty cycle and output level 4.
Wash the tip of the sonicator and repeat step 11 two more times (3 total) (see Note 14).
Add 1260 μL of lysis buffer and vortex briefly.
Apply sonication for 1 min at the same settings as in step 11.
Transfer the contents equally to two pre-chilled 2 mL tubes (i.e., 1200 μL/tube) (see Note 15).
Spin down the sample at 16,500× g for 10 min at 4°C.
You may collect and flash freeze the lysates in liquid nitrogen at this step. From the 2 mL tubes remove the supernatant carefully without disturbing the cell pellet, place the supernatant in a 5 mL tube, flash freeze, and store in −80 °C freezer. All samples that are to be compared within a given pull-down need to go through the processes on day 2 at the same time for consistency. Freeze lysates at this step if you are waiting on other samples.
During centrifugation in step 16, add 1 mL of lysis buffer and 50 μL of sepharose gelatin beads to a new 1.5 mL tube.
Mix gently and centrifuge for 3 min at 80 × g.
Remove supernatant and resuspend/wash beads in 1 mL of lysis buffer.
Repeat step 18.
After sample centrifugation (step 16), transfer the supernatant to a new 5 mL tube.
Once sepharose gelatin beads have been washed twice with lysis buffer to remove residual ethanol (steps 17–20), remove and discard the supernatant.
Transfer 1 mL of sample supernatant to the 1.5 mL tube with beads and resuspend by gentle pipetting.
Transfer the sample/bead mixture back to the 5 mL tube containing the rest of the sample.
Incubate the tube on rotator at 4 °C for 2 h (see Note 16).
Centrifuge samples at 4 °C for 5 min at 800 × g.
During step 26, wash 50 μL of sepharose streptavidin beads to remove residual ethanol as described in steps 17–20.
After sample centrifugation, transfer sample lysate to a new 5 mL tube, careful not to disturb gelatin bead pellet.
Follow steps 23 and 24 to resuspend streptavidin beads in sample lysate.
Incubate the tube on rotator at 4 °C overnight.
3.2.2. Day 2 (Bead Wash)
Centrifuge 5 mL sample tubes from step 31 at 4°C for 5 min at 800 × g.
Remove the supernatant gently by pipetting. Avoid disturbing the beads.
Add 1 mL of wash buffer to the tube; resuspend the beads gently by pipetting and transfer to a new 1.5 mL tube.
Place the tube on rotator and wash for 8 min at 25 °C.
Centrifuge at 25 °C for 3 min at 800 × g.
Remove supernatant and resuspend beads in 1 mL wash buffer.
Repeat steps 4–6 three more times (four total washes).
After the four washes remove supernatant.
Resuspend samples in 1 mL wash buffer and transfer 100 μL (10% of total) to a new 1.5 mL tube for further analysis by western blot (Fig. 2b).
Centrifuge the tube containing 100 μL of the sample at room temperature for 3 min at 800 × g.
Remove the supernatant from the tube.
Resuspend the beads using 100 μL in 50 mM Tris (pH 7.4).
Repeat step 10.
Remove the supernatant and add 100 μL of 1 SDS-PAGE sample buffer. Heat samples at 98 °C for 5 min. Samples can be stored at−20 °C for further analysis by immunoblot.
Collect the beads from the remaining 900 μL tube (step 9) by centrifugation at 25 °C for 3 min at 800 × g.
After removing the supernatant, resuspend the beads using 50 μL of 50 mM ammonium bicarbonate containing 1 mM of biotin (see Note 17) for shipment for MS analysis.
Flash freeze samples and place them in the −80 °C freezer until shipment to MS facility.
3.2.3. Analyzing Mass Spectrometry Data
4. Notes
BioID/BioID2 plasmids are available from the nonprofit repository Addgene at https://www.addgene.org/Kyle_Roux.
Methanol fixation should be avoided for IFs in which a streptavidin-conjugated Alexa Fluor is utilized as high mitochondrial staining is often detected resulting in lower signal-to-noise ratio.
Each BioID pull-down will require a BioID-only control as a positive control during fusion protein and stable cell line validation, as well as a background control necessary for deciphering proteomic data. To make an organelle-specific control, fuse a targeting peptide sequence to the BioID ligase.
Two-plate minimum is an estimate based on average 8–10 million cells per plate. Adjust the number of plates as necessary.
To minimize keratin contamination, use autoclaved DNase/ RNase-free tubes, and a tube cap opener when possible, and wear gloves.
The BioID fusion should not interrupt protein function or localization. Previous functional fusion with GFP or other similarly sized fusion protein can be a promising indicator of whether BioID should be N-terminal or C-terminal. If unsure, make and test both versions.
For transient expression for the sake of validation highly active promoters are acceptable. However, for stable cell line generation, use a vector with a low-level promoter such as viral LTR rather than a highly active promoter such as CMV to decrease the potential for false positives due to overexpression.
Addition of biotin enables promiscuous biotinylation by the BioID fusion protein.
Use PFA fixation rather than methanol to avoid a strong mitochondrial signal. Permeabilize with PBST. Use DNA-labeling reagent (e.g., Hoechst, DAPI) and streptavidin-Alexa Fluor at 1:1000 to examine biotinylation localization.
Streptavidin-HRP concentration may need to be optimized depending on the manufacturer. If a blocking step is desired, use BSA or other protein sources that lack biotin which will inactivate streptavidin’s ability to bind to biotinylated proteins on the membrane.
The biotin-streptavidin interaction is of extreme high affinity; therefore, conventional stripping methods are not appropriate for this application. Quenching with H2O2 permanently inactivates the HRP, allowing further probing with additional antibodies. To verify successful quenching, reapply ECL reagent to membrane following incubation with H2O2 and brief PBS wash.
Use an anti-tag antibody (e.g., HA or myc) or anti-BioID/ BioID2 antibody (BioFront Technologies). Block and probe diluted in ABS or other blocking buffer.
Remember to add 50 μM biotin the day before IF or WB to visualize biotinylation.
The breaks in sonication are necessary to avoid sample heating. If the sample is still viscous and cloudy after sonication, apply another session of sonication.
2 mL tubes are suggested as pellets tend to be more visible as compared to in 1.5 mL tubes.
This step functions to preclear sample lysate of proteins that nonspecifically bind to sepharose beads.
Excess biotin in the ammonium bicarbonate is included to prevent biotinylated peptides from reattaching to the streptavidin beads during and after on-bead trypsinization.
Detection of a protein via MS does not necessarily inform on biological relevance. Low-abundance proteins may be interesting candidates, especially if abundance levels differ in comparative BioID experiments. There are several proteins often identified by BioID only and BioID fusion proteins which could be considered background including AHNAK, FLNA, PARP1, EEF1A1, PRKDC, TOP1, and PKM. Histones and ribosomal subunits are also highly detected in all BioID samples, so one must be careful when pursuing such “candidates” for PPI relevance. Endogenously biotinylated carboxylases should also be considered as background.
BioID pull-downs can be performed at minimum in triplicate to enable assignment of statistical values to each protein identified.
References
- 1.Roux KJ, Kim DI, Raida M, Burke B (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196(6):801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY (2014) Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell 55(2):332–341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi-Rhee E, Schulman H, Cronan JE (2004) Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci 13 (11):3043–3050. 10.1110/ps.04911804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cronan JE (2005) Targeted and proximity- dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. J Nutr Biochem 16(7):416–418. 10.1016/j.jnutbio.2005.03.017 [DOI] [PubMed] [Google Scholar]
- 5.Kwon K, Beckett D (2000) Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci 9(8):1530–1539. 10.1110/ps.9.8.1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ (2014) Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 111(24):E2453–E2461. 10.1073/pnas.1406459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ (2016) An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27 (8):1188–1196. 10.1091/mbc.E15-12-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2017) Directed evolution of TurboID for efficient proximity labeling in living cells and organisms. bioRxiv. 10.1101/196980 [DOI] [Google Scholar]
- 9.Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, Roth JG, Gai H, Kovalski JR, Siprashvili Z, Palmer TD, Carette JE, Khavari PA (2018) RNA-protein interaction detection in living cells. Nat Methods 15 (3):207–212. 10.1038/nmeth.4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birendra K, May DG, Benson BV, Kim DI, Shivega WG, Ali MH, Faustino RS, Campos AR, Roux KJ (2017) VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol Biol Cell 28 (17):2241–2250. 10.1091/mbc.E17-03-0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, Warren G (2013) Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell 12(2):356–367. 10.1128/EC.00326-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DI, Roux KJ (2016) Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol 26(11):804–817. 10.1016/j.tcb.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opitz N, Schmitt K, Hofer-Pretz V, Neumann B, Krebber H, Braus GH, Valerius O (2017) Capturing the Asc1p/receptor for activated C kinase 1 (RACK1) microenvironment at the head region of the 40S ribosome with quantitative BioID in yeast. Mol Cell Proteomics 16(12):2199–2218. 10.1074/mcp.M116.066654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen AL, Kim EW, Toh JY, Vashisht AA, Rash-off AQ, Van C, Huang AS, Moon AS, Bell HN, Bentolila LA, Wohlschlegel JA, Bradley PJ (2015) Novel components of the toxoplasma inner membrane complex revealed by BioID. mBio 6(1): e02357–02314. 10.1128/mBio.02357-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batsios P, Meyer I, Graf R (2016) Proximity-dependent biotin identification (BioID) in Dictyostelium amoebae. Methods Enzymol 569:23–42. 10.1016/bs.mie.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 16.Schnider CB, Bausch-Fluck D, Bruhlmann F, Heussler VT, Burda PC (2018) BioID reveals novel proteins of the plasmodium parasitophorous vacuole membrane. mSphere 3(1). 10.1128/mSphere.00522-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Q, Zhou Z, Luo W, Fang M, Li M, Li H (2017) Screening of proximal and interacting proteins in rice protoplasts by proximity- dependent biotinylation. Front Plant Sci 8:749 10.3389/fpls.2017.00749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brudvig JJ, Cain JT, Schmidt-Grimminger GG, Stumpo DJ, Roux KJ, Blackshear PJ, Weimer JM (2018) MARCKS is necessary for netrin-DCC signaling and corpus callosum formation. Mol Neurobiol. 10.1007/s12035-018-0990-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan PK, Srikumar T, Dingar D, Kalkat M, Penn LZ, Raught B (2014) BioID data of c-MYC interacting protein partners in cultured cells and xenograft tumors. Data Brief 1:76–78. 10.1016/j.dib.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH (2016) Identification of an elaborate complex mediating postsynaptic inhibi- tion. Science 353(6304):1123–1129. 10.1126/science.aag0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, Choi H, Gingras AC (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal 6(302):rs15 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- 22.Firat-Karalar EN, Stearns T (2015) Probing mammalian centrosome structure using BioID proximity-dependent biotinylation. Methods Cell Biol 129:153–170. 10.1016/bs.mcb.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng YS, Seibert O, Kloting N, Dietrich A, Strassburger K, Fernandez-Veledo S, Vendrell JJ, Zorzano A, Bluher M, Herzig S, Berriel Diaz M, Teleman AA (2015) PPP2R5C couples hepatic glucose and lipid homeostasis. PLoS Genet 11(10):e1005561 10.1371/journal.pgen.1005561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coyaud E, Mis M, Laurent EM, Dunham WH, Couzens AL, Robitaille M, Gingras AC, Angers S, Raught B (2015) BioID-based identification of Skp cullin F-box (SCF)beta- TrCP1/2 E3 ligase substrates. Mol Cell Proteomics 14(7):1781–1795. 10.1074/mcp.M114.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, Stewart C, Burke B (2016) A-type lamins form distinct filamentous net- works with differential nuclear pore complex associations. Curr Biol 26(19):2651–2658. 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- 26.Firat-Karalar EN, Rauniyar N, Yates JR 3rd, Stearns T (2014) Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol 24(6):664–670. 10.1016/j.cub.2014.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer I, Peter T, Batsios P, Kuhnert O, Kruger-Genge A, Camurca C, Graf R (2017) CP39, CP75 and CP91 are major structural components of the Dictyostelium centrosome’s core structure. Eur J Cell Biol 96 (2):119–130. 10.1016/j.ejcb.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 28.De Munter S, Gornemann J, Derua R, Lesage B, Qian J, Heroes E, Waelkens E, Van Eynde A, Beullens M, Bollen M (2017) Split- BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett 591(2):415–424. 10.1002/1873-3468.12548 [DOI] [PubMed] [Google Scholar]
- 29.Schopp IM, Amaya Ramirez CC, Debeljak J, Kreibich E, Skribbe M, Wild K, Bethune J (2017) Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat Commun 8:15690 10.1038/ncomms15690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long S, Brown KM, Sibley LD (2018) CRISPR-mediated Tagging with BirA Allows Proximity Labeling in Toxoplasma gondii. Bio Protoc 8(6). 10.21769/BioProtoc.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DI, Cutler JA, Na CH, Reckel S, Renuse S, Madugundu AK, Tahir R, Goldschmidt HL, Reddy KL, Huganir RL, Wu X, Zachara NE, Hantschel O, Pandey A (2018) BioSITe: a method for direct detection and quantitation of site-specific biotinylation. J Proteome Res 17(2):759–769. 10.1021/acs.jproteome.7b00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Udeshi ND, Pedram K, Svinkina T, Fereshetian S, Myers SA, Aygun O, Krug K, Clauser K, Ryan D, Ast T, Mootha VK, Ting AY, Carr SA (2017) Antibodies to biotin enable large-scale detection of biotinylation sites on proteins. Nat Methods 14 (12):1167–1170. 10.1038/nmeth.4465 [DOI] [PMC free article] [PubMed] [Google Scholar]