Summary
Patch clamp electrophysiology is the standard technique used for the high resolution functional measurements on ion channels. While studies using patch clamp are commonly carried out following ion channel expression in a heterologous system such as Xenopus oocytes or tissue culture cells, these studies can also be carried out using ion channels reconstituted into lipid vesicles. In this chapter, we describe the methodology for reconstituting the ion channels into liposomes and the procedure for generation of unilamellar blisters from these liposomes that are suitable for patch clamp. Here we focus on the bacterial K+ channel KcsA, although the methodologies described in this chapter should be applicable for the functional analysis of other ion channels.
Keywords: GUV recordings, KcsA, lipid vesicles, blisters, patch clamp
Introduction
Ion channels are ubiquitously present integral membrane proteins that provide pathways for the movement of ions through biological membranes [1]. Ion channels play essential roles in a number of physiological processes such as neuronal signaling, cardiac excitability, insulin release, and fluid secretion. Functional studies on ion channels are carried out by the electrophysiological techniques of voltage clamp or patch clamp generally following the heterologous expression of the ion channels in Xenopus oocytes or HEK cells [2, 3]. Alternately, functional studies on ion channels can be carried out using reconstituted systems, which consist of the purified ion channel proteins incorporated into lipid bilayers [4]. Studies using reconstituted systems are required when the ion channels either do not express or do not properly fold or traffic to the cell membrane in heterologous expression systems. This situation is commonly encountered during heterologous expression of bacterial and archaeal ion channels. Another complication in using a heterologous expression system is that the endogenous proteins or lipids present may alter the function of the ion channel in a non-native manner. This complication is not encountered in studies on reconstituted ion channels as the all the components present are biochemically defined. A challenge with using reconstituted systems is the requirement of the purified ion channel protein. However, recent advances in membrane protein biochemistry have made overexpression and purification of many ion channel proteins very feasible.
Purification of ion channels requires the use of detergents for extraction from the cellular membranes and for stabilizing these proteins during purification. Studies to measure the equilibrium binding of ions, ligands, or inhibitors can be carried out using the purified ion channels in detergent, but the electrophysiological studies to characterize the detailed functional properties require reconstitution of the purified ion channel into a lipid bilayer [4]. Ion channels have been reconstituted into a variety of lipid vesicles that range in size from small unilamellar vesicles (SUVs, tens of nm) [5] to giant unilamellar vesicles (GUVs, 1–100 μm), roughly the size of biological cells [6]. Functional studies on ion channels reconstituted into SUVs are carried out using flux assays, which measure the movement of ions into or out of many vesicles [4]. These flux assays provide the bulk functional properties of the ion channel population but do not allow for easy determination of the single channel properties such as the conductance or the open probability of the ion channel. These assays also do not allow for rapid control of membrane potential or for rapid exchange of solutions surrounding the membrane without highly specialized stopped-flow equipment. SUVs containing ion channels can also be fused with planar lipid bilayers and electrophysiological methods can be used for measurement of ion channel activity [4]. A limitation of the planar lipid bilayer recording setups that are presently available is that they do not allow for rapid exchange of solutions. Further, the size of the membranes formed in a planar lipid bilayer setup make them more prone to electrical and mechanical noise which interfere with the ion channel recordings. The membrane size also results in large capacitances which cause for a slowing of the voltage response and therefore fast channel events (rapid activation, for example) cannot be investigated.
Ion channels can also be reconstituted into GUVs. GUVs are advantageous compared to SUVs since their large size allows them to be visualized by optical or fluorescence microscopy, thus allowing them to be used for patch-clamp electrophysiological recordings of ion channels. Ion channels that have been functionally characterized in GUVs include the proton-gated K+ channel KcsA [7, 8, 9 ], the voltage gated K+ channel KvAP [10], the mechanosensitive ion channels MscS and MscL [11–13], the bacterial porins OmpC and OmpF [14, 15], and the eukaryotic ion channel TrpV1 [16]. In addition to studies on ion channels, GUVs have also been used as model membranes to study characteristics of lipid domains [17], membrane-protein interactions [18], DNA-membrane interactions [19] and vesicle budding and fission [20, 21].
The techniques commonly used for forming GUVs include spontaneous swelling and electroformation [22–26]. In the spontaneous swelling method, SUVs are dried down, forming a lipid film which consists of the lipid bilayers organized roughly in stacks [27]. Rehydration causes the bilayers to swell as the hydrophilic head groups attract water and results in forming GUVs as water seeps into the interlamellar space. At physiological ionic strength, negatively charged lipids are crucial for the electrostatic repulsion necessary to promote swelling [28]. Neutral lipids may be used in the presence of high concentrations of divalent cations [29], at low ionic strength (< 1 mM) [28] or with the addition of non-electrolytic monosaccharides such as glucose, mannose, or fructose [30] , that drive water towards the interlamellar space as a result of osmotic pressure. Electroformation works similarly to the spontaneous swelling method, except that an alternating current (AC)-electric field is applied during the rehydration step. The alternating electric field causes the vesicles to become more ordered and also to vibrate, aiding in their ability to fuse and then detach from the surface to form GUVs [31]. While electroformation generally results in a more homogenous GUV population, the drawbacks of electroformation are that it works best under low ionic concentrations that are not physiological, is hampered by the presence of charged lipids, and requires specialized equipment [22].
We have adapted a technique similar to the dehydration/rehydration method for the functional characterization of ion channels. In this method, proteoliposomes containing the ion channels are dehydrated on a glass cover slip, and then rehydrated in the presence of magnesium to induce the formation of large unilamellar blisters [32]. We have found it easier to identify these unilamellar blisters and to obtain gigaohm seals from these blisters than compared to the spherical GUVs obtained by electroformation. Further this method relies on a regular patch clamp setup that is commonly available and therefore provides a relatively easy way for functional studies on reconstituted ion channels. In this chapter, we describe how this approach is used for the functional characterization of the bacterial potassium channel KcsA.
2. Materials
2.1. Reconstitution of the ion channel into liposomes
Soybean polar lipid extract in chloroform (Avanti Polar Lipids)
Borosilicate glass tubes, 20×150 mm with lids (Fisher Scientific)
Rotator
Reconstitution buffer: 200 mM KCl, 5 mM MOPS-KOH, pH 7.0 (see Note 1)
Tris(2-carboxyethyl)phosphine (TCEP)
High power bath sonicator (Laboratory Supplies, Inc)
Liquid nitrogen
n-Dodecyl-β-D-maltopyranoside (DDM)
Purified ion channel in detergent
Bio-Beads SM2 (Bio-Rad)
Ultracentrifuge and rotor (such as Beckman Optima TLX Ultracentrifuge with rotor TLA 100.4)
Ultracentrifuge tubes
Microcentrifuge tube homogenizers
2.2. Formation of blisters
Glass cover slips
Desiccator at 4°C
Petri dishes (35×10 mm)
Phase contrast, DIC, or fluorescent microscope (we use a Nikon Eclipse TS100 with a 40X objective and a 10X eye piece)
Rehydration buffer (200 mM KCl, 10 mM MOPS-KOH, pH 7.0)
Blister formation buffer (200 mM KCl, 40 mM MgCl2, 0.5 mM CaCl2, 10 mM MOPS-KOH pH 7.0)
2.3. Patching of blisters
Patch Amplifier (Molecular Devices)
Analog to digital converter (Molecular Devices)
Borosilicate glass pipettes (we use Sutter SF150–86-10, OD 1.5 mm, ID 0.86 mm, 10 cm length)
Rapid solution changer (BioLogic Science Instruments)
Pipette solution (200 mM KCl, 10 mM succinate-KOH, 0.5 mM CaCl2 pH 4.0 for typical wild-type KcsA recordings)
Solutions for rapid solution changer (200 mM KCl, 10 mM MOPS-KOH pH 8.0 and 200 mM KCl, 10 mM succinate pH 3.0 for typical wild-type KcsA recordings)
3. Methods
3.1. Reconstitution of the ion channel into liposomes
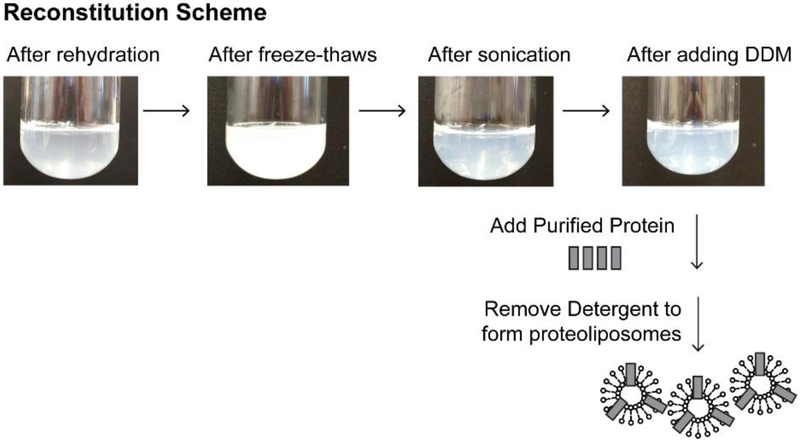
Our protocol is based on the procedure used by Cao et al. for the reconstitution of the TrpV1 channel [16]. In this protocol, lipid vesicles are gently solubilized by detergent, purified ion channel protein is added, and the detergent is slowly removed by Bio-Beads for incorporation of the ion channel proteins into the liposomes (Figure 1).
Figure 1. Reconstitution of the KcsA channel into lipid vesicles.
Pictures of lipids at various stages during the preparation of solubilized lipids for ion channel reconstitution, along with a cartoon illustrating the incorporation of KcsA channels into the liposomes.
Dry 2.5–10 mg of soybean polar lipid extract (in a chloroform solution) in a round bottom glass tube with lid using a gentle stream of argon. During drying, the tube is gently rotated to get a thin layer of lipid. After the chloroform has evaporated, dry under argon for an additional 5 minutes and then place under vacuum in a desiccator for 2–18 hours at room temperature (see Note 2).
Rehydrate lipids at 5 mg/mL in 200 mM KCl, 5 mM MOPS-KOH, 2 mM TCEP pH 7.0. Rotate gently for 30 minutes at room temperature.
Freeze-thaw the lipids ten times, alternating between a liquid nitrogen and a warm water bath.
Sonicate lipids in a bath sonicator for 3–5 minutes, until the lipid solution become translucent (see Note 3).
Add n-Dodecyl β-D-maltoside (DDM, solid) to 4 mM and rotate lipids for 30 min at room temperature.
Add the purified protein to lipids at the desired protein: lipid ratio (we have used a 1: 20–200 ratio for macroscopic and a 1: 10,000 ratio for single channel KcsA recordings). Bring total volume to 1 mL with reconstitution buffer. Rotate for 30 min at room temperature (see Note 4).
Remove detergent using Bio-Beads. Add Bio-Beads in four aliquots (30 mg, 30 mg, 50 mg, and 100 mg respectively), one aliquot per hour and continue rotating at room temperature. Add 1 mL of reconstitution along with each of the first three additions of Bio-Beads thereby diluting the reconstitution solution to a final volume of 4 mL. Following the additions of Bio-Beads, rotate the reconstitution mixture overnight at 4 °C. To ensure complete removal of detergent, add an additional 100 mg of Bio-Beads the following morning and rotate the mixture at room temperature an additional 1–3 hours (see Notes 5 and 6).
To harvest the liposomes formed, centrifuge the reconstitution mixture at 100,000 g for 1 hour at 4 °C
Resuspend the liposome pellet at ~50 mg/mL of lipid in 200 mM KCl, 10 mM MOPS-KOH pH 8.0. High concentrations of liposomes are necessary to promote GUV formation (see Note 7).
Divide liposomes into ~7 μL aliquots and freeze at −80°C.
3.2. Formation of blisters
Dry 7 μL of liposomes on a cover slip in a desiccator for 12– 36 hours at 4°C (see Note 8).
Rehydrate liposomes by adding 60 μL rehydration buffer directly on top of the dried liposomes. Incubate overnight at 18 °C or ≥2 hours at room temperature. If rehydrating overnight, put the cover slip into a container containing a paper towel soaked in rehydration buffer to prevent the drop from drying out.
Using a P2 pipette tip, gently mix the rehydrated liposomes. The solution should contain many small white chunks.
Fill 3– 4 small petri dishes with 4 mL of the blister formation solution. Using a P2 pipette, gently distribute very small amounts of rehydrated liposomes around the dish. For this step, pipet up 2 μL of the liposome solution and release in 10–20 small drops around the dish, repeating until the dish contains a total of 15–20 μL of rehydrated liposomes. Mg2+ is required for blister formation.
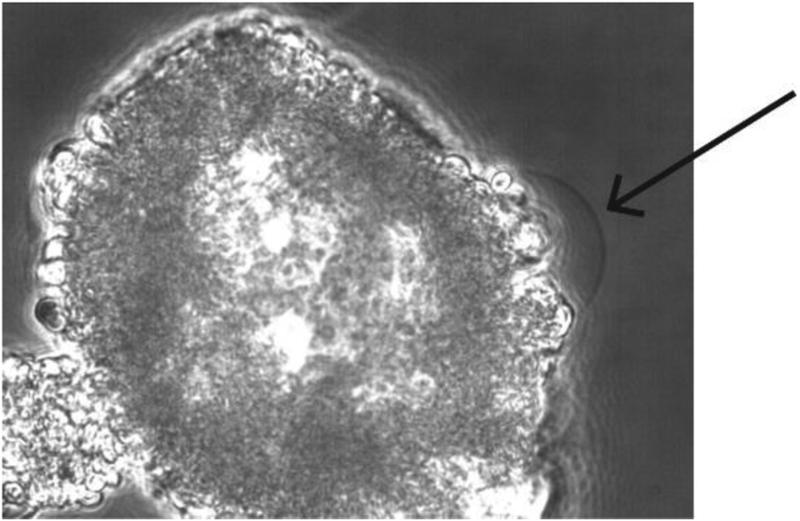
After ~1 hour in the Mg2+-containing solution, blisters are observed using a phase-contrast, DIC, or fluorescent microscope (Figure 2). Locating blisters on the dish typically requires much patience, but they are found hanging off of the sides of clumps of lipids and there are typically several blisters per dish.
Figure 2. A unilamellar blister used for patching.
A blister attached to a clump of collapsed liposomes observed through a phase contrast microscope is shown. Unilamellar blisters can be recognized from their thin and transparent outline (see arrow).
3.3. Patching of blisters
Use patch pipettes with 1.5–5 megaohm resistance to form gigaohm seals on blisters. Fill pipettes with 200 mM KCl, 0.5 mM CaCl2, 10 mM succinate-KOH, pH 4.0 solution for typical KcsA recordings.
Apply slight outward pressure once the pipette is in solution to prevent the tip of the pipette from getting clogged.
As you approach the blister, remove outward pressure just before the tip of the pipette touches the blister. Once the pipette is lightly touching the blister, apply inward pressure to form a gigaohm seal. Typically, seals form quickly and very little inward pressure is required. Once a seal is obtained, slowly pull back the pipette to get an inside-out patch (see Note 9).
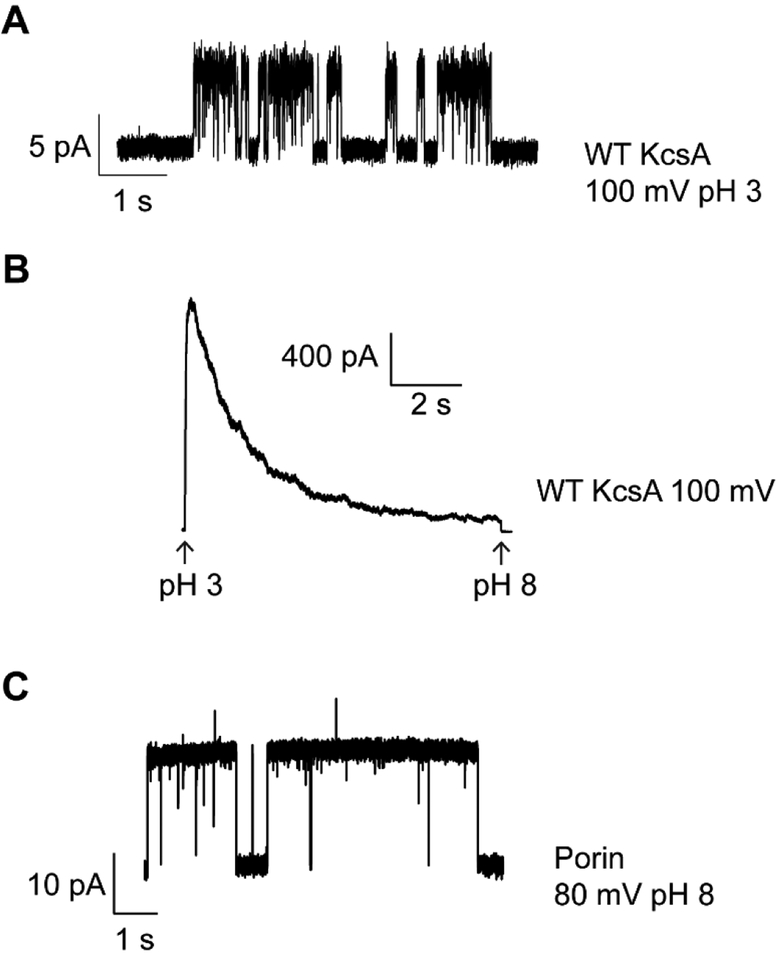
To observe activity of the KcsA channel, place the pipette in front of rapid solution changer tube flowing the pH 8.0 solution. Rapidly switch the pipette to the front of low-pH solution tube to observe activation and inactivation of KcsA. Figure 3a shows a single KcsA channel at 100 mV with the bath solution at pH 3.0. Figure 3b shows a macroscopic KcsA recording at 100 mV (see Notes 10 and 11).
Figure 3.
Electrophysiological recordings of ion channels.(A) Single channel recording of KcsA at 100 mV with 200 mM KCl, 10 mM succinate pH 3.0 in the bath and 200 mM KCl, 0.5 mM CaCl2, 10 mM succinate-KOH pH 4.0 in the pipette. (B) Macroscopic recording of KcsA at 100 mV. Channels were activated by rapid perfusion of the bath solution from 200 mM KCl, 10 mM MOPS-KOH pH 8.0 to 200 mM KCl, 10 mM succinate pH 3.0. On the change in pH, the channels rapidly activate and then inactivate. (C) Representative porin single channel activity with 200 mM KCl in the recording solutions. Porins are a common contaminant that can easily incorporate into blisters.
4. Notes
In electrophysiological recordings using purified proteins, one must always watch out for contamination by porins. The porins observed either co-purify with the ion channels or originate from small amounts of bacterial contamination in the solutions used. Figure 3c shows an example of a recording showing porin activity. To minimize porin contamination, all the solutions used for protein purification, reconstitution, and recordings should be made fresh starting from powder, stored at 4 °C, and used with one week.
When drying down the soybean polar lipid solution, it is important for good reconstitutions that the lipid layer formed be as thin as possible. A rotary evaporator can be used for this step but we have found that steady manual rotation under a gentle stream of argon works sufficiently.
Formation of unilamellar vesicles is required before protein addition. Vesicle formation is carried out by sonication and the formation of unilamellar vesicles is indicated by the solution turning from cloudy to translucent. If the lipid solution does not look translucent, then additional sonication steps should be used. Care should be taken during the sonication steps to prevent heating as an increase in temperature can cause spoiling of the lipid vesicles. Sonication is typically done in 30 s intervals with an intervening 30– 60 s rest period. The rest period between rounds of sonication allows the lipids to cool to prevent overheating.
The protein to lipid ratio used during reconstitution is dependent upon the specific channel being investigated and the desired number of channels in the patch. Too much protein will make blister formation challenging and give leaky patches while too little protein will result in too many patches with no channel activity.
Bio-Beads should be washed three times each with methanol, water, and reconstitution buffer before use.
The amount of Bio-Beads to use will depend upon the detergent concentration in the purified protein sample. Since proteins are typically concentrated in a manner where the exact detergent concentration in the sample is not known, a trial and error approach has to be used. When detergent present has not been fully removed by the Bio-Beads, we often observe extra bubbles in the rotating protein/lipid mixture. In these cases, an additional aliquot of Bio-Beads is added to remove the excess detergent. If the detergent is not completely removed, then the blisters observed do not give the stable gigaohm seals that are necessary for electrophysiological recordings.
To resuspend the liposome pellet obtained following ultracentrifugation, we initially use a pipette tip to break up the pellet into smaller chunks. The solution with the chunks is then transferred into a micro-centrifuge tube and homogenized. Only gentle force must be used during the homogenization step as we have found that using too much force during homogenization can inhibit good blister formation. Gentle homogenization is followed by brief bath sonication to ensure the formation of a uniform solution before the liposomes are aliquoted and frozen.
Some ion channels require the addition of sucrose or ethylene glycol to prevent damage during the dehydration step [33–35]. The requirement for these additives has to be determined for the specific protein. We have determined that these additives are not necessary for maintaining the activity of the wild type KcsA channel during dehydration.
Patching the blisters is challenging as they tend to move slightly in the solution along with the lipid chunks that the blisters grow from. Positioning the pipette to touch the moving blister for patching can be done with patience and practice.
When recording KcsA currents, rapid perfusion is essential for obtaining accurate rates of activation and inactivation. It is important to make sure the solution lines in the rapid perfusion set-up are free of air bubbles as air bubbles will cause the patch to break.
Proteins typically reconstitute into lipid bilayers in both orientations and therefore the patch obtained will contain channels oriented both ways. For the KcsA channel, we use pH 4 in the pipette to fully inactivate and thereby silence the channels that are oriented with their intracellular side facing the pipette. For other channels, a specific blocker can be added, either to the pipette or the bath solution, to silence channel in a specific orientation.
Acknowledgement:
This research was supported by a grant from the NIH (GM087546) to FIV.
References
- 1.Hille B, Ion Channels of Excitable Membranes. 2001, Sunderland, MA: Sinauer [Google Scholar]
- 2.Stuhmer W, Electrophysiologic recordings from Xenopus oocytes. Methods Enzymol, 1998. 293: p. 280–300. [DOI] [PubMed] [Google Scholar]
- 3.Thomas P and Smart TG, HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods, 2005. 51(3): p. 187–200. [DOI] [PubMed] [Google Scholar]
- 4.Stockbridge RB and Tsai MF, Lipid reconstitution and recording of recombinant ion channels. Methods Enzymol, 2015. 556: p. 385–404. [DOI] [PubMed] [Google Scholar]
- 5.Lin CM, et al. , Size-dependent properties of small unilamellar vesicles formed by model lipids. Langmuir, 2012. 28(1): p. 689–700. [DOI] [PubMed] [Google Scholar]
- 6.Walde P, et al. , Giant vesicles: preparations and applications. Chembiochem, 2010. 11(7): p. 848–65. [DOI] [PubMed] [Google Scholar]
- 7.Yanagisawa M, et al. , Oriented reconstitution of a membrane protein in a giant unilamellar vesicle: experimental verification with the potassium channel KcsA. J Am Chem Soc, 2011. 133(30): p. 11774–9. [DOI] [PubMed] [Google Scholar]
- 8.Matulef K, et al. , Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation. Structure, 2016. 24(5): p. 750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrapani S, Cordero-Morales JF, and Perozo E, A quantitative description of KcsA gating I: macroscopic currents. J Gen Physiol, 2007. 130(5): p. 465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aimon S, et al. , Functional reconstitution of a voltage-gated potassium channel in giant unilamellar vesicles. PLoS One, 2011. 6(10): p. e25529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinac B, et al. , Bacterial mechanosensitive channels: models for studying mechanosensory transduction. Antioxid Redox Signal, 2014. 20(6): p. 952–69. [DOI] [PubMed] [Google Scholar]
- 12.Martinac B, et al. , Studying mechanosensitive ion channels using liposomes. Methods Mol Biol, 2010. 606: p. 31–53. [DOI] [PubMed] [Google Scholar]
- 13.Battle AR, et al. , Rapid and improved reconstitution of bacterial mechanosensitive ion channel proteins MscS and MscL into liposomes using a modified sucrose method. FEBS Lett, 2009. 583(2): p. 407–12. [DOI] [PubMed] [Google Scholar]
- 14.Mahendran KR, et al. , Permeation of antibiotics through Escherichia coli OmpF and OmpC porins: screening for influx on a single-molecule level. J Biomol Screen, 2010. 15(3): p. 302–7. [DOI] [PubMed] [Google Scholar]
- 15.Kreir M, et al. , Rapid screening of membrane protein activity: electrophysiological analysis of OmpF reconstituted in proteoliposomes. Lab Chip, 2008. 8(4): p. 587–95. [DOI] [PubMed] [Google Scholar]
- 16.Cao E, et al. , TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron, 2013. 77(4): p. 667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahya N, et al. , Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J Biol Chem, 2003. 278(30): p. 28109–15. [DOI] [PubMed] [Google Scholar]
- 18.Nikolaus J, et al. , Hemagglutinin of influenza virus partitions into the nonraft domain of model membranes. Biophys J, 2010. 99(2): p. 489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angelova MI and Tsoneva I, Interactions of DNA with giant liposomes. Chem Phys Lipids, 1999. 101(1): p. 123–37. [DOI] [PubMed] [Google Scholar]
- 20.Kovacic J, Bozic B, and Svetina S, Budding of giant unilamellar vesicles induced by an amphitropic protein beta2-glycoprotein I. Biophys Chem, 2010. 152(1–3): p. 46–54. [DOI] [PubMed] [Google Scholar]
- 21.Staneva G, et al. , Detergents induce raft-like domains budding and fission from giant unilamellar heterogeneous vesicles: a direct microscopy observation. Chem Phys Lipids, 2005. 136(1): p. 55–66. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen IL, Kemmer GC, and Pomorski TG, Membrane protein reconstitution into giant unilamellar vesicles: a review on current techniques. Eur Biophys J, 2016. [DOI] [PubMed] [Google Scholar]
- 23.Varnier A, et al. , A simple method for the reconstitution of membrane proteins into giant unilamellar vesicles. J Membr Biol, 2010. 233(1–3): p. 85–92. [DOI] [PubMed] [Google Scholar]
- 24.Garten M, et al. , Reconstitution of a transmembrane protein, the voltage-gated ion channel, KvAP, into giant unilamellar vesicles for microscopy and patch clamp studies. J Vis Exp, 2015(95): p. 52281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manley S and Gordon VD, Making giant unilamellar vesicles via hydration of a lipid film. Curr Protoc Cell Biol, 2008. Chapter 24: p. Unit 24 3. [DOI] [PubMed] [Google Scholar]
- 26.Collins MD and Gordon SE, Giant liposome preparation for imaging and patch-clamp electrophysiology. J Vis Exp, 2013(76). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeves JP and Dowben RM, Formation and properties of thin-walled phospholipid vesicles. J Cell Physiol, 1969. 73(1): p. 49–60. [DOI] [PubMed] [Google Scholar]
- 28.Akashi K, et al. , Preparation of giant liposomes in physiological conditions and their characterization under an optical microscope. Biophys J, 1996. 71(6): p. 3242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akashi K, et al. , Formation of giant liposomes promoted by divalent cations: critical role of electrostatic repulsion. Biophys J, 1998. 74(6): p. 2973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsumoto K, et al. , Efficient formation of giant liposomes through the gentle hydration of phosphatidylcholine films doped with sugar. Colloids Surf B Biointerfaces, 2009. 68(1): p. 98–105. [DOI] [PubMed] [Google Scholar]
- 31.Wesolowska O, et al. , Giant unilamellar vesicles - a perfect tool to visualize phase separation and lipid rafts in model systems. Acta Biochim Pol, 2009. 56(1): p. 33–9. [PubMed] [Google Scholar]
- 32.Delcour AH, et al. , Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys J, 1989. 56(3): p. 631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doeven MK, et al. , Distribution, lateral mobility and function of membrane proteins incorporated into giant unilamellar vesicles. Biophys J, 2005. 88(2): p. 1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller BU, et al. , Single channel recordings of reconstituted ion channel proteins: an improved technique. Pflugers Arch, 1988. 411(1): p. 94–100. [DOI] [PubMed] [Google Scholar]
- 35.Riquelme G, et al. , Giant liposomes: a model system in which to obtain patch-clamp recordings of ionic channels. Biochemistry, 1990. 29(51): p. 11215–22. [DOI] [PubMed] [Google Scholar]