Abstract
Genetically modified mice are commonly generated by the microinjection of pluripotent embryonic stem (ES) cells into wild-type host blastocysts1, producing chimeric progeny that require breeding for germline transmission and homozygosity of modified alleles. As an alternative approach and to facilitate studies of the immune system, we previously developed RAG2-deficient blastocyst complementation2. Because RAG2-deficient mice cannot undergo V(D)J recombination, they do not develop B or T lineage cells beyond the progenitor stage2: injecting RAG2-sufficient donor ES cells into RAG2-deficient blastocysts generates somatic chimaeras in which all mature lymphocytes derive from donor ES cells. This enables analysis, in mature lymphocytes, of the functions of genes that are required more generally for mouse development3. Blastocyst complementation has been extended to pancreas organogenesis4, and used to generate several other tissues or organs5–10, but an equivalent approach for brain organogenesis has not yet been achieved. Here we describe neural blastocyst complementation (NBC), which can be used to study the development and function of specific forebrain regions. NBC involves targeted ablation, mediated by diphtheria toxin subunit A, of host-derived dorsal telencephalic progenitors during development. This ablation creates a vacant forebrain niche in host embryos that results in agenesis of the cerebral cortex and hippocampus. Injection of donor ES cells into blastocysts with forebrain-specific targeting of diphtheria toxin subunit A enables donor-derived dorsal telencephalic progenitors to populate the vacant niche in the host embryos, giving rise to neocortices and hippocampi that are morphologically and neurologically normal with respect to learning and memory formation. Moreover, doublecortin-deficient ES cells-generated via a CRISPR-Cas9 approach-produced NBC chimaeras that faithfully recapitulated the phenotype of conventional, germline doublecortin-deficient mice. We conclude that NBC is a rapid and efficient approach to generate complex mouse models for studying forebrain functions; this approach could more broadly facilitate organogenesis based on blastocyst complementation.
The complexity of the cerebral cortex is a longstanding obstacle to elucidating key questions about its development and function. Experiments on cultured neurons are limited in interpretability, as they often do not recapitulate normal physiological conditions11, including specific cues involved in regional patterning during development, activity-dependent synaptic pruning, and experience-dependent plasticity. In vitro 3D brain organoid approaches can be problematic for similar reasons. Also, in vitro approaches do not allow neurobehavioral testing. Consequently, genetically engineered transgenic and germline mutant mouse models designed to interrogate specific gene functions in the cerebral cortex and hippocampus have been critically important for the field. However, generation of such mutant mice using conventional methods requires breeding of chimeras for germline transmission of the modified allele (Fig. 1a), and is time-consuming, costly, and limited to genes not required for early mouse development. Germline gene knockout approaches can also be impractical with respect to analyzing complex genetic models that involve independently segregating alleles12. Thus, we sought to develop a blastocyst complementation approach to facilitate neurobiological studies of the mouse forebrain, by creating and repopulating a vacant forebrain niche during neurodevelopment.
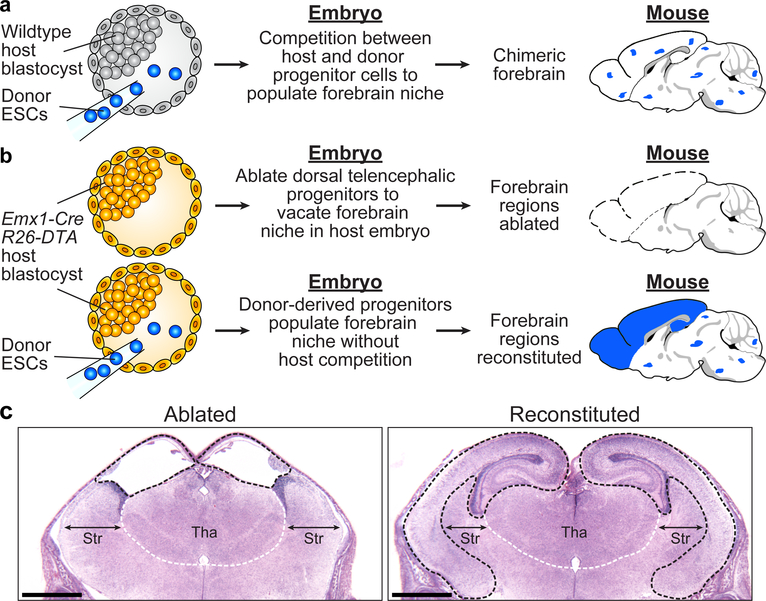
Figure 1. Generation of NBC chimeras with donor ESC-derived forebrain regions.
a, Schematic of the conventional approach to generate mouse models. Competition between host and donor cells generates mice with chimeric brains (blue dots indicate donor contribution). b, Schematic of the NBC approach. Upper: Emx1-Cre activation of DTA expression at E9.5 causes dorsal telencephalic progenitor ablation in host embryos, which develop into mice that lack forebrain regions (dotted lines). Lower: Injection of donor ESCs into host blastocysts yields mice with reconstituted, donor-derived forebrain regions (blue). Mouse brain schematics in a and b were generated based on a reference image from the Allen Mouse Brain Atlas. c, Representative H&E-stained coronal brain sections (E18.5) from 42 mice generated from non-ESC-injected (left) or ESC-injected (right) Emx1-Cre; R26-DTA blastocysts (n=3 from ESC-injected Emx1-Cre; R26-DTA blastocysts; n=38 non-ESC-injected Emx1-Cre; R26-DTA blastocysts). Ablated (left) or reconstituted (right) forebrain regions are demarcated by black dotted lines. White dotted lines demarcate the approximate boundary of the thalamus (Tha); double-headed arrows indicate width of the striatum (Str). Scale bars,1 mm. See also Extended Data Figure 1.
To ablate dorsal telencephalic progenitors (DTPs) in mice, we crossed mice that express Cre recombinase from an Emx1 promoter with mice carrying a floxed STOP cassette upstream of an attenuated diphtheria toxin subunit A (DTA) that was introduced into the Rosa26 locus13,14. Emx1 is a homeobox transcription factor expressed at E9.5 in DTPs, which give rise to neocortical, hippocampal, and olfactory bulb structures14,15. We predicted that Emx1-Cre; R26-DTA mice would lack neocortices and hippocampi, based on findings of prior studies that tested Emx1-Cre-mediated deletion of factors required for neuronal survival16,17, and the known ability of directed DTA expression to ablate specific cell lineages in mice13,18,19 (Fig. 1b). Following transfer into foster females, pups derived from Emx1-Cre; R26-DTA blastocysts survived birth, acquired a characteristic pink coloration (Extended Data Fig. 1a), and vocalized, indicating normal respiratory and cardiovascular systems, as well as functionally intact brainstems20. However, histological analysis of serial brain sections of the Emx1-Cre; R26-DTA pups revealed extensive ablation of hippocampal, neocortical, and olfactory bulb structures, resulting in fusion and dilation of the lateral ventricles over the thalamus (Fig. 1c). Thalamus, midbrain, hindbrain and cerebellum appeared normal, in line with the specificity of Emx1-Cre recombination14. However, the mice exhibited incomplete jaw development likely due to off-target recombination (Extended Data Fig. 1a), consistent with a similar defect documented in an independent mouse model using the same Emx1-Cre strain19.
We sought to repopulate the vacant dorsal telencephalic niche in Emx1-Cre; R26-DTA mice by injecting wild-type (WT) donor ESCs into Emx1-Cre; R26-DTA NBC host blastocysts, followed by transfer into foster females to generate NBC chimeras (Fig. 1b). Histological analysis of NBC chimeras revealed complete reconstitution of hippocampal and neocortical structures (Fig. 1c), and rescue of the jaw malformation (Extended Data Fig. 1a). Reconstituted NBC chimeras displaying a normal jaw phenotype were viable, developed normally after birth, and showed no increased mortality up to 24 months of age (Extended Data Fig. 1a–d). Both male and female NBC chimeras were fertile, and female chimeras displayed normal nurturing behavior towards their pups.
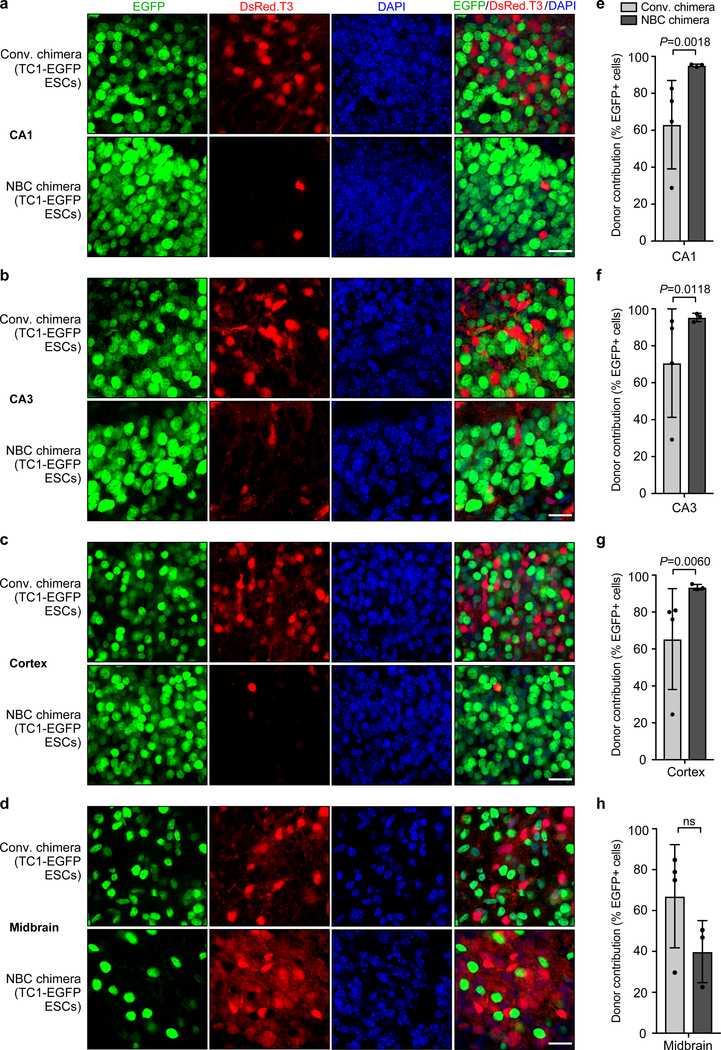
To directly assess and quantify donor and host cell contribution in NBC chimeras, we generated and injected H2B-EGFP-expressing donor ESCs (“TC1-EGFP”) into blastocysts derived from Emx1-Cre; R26-DTA mice crossed with CAG-DsRed.T3 transgenic mice (see Methods and Extended Data Fig. 2). The DsRed.T3 transgene mediates widespread constitutive expression of the red fluorescent protein variant DsRed.T3 driven by the chicken beta actin promoter coupled with the cytomegalovirus immediate early enhancer. Thus, this strategy generates dual-labeled chimeras in which donor ESC-derived cells are EGFP-positive (green) and host blastocyst-derived cells are DsRed.T3-positive (red). Postnatal day 0 (P0) NBC chimeras generated from TC1-EGFP ESC-injected Emx1-Cre; R26-DTA; CAG-DsRed.T3 blastocysts had nearly 100 percent EGFP-positive cells in hippocampus and cerebral cortex, indicating reconstitution of the vacant forebrain niche (Fig. 2a–c and e–g). In contrast, conventional P0 chimeras generated by injection of TC1-EGFP donor ESCs into CAG-DsRed.T3 blastocysts consistently showed substantially lower, and much more variable, percentages of EGFP-positive cells, due to host-donor competition (Fig. 2a–c and e–g). In the midbrain, where there is no ablation and thus no host-donor competition, both NBC and conventional chimeras showed lower numbers of EGFP-positive cells and large chimera-to-chimera variability (Fig. 2d and h), similar to other non-ablated tissues (Extended Data Fig. 3). We note that the small percentage of DsRed.T3-positive cells in the forebrain regions of NBC chimeras likely corresponds to non-dorsal telencephalon-derived cells of host origin (e.g., interneurons that migrate tangentially into the cortex, microglia that derive from resident macrophages, or endothelial cells from blood vessels).
Figure 2. NBC chimeras show consistently high donor contribution in forebrain regions as compared to conventional chimeras.
a-d, Representative images showing extent of TC1-EGFP donor ESC contribution to the indicated brain regions in conventional (conv.) and NBC chimeras at P0, depicting donor-derived cells in green and host-derived cells in red. Nuclei were DAPI stained (blue). CA, cornu ammonis subfields of the hippocampus. Scale bars, 10 μm. e-h, Quantification of donor ESC contribution in conventional (n=4) and NBC chimeras (n=3). Data are mean ± SD. Variance was significantly different between conventional and NBC chimeras for CA1, CA3, and cortex (F test for equality of variances), reflecting the wide variation in donor contribution among individual conventional chimeras, in contrast to the consistently high donor contribution in NBC chimeras. No difference in variance was observed for midbrain (P = 0.5534; ns, not significant; F test for equality of variances), consistent with random donor contribution to this non-ablated brain region in both types of chimeras. See also Extended Data Figures 2 and 3.
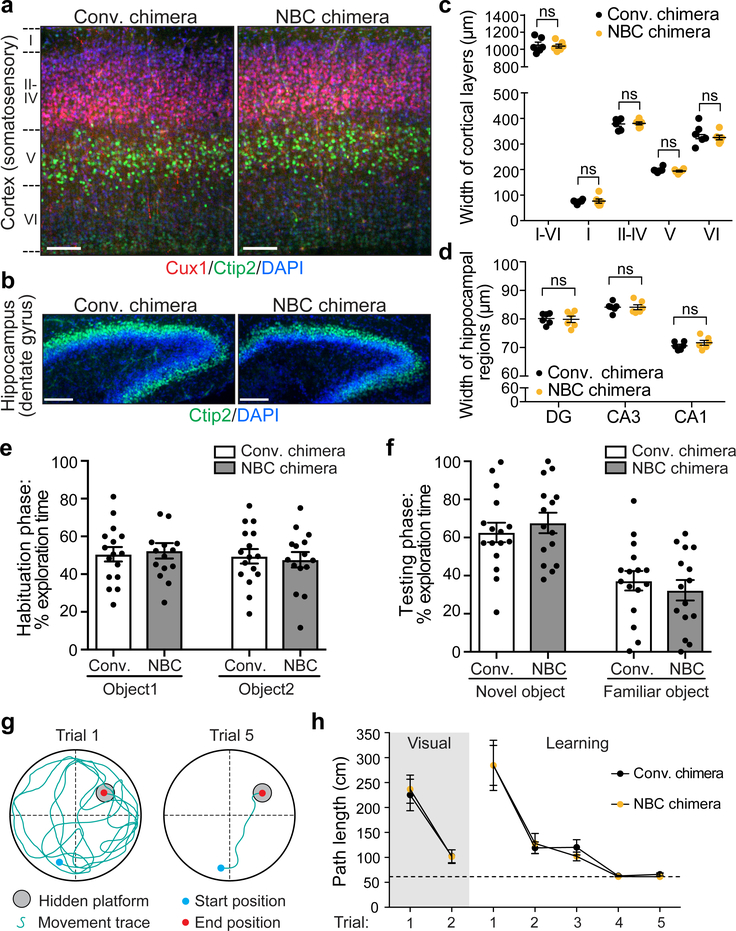
To assess the cytoarchitecture and lamination of the neocortex and hippocampus in P7 WT NBC chimeras, we performed immunofluorescence staining for layer-specific markers21 (Fig. 3a–d and Extended Data Fig. 4). The reconstituted brains of P7 WT NBC chimeras were morphologically normal, with layer widths and cellular densities indistinguishable from conventional chimera controls (Fig. 3a–d and Extended Data Fig. 4). Numbers of oligodendrocytes, astrocytes, and microglia were similar to controls, and there were no signs of neuroinflammation (Extended Data Fig. 5). Correspondingly, basic neurological functions were intact in the two- to three-month-old WT NBC chimeras (Supplementary Table 1). To assess higher cognitive functions, we tested the behavior of adult (two-month-old) chimeras in the novel object recognition (NOR) and Morris water maze (MWM) paradigms, two behavioral tasks that are dependent on intact forebrain function22,23. Adult WT NBC chimeras performed indistinguishably from controls in both paradigms, demonstrating that learning and memory are intact (Fig. 3e–h and Extended Data Fig. 6). Collectively, these studies indicate that NBC mice are indistinguishable from normal mice with respect to brain structure and function.
Figure 3. Brains of NBC mice are structurally and functionally normal.
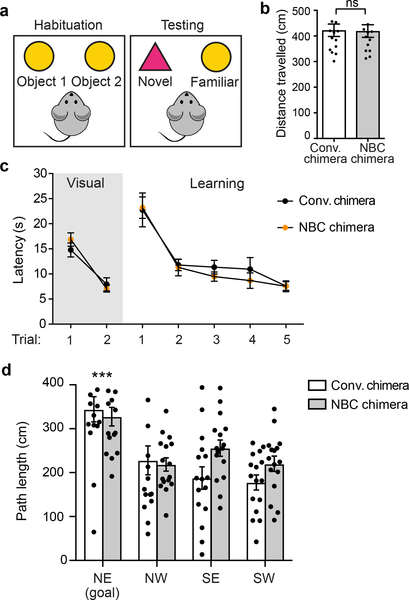
a, Representative images of somatosensory cortex sections from six independent, P7 NBC and conventional chimeras. Layers (L) II-IV (Cux1; red), L V (Ctip2; green), and nuclei (DAPI; blue) are shown. Scale bars, 100 μm. b, Representative images of hippocampal sections from six independent, P7 NBC and conventional chimeras stained for Ctip2 (green) and DAPI (blue). Scale bars, 100 μm. c, Somatosensory cortex layer widths in P7 conventional (black dots) or NBC (orange dots) chimeras (n=6 mice each). d, Widths of P7 dentate gyrus (DG), CA3, and CA1 regions in conventional or NBC chimeras (n=6 mice each). Horizontal bars in c and d indicate mean ± SEM; ns, not significant, P>0.05 (unpaired, two-tailed t test). See also Extended Data Figure 4. e, No significant difference in percentage of total exploration time spent on object 1 vs. object 2 during habituation phase of novel object recognition (NOR) task in conventional and NBC chimeras. f, Significant preference for the novel object in male 2-month-old conventional (n=15) and NBC (n=14) chimeras. Data represent mean ± SEM; conventional chimera: novel vs. familiar, P=0.0051; NBC chimera: novel vs. familiar, P<0.0001 (one-way ANOVA, Tukey’s post hoc correction). g, Schematic of MWM task. h, Mean path length (± SEM) to platform for conventional (black dots) or NBC (orange dots) chimeras in visual and learning trials. Dotted line indicates minimum possible path length to platform. No significant difference between male 2-month-old conventional and NBC chimeras (n=16 mice each) in visual [F(1,30)=0.052, P=0.822] or learning trials [F(1,30)=0.041, P=0.841] (two-way mixed model ANOVA). Path length across learning trials was significantly different indicating both groups had learned the task [F(1.393,41.787)=34.57, P<0.0005] (two-way mixed model ANOVA with Greenhouse-Geisser correction for violation of sphericity). See also Extended Data Figure 6.
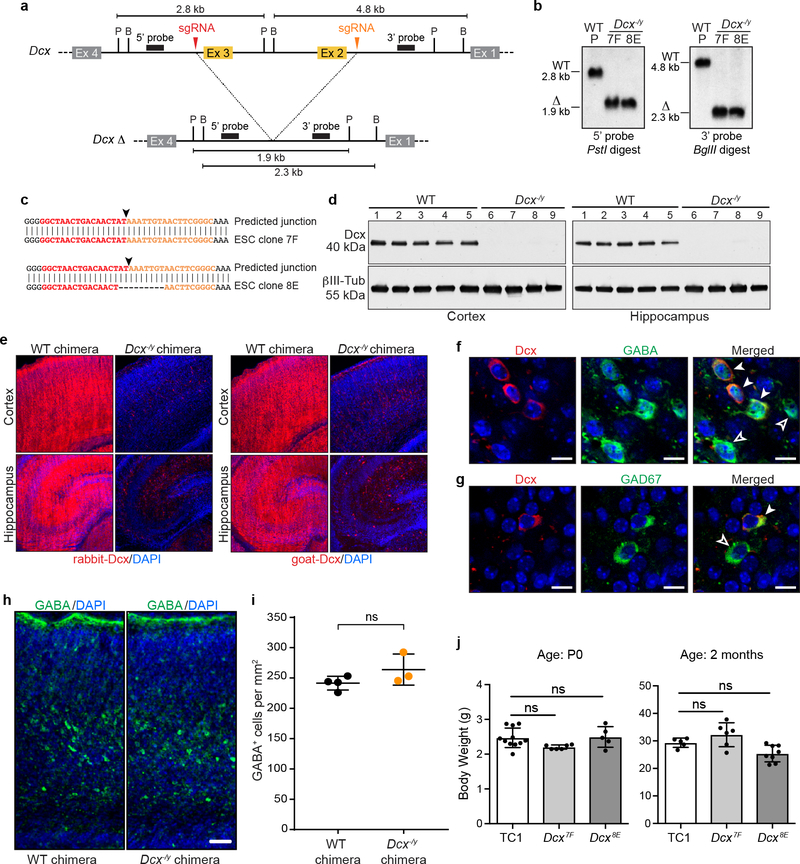
To further evaluate NBC, we tested the ability of this approach to rapidly assess a brain phenotype previously established by a conventional germline knockout approach24,25. Doublecortin (Dcx) lies on the mouse X chromosome and encodes a microtubule-associated protein that functions in neuronal migration26,27. Male mice in which the single Dcx copy is functionally abrogated show disrupted hippocampal lamination with a normal neocortex from age E17.5 on (including at P0 and two months of age)24,25. We targeted male mouse ESCs via CRISPR/Cas9 to delete Dcx exons 2 and 3 (Extended Data Fig. 7a–b), which encode the tubulin-binding domains essential for Dcx function28. Two Dcx−/y ESC clones, generated from independent deletion events (Extended Data Fig. 7c), were injected into NBC blastocysts to create NBC chimeras. Resulting P0 Dcx−/y NBC chimeras showed a dramatic reduction in Dcx in protein lysates from neocortical and hippocampal tissue, relative to control NBC chimeras generated from the parental WT ESC line (Extended Data Fig. 7d). In addition, immunofluorescence analyses with two different antibodies revealed that Dcx was absent in most cells of the neocortex and hippocampus of P0 Dcx−/y NBC chimeras (Extended Data Fig. 7e). The remaining Dcx-positive neurons expressed markers of GABAergic neurons (Extended Data Fig. 7f and g) and are likely immature interneurons that originate in the ventral telencephalon where Emx1 is not expressed14. Overall positioning and cell density of GABA-positive interneurons in in the somatosensory cortex were similar in WT and Dcx−/y chimeras (Extended Data Figure 7h and i). We did not observe decreased postnatal viability of reconstituted Dcx−/y chimeras; all mice that were not euthanized for experiments at P0 survived to adulthood (i.e., at least two months of age; n=14, generated from 2 different Dcx−/y ESC clones; Extended Data Figure 7j). Further, body weights of WT (TC1) and Dcx−/y chimeras were similar at P0 and two months of age (Extended Data Figure 7j).
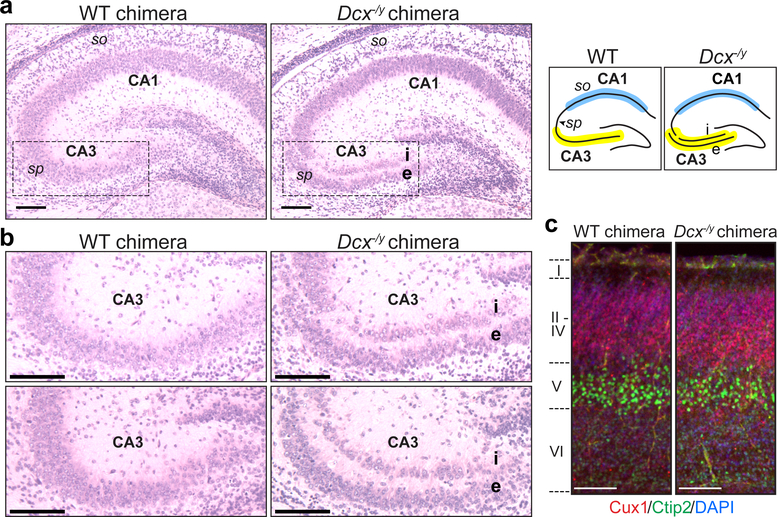
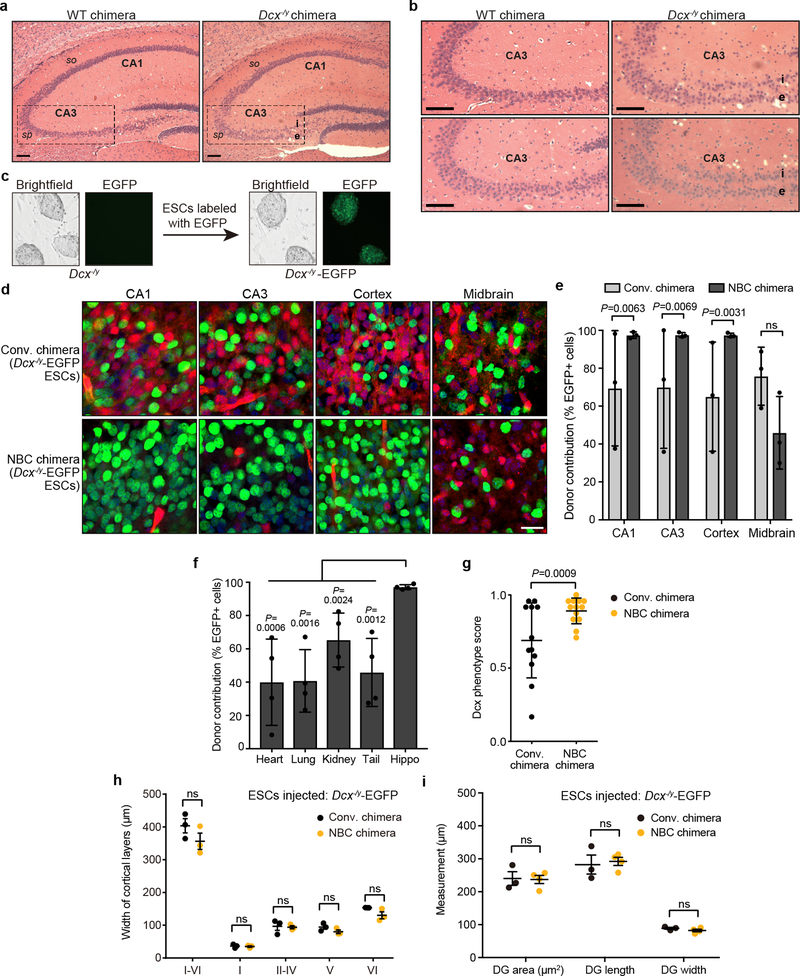
Dcx−/y NBC chimeras accurately phenocopied the hippocampal dysplasias observed in P0 and two-month-old germline Dcx−/y mice24,25. At P0, we observed an abnormal division of the stratum pyramidale of the cornu ammonis (CA) 3 region into an internal and external layer, resulting in a distinct band of heterotopic neurons (Fig. 4a, b). We also noted a less pronounced band of heterotopic neurons in the stratum oriens of the CA1 region (Fig. 4a). When Dcx−/y NBC chimeras were examined at 2 months of age, the heterotopic band of CA3 neurons was still present; however, the CA1 heterotopias appeared to have resolved (Extended Data Fig. 8a, b). The mutant hippocampal phenotype was reproducibly observed in all NBC chimeras from both Dcx−/y ESC clones, demonstrating full recapitulation of the brain phenotype of germline Dcx knockout mice at both ages examined24,25. As in germline Dcx−/y mice, cortical layering in P0 Dcx−/y NBC chimeras was intact (Fig. 4c, Extended Data Fig. 8h), providing an internal control to verify the absence of non-specific neurodevelopmental defects in the Dcx−/y NBC chimeras.
Figure 4. Dcx−/y NBC chimeras recapitulate the phenotype of germline Dcx−/y mice.
a, Representative images of H&E-stained coronal brain sections showing hippocampal defects in P0 Dcx−/y NBC chimeras (right) but not wild-type (WT) chimeras (left). Dashed lines highlight the CA3 region, where the stratum pyramidale (sp) is present as a single layer in WT chimeras (left) but is abnormally divided into an internal (i) and external (e) layer in the Dcx−/y chimeras (right). This phenotype is diagrammed on the right, with the CA3 (yellow) and CA1 (blue) regions highlighted. so, stratum oriens. b, Enlarged, representative images of the CA3 region in two P0 WT (left) and Dcx−/y (right) chimeras. c, Representative images of Cux1 (red), Ctip2 (green), and DAPI (blue) co-stained, coronal somatosensory cortex sections from P0 WT (left) and Dcx−/y (right) chimeras. Approximate location of cortical layers I-VI is indicated on the left. Scale bars, 100 μm. Images in a-c are representative of experiments performed on 21 mice (n=6 for Dcx−/y ESC clone 7F; n=4 for Dcx−/y ESC clone 8E; n=11 for wild-type ESCs). Mice were generated and characterized as depicted in Extended Data Figures 7–8.
We further generated H2B-EGFP-expressing Dcx−/y ESCs (Dcx−/y-EGFP) and injected them into either Emx1-Cre; R26-DTA; CAG-DsRed.T3 blastocysts or CAG-DsRed.T3 blastocysts to generate, respectively, Dcx−/y-EGFP NBC chimeras or Dcx−/y-EGFP conventional chimeras (Extended Data Fig. 8d–i). Consistent with our findings with NBC and conventional chimeras generated with WT TC1-EGFP ESCs (Fig. 2), P0 Dcx−/y-EGFP NBC chimeras showed significantly higher donor contribution and lower variability in forebrain regions, but not in non-ablated tissues, than conventional Dcx−/y-EGFP chimeras (Extended Data Fig. 8d–f). Moreover, all P0 Dcx−/y NBC chimeras showed the well-characterized Dcx knockout phenotype with high consistency, allowing clear-cut experimental analyses and conclusions (Extended Data Fig. 8g). In contrast, Dcx−/y conventional chimeras showed highly variable penetrance of the Dcx knockout phenotype (Extended Data Fig. 8g). Thus, this experiment clearly reveals the advantage of using the NBC system in which competition between donor Dcx−/y ESCs-derived progenitors and host blastocyst-derived progenitors does not occur. Beyond further establishing the utility of the NBC system for analysis of the known relatively subtle and specific Dcx−/y phenotype, these experiments also revealed that Dcx−/y ESC-derived neural progenitors are not at a major competitive disadvantage as compared to WT progenitors during neurodevelopment.
The NBC approach we describe provides a blastocyst complementation system that should greatly facilitate many in vivo studies of the cerebral cortex and hippocampus. Blastocyst complementation has been used to reconstitute several tissues and organs, including the mature adaptive immune system, ocular lens, pancreas, thymus, kidney, and heart2–10. Until now, however, blastocyst complementation has never been reported for the brain. NBC is based on ESC complementation of blastocysts that, due to programmed ablation of blastocyst-derived DTPs, cannot generate major forebrain structures. The approach works extremely well, as we found consistent pup survival rates, recovery rates, and litter sizes across a large number of experiments (Extended Data Fig. 9). Moreover, use of normal ESCs for NBC complementation leads to the generation of NBC chimeras with donor-derived forebrain structures that are structurally and functionally indistinguishable from those of conventional chimeras (Figs. 1–3). Thus, use of normal ESCs for NBC can reliably generate key structures of the brain, arguably the most complex organ in vertebrates. Moreover, use of Dcx−/y ESCs for NBC complementation fully recapitulated the reported brain phenotypes of Dcx germline knockout mice (Fig. 4). Thus, the NBC approach is anticipated to substantially accelerate studies of the function of genes and other genomic elements in cortical and hippocampal development and physiology in mice.
Prior blastocyst complementation approaches relied on creation of cellular niches by inactivation of genes critical for organ development in blastocyst-derived progenitor cells2–6,8–10. In contrast, NBC abrogates organogenesis by selective progenitor cell ablation via targeted DTA expression. The general DTA ablation strategy we use for NBC should also be applicable for blastocyst complementation approaches in the context of other brain regions and other organ systems. Moreover, compared to traditional methods used to generate genetically modified mice via germline genetic modifications, NBC should offer many of the same advantages for brain studies that have been extensively documented for RDBC in the context of immune system studies. Thus, as documented by our studies of the Dcx mutation, genetic modifications introduced into ESCs by CRISPR/Cas9 approaches can be rapidly and accurately assessed via NBC with respect to causing even a subtle brain phenotype. Moreover, as extensively documented via RDBC analyses of the immune system12,29–30, multiple genetic modifications can be introduced into ESCs, which then could be directly analyzed for effects on forebrain development, function, or disease in NBC chimeras, eliminating the time-consuming and often prohibitive task of intercrossing of multiple, segregating alleles in mutant mice12,29. Finally, recent studies have focused on analyses of interspecies chimeras4,8–10; the NBC approach could provide an ideal platform for such studies in the context of forebrain development and function.
METHODS
No statistical methods were used to predetermine sample size. Samples for image acquisition and image analysis, and mice for neurobehavioral tests were randomized, and investigators were blinded to allocation during the experiment and outcome assessment.
Mice and Blastocyst Injections
All studies were in compliance with ethical regulations established by the Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC) and Institutional Biosafety Committee (IBC). Animals were maintained on a 12-h light-dark schedule in a temperature-controlled environment, with food and water provided ad libitum. Behavioral testing was conducted during the light phase. Emx1-Cre (stock no.: 005628; C57BL/6) and R26-DTA mice (stock no.: 010527; 129Sv:C57BL/6) were purchased from the Jackson Laboratory and maintained as homozygous breeding pairs. Tg(CAG-DsRed*MST)1Nagy/J mice (Jackson Laboratory stock no.: 006051; C57BL/6)31 were a kind gift from Carla F. Kim (Boston Children’s Hospital) (see Supplementary Table 2 for genotyping information). Blastocyst injections were performed as previously described3. Briefly, 10–12 ESCs were injected per blastocyst (harvested at dpc 3.5 from superovulated R26-DTA females crossed to Emx1-Cre or Emx1-Cre; DsRed.T3 stud males), and 10–12 blastocysts were implanted per pseudo-pregnant CD-1 foster females (Charles River Laboratories). For a detailed protocol of the above methods, see Protocol Exchange32. For all studies, NBC chimeras with normally developed jaws were analyzed. The following parental ESC lines were used for blastocyst injections: TC1 (a gift from P. Leder, Harvard Medical School; 129Sv; used for analysis of brain morphology, donor vs. host contribution, and Dcx mutation), EF1 (derived in house from a 129Sv:C57BL/6 F1 hybrid mouse; used for analysis of brain morphology, and behavior experiments), R26-lacZ (TC1-derived; a gift from S. H. Orkin, Boston Children’s Hospital; 129Sv; used for brain morphology analysis). All cell lines were confirmed to be mycoplasma free.
ES Cell Culture, CRISPR/Cas9 Gene Editing, Fluorescent Labeling of Cells and Generation of Dcx−/y mice
ESCs were maintained on a monolayer of γ-irradiated mouse embryonic fibroblast feeder cells and passaged every 2–3 days. Cells were cultured at 37°C and 5% CO2 in DMEM medium supplemented with 15% (v/v) fetal bovine serum, 20 mM HEPES (pH 7.4), 2 mM L-glutamine, 50 U/mL penicillin/streptomycin, 0.1 mM MEM non-essential amino acids, 50 μM β-mercaptoethanol, and 500 U/mL recombinant mouse leukemia inhibitory factor. To generate fluorescently labeled ESC lines, wild-type TC1 and Dcx mutant ESCs (described below) were transduced with the lentiviral plasmid FUHGW (hUbc-driven H2B-EGFP)33, a kind gift from Guo-Liang Xu (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences). Two Cas9 sgRNAs targeting the Dcx locus (see Supplementary Table 2 for nucleotide sequences) were cloned into the pX330 vector (Addgene plasmid 42230), and nucleofected into wild-type TC1 ESCs (1 μg each pX330 vector / 2 × 106 cells) per the Amaxa 4D-nucleofector protocol (solution P3, CG-104, Lonza). A puromycin resistance plasmid was co-transfected together with the sgRNAs to allow positive clones to be identified by puromycin selection. ESC clones with Dcx deletion were identified by PCR screening, Southern blotting (see below), and nucleotide sequencing of the deletion junction (see Supplementary Table 2 for details and oligonucleotide sequences). The two independent ESC clones (Dcx7F and Dcx8E) used for blastocyst injections were confirmed to have a normal karyotype by metaphase analysis. Clones were also confirmed by Southern blotting to be negative for genomic integration of the pX330 plasmid backbone or the puromycin resistance plasmid backbone. For a detailed protocol of the above methods, see Protocol Exchange32. As Dcx is developmentally regulated, mice were analyzed at postnatal day (P) 0 for Dcx expression. For assessment of hippocampal and cortical phenotypes, two differently aged cohorts of NBC chimeras (Dcx7F or Dcx8E ESCs into Emx1-Cre; R26-DTA blastocysts) and control chimeras (wild-type parental TC1 ESCs into Emx1-Cre; R26-DTA blastocysts) were analyzed. For the P0 cohort, n=6 for Dcx7F; n=4 for Dcx8E; n=11 for wild-type controls. For the 2-month-old cohort, n=6 for Dcx7F; n=8 for Dcx8E; n=5 for wild-type controls. Additional mouse cohorts were generated using Dcx−/y-EGFP ESCs and analyzed at P0; n=3 for Dcx−/y-EGFP NBC chimeras (Dcx−/y-EGFP ESCs into Emx1-Cre; R26-DTA; DsRed.T3 blastocysts), and n=3 for control chimeras (Dcx−/y-EGFP ESCs into R26-DTA; DsRed.T3 blastocysts). In all cases, sample order was randomized, and investigators were blinded to genotype.
Histology and Immunofluorescence Analysis
Mice were anesthetized with ketamine/xylazine, and transcardially perfused with ice-cold PBS prior to brain removal and fixation. For histological analyses, hemibrains were fixed overnight in Bouin’s solution (Sigma) at 4°C with gentle agitation, embedded in paraffin, and sectioned at 5 μm. Hematoxylin and eosin (H&E) staining was performed on coronal brain sections per standard protocols. For Nissl staining, brains were fixed in 4% (w/v) paraformaldehyde at 4°C for 24 hours with gentle agitation, cryoprotected in 30% (w/v) sucrose in 0.1 M sodium phosphate buffer overnight at 4°C, embedded in optimal cutting temperature (OCT) compound, and cryosectioned at 35 μm. Coronal sections were mounted onto glass slides and dried. Slides were rehydrated in TBS, stained with thionin solution (0.25% (w/v) thionin, 1.2% (v/v) glacial acetic acid, 0.144% (w/v) sodium hydroxide) for 1 min, dehydrated by incubation in a series of increasing ethanol concentrations, cleared with xylene, and coverslipped (Permount mounting medium; Electron Microscopy Sciences). Images were acquired with an Olympus BX51 microscope and a Micropublisher 5.0 RTV camera (QImaging) and processed with the QCapture software (QImaging).
For immunofluorescence analyses, hemibrains were fixed overnight in 4% (w/v) paraformaldehyde at 4°C with gentle agitation. For use with GABA and GAD67 antibodies (see below), 0.2% (v/v) glutaraldehyde was added during this fixation step. Fixed brains were cryoprotected, embedded, and cryosectioned at 35 μm. Floating coronal sections were rinsed in TBS, quenched in 50 mM NH4Cl, permeabilized with 0.4–1% (w/v) Triton X-100, blocked (5% (v/v) serum from the species used to generate the secondary antibodies, 0.5% (w/v) fish gelatin, 0.2% (w/v) Tween-20), and incubated with primary antibody at 4°C overnight. The following primary antibodies were used: Cux1 (CDP; rabbit polyclonal, 1:250; sc-13024 [M-222], Santa Cruz Biotechnology), Ctip2 (rat monoclonal [clone 25B6], 1:100, ab18465, Abcam), GFAP (rabbit polyclonal, 1:1,000; Z0334, DAKO), Olig2 (rabbit polyclonal, 1:1,000; AB9610, Millipore), Iba1 (rabbit polyclonal, 1:1,000; 019–19741, Wako chemicals), Dcx (rabbit polyclonal, 1:1,000; ab18723, Abcam; or goat polyclonal, 1:1,000; sc-8066 [C-18], Santa Cruz Biotechnology), GABA (rabbit polyclonal, 1:1,000; A2052, Sigma), GAD67 (mouse monoclonal [clone 1G10.2], 1:500; MAB5406, Millipore), Satb2 (mouse monoclonal [clone SATBA4B10], 1:500; ab51502, Abcam), Foxp2 (rabbit polyclonal, 1:1,000; ab16046, Abcam). Sections were washed three times with TBS or TBS with 0.05–0.1% (w/v) Triton X-100 and incubated with secondary antibody (1:500; Alexa Fluor 488, 594, or 647; ThermoFisher Scientific) for two hours at room temperature. Sections were washed twice with TBS or TBS with 0.05–0.1% (w/v) Triton X-100, mounted on SuperFrost Plus glass slides, and coverslipped with DAPI Fluoromount-G (SouthernBiotech). Non-brain tissues (heart, lung, kidney, tail) were fixed and sectioned similarly to the brains.
For image acquisition and analysis, sample order was randomized, and investigators were blinded to genotype. Images were acquired with a Zeiss confocal spinning disk microscope (Yokogawa CSU-X1, 3i Intelligent Imaging Innovations) and a QuantEM:512SC EMCCD camera (Photometrics) and processed with SlideBook6 software (3i Intelligent Imaging Innovations). To assess donor and host contribution, brain slices from P0 NBC chimeras with normal jaw morphology indicative of successful ESC contribution (Dcx−/y-EGFP or TC1-EGFP ESCs into Emx1-Cre; R26-DTA; DsRed.T3 blastocysts) and control chimeras (Dcx−/y-EGFP or TC1-EGFP ESCs into R26-DTA; DsRed.T3 blastocysts) were analyzed by fluorescence imaging. Graphed data points represent the average cell counts from three coronal sections per mouse.
For analysis of layer widths and cell densities, brain slices from P7 NBC chimeras (TC1-derived ESCs or EF1 ESCs injected into Emx1-Cre; R26-DTA blastocysts) and conventional chimera controls (TC1-derived ESCs or EF1 ESCs injected into wild-type strain background-matched blastocysts) were analyzed. Width of layer (L) I-VI, L II-IV and L V was determined by DAPI, Cux1, and Ctip2 immunostaining, respectively. L I and VI were determined by the boundary of cortex and adjacent layer markers. Width of cortical layers in Dcx−/y-EGFP chimeras were analyzed similarly using brain slices from P0 NBC chimeras (Dcx−/y-EGFP injected into Emx1-Cre; R26-DTA; DsRed.T3 blastocysts) and control chimeras (Dcx−/y-EGFP ESCs injected into R26-DTA; DsRed.T3 blastocysts) and the width of layer II-IV was determined by DAPI staining. Graphed data points represent the average measurements or cell counts from twelve coronal step-sections per mouse. For each section, layer width in the hippocampus was estimated from seven measurements across each hippocampal region, and layer width in the cortex was estimated from three measurements across the somatosensory cortex (S1). Quantification of DG measurements was performed in at least three hippocampal sections per mouse. Numbers of mice analyzed are stated in the figure legends. DG area was measured by outlining the DG region. DG length and width, respectively, were assessed by determining the length of the granule cell layer or the distance between the ends of the granule cell layer.
Southern Blotting
Genomic DNA was prepared by lysing cells at 56°C overnight in lysis buffer (200 mM NaCl, 100 mM Tris-Cl pH 8.0, 5 mM EDTA, 0.2% SDS), supplemented with 0.2 – 0.4 mg/mL proteinase K, followed by precipitation and purification by phenol-chloroform-isoamyl alcohol extraction per standard protocols. DNA was digested with the indicated restriction enzymes, genomic fragments were separated by agarose gel electrophoresis, transferred to nylon membranes (Zeta-Probe GT, Bio-Rad), and hybridized with radiolabeled DNA probes per standard protocols. Details of Dcx probes are listed in Supplementary Table 2.
Protein Extraction and Immunoblotting
Whole cell extracts were prepared from microdissected cortical and hippocampal tissue in ice-cold THB buffer (250 mM sucrose, 20 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA), supplemented with protease inhibitors (cOmplete EDTA-free protease inhibitor cocktail, Roche) and 0.5 mM PMSF. Tissue was homogenized by 30 strokes of a Teflon pestle in a Potter-Elvehjem tissue grinder (#19; Kontes). Protein concentration was determined (Bio-Rad Protein DC assay), samples were briefly sonicated, 6× SDS sample buffer (0.5 M Tris-Cl pH 6.8, 30% (w/v) glycerol, 10% (w/v) SDS, 0.6 M dithiothreitol, 0.012% (w/v) bromophenol blue) was added to a final concentration of 1×, and samples were denatured by incubation at 95°C for three minutes. Equal amounts of protein were separated by SDS-PAGE, followed by immunoblotting with primary antibodies against Dcx (rabbit polyclonal, 1:10,000; ab18723, Abcam) and βIII-tubulin (rabbit polyclonal, 1:20,000; ab18207, Abcam).
Neurobehavioral Analysis
Cohorts of two- to three-month-old male mice generated by injection of two different ESC lines were tested: a larger cohort generated from wild-type EF1 donor ESCs and a smaller cohort generated from wild-type TC1 donor ESCs. Cohorts of age-matched, male conventional chimeras generated from the same donor ESC lines were used as controls. Mice were randomized for testing, and investigators administering the tests were blinded to genotype. Prior to behavioral testing, mice were transferred to the testing room and allowed to acclimatize for at least 30 minutes. Basic autonomic, neurological, and sensory functions were assessed using a modified SHIRPA (SmithKline Beecham, Harwell, Imperial College, Royal London Hospital, phenotype assessment) protocol34. Parameters were scored in accordance with guidelines from the International Mouse Phenotyping Resource of Standardised Screens (IMPReSS) developed by the International Mouse Phenotyping Consortium (IMPC).
Novel Object Recognition (NOR)
The NOR task was administered in a square arena (30 × 30 cm) maintained at 30 lux throughout the experiment. During the habituation phase, mice were allowed to explore the arena and two identical objects freely, and the trial was stopped once the mouse had explored both objects for a combined total of 20 seconds. The mice were then returned to the home cage for a 10-minute intersession interval. During the testing phase, one of the objects was replaced with a novel object, and mice were allowed to explore freely for 5 minutes. The position of the novel object was randomized for each trial. Mice were monitored by automated, multiple body point video tracking with an overhead camera (Ethovision XT 11.5, Noldus Information Technology), and novel object discrimination was calculated as the percentage of total exploration time spent exploring the novel or familiar object. Locomotor activity (distance traveled in cm) was determined by automated video tracking. Nose-point heat maps, track maps, and videos were generated for visualization purposes.
Morris Water Maze (MWM)
Mice were placed in a circular pool (137 cm diameter) filled with room temperature water, divided into quadrants marked with visual cues for spatial orientation. Swim patterns were recorded by automated video tracking (Ethovision XT 11.5, Noldus Information Technology). Track maps for each mouse were visually inspected, and a total of three conventional chimeras were excluded due to technical tracking issues. In the visual phase (Day 1), mice were given 90 seconds to swim to a visible platform elevated 0.5 cm above the water and marked with a flag. The position of the platform was kept constant, but the starting quadrant varied. Four starts (each from one of the four quadrants) were administered in one trial, and two visual trials were administered during Day 1. In the learning phase (Days 2 and 3), the platform was moved to the opposite quadrant, submerged 0.5 cm below the water, and the flag removed. Mice were given 90 seconds to find the hidden platform and left on the platform for 5 seconds to orientate themselves. Mice that did not find the platform within 90 seconds were guided to it. Each mouse completed three learning trials on Day 2 (Trials 1 to 3), and two learning trials on Day 3 (Trials 4 and 5), with each trial comprising four starts each. In the probe trial (Day 4), the platform was removed. Mice were placed in the quadrant opposite the quadrant that had previously contained the platform (NE), and given 60 seconds to swim. Time spent, and path length traversed, in each of the four quadrants was recorded.
Statistical Analysis
Graphs were generated using GraphPad Prism 6.05. Data from the quantification of donor ESC contribution were analyzed with an unpaired Welch’s t test, and F test for equality of variances. Data from the quantification of cortical and hippocampal widths and cell densities were analyzed with an unpaired, two-tailed t test. Data from the NOR testing were analyzed with a one-way ANOVA with Tukey’s post hoc correction. MWM data for two-way mixed model ANOVA analysis were first tested for normality and sphericity, using the Shapiro-Wilks test in GraphPad Prism and Mauchly’s test in SPSS Statistics, respectively. MWM data were found to be normally distributed, and in cases where the data were found to violate the assumption of sphericity, the Greenhouse-Geisser correction was used to correct the F statistic and assess statistical significance. Data were then analyzed in SPSS Statistics with a two-way mixed model ANOVA, with genotype (NBC chimera and conventional chimera) as the between-subject factor, and path length or latency to platform (visual and learning trials), or path length within each quadrant (probe trial) as the within-subject factor. Bonferroni post hoc correction was applied in cases where multiple pairwise comparisons were performed.
Data Availability
All data generated or analyzed during this study are included in this manuscript (and its supplementary information files).
Extended Data
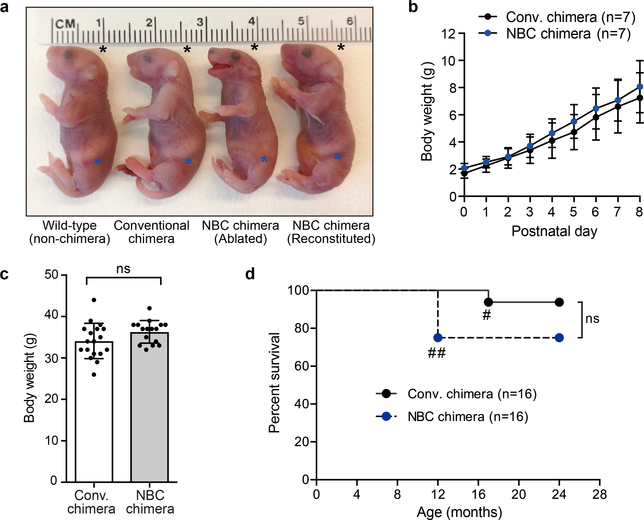
Extended Data Figure 1. NBC chimeras have normal overall appearance, postnatal growth, body weights, and survival rates.
a, Representative photograph of newborn mice. From left to right: (1) Wild-type (WT) non-chimeric pup from a normal mouse breeding cross; (2) Conventional chimera generated by injecting donor ESCs into a WT host blastocyst; (3) NBC chimera without donor ESC reconstitution of the forebrain (“Ablated”); (4) NBC chimera with donor ESC reconstitution of the forebrain (“Reconstituted”). Note the abnormal head curvature of the ablated NBC chimera relative to the other mice (black asterisks), consistent with the lack of a forebrain. Also note the lack of a milk spot (blue asterisks), consistent with the inability to suckle as a result of incomplete jaw development. At least three mice per group from independent experiments were analyzed with similar results. b, Body weight gain of conventional and NBC chimeras with normal jaws (n=7 each) in the first postnatal week. Data represent mean ± SD; not significant, P>0.05 (multiple unpaired, two-tailed t tests, Holm-Sidak post hoc correction) for all data points. c, Body weights of 2- to 3-month-old male conventional (n=18) and NBC chimeras (n=16). Data represent mean and SD; ns, not significant, P>0.05 (unpaired, two-tailed t test). d, Postnatal survival rate of conventional (n=16 males) and NBC chimeras (n=16 males); ns, not significant; P>0.05 (two-sided Log-Rank, Mantel-Cox test. #One conventional chimera was euthanized because of a genital mass; ##four NBC chimeras died because their cage flooded; all other mice were euthanized at 24 months of age.
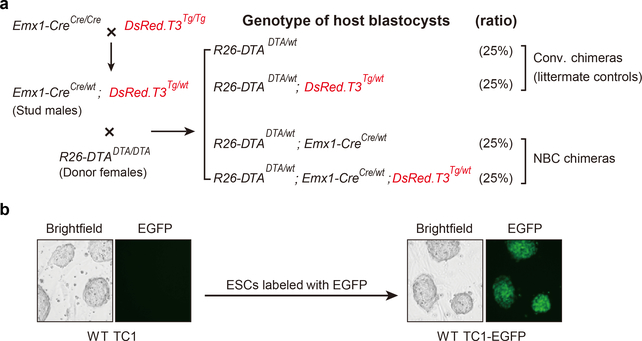
Extended Data Figure 2. Generation of Emx1-Cre; R26-DTA; DsRed.T3 NBC host blastocysts and EGFP-labeled donor ESCs.
a, Schematic of the breeding crosses used to generate DsRed.T3-labeled NBC host blastocysts. Tg, transgene. b, Representative bright field and fluorescence microscopy images of wild-type TC1 ESCs before and after lentiviral integration of H2B-EGFP. The ESCs shown stably express H2B-EGFP and were repeatedly imaged during the study with similar results.
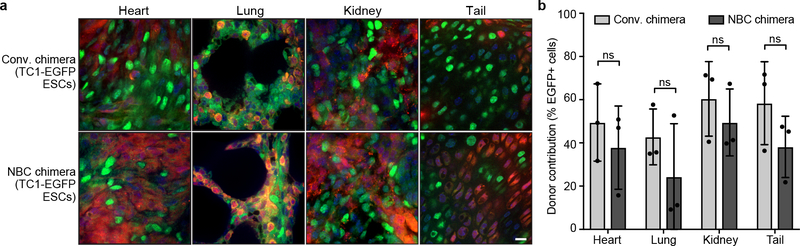
Extended Data Figure 3. NBC chimeras show variable donor contribution in non-brain tissues similar to conventional chimeras.
a, Representative images showing the extent of TC1-EGFP donor ESC contribution to the indicated non-brain tissues in conventional (conv.) and NBC chimeras at P0, depicting donor-derived cells in green and host-derived cells in red. Nuclei were visualized by DAPI (blue). Scale bar, 10 μm. These experiments were performed on six mice (n=3, NBC chimeras; n=3, conventional chimeras). b, Quantification of donor ESC contribution in indicated tissues of conventional and NBC chimeras (n=3 mice each). Data represent mean and SD. The variance in donor contribution was not significantly different between conventional and NBC chimeras for all tissues examined, consistent with competition between donor and host cells in these non-neuronal tissues in both types of chimeras (ns, not significant, P>0.05; F test for equality of variances). See also Figure 2.
Extended Data Figure 4. Brains of NBC mice are structurally normal.
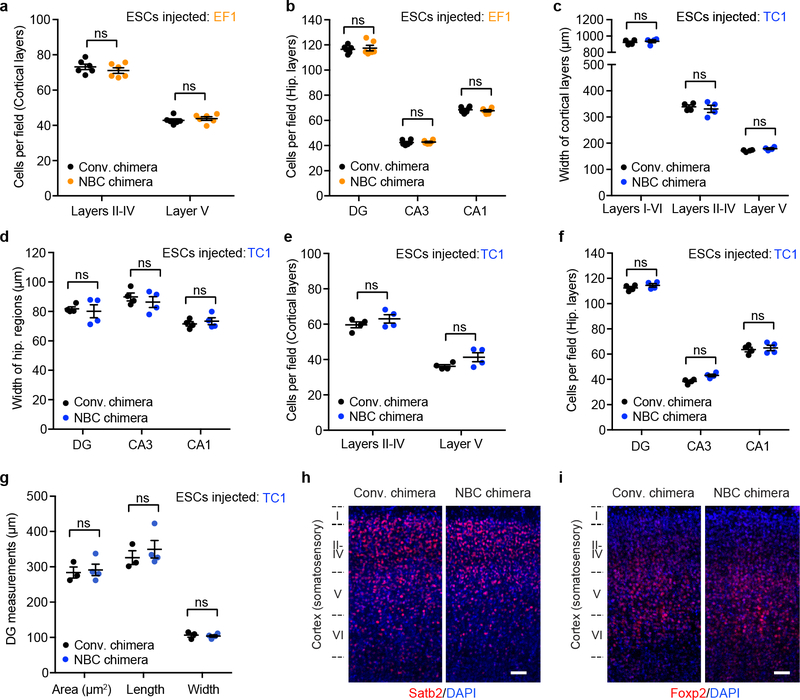
a, b, Cell numbers in the indicated somatosensory cortical layers (a) and hippocampal regions (b) in P7 conventional (black dots) and NBC chimeras (orange dots) (n=6 mice each) generated by blastocyst injection of EF1 ESCs. Cortical layers (L) II-IV and V were identified by Cux1 and Ctip2 immunoreactivity, respectively. Nuclei were visualized with DAPI. Hip, hippocampus; DG, dentate gyrus. c, d, Width of the indicated somatosensory cortical layers (c) and hippocampal regions (d) in P7 conventional (black dots) and NBC chimeras (blue dots) generated by blastocyst injection of TC1-derived ESCs (n=4 mice each), analyzed as in Figure 3. e, f, Cell numbers in the indicated somatosensory cortical layers (e) and hippocampal regions (f) in P7 conventional (black dots) and NBC chimeras (blue dots) generated by blastocyst injection of TC1-derived ESCs (n=4 mice each), analyzed as in a, b. g, DG measurements in P0 conventional (conv.) chimeras (n=3; black dots) and NBC chimeras (n=4; blue dots) generated by blastocyst injection of TC1-EGFP ESCs. For panels a-g, horizontal bars indicate mean; error bars denote SEM; ns, not significant, P>0.05 (unpaired, two-tailed t test). h, i, Representative images of coronal somatosensory cortex sections at P7 stained for Satb2 (h, red), Foxp2 (i, red) and DAPI (blue). Scale bars, 50 μm. These experiments were repeated on six mice (n=3, NBC chimeras; n=3, conventional chimeras).
Extended Data Figure 5. NBC mice have normal proportions of non-neuronal cells and do not show signs of neuroinflammation.
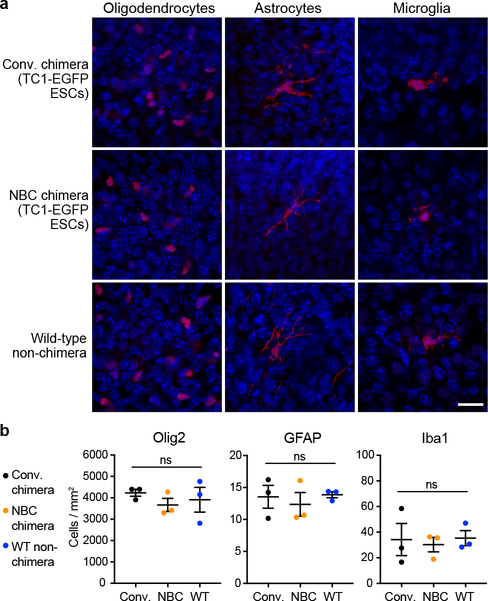
a, Representative immunofluorescence images of oligodendrocytes (Olig2), astrocytes (GFAP), and microglia (Iba1) in cortical brain sections of conventional (conv.) and NBC chimeras, with a wild-type, non-chimeric mouse for comparison. The non-neuronal cells are shown in red; DAPI-stained nuclei are in blue. Scale bar, 10 μm. These experiments were repeated on 9 mice (n=3, NBC chimeras; n=3, conventional chimeras; n=3, non-chimeric wild-type mice). b, Quantification Olig2-, GFAP-, or Iba1-positive cells in cortical brain sections of conventional chimeras, NBC chimeras, and wild-type non-chimeric mice (n=3 each). Data represent mean and SEM; ns, not significant, P>0.05 (one-way ANOVA, Tukey’s post hoc correction for multiple comparisons).
Extended Data Figure 6. Brains of NBC mice are functionally normal.
a, Schematic of the NOR task. b, Locomotor activity of male, 2-month-old conventional (n=15) or NBC chimeras (n=14) during the habituation phase of the NOR paradigm. Data represent mean and SEM; ns, P>0.05 (unpaired two-tailed t test). c, Latency to platform for conventional (black dots) or NBC (orange dots) chimeras in the indicated visual and learning trials of the MWM task. Male 2-month-old conventional and NBC chimeras (n=16 mice each) were analyzed. Data represent mean and SEM; no significant difference was observed between conventional and NBC chimeras across the visual trials [F(1,30) = 0.223, P = 0.640] or learning trials [F(1,30) = 0.296, P = 0.590] (two-way mixed model ANOVA). A significant difference was observed in latency over the learning trials, indicating that both groups of mice are able to learn [F(1.627,48.815) = 29.426, P<0.0005] (two-way mixed model ANOVA with Greenhouse-Geisser correction for violation of sphericity). d, Path length traversed in each quadrant by conventional and NBC chimeras during the probe trial. Male 2-month-old conventional and NBC chimeras (n=16 mice each) were analyzed. Data represent mean and SEM; no significant difference between conventional and NBC chimeras [F(1,30) = 1.433, P = 0.241] (two-way mixed model ANOVA). Significant preference was observed for the NE quadrant, where the platform had previously been located, indicating memory retention of the prior platform location; ***, P<0.001 (multiple pairwise comparisons with Bonferroni post hoc correction).
Extended Data Figure 7. Generation of Dcx−/y NBC chimeras.
a, Upper, Schematic of Dcx inactivation. sgRNA target sequences (red and orange arrowheads) flanking exon 2 (Ex 2) and exon 3 (Ex 3) are indicated. Probes (black rectangles) and restriction enzyme sites (P, PstI; B, BglII) used for Southern blotting are indicated. Predicted restriction fragment sizes are indicated in kb. Lower, predicted Dcx exon 2/3 deletion (Δ) locus. b, Southern blot analysis of genomic DNA from parental Dcx+/y (WT) ESC line and Dcx−/y ESC clones 7F and 8E using the probes and digests depicted in a. Southern blot analysis was performed five times. c, Sequence analysis of Dcx deletion junctions (black arrowheads); sgRNA target sequences flanking the deletion junction are denoted in red and orange. Both Dcx−/y ESC clones have unique junction sequences, suggesting each derived from an independent deletion event. d, Western blotting of whole cell lysates from cortex (left) or hippocampus (right) of WT or Dcx−/y NBC chimeras at P0 with antibodies to Dcx (rabbit anti-Dcx) and βIII-tubulin as a loading control. Each lane (1–9) depicts cortex or hippocampus whole cell lysate from one mouse; experiments were performed on nine mice (n=4, Dcx−/y ESC clone 8E; n=5, wild-type ESCs) e, Representative immunofluorescence images of Dcx expression in cortex (upper) and hippocampus (lower) of P0 WT or Dcx−/y chimeras. 21 mice (n=6, Dcx−/y ESC clone 7F; n=4 Dcx−/y ESC clone 8E; n=11 wild-type ESCs) were analyzed. Dcx-expressing cells (red) were detected with rabbit (left) or goat (right) Dcx antibodies. Nuclei were DAPI stained (blue). f, Representative images of somatosensory cortical brain sections (acquired between layers IV-V) from P0 Dcx−/y chimeras co-stained Dcx (red, left) and GABA (green, middle) antibodies. Merged images are shown on right. Nuclei were DAPI stained (blue). Solid white arrowheads indicate Dcx/GABA double-positive cells (host-derived immature interneurons). Hollow arrowheads indicate GABA single-positive cells (either donor-derived mature interneurons, donor-derived immature interneurons, or host-derived mature interneurons). g, Representative images of somatosensory cortical brain sections (acquired between layers IV-V) from P0 Dcx−/y chimeras co-stained with Dcx (red, left) and GAD67 (green, middle) antibodies. Merged images are shown on right. Nuclei were DAPI stained (blue). Solid white arrowheads indicate host-derived immature interneurons, whereas hollow arrowheads indicate cells that are either donor-derived mature interneurons, donor-derived immature interneurons, or host-derived mature interneurons. Scale bar, 10 μm. For experiments in f and g, nine mice (n=4, Dcx−/y ESC clone 8E; n=5 wild-type ESCs) were analyzed. h, Representative images of somatosensory cortical brain sections from P0 WT and Dcx−/y chimeras stained GABA antibodies (green). Nuclei DAPI stained (blue). Scale bar, 50 μm. Seven mice were analyzed (n=3, NBC chimeras; n=4, conventional chimeras). i, Cell density of GABA-positive interneurons in somatosensory cortex from control chimeras (n=4 mice) and Dcx−/y chimeras (n=3 mice). Center denotes mean, error bars indicate SEM. ns, not significant, P>0.05 (unpaired, two-tailed t test). j, Mean (± SD) body weights of WT (TC1) or Dcx−/y chimeras; ns, not significant, P>0.05 (one-way ANOVA, Tukey’s post hoc correction). For P0 cohort, n=6 for Dcx7F; n=5 for Dcx8E; n=11 for WT TC1 controls. For two-month-old cohort, n=6 for Dcx7F; n=8 for Dcx8E; n=5 for WT TC1 controls.
Extended Data Figure 8. Dcx−/y NBC chimeras show a mutant hippocampal phenotype.
a, Representative images of H&E-stained coronal sections of hippocampus from 2-month-old wild-type (WT) chimeras and Dcx−/y chimeras. Dashed lines highlight the CA3 region, where the stratum pyramidale (sp) is present as a single layer in WT chimeras (left) but is abnormally divided into an internal (i) and external (e) layer in the Dcx−/y chimeras (right). Refer to Figure 4a for a schematic of the hippocampus. so, stratum oriens. b, Enlarged, representative images of the CA3 region in two 2-month-old WT and Dcx−/y chimeras. Scale bars, 100 μm. For experiments in a and b, 14 two-month-old Dcx−/y chimeras, generated from two different Dcx−/y ES clones (n=6 from clone Dcx7F; n=8 from clone Dcx8E), and five WT chimeras were analyzed. All 14 Dcx−/y chimeras showed the hippocampal mutant phenotype. c, Representative bright field and fluorescence microscopy images of Dcx−/y ESCs before and after lentiviral integration of H2B-EGFP. d, Representative images from experiments performed on 6 mice (n=3, NBC chimeras; n=3, conventional chimeras) showing extent of Dcx−/y-EGFP donor ESC contribution to indicated brain regions in P0 conventional and NBC chimeras (green, donor ESC-derived cells; red, host-derived cells; blue, DAPI-stained nuclei). Scale bar,10 μm. e, Quantification of Dcx−/y-EGFP donor ESC contribution in conventional and NBC chimeras (n=3 mice each). Data are mean ± SD. Variance was significantly different between conventional and NBC chimeras for CA1, CA3, and cortex (F test for equality of variances), reflecting the wide variation in donor contribution among individual conventional chimeras in contrast to consistently high donor contribution in NBC chimeras. No difference was observed for midbrain (ns, not significant; P = 0.7812), consistent with competition between donor and host cells in this non-ablated brain region in both types of chimeras. f, Quantification of variance in Dcx−/y-EGFP donor ESC contribution in non-brain tissues relative to hippocampus in NBC chimeras (n=4). Donor contribution in the hippocampus was calculated as the average of CA1 and CA3 values. Data are mean ± SD (F test for equality of variances). g. Quantification of Dcx mutant hippocampal phenotype in conventional chimeras (n=12) and NBC chimeras (n=13) at P0. Dcx phenotype scores were determined by reviewing a series of coronal brain step-sections from each chimera and determining the number of sections that show a clear CA3 double layer (Score = 1), some degree of CA3 disorganization (Score = 0.5), or normal CA3 layer morphology (Score = 0). Mean Dcx phenotype scores across all sections of a given chimera are plotted. Each data point corresponds to one chimera. Horizontal lines indicate mean phenotype score (± SD) across all chimeras per group. Conventional chimeras show significant variation in the severity of the mutant phenotype, in contrast to the NBC chimeras (F test for equality of variances). h, Somatosensory cortex layer widths in P0 Dcx−/y-EGFP ESC-injected conventional (n=3, black dots) or NBC (n=3, orange dots) chimeras determined by DAPI and Ctip2 staining. Center denotes mean, error bars indicate SEM. ns, not significant, P>0.05 (two-way ANOVA with Bonferroni post hoc correction for multiple comparisons). i, Dentate gyrus (DG) measurements in conventional chimeras (n=3; black dots) and NBC chimeras (n=4; orange dots) at P0, generated by blastocyst injection of Dcx−/y-EGFP ESCs. Horizontal bars indicate mean; Error bars denote SEM; ns, not significant, P>0.05 (unpaired, two-tailed t test).
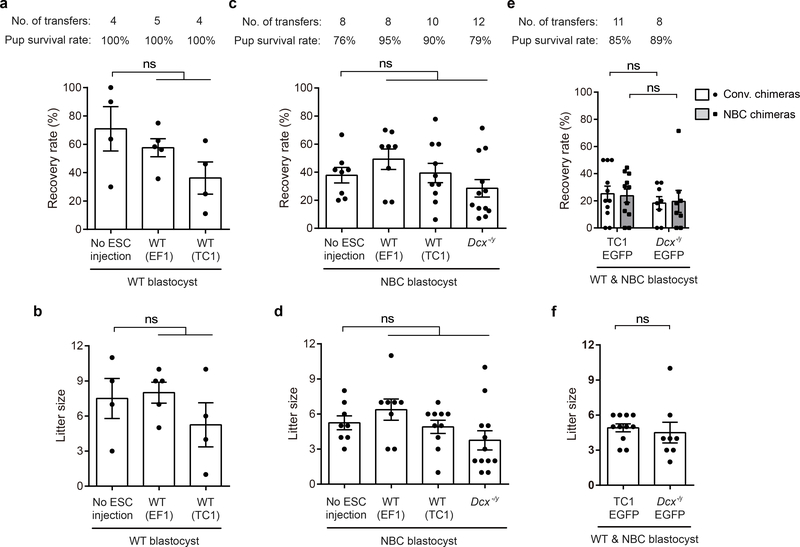
Extended Data Figure 9. Rates of recovery, survival, and litter sizes across multiple pregnancies and ESC lines.
a, Average recovery rate (± SEM) of liveborn pups from WT blastocysts without ESC injection or injected with WT ESCs (EF1, TC1). ns, not significant, P>0.05 (one-way ANOVA with Tukey’s post hoc correction for multiple comparisons). Number of transfers (foster pregnancies) and pup survival rate at P0 is listed above graph. b, Mean litter size (± SEM) per foster recipient of WT blastocysts without ESC injection or injected with WT ESCs (EF1, TC1); ns, not significant, P>0.05 (one-way ANOVA, Tukey’s post hoc correction for multiple comparisons). c, Average recovery rate (± SEM) of liveborn pups from NBC blastocysts without ESC injection or injected with WT (EF1, TC1) or Dcx−/y ESCs. ns, not significant, P>0.05 (one-way ANOVA with Tukey’s post hoc correction for multiple comparisons). Number of transfers (foster pregnancies) and pup survival rate at P0 is listed above graph. d, Mean litter size (± SEM) per foster recipient of NBC blastocysts without ESC injection or injected with WT (EF1, TC1) or Dcx−/y ESCs; ns, not significant, P>0.05 (one-way ANOVA, Tukey’s post hoc correction for multiple comparisons). e, Average recovery rate (± SEM) of mice from mixed WT/NBC blastocyst transfers; ns, not significant, P>0.05 (unpaired, two-tailed t test). f, Mean litter size (± SEM) per foster recipient; ns, not significant, P>0.05 (unpaired, two-tailed t test). For experiments in e and f, mice were intercrossed to simultaneously generate WT or NBC blastocysts with or without DsRed.T3 expression (see Extended Data Figure 2a) and were injected with either TC1-EGFP or Dcx−/y-EGFP ESCs followed by transfer into fosters. Pups were then genotyped to determine whether they were WT or NBC blastocyst-derived.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Alt lab and Christopher A. Walsh for stimulating discussions, Pei-Yi Huang for help with blastocyst injections, Hwei-Ling Cheng for advice and help with ESC culture, and Sophie V. Griswold and Trishala Chari for assistance with behavioral experiments. Behavioral testing was carried out at the Boston Children’s Hospital (BCH) Neurodevelopmental Behavior Core (CHB IDDRC, 1U54HD090255). This work was supported by the Howard Hughes Medical Institute, the BCH Department of Medicine (DOM) Support Fund, and the BCH DOM Anderson Porter Fund and a major grant from the Charles H. Hood Foundation, Inc., Boston, MA. B.S. is a Kimmel Scholar of The Sidney Kimmel Foundation, supported by NIA/NIH grant AG043630, the UCSF Brain Tumor SPORE Career Development Program, the American Cancer Society, the Andrew McDonough B+ Foundation, the Shurl and Kay Curci Foundation, and a Martin D. Abeloff V Scholar award of The V Foundation for Cancer Research. B.S. also holds the Suzanne Marie Haderle and Robert Vincent Haderle Endowed Chair, UCSF. H-Q.D. is a fellow of the Cancer Research Institute of New York. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
REFERENCES
- 1.Capecchi MR Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nature reviews. Genetics 6, 507–512, doi: 10.1038/nrg1619 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Lansford R, Stewart V, Young F & Alt FW RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proceedings of the National Academy of Sciences of the United States of America 90, 4528–4532 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J et al. Generation of normal lymphocyte populations by Rb-deficient embryonic stem cells. Current biology : CB 3, 405–413 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142, 787–799, doi: 10.1016/j.cell.2010.07.039 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Liegeois NJ, Horner JW & DePinho RA Lens complementation system for the genetic analysis of growth, differentiation, and apoptosis in vivo. Proceedings of the National Academy of Sciences of the United States of America 93, 1303–1307 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanger BZ, Tanaka AJ & Melton DA Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature 445, 886–891, doi: 10.1038/nature05537 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Usui J et al. Generation of kidney from pluripotent stem cells via blastocyst complementation. The American journal of pathology 180, 2417–2426, doi: 10.1016/j.ajpath.2012.03.007 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Wu J et al. Stem cells and interspecies chimaeras. Nature 540, 51–59, doi: 10.1038/nature20573 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Wu J et al. Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 168, 473–486.e415, doi: 10.1016/j.cell.2016.12.036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi T et al. Interspecies organogenesis generates autologous functional islets. Nature 542, 191–196, doi: 10.1038/nature21070 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Conti L & Cattaneo E Neural stem cell systems: physiological players or in vitro entities? Nature reviews. Neuroscience 11, 176–187, doi: 10.1038/nrn2761 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Ba Z et al. A Rapid Embryonic Stem Cell-Based Mouse Model for B-cell Lymphomas Driven by Epstein-Barr Virus Protein LMP1. Cancer immunology research 3, 641–649, doi: 10.1158/2326-6066.cir-15-0058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Wu Y & Capecchi MR Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development 133, 581–590, doi: 10.1242/dev.02236 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Gorski JA et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 6309–6314, doi:20026564 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simeone A, Acampora D, Gulisano M, Stornaiuolo A & Boncinelli E Nested expression domains of four homeobox genes in developing rostral brain. Nature 358, 687–690, doi: 10.1038/358687a0 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Kim S et al. The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron 66, 69–84, doi: 10.1016/j.neuron.2010.03.019 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y et al. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nature neuroscience 15, 819–826, doi: 10.1038/nn.3097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmiter RD et al. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell 50, 435–443 (1987). [DOI] [PubMed] [Google Scholar]
- 19.Yoshida M, Assimacopoulos S, Jones KR & Grove EA Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development 133, 537–545, doi: 10.1242/dev.02209 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Turgeon B & Meloche S Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases. Physiological reviews 89, 1–26, doi: 10.1152/physrev.00040.2007 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Molyneaux BJ, Arlotta P, Menezes JR & Macklis JD Neuronal subtype specification in the cerebral cortex. Nature reviews. Neuroscience 8, 427–437, doi: 10.1038/nrn2151 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Leger M et al. Object recognition test in mice. Nature protocols 8, 2531–2537, doi: 10.1038/nprot.2013.155 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Vorhees CV & Williams MT Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nature protocols 1, 848–858, doi: 10.1038/nprot.2006.116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbo JC et al. Doublecortin is required in mice for lamination of the hippocampus but not the neocortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 7548–7557 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kappeler C et al. Magnetic resonance imaging and histological studies of corpus callosal and hippocampal abnormalities linked to doublecortin deficiency. The Journal of comparative neurology 500, 239–254, doi: 10.1002/cne.21170 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Gleeson JG, Lin PT, Flanagan LA & Walsh CA Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 23, 257–271 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Francis F et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 23, 247–256 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Taylor KR, Holzer AK, Bazan JF, Walsh CA & Gleeson JG Patient mutations in doublecortin define a repeated tubulin-binding domain. J Biol Chem 275, 34442–34450 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Tian M et al. Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires. Cell 166, 1471–1484.e1418, doi: 10.1016/j.cell.2016.07.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeap LS et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell 163, 1124–1137, doi: 10.1016/j.cell.2015.10.042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
ADDITIONAL REFERENCES CITED IN METHODS
- 31.Vintersten K et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis 40, 241–246, doi: 10.1002/gene.20095 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Schwer B et al. CRISPR/Cas9-mediated genome editing in mouse embryonic stem cells and direct analysis of brain phenotypes via neural blastocyst complementation. Protoc. Exch, https://doi.org/xxxxxxx (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14, 512–522, doi: 10.1016/j.stem.2014.01.001 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Masuya H et al. Implementation of the modified-SHIRPA protocol for screening of dominant phenotypes in a large-scale ENU mutagenesis program. Mammalian Genome 16, 829–837 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript (and its supplementary information files).