Arabidopsis cytochrome b5 isoform D physically interacts with monolignol biosynthetic cytochrome P450s and is specifically required for syringyl lignin biosynthesis in the cell wall.
Abstract
Angiosperms have evolved the metabolic capacity to synthesize p-hydroxyphenyl, guaiacyl (G), and syringyl (S) lignin subunits in their cell walls to better adapt to the harsh terrestrial environment. The structural characteristics of lignin subunits are essentially determined by three cytochrome P450-catalzyed reactions. NADPH-dependent cytochrome P450 oxidoreductase (CPR) is commonly regarded as the electron carrier for P450-catalyzed reactions during monolignol biosynthesis. Here, we show that cytochrome b5 isoform D (CB5D) is an indispensable electron shuttle protein specific for S-lignin biosynthesis. Arabidopsis (Arabidopsis thaliana) CB5D localizes to the endoplasmic reticulum membrane and physically associates with monolignol P450 enzymes. Disrupting CB5D in Arabidopsis resulted in a >60% reduction in S-lignin subunit levels but no impairment in G-lignin formation compared with the wild type, which sharply contrasts with the impaired G- and S-lignin synthesis observed after disrupting ATR2, encoding Arabidopsis CPR. The defective S-lignin synthesis in cb5d mutants was rescued by the expression of the gene encoding CB5D but not with mutant CB5D devoid of its electron shuttle properties. Disrupting ATR2 suppressed the catalytic activity of both cinnamic acid 4-hydroxylase and ferulate 5-hydroxylase (F5H), but eliminating CB5D specifically depleted the latter’s activity. Therefore, CB5D functions as an obligate electron shuttle intermediate that specifically augments F5H-catalyzed reactions, thereby controlling S-lignin biosynthesis.
INTRODUCTION
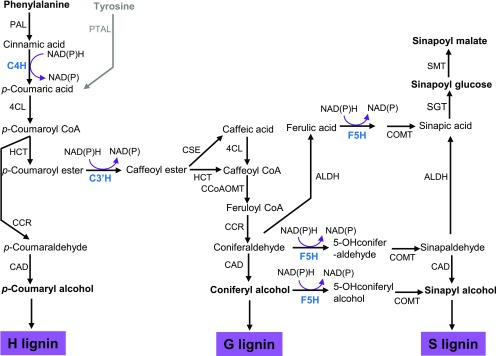
Vascular plants produce lignin to sustain the mechanical strength of their cell walls and to confer hydrophobicity to the vasculature for water conductance. Lignin, a major structural polymer of the cell wall, is primarily formed by the oxidative polymerization of three monolignols: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. After their incorporation into lignin, those monolignols give rise to the p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) subunits, respectively (Figure 1; Boerjan et al., 2003; Vanholme et al., 2010; Liu, 2012). While the cell walls of gymnosperms are predominantly composed of G-lignin with a small proportion of H-lignin subunits, angiosperms also build S-lignin structural subunits in their walls. The methoxylated structural characteristics of monolignols are determined by ring-hydroxylation, which is catalyzed by three endoplasmic reticulum (ER)-resident cytochrome P450 monooxygenases (CYPs) and by the subsequent methylation reactions (Figure 1). In Arabidopsis (Arabidopsis thaliana), three P450 enzymes involved in monolignol biosynthesis are cinnamic acid 4-hydroxylase (C4H; CYP73A5), p-coumaroyl ester 3′-hydroxylase (C3′H; CYP98A3), and ferulate 5-hydroxylase/coniferaldehyde 5-hydroxylase (F5H; CYP84A1). C4H catalyzes 4-hydroxylation of the phenyl ring of cinnamic acid, the first aromatic compound in the general phenylpropanoid pathway, producing p-coumaric acid, which serves as a common precursor for the formation of a whole set of phenolic metabolites, including monolignols/lignin, flavonoids, and phenolic esters. C3′H hydroxylates p-coumaroyl ester derivatives at their phenyl ring 3-positions and diverts general phenylpropanoid precursors to the synthetic branch for both G- and S-monolignols. F5H, which acts as the key branch point enzyme, hydroxylates coniferyl alcohol/coniferaldehyde (the G-lignin precursor) at its C-5 position, leading to the formation of S-monolignol in angiosperms (Figure 1; Boerjan et al., 2003; Vanholme et al., 2010; Liu, 2012). In the Brassicaceae family, the activity of F5H also leads G-lignin precursors en route to the methanolic soluble phenolics sinapoyl esters (Figure 1); these compounds typically act as photoprotectants, protecting plants from UV light damage (Liu, 2010; Fraser and Chapple, 2011).
Figure 1.
Simplified Scheme of the Phenylpropanoid-Lignin Biosynthetic Pathway, Illustrating Three P450 Enzyme-Catalyzed Hydroxylation Reactions in Monolignol Biosynthesis.
ALDH, aldehyde dehydrogenase; CAD, (hydroxy)cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; CCR, cinnamoyl CoA reductase; 4CL, 4-hydroxycinnamoyl CoA ligase; COMT, caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase; CSE, caffeoyl shikimate esterase; HCT, hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase; PAL, phenylalanine ammonia lyase; PTAL, phenylalanine and tyrosine ammonia lyase; SGT, sinapate glucosyltransferase; SMT, sinapoylglucose:malate sinapoyltransferase. Gray letters and arrows indicate that the pathway occurs in grass.
P450 enzyme catalysis depends on two single-electron transfer steps per cycle, in which electron donor protein(s) are required to shuttle two electrons from the reducing power (NADPH or NADH) to the heme iron center of P450 to activate molecular oxygen (Supplemental Figure 1; Hannemann et al., 2007). The microsomal P450 system of eukaryotic cells is generally composed of two integral membrane proteins, P450 itself and its redox partner, the flavin adenine dinucleotide and flavin mononucleotide domains-containing NADPH-dependentcytochrome P450 oxidoreductase (CPR; Jensen and Møller, 2010). CPR is required for C4H catalytic activity, as revealed in a yeast heterologous expression system (Urban et al., 1997). The expression of one of the two Arabidopsis CPR homologous genes (i.e., ATR2) appears to be closely correlated with the expression of lignin biosynthetic genes and is induced during stress responses and lignin biosynthesis (Sundin et al., 2014). Moreover, the disruption of ATR2 resulted in an ∼6% reduction in total lignin content in inflorescence stems as well as a compositional shift of the remaining lignin, suggesting that CPR indeed serves as the electron carrier for lignin formation (Sundin et al., 2014). Nevertheless, the modest reduction in stem lignin content upon ATR2 disruption suggests the potential existence of additional redox partner(s) for monolignol biosynthesis.
In fact, the ER membrane of eukaryotic cells possesses additional electron shuttle components that form two interconnected electron transfer systems: the NADPH-CPR electron transfer chain and the NADH-cytochrome b5 reductase (CBR)-cytochrome b5 (CB5) chain (Supplemental Figure 1; Porter, 2002; Schenkman and Jansson, 2003). Notably, in yeast and mammalian cells, CB5 can be reduced both by CBR (Oshino et al., 1971) and CPR (Dailey and Strittmatter, 1979; Enoch and Strittmatter, 1979). Therefore, CB5 is involved in two electron transfer chains, either of which delivers two electrons from CBR or the second electron from CPR to a number of oxidative reactions that are catalyzed by non-heme-containing desaturases or the heme-containing cytochrome P450s during biological processes such as anabolic metabolism of fats and steroids and catabolism of xenobiotics, drugs, and compounds involved in endogenous metabolism (Supplemental Figure 1; Jansson and Schenkman, 1977; Noshiro et al., 1979; Porter, 2002; Kandel and Lampe, 2014). CB5 is a heme binding, tail-anchored membrane protein with a molecular mass of ∼15 kD. This protein resides on the cytoplasmic face of the ER membrane via its hydrophobic C-terminal domain, and its hydrophilic N-terminal domain protrudes into the cytosol (Porter, 2002; Schenkman and Jansson, 2003). In contrast to the single copy of CB5 in mammals and yeast (Porter, 2002; Kandel and Lampe, 2014), flowering plants including Arabidopsis have evolved multiple CB5 genes (Smith et al., 1992; Napier et al., 1995; Fukuchi-Mizutani et al., 1999; Hwang et al., 2004; Kumar et al., 2006, 2012; Maggio et al., 2007). Six CB5 family members are annotated from the Arabidopsis genome: AtCB5A, AtCB5B, AtCB5C, AtCB5D, AtCB5E, and AtCB5F. Among these, AtCB5B, AtCB5C, AtCB5D, and AtCB5E share high sequence similarity at the amino acid level and were proven or predicted to be localized to the ER membrane (Hwang et al., 2004; Maggio et al., 2007). AtCB5F has different N and C termini from the others and is defined as a CB5-like protein (Hwang et al., 2004; Maggio et al., 2007). Similar to mammals and yeast, the CB5s in flowering plants also function as electron shuttle proteins in lipid and steroid modification reactions, such as the desaturation of fatty acids of microsomal membranes in developing safflower (Carthamus tinctorius) cotyledons (Smith et al., 1990), the hydroxylation of oleate in castor bean (Ricinus communis) seeds (Smith et al., 1992), and the desaturation of sterol precursors in maize (Zea mays; Rahier et al., 1997). Therefore, CB5 in flowering plants is generally regarded as a redox component involved in lipid and steroid biosynthetic reactions.
Cytochrome P450 enzymes physically and stochastically associate with their redox partner CPR or CB5 via electrostatic interactions to form functional 1:1 complexes (Vergéres and Waskell, 1995; Backes and Kelley, 2003; Im and Waskell, 2011; Scott et al., 2016). Such redox partner-enzyme complex formation is essential for the roles of P450 enzymes in xenobiotic detoxification and drug metabolism in mammals (Kandel and Lampe, 2014). In this study, using coimmunoprecipitation (Co-IP)-mass spectrometry analysis, we systematically explored the proteins that potentially associate with monolignol biosynthetic P450 enzymes in lignifying stem tissues of Arabidopsis (Gou et al., 2018). Among the list of recognized proteins, we uncovered the ER-resident CB5 isoform, CB5D. Disrupting the gene encoding CB5D specifically impaired F5H catalytic activity and S-lignin subunit deposition, while C4H activity and G-lignin subunit levels remained unchanged or even slightly increased in the stems of mutant plants. This phenomenon sharply contrasts with the results of disrupting ATR2, encoding the electron donor CPR, which reduced both G- and S-lignin synthesis. These findings demonstrate that CB5D functions as an electron shuttle protein specifically for F5H-catalyzed reactions and is responsible for S-lignin biosynthesis in Arabidopsis. Interestingly, while disrupting either CB5D or ATR2 led to reduced S-lignin levels, the deficiency of CBR did not obviously affect lignin synthesis, suggesting that both CB5D and ATR2, but not CBR, are required for S-lignin formation. Therefore, CB5D likely participates in the NADPH-CPR electron transfer chain but not in the NADH-CBR-mediated electron transfer system to augment F5H activity.
RESULTS
Cytochrome b5 Isoform D Interacts with Monolignol P450s
Monolignol biosynthetic P450 enzymes are thought to function as anchor proteins that associate with other enzymes/proteins to regulate lignin biosynthesis (Dixon et al., 2001; Jørgensen et al., 2005). In an attempt to comprehensively explore the components associated with the three monolignol P450s involved in phenylpropanoid-lignin biosynthesis, we expressed hemagglutinin (HA)-PreScission-Biotin (HPB)-tagged C4H, C3′H, and F5H driven by the AtC4H promoter in Arabidopsis. We then affinity-purified the HPB-tagged C4H, C3′H, and F5H proteins isolated from membrane fractions, particularly from the lignifying inflorescence stems of transgenic plants (Gou et al., 2018). The eluted proteins were resolved via liquid chromatography-mass spectrometry (LC-MS) in two sets of independent experiments. Examining the resolved peptides revealed a number of peptides corresponding to electron carrier proteins (Table 1). In the experiment in which the eluted proteins were resolved by SDS-PAGE and thegel slices were subjected to trypsin digestion, two CB5 isoforms, CB5D and CB5E, coeluted with the monolignol P450s. CB5D appeared in the set of proteins that coeluted with the three monolignol P450s, while CB5E was found only in the C4H coeluted proteins (Table 1). Among the eluted proteins on affinity binding beads that were directly subjected to trypsin digestion, cytochrome P450 reductases ATR1 and ATR2, cytochrome b5 reductase CBR1, and the cytochrome b5 isoforms CB5C, CB5D, and CB5E were all detected (Table 1). These results suggest that the detected electron transfer components might be in close proximity to, if not physically interact with, the monolignol biosynthetic P450s in the endosomal membrane.
Table 1. The potential electron transfer components identified from affinity purification-mass spectrometry analysis of monolignol P450 enzymes.
| Bait Protein | Protein Identified | Spectral Counts | Unique Peptides | Percentage Coverage |
|---|---|---|---|---|
| C4H | CPR1 | 5 | 3 | 6.8 |
| CPR2 | 2 | 1 | 2.1 | |
| CBR1 | 7 | 4 | 18 | |
| CB5D | 7 (4) | 4 (3) | 41 (36) | |
| CB5C | 4 | 2 | 17 | |
| CB5E | 1 (2) | 1 (2) | 10 (28) | |
| C3′H | CPR1 | 4 | 3 | 7.4 |
| CPR2 | 1 | 1 | 2.3 | |
| CBR1 | 5 | 3 | 15 | |
| CB5D | 7 (1) | 4 (1) | 41 (9.3) | |
| CB5C | 2 | 1 | 7.6 | |
| CB5E | 3 | 2 | 16 | |
| F5H | CPR1 | 4 | 3 | 6.9 |
| CPR2 | 1 | 1 | 2.3 | |
| CBR1 | 5 | 4 | 18 | |
| CB5D | 4 (2) | 2 (2) | 25 (24) | |
| CB5C | 4 | 2 | 17 | |
| CB5E | 2 | 1 | 10 |
Purified proteins were subjected to SDS-PAGE, and the sliced gel pieces were processed for in-gel digestion and peptide elution (for data in parentheses), or the proteins were digested and eluted directly from protein-bound beads. The eluted peptides were injected for LC-MS analysis. The MS/MS data was searched with GPM X!Tandem software against the UniProt Arabidopsis database with P = 0.01.
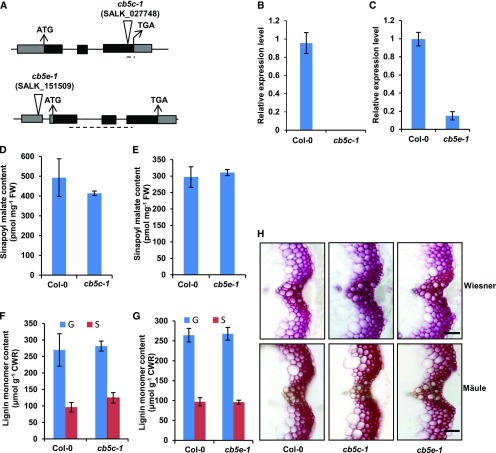
Among the recognized CB5 isoforms, homozygous mutant plants deficient in CB5C (cb5c-1; SALK_027748) and CB5E (cb5e-1; SALK_151509) did not exhibit significant effects on lignin deposition or the accumulation of soluble sinapoyl esters, although CB5C expression level was completely diminished in cb5c-1 and CB5E expression level was suppressed up to 85% in cb5e-1 (Figure 2). By contrast, plants deficient in CB5D exhibited drastic defects in sinapoyl ester accumulation and lignin formation (see below for details). We therefore primarily focused on the CB5D isoform for further characterization. Similar to the typical ER localization pattern of C4H-, C3′H-, and F5H-GFP (Figure 3A), when transiently expressing CB5D-GFP (green fluorescent protein) in wild tobacco (Nicotiana benthamiana) leaf epidermal cells, its fluorescence distribution exhibited a typical ER network pattern, which resembles the behavior of the ER marker protein SP-GFP-HDEL, which harbors a signal peptide of pumpkin 2S albumin and GFP followed by a 12-amino acid sequence including an ER retention signal, HDEL (Matsushima et al., 2002), but clearly differs from that of free GFP, which was distributed throughout the cytoplasm (Figure 3A). These data confirm the notion that CB5D, like monolignol P450 enzymes, localizes to the ER membrane.
Figure 2.
Characterization of the cb5c-1 and cb5e-1 Mutants.
(A) Diagrams of the T-DNA insertion mutants of CB5C and CB5E. The triangles indicate the T-DNA insertion sites in CB5D and CB5E. The dashed lines indicate the regions used for qRT-PCR analysis of gene expression.
(B) and (C) qRT-PCR analysis of the relative expression levels of CB5C and CB5E in Col-0 wild-type and cb5c (B) or cb5e (C) seedlings. Approximately 12 2-week-old seedlings were grouped as one biological replicate for RNA extraction. qRT-PCR was performed with three biological replicates, each with four technical repeats. The data represent means ± sd of three biological replicates.
(D) and (E) Sinapoyl malate content in 2-week-old seedlings of the Col-0 wild type and cb5c-1 (D) or the Col-0 wild type and cb5e-1 (E). The data represent means ± sd of three biological repeats; each replicate is composed of 0.5 g fresh weight (FW) of pooled seedlings.
(F) and (G) Quantification of thioacidolytic lignin monomers in the cell walls of cb5c-1 (F) and cb5e-1 (G). Inflorescence stems from 12-week-old plants were used in the analysis. Stems from at least six plants were pooled as one biological replicate. The data represent means ± sd of three biological repeats. CWR, cell wall residues.
Statistical analysis of the data in (D) to (G) was conducted with two-tailed Student’s t tests. No significant differences were observed between the wild type and mutants.
(H) Histochemical observation of lignin in stem cross sections (1 cm from the bottom, 80 µm thick) of 7-week-old Col-0 wild-type, cb5c-1, and cb5e-1 plants with Wiesner staining for total lignin (top panels) and Mӓule staining for S-lignin (bottom panels). Bars = 30 µm.
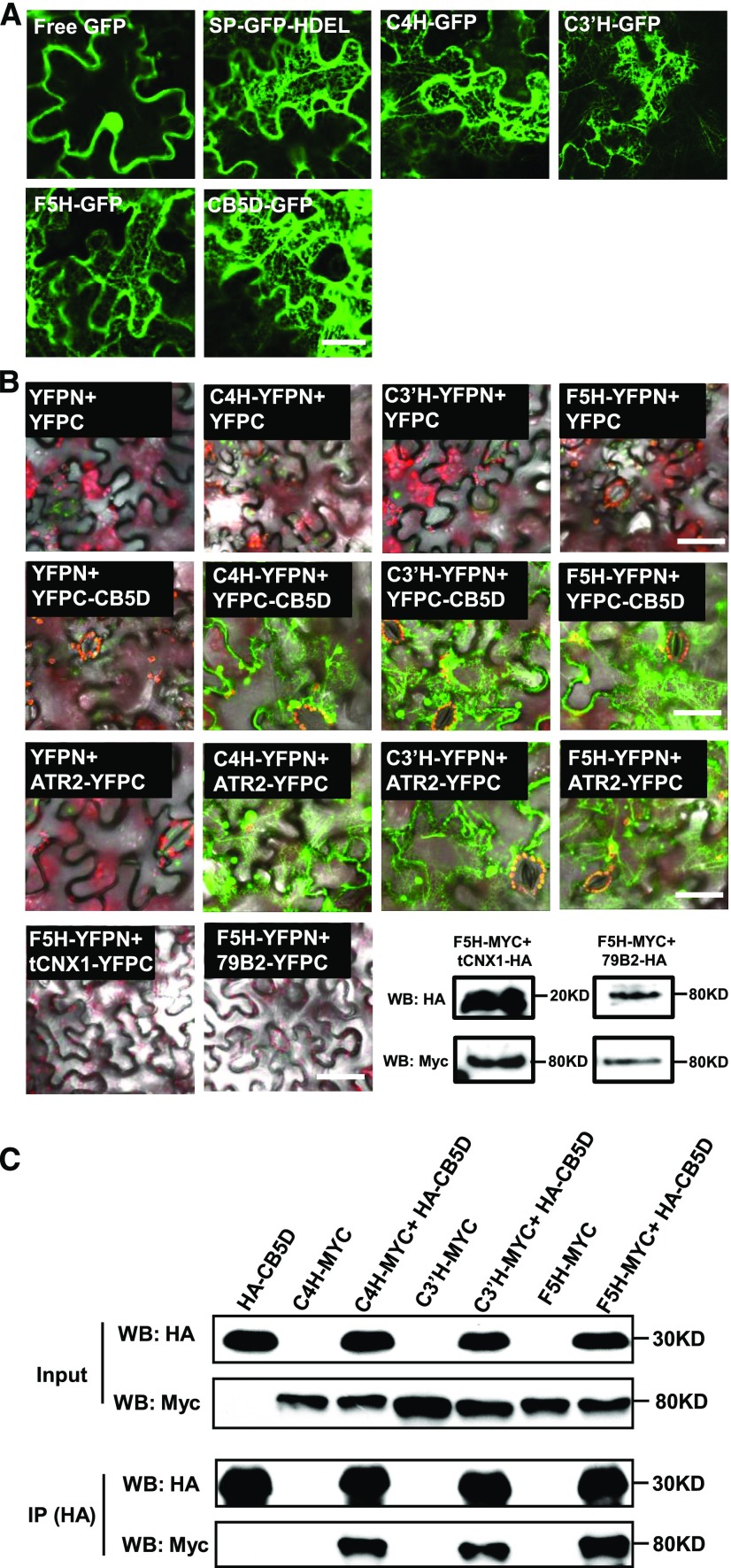
Figure 3.
Colocalization and Interaction of Monolignol Biosynthetic P450s and CB5D on the ER Membrane.
(A) Fluorescence distribution patterns of C4H-, C3′H-, F5H-, and CB5D-GFP transiently expressed in wild tobacco leaves. Free GFP and SP-GFP-HDEL were used as controls. Bar = 25 µm.
(B) Pair-wise BiFC assay of C4H, C3′H, and F5H with CB5D and ATR2. F5H was coexpressed with a truncated ER membrane-localized CNX1 (tCNX1) protein containing its C-terminal transmembrane domain and with another P450 CYP79B2 as the negative controls. The bottom right panel shows immunoblots of the expressed tCNX1-HA with F5H-Myc and CYP79B2-HA with F5H-Myc as negative controls with anti-HA and anti-Myc antibodies. WB indicates immunoblot (protein gel blot). Bars = 50 µm.
(C) Co-IP assay of C4H, C3′H, and F5H with CB5D using proteins expressed in the BiFC assay. The proteins were immunoprecipitated (IP) with anti-HA antibody and probed with anti-HA and anti-Myc antibodies.
We then validated the potential interactions of the three monolignol biosynthetic P450s with CB5D via bimolecular fluorescence complementation (BiFC) assay and/or Co-IP-immunoblot detection. In the BiFC assay, strong chimeric fluorescence was observed in wild tobacco leaves coexpressed with C4H, C3′H, or F5H fused with the N-terminal half of yellow fluroesent protein (YFPN) and CB5D fused with the complementary C-terminal half of YFP (YFPC). Similarly, when we coexpressed P450-YFPN with ATR2-YFPC in wild tobacco cells, substantial chimeric fluorescence was also observed (Figure 3B). By contrast, no fluorescent signal was detected in the negative controls (Figure 3B). The control experiments included wild tobacco leaf cells coinfiltrated with agrobacteria harboring BiFC empty vectors, or an empty BiFC vector with a P450-YFPN or YFPC-CB5D construct, and cells coexpressing F5H-YFPN and either YFPC fused with the transmembrane domain from calnexin1 (tCNX1), a highly conserved ER chaperone protein (Liu et al., 2017), or with another P450 enzyme, CYP79B2, which is involved in auxin/glucosinolate biosynthesis.
We then examined fusion proteins of F5H, tCNX1, and CYP79B2 in the negative control sets via immunoblot analysis (Figure 3B), which validated the proper expression and accumulation of the fusion proteins, thus excluding the possibility that the failure to generate chimeric fluorescence signals was due to the lack of expressed proteins in the wild tobacco cells. Taken together, these data indicate that the BiFC assay is a relatively reliable approach to monitoring proteins on the same membrane that are either in close proximity or physically interacting. These data also suggest that CB5D as well as other electron transfer components might be close to or directly interact with monolignol P450s in planta. We further validated the interactions of the P450s with CB5D via Co-IP-immunoblot assays, in which we took advantage of the HA and Myc tags in each of the BiFC vectors and immunoprecipitated HA-CB5D using anti-HA antibody and then detected the pulled-down proteins with anti-Myc antibody for the C4H-, C3′H-, and F5H-Myc fusions. While no P450-Myc signal was detected when HA-CB5D was expressed alone in wild tobacco cells, three P450s were readily detected in the immunoprecipitated proteins from cells coexpressing HA-CB5D and P450-Myc (Figure 3C). Both BiFC and the Co-IP experiments verify the spatial association of CB5D with monolignol biosynthetic P450s C4H, C3′H, and F5H in planta.
CB5D Is Expressed in Lignifying Tissues of Arabidopsis
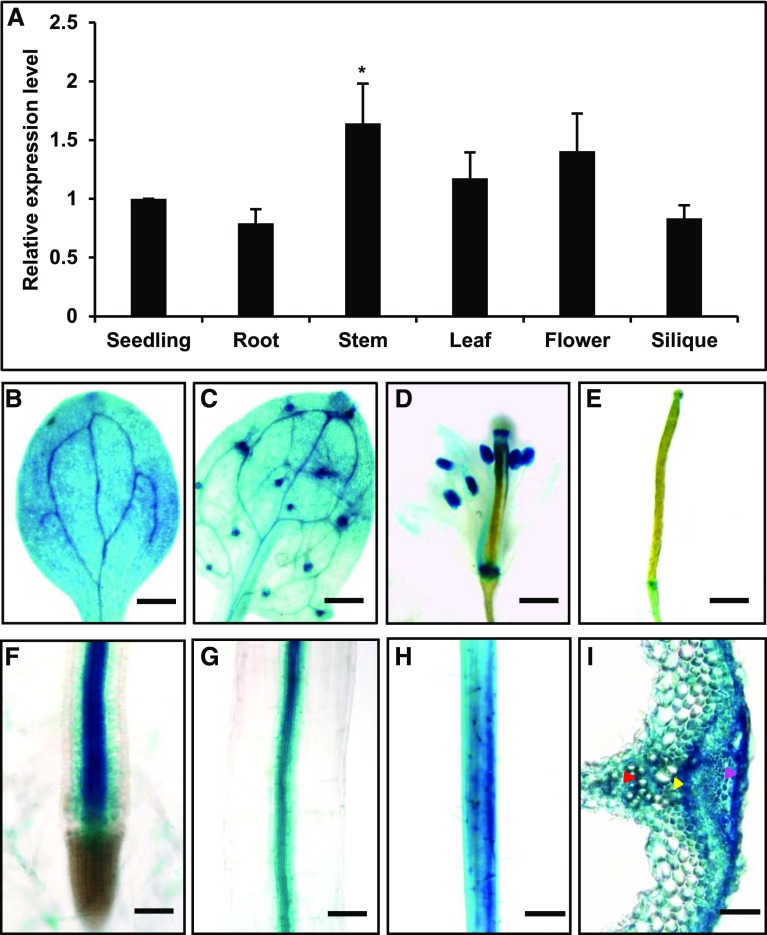
We then examined the gene expression pattern of CB5D using qRT-PCR. CB5D transcripts were detected in all tissues examined, including root, stem, leaf, flower, and silique tissue, but the highest expression level was observed in stem (Figure 4A). In agreement with the qRT-PCR data, an assay using GUS driven by the native CB5D promoter revealed strong GUS staining in hypocotyl, cotyledon, root, stem, leaf, and flower tissue (Figures 4B to 4H), with the stronger staining appearing in the vascular tissues of roots and hypocotyls (Figures 4F and 4G), the vein cells of cotyledons and leaves (Figures 4B and 4C), or the anthers of flowers (Figure 4D). In stem cross sections, high-intensity GUS staining was detected in xylem, cambium, and epidermal cells (Figure 4I), where lignin or soluble phenolic biosynthesis typically occurs. These gene expression patterns point to a potential functional association of CB5D with lignin biosynthesis.
Figure 4.
Expression Pattern of CB5D in Planta.
(A) qRT-PCR analysis of relative CB5D transcript levels in different tissues of the Col-0 wild type. The roots and leaves from 30-d-old plants, the stems and flowers from ∼40-d-old plants, and the siliques from ∼50-d-old plants were collected. Tissues from at least five individual plants or ∼12 2-week-old seedlings were pooled as one biological replicate. The data represent means ± sd from three biological replicates; each replicate represents the average of three technical repeats. The asterisk indicates a significant difference compared with the seedling samples (Student’s t test; P < 0.05).
(B) to (I) GUS staining of different tissues of pCB5D:GUS transgenic plants in the Col-0 wild-type background.
(B) Cotyledon. Bar = 500 µm.
(C) True leaf. Bar = 300 µm.
(D) Flower. Bar = 300 µm.
(E) Young silique. Bar = 500 µm.
(F) Root. Bar = 150 µm.
(G) Hypocotyl. Bar = 75 µm.
(H) Stem. Bar = 300 µm.
(I) Cross section of stem. The red arrowhead points to xylem cells, the yellow arrowhead points to cambium cells, and the magenta arrowhead points to epidermal cells. Bar = 75 µm.
Disruption of CB5D Compromises Sinapoyl Ester, α-Pyrone, and S-Lignin Accumulation
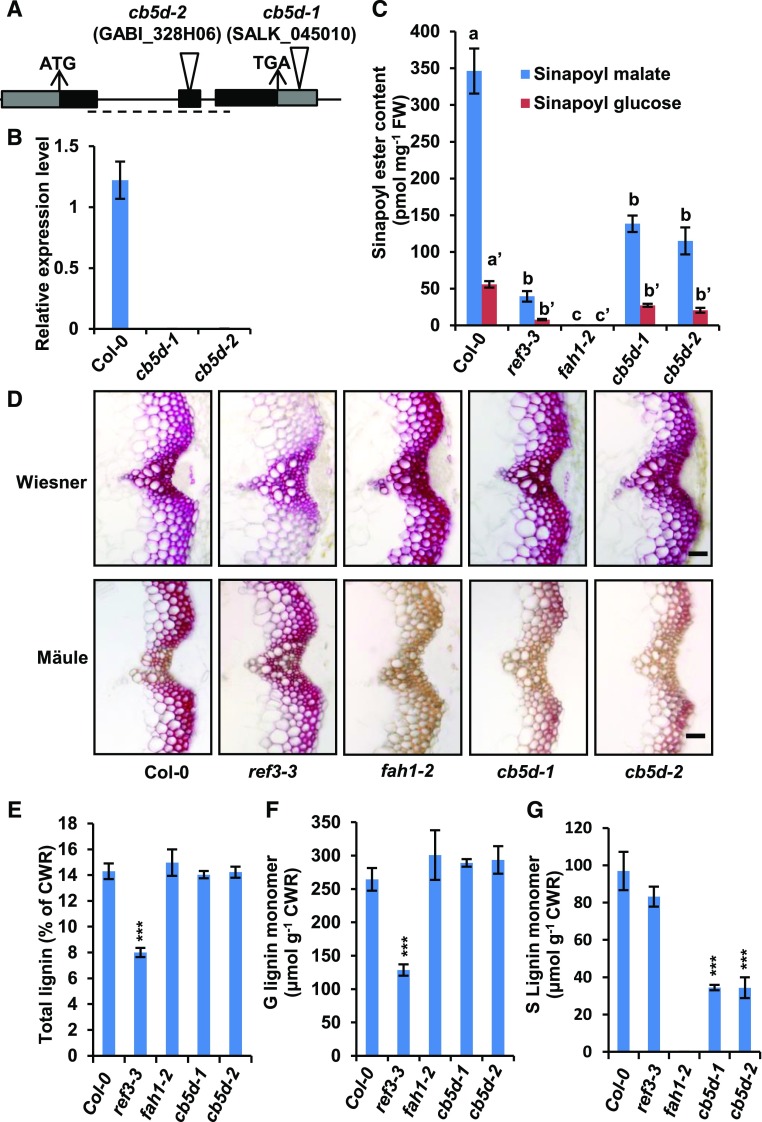
We then measured the contents of soluble phenolics and lignin in two T-DNA insertion mutants of CB5D, designated as cb5d-1 (SALK_045010) and cb5d-2 (GABI-328H06), where the T-DNA was inserted in the 3′ untranslated region and the second exon of CB5D, respectively (Figure 5A). The homozygosity of both mutants was validated prior to subsequent analysis. The T-DNA insertion resulted in barely detectable CB5D expression in both lines (Figure 5B); they therefore represent null mutants. Both cb5d-1 and cb5d-2 were morphologically similar to the Col-0 wild type, although a slight decrease in the biomass of cb5d-2 was detected (Supplemental Figure 2). However, when soluble phenolics were examined, we noticed that the levels of both sinapoyl malate and sinapoyl glucose in cb5d-1 and cb5d-2 were reduced by ∼40 to 50% compared with wild-type levels (Figure 5C). When the basal stems of 6-week-old mutant plants were cut into cross sections and stained to monitor total lignin content (by phloroglucinol-HCl or Wiesner staining) and lignin composition (by Mäule staining), the C4H mutant ref3-3 (reduced epidermal fluorescence3-3; as a control) showed a substantial reduction in total lignin content in its interfascicular fiber cells but a minor impairment of S-lignin accumulation, as indicated by much weaker violet-red coloration after Wiesner staining compared with the wild type but slightly weaker color intensity after Mäule staining, which specifically probes S-lignin subunits (Figure 5D). These results are consistent with the previous characterization of the ref3-3 mutant (Schilmiller et al., 2009) and suggest that a partially complementary pathway might bypass C4H and lead to the formation of S-lignin. In sharp contrast, when cross sections of cb5d-1 and cb5d-2 were stained with both Wiesner and Mäule reagents, compared with the wild type, the intensity of Wiesner staining for total lignin was not compromised at all (or it was even strengthened), but a dramatic reduction in red coloration after Mäule staining occurred in both the xylem bundle and interfascicular fiber cells (Figure 5D). These histochemical reactions phenocopied those of the F5H mutant fah1-2 (Figure 5D), suggesting that CB5D, like F5H, is more functionally associated with the biosynthesis of S-lignin.
Figure 5.
Characterization of cb5d-Related Mutants.
(A) Diagram of the T-DNA insertion mutants of CB5D. The triangles indicate the T-DNA insertion sites in CB5D. The dashed line shows the region used for qRT-PCR analysis of CB5D gene expression.
(B) qRT-PCR analysis of the relative expression levels of CB5D in the Col-0 wild type, cb5d-1, and cb5d-2. Each of ∼12 2-week-old seedlings was grouped as one biological replicate for RNA extraction. Three biological replicates, each with four technical repeats, were performed in qRT-PCR analysis. The data represent means ± sd of three biological repeats.
(C) Quantification of sinapoyl ester content in 2-week-old seedlings of the indicated genotypes. The data represent means ± sd of four biological repeats; each replicate is composed of 0.5 g of pooled seedlings. Different letters indicate significant differences (P < 0.05; one-way ANOVA, Tukey’s HSD test) between the different genotypes. FW, fresh weight.
(D) Histochemical observation of cross sections of 7-week-old stems (1 cm from the bottom, 50 µm thick) of the indicated genotypes with Wiesner staining for total lignin (top panels) and Mӓule staining for S-lignin (bottom panels). Bars = 30 µm.
(E) Quantification of total acetyl bromide lignin content in the cell walls of the indicated genotypes.
(F) Quantification of thioacidolytic G-lignin monomer in the cell walls of the indicated genotypes.
(G) Quantification of thioacidolytic S-lignin monomer in the cell walls of the indicated genotypes.
In (E) to (G), the data represent means ± sd of three biological replicates of stems from six fully mature (14-week-old) plants per replicate. CWR, cell wall residues. Asterisks indicate significant differences compared with the seedling samples (Student’s t test; ***, P < 0.001).
We then conducted chemical measurements of the total acetyl bromide lignin content in the cell walls of 14-week-old fully mature stems. In agreement with the results of histochemical analysis, the data revealed no significant difference between cb5d-1, cb5d-2, fah1-2, and the wild type, whereas ref3-3 showed a >40% reduction in total lignin content (Figure 5E). However, when we analyzed lignin composition via diagnostic thioacidolysis, which quantifies lignin monomers released from β-O-4-linked lignin substructures upon chemical degradation (Lapierre et al., 1985), we detected a dramatic reduction in the levels of released S-monomer (up to 65% of wild-type levels) in the lignin of cb5d-1 and cb5d-2 (Figure 5G). By contrast, the level of released G-monomers remained unchanged in these mutants or even slightly increased compared with the wild type (although such an increase was not statistically significant due to the large variation among wild-type samples; Figure 5F). Consistent with a previous report (Meyer et al., 1996), fah1-2 accumulated no S-lignin monomer, whereas its G-monomer level was similar or slightly higher than that of the wild type (Figures 5F and 5G). By contrast, ref3-3 exhibited a >50% reduction in G-lignin monomer levels but little or no reduction in S-lignin content, as previously reported (Figures 5F and 5G; Schilmiller et al., 2009). These results of chemical analysis confirm the notion that CB5D is primarily involved in the biosynthesis of S-lignin subunits. These data also point to a potential functional association of CB5D with F5H but not C4H activity.
F5H (CYP84A1) in Arabidopsis has a close paralog, CYP84A4 (At5G04330), which is responsible for the biosynthesis of α-pyrones (Weng et al., 2012). We analyzed aqueous extracts from stems of both cb5d-1 and cb5d-2 and found that the contents of the major α-pyrones arabidopyl alcohol and iso-arabidopic acid were reduced by ∼80% in both mutant lines compared with the wild-type control (Supplemental Figure 3A), suggesting that CB5D is also functionally required for the CYP84A4-catalyzed formation of arabidopyrones. However, when phenolic extracts from seeds were analyzed, the levels of the major flavonol, quercetin, did not significantly differ between the cb5d mutants and the wild type, whereas the levels of sinapoyl esters in the seeds of both cb5d mutants were reduced by up to 70% (Supplemental Figures 3B and 3C). The synthesis of quercetin requires flavonoid 3′-hydroxylase (F3′H; CYP75B), an evolutionarily close homolog of F5H (Weng et al., 2008). These data suggest that CB5D is specifically required for reactions catalyzed by CYP84A subfamily members in Arabidopsis.
Complementation of the cb5d Mutants with the CB5D Transgene
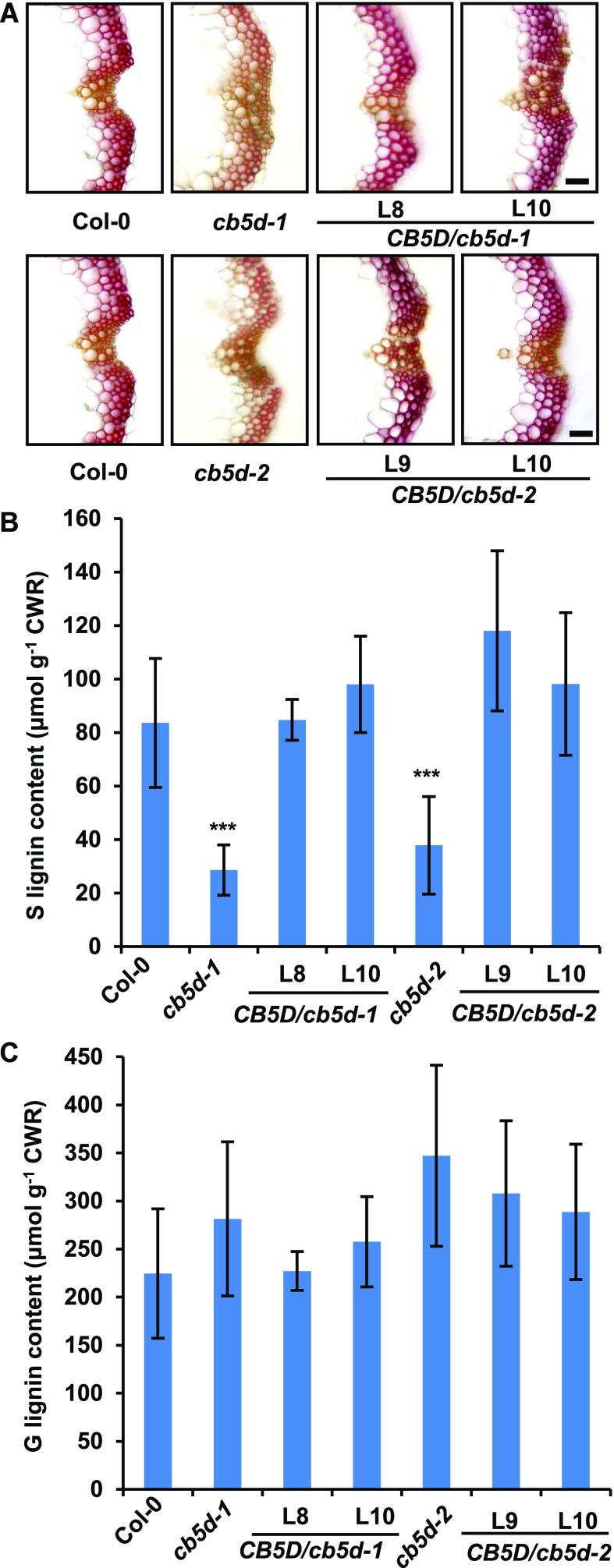
To confirm that the defects in S-lignin deposition and the accumulation of soluble phenolics in cb5d are indeed due to the loss of CB5D function, we generated two sets of complementation transgenic lines. One set of lines was transformed with a genomic DNA fragment spanning the native CB5D promoter and its coding region into the cb5d-1 and cb5d-2 homozygous mutant backgrounds, and in the other set, since the C4H promoter effectively drives F5H overexpression in lignifying tissues (Meyer et al., 1998), a pC4H:CB5D expression cassette was constructed and transformed into homozygous cb5d-1 and cb5d-2 lines. In the resulting T1 transgenic lines, the defective sinapoyl ester accumulation in cb5d-1 and cb5d-2 was rescued to close to wild-type levels (Supplemental Figure 4). Histochemical staining of stem cross sections of the representative T2 complementation lines of pC4H:CB5D in both the cb5d-1 and cb5d-2 backgrounds revealed a clear restoration of S-lignin deposition in the vasculature of the transgenic plants (Figure 6A). Chemical composition analysis confirmed that the contents of both G- and S-lignin monomers in the cell walls of the complementation lines were restored to wild-type levels (Figures 6B and 6C). These results confirm the notion that the loss of function of CB5D indeed is responsible for the defective biosynthesis of S-lignin and related sinapoyl esters in cb5d mutant plants.
Figure 6.
Complementation of S-Lignin Synthesis in cb5d Mutants by CB5D Expression.
(A) Histochemical observation of S-lignin (Mӓule staining) in the Col-0 wild type, cb5d-1, cb5d-2, and two representative T2 transgenic lines of pC4H:CB5D in the cb5d-1 and cb5d-2 homozygous backgrounds. Cross sections of 7-week-old stems (1 cm from the bottom, 50 µm thick) were used. Bars = 30 µm.
(B) and (C) Quantification of S-lignin (B) and G-lignin (C) monomer in the cell walls of the indicated genotypes. Stems of 14-week-old Col-0 wild type, cb5d-1, cb5d-2, and two T2 transgenic lines of pC4H:CB5D in the cb5d-1 and cb5d-2 backgrounds were used in the analysis. Stems from at least six plants were pooled as one replicate. The data represent means ± sd of three biological replicates. Asterisks indicate significant differences compared with the Col-0 wild type (two-tailed Student’s t test; ***, P < 0.001). No significant differences were observed for G-lignin in each genotype compared with the Col-0 wild type. CWR, cell wall residues.
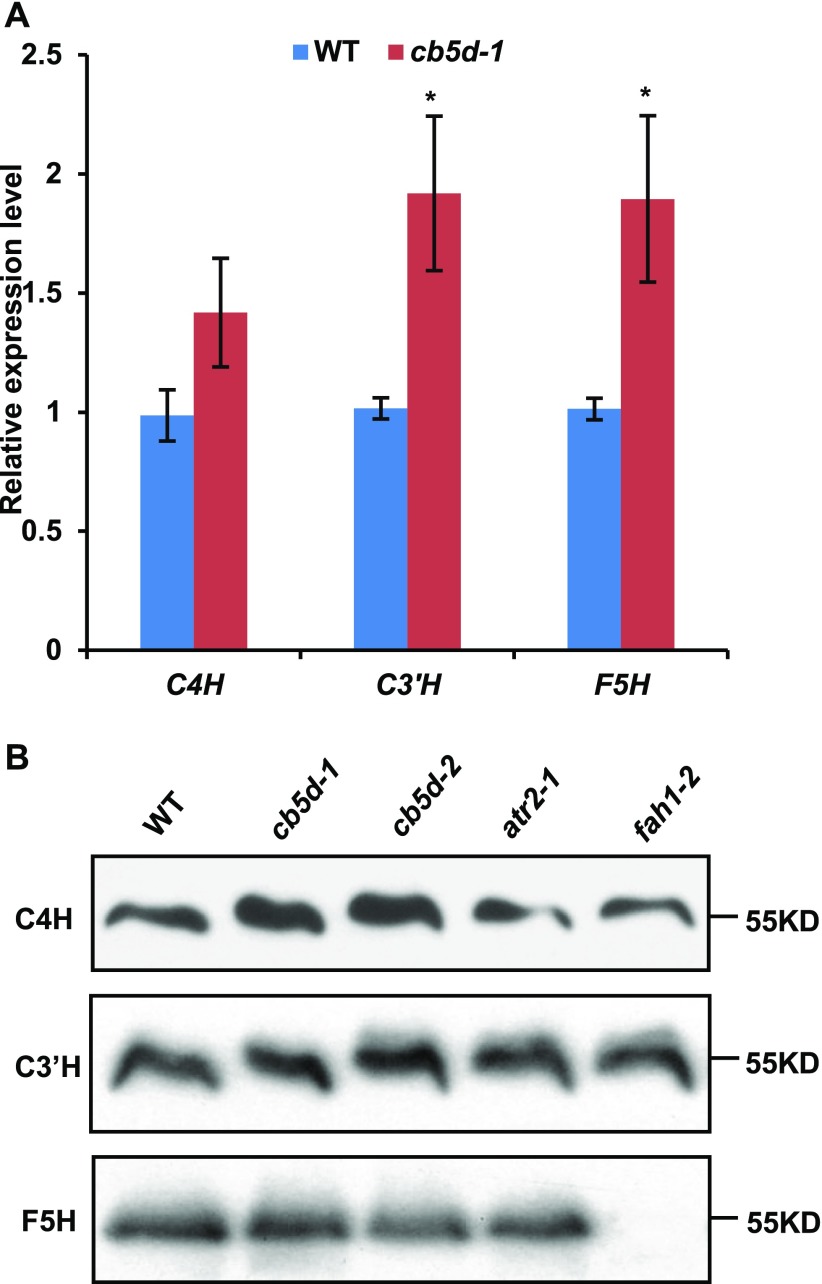
It was still possible that the defective S-lignin synthesis in cb5d was caused by nonspecific or secondary effects of CB5D disruption on gene expression or protein stability of the monolignol biosynthetic P450s. We therefore measured the transcript and protein levels of C4H, C3′H, and F5H in the cb5d mutants. qRT-PCR revealed that the transcript levels of C4H, C3′H, and F5H in both mutants were actually slightly higher than those of the wild type (Figure 7A). Immunoblot analysis further verified that the protein levels of C3′H and F5H in both cb5d mutants were nearly the same as those of the wild type, while C4H protein levels appeared even slightly higher in the mutants than in the wild type (Figure 7B). These data indicate that the defective S-lignin biosynthesis in the mutants, along with their CB5D deficiency, did not result from the depletion of the transcripts and/or cellular protein concentrations of monolignol biosynthetic P450 enzymes.
Figure 7.
Transcript Abundance and Protein Levels of Three P450s in cb5d Mutant Lines.
(A) qRT-PCR analysis of the relative expression levels of C4H, C3′H, and F5H in the Col-0 wild type and cb5d-1. Two-week-old seedlings were used for RNA extraction. Each of ∼12 seedlings were grouped as a biological replicate. Three biological repeats, each with four technical repeats, were performed in the analysis. The data represent means ± sd of three biological repeats. Asterisks indicate significant differences compared with wild-type samples (Student’s t test; *, P < 0.05).
(B) Immunoblots of C4H, C3′H, and F5H protein levels in 2-week-old seedlings of the Col-0 wild type, cb5d-1, cb5d-2, atr2-1, and fah1-2. Equal amounts of microsomal proteins were loaded in each lane.
CB5D Functions as an Electron Carrier for S-Lignin Synthesis
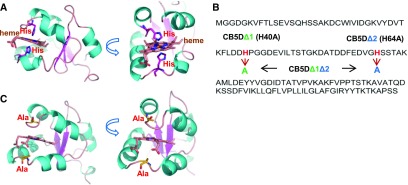
Studies on mammalian and human P450 systems suggest that the electron transfer function of CB5 relies on its iron center of bound heme molecule (Schenkman and Jansson, 2003). Sequence analysis and protein homology modeling of Arabidopsis CB5D indicated that this protein, like other cytochrome b5 family members, shares a conserved heme binding domain and two highly conserved His residues, His-40 and His-64, which serve as the critical axial ligands for heme iron coordination and for electron transfer in CB5 family members (Figure 8A; Cowley et al., 2002; Schenkman and Jansson, 2003). To explore whether the effect of CB5D on S-lignin biosynthesis is due to its electron carrier properties, we generated CB5D mutant variants by substituting either one or two conserved His residues with Ala (Figure 8B), therefore impairing the coordination with heme iron and thus depleting its intramolecular electron transfer properties (Cowley et al., 2002; Schenkman and Jansson, 2003). Both substitutions occurred on the flexible coil regions connecting two helices and thus likely would not impair protein folding or overall structure. This was demonstrated by homology modeling analysis, in which the mutant variants exhibited identical tertiary structures to the parental protein, even after the models went through intensive simulation and energy minimization during the homology modeling process (Figure 8C).
Figure 8.
Creation of CB5D Variants.
(A) Homology model of AtCB5D based on the crystal structure of human CB5B (PDB ID: 3NER). The bound heme and its interacting His residues are presented as sticks.
(B) Diagram of the substitutions (His to Ala) in AtCB5D. Mutants with substitutions of His-40, or His-64, or both with Ala are designated as CB5DΔ1, CB5DΔ2, and CB5DΔ1Δ2, respectively.
(C) Homology model of CB5DΔ1Δ2, showing its identical overall structure to the parental protein.
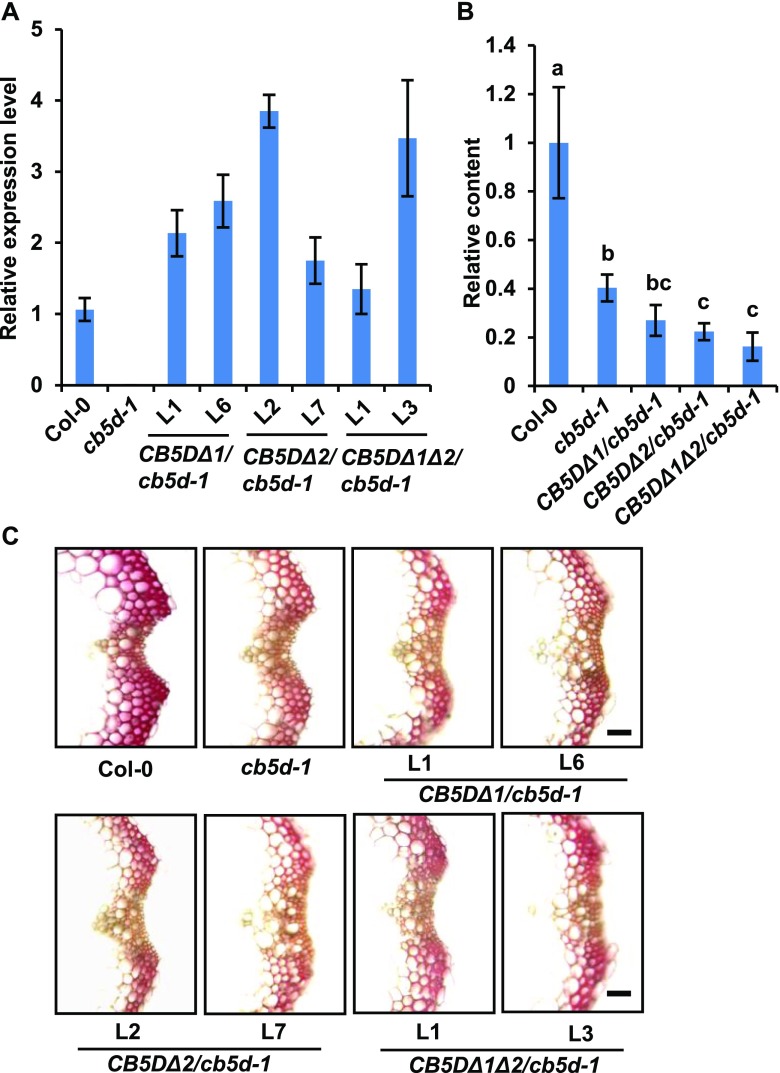
We then transformed Arabidopsis plants in the homozygous cb5d-1 background with the genes encoding these mutant variants, including CB5DΔ1 (H40A), CB5DΔ2 (H64A), and CB5DΔ1 Δ2 (H40A/H64A), driven by the C4H promoter. While transcripts of the CB5DΔ transgenes were detected in the resulting transgenic lines (Figure 9A), in contrast to the expression pattern of pC4H:CB5D in cb5d-1 (Figure 6), none of the transgenic lines harboring the CB5DΔ variants showed a restoration of sinapoyl ester accumulation (Figure 9B). In fact, the levels of sinapoyl esters in transgenic lines harboring the CB5DΔ variants, primarily CB5DΔ1Δ2, were even lower than those of the cb5d-1 parental line (Figure 9B). These results point to the possible dominant suppression of the overexpressed CB5DΔ variants over the endogenous P450 reactions in planta, presumably due to the competition of the expressed mutant variants for other endogenous electron donor proteins in the electron transfer chain. When S-lignin deposition was monitored by Mäule staining of stem cross sections of the representative T2 lines expressing the CB5DΔ variants, the intensity of color staining in these lines was similar to that of the cb5d-1 parental lines and drastically weaker than that of the wild type (Figure 9C), indicating the failure of the CB5D variants to restore S-lignin synthesis. Taken together, these findings demonstrate that heme binding and, thus, the electron transfer properties of CB5D are essential for regulating sinapoyl ester and S-lignin biosynthesis.
Figure 9.
Complementation Test Using Genes Encoding Mutant Variants of CB5D.
(A) qRT-PCR analysis of gene expression levels in T2 transgenic lines expressing pC4H:CYB5DΔ1, pC4H:CYB5DΔ2, or pC4H:CYB5DΔ1Δ2 in the cb5d-1 background. Two-week-old T2 seedlings were used for RNA extraction. Approximately 12 seedlings were pooled as one biological replicate. The data represent means ± sd of three biological replicates, each with three technical repeats.
(B) Relative content of sinapoyl malate in 4-week-old rosette leaves of the Col-0 wild type, cb5d-1, and T1 transgenic lines harboring pC4H:CYB5DΔ in the cb5d-1 background. The data represent means ± sd of all biological replicates and are expressed as relative values compared with the Col-0 wild type. The data for the Col-0 wild type and cb5d-1 were obtained from three independent plants representing three biological replicates; the data for pC4H:CYB5DΔ transgenic plants were obtained from an average of 10 T1 independent transgenic lines per transgene. Different letters indicate significant differences (P < 0.05; one-way ANOVA, Tukey’s HSD test) between the different genotypes.
(C) Histochemical observation of S-lignin in the Col-0 wild type, cb5d-1, and two independent T2 transgenic lines of pC4H:CYB5DΔ1, pC4H:CYB5DΔ2, and pC4H:CYB5DΔ1Δ2 in the cb5d-1 background with Mӓule staining. Cross sections of 7-week-old stems (1 cm from the bottom, 50 µm thick) were used. Bars = 30 µm.
Contribution of CB5D, ATR2, and CBR1 to Lignin Biosynthesis
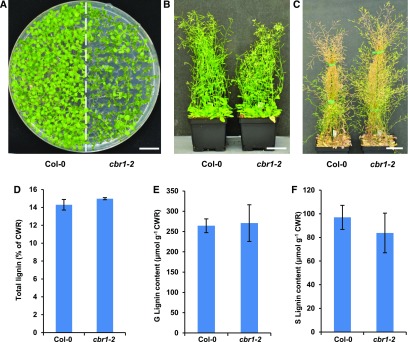
CB5 can receive electrons from either CBR or CPR and shuttle them to the oxidative enzymes. To further evaluate the involvement of CB5D-mediated electron transfer chain(s) in lignin biosynthesis, we examined the effects of Arabidopsis CBR and CPR on lignin deposition. Arabidopsis has two annotated CBR homologs, but only CBR1 has been demonstrated to be part of the ER electron transport system. Disrupting CBR1 affects fatty acid biosynthesis and plant fertility (Kumar et al., 2006; Wayne et al., 2013). The second member, CBR2, is localized to the inner mitochondrial membrane, and there is no evidence showing its involvement in the ER electron transport system (Heazlewood et al., 2004). Consistent with a previous study (Wayne et al., 2013), disrupting CBR1 (in the cbr1-2 homozygous mutant background) resulted in delayed germination and retarded growth, with most plants exhibiting smaller statures than the wild type at the seedling stage, although a few seedlings were phenotypically similar to the wild type (Figure 10A). The growth of the cbr1-2 mutants eventually caught up to that of the wild type, with similar heights when they approached full maturation (Figures 10B and 10C). When measured at the fully mature stage, the total lignin content and levels of both G- and S-lignin monomers released from the cell walls of cbr1-2 plants were essentially the same as those of the wild type, although there appeared to be a slight (but not statistically significant) reduction in the levels of S-monomers (Figures 10D to 10F). These results suggest that the NADH-CBR-CB5 electron transport system has little or no role in lignin biosynthesis or that it plays a redundant role with other electron transfer systems and its defect is compensated for by other electron transport components.
Figure 10.
Characterization of the cbr1-2 Mutant.
(A) Growth phenotypes of 2-week-old seedlings of the Col-0 wild type and cbr1-2 on 0.5× Murashige and Skoog plates. Bar = 1 cm.
(B) Growth phenotypes of 7-week-old plants of the Col-0 wild type and cbr1-2. Bar = 5 cm.
(C) Growth phenotypes of 12-week-old mature plants of the Col-0 wild type and cbr1-2. Bar = 5 cm.
(D) Quantification of total acetyl bromide lignin content in the cell walls of 14-week-old plants of the Col-0 wild type and cbr1-2.
(E) Quantification of thioacidolytic G-lignin monomer in the cell walls of 14-week-old plants of the Col-0 wild type and cbr1-2.
(F) Quantification of thioacidolytic S-lignin monomer in the cell walls of 14-week-old plants of the Col-0 wild type and cbr1-2.
Data in (D) to (F) represent means ± sd of three biological replicates of pooled stems from at least six fully mature plants per replicate. There was no significant difference in total lignin or G- or S-lignin monomer contents in cbr1-2 compared with the Col-0 wild type (two-tailed Student’s t test). CWR, cell wall residues.
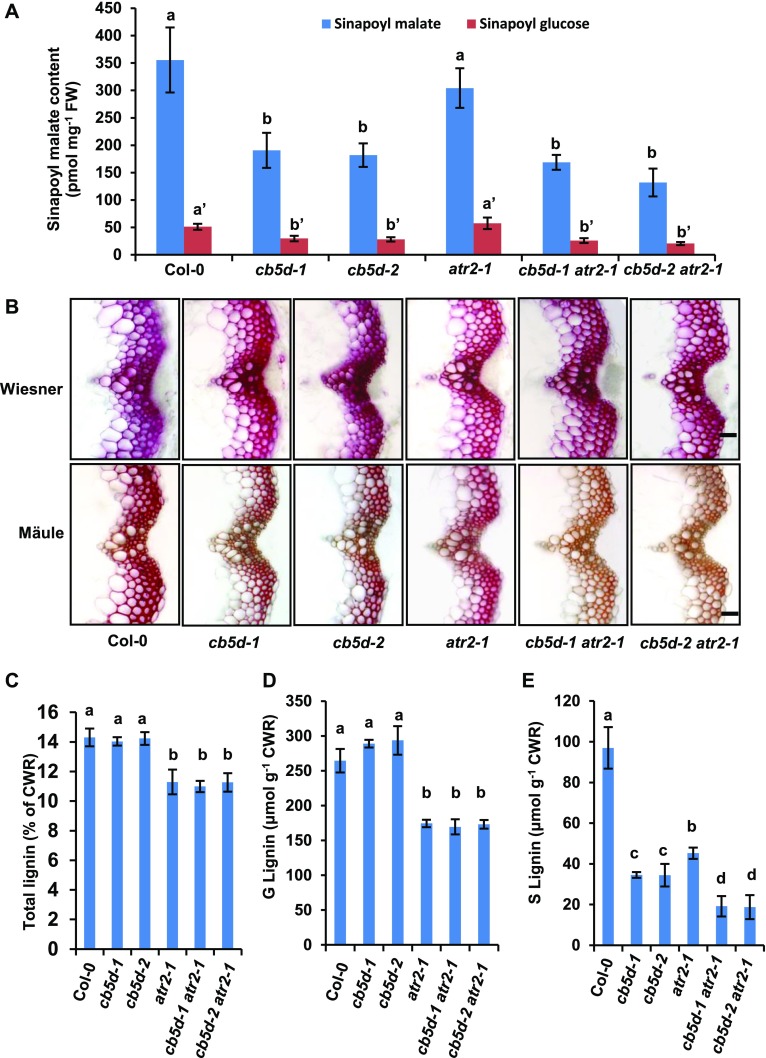
The Arabidopsis cytochrome P450 reductase ATR2 was previously shown to be involved in lignin biosynthesis (Sundin et al., 2014). We reexamined the effects of ATR2 on lignin deposition and compared them with those of CB5D under our growth conditions. Disrupting ATR2 (in the atr2-1 homozygous mutant background) did not result in a significant impairment in sinapoyl malate or sinapoyl glucose accumulation compared with the wild type, in strong contrast to the significant reduction in sinapoyl ester levels in cb5d-1 and cb5d-2 (Figure 11A). Moreover, unlike cb5d-1 and cb5d-2, which showed no depletion of total lignin deposition but a specific impairment of S-lignin accumulation (Figures 5 and 11), disrupting ATR2 reduced the total lignin content by ∼20% and compromised the biosynthesis of both G- and S-lignin monomers, which was monitored by thioacidolysis, with an ∼34% reduction for G-subunits and an ∼53% reduction for S-subunits in atr2-1 compared with wild-type levels under our growth conditions (Figures 11B to 11E).
Figure 11.
Characterization of CB5D and CPR Mutants.
(A) Quantification of sinapoyl ester content in 2-week-old seedlings of the indicated genotypes. The data represent means ± sd of four biological repeats. FW, fresh weight.
(B) Histochemical observation of cross sections of 7-week-old stems (1 cm from the bottom, 80 µm thick) of the indicated genotypes with Wiesner staining for total lignin (top panels) and Mӓule staining for S-lignin (bottom panels). Bars = 30 µm.
(C) Quantification of total acetyl bromide lignin content in the cell walls of the indicated genotypes.
(D) Quantification of thioacidolytic G-lignin monomer in the cell walls of the indicated genotypes.
(E) Quantification of thioacidolytic S-lignin monomer in the cell walls of the indicated genotypes.
In (C) to (E), the data represent means ± sd of three biological replicates of pooled stems from at least six fully mature plants per replicate. CWR, cell wall residues. In (A) and (C) to (E), different letters indicate significant differences (P < 0.05; one-way ANOVA, Tukey’s HSD test) between the different genotypes.
To further assess whether CB5D and ATR2 share functionally redundant roles in regulating S-lignin biosynthesis, we generated cb5d-1 atr2-1 and cb5d-2 atr2-1 double mutants. Homozygosity of the resulting double mutant was confirmed prior to detailed chemical analysis. Similar to atr2-1, the double mutants showed reduced height and biomass compared with the wild type (Supplemental Figure 2). Simultaneously knocking out both CB5D and ATR2 resulted in a reduction in total lignin content and the levels of both G- and S-lignin monomers, as observed in the atr2-1 single mutant (Figures 11C and 11D). Notably, there was a greater reduction in S-lignin content in the double mutant, with a >80% reduction compared with the wild type versus an ∼53% reduction in atr2-1 and an ∼65% reduction in cb5d-1 and cb5d-2 (Figure 11E). Such an enhanced reduction in S-lignin content was also visually discernible via Mӓule staining of cross sections of the stems of double mutant plants, particularly compared with atr2-1 (Figure 11B). These data indicate that ATR2 and CB5D have an additive effect on the synthesis of S-lignin.
Furthermore, we reciprocally expressed ATR2 and CB5D in cb5d and atr2 knockout mutant lines. When ATR2 was expressed in cb5d-1 driven by the C4H promoter, the defective sinapoyl ester synthesis and S-lignin deposition in the mutant line were not restored (Supplemental Figure 5). Conversely, when CB5D driven by the C4H promoter was expressed in the atr2-1 background, although an apparently higher level of sinapoyl ester was detected in the resulting T1 transgenic lines compared with atr2-1, the increase was not statistically significant (Supplemental Figure 6A). Furthermore, the defective G- and S-lignin synthesis in atr2-1 was not rescued, as measured chemically and histochemically in the stem cell walls of the corresponding T2 transgenic lines (Supplemental Figures 6B to 6D). These data indicate that both ATR2 and CB5D are indispensable components for lignin biosynthesis.
Both CB5D and ATR2 Are Required to Augment the F5H-Catalyzed Reaction
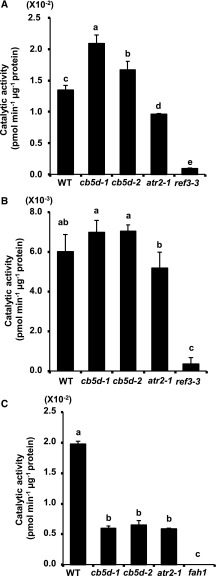
The specific effect of CB5D on S-lignin monomer synthesis suggests that this electron transfer protein might functionally prefer to associate with F5H, the branch point enzyme leading to S-lignin monomer synthesis, compared with C4H and/or C3′H. To test this hypothesis, we prepared Arabidopsis microsomes from wild-type, cb5d, and atr2-1 plants. After monitoring F5H, C4H, and C3′H protein levels (Figure 7B), we measured both F5H and C4H activity in the presence of NADPH or NADH as a reductant; the reaction products were determined and quantified with the aid of LC-MS (Supplemental Figures 7 and 8). Due to the lack of available substrate for C3′H, it was not included in the activity measurements. Rather, we chose C4H as the representative enzyme. When NADPH was used as a reductant, as expected, C4H activity was severely compromised in the C4H mutant ref3-3 and was also reduced in atr2-1 (Figure 12A). By contrast, C4H activity was not reduced (it actually increased) in the cb5d mutants compared with the wild type (Figure 12A; Supplemental Figure 7), indicating that a defect in CB5D had no detrimental effect on C4H catalytic activity. The slight increase in C4H activity is likely due to the discernible increase in C4H protein levels in microsomes prepared from the cb5d mutants (Figure 7B). When NADH was used as the reducing reagent, the C4H activity in all microsomal samples was lower than those measured when we used NADPH as a reductant, and there were no significant differences between cb5d (or atr2-1) and the wild type (Figure 12B). These results further confirm the notion that disrupting CB5D does not affect C4H enzymatic activity. Interestingly, when we measured F5H activity, it was only detectable when NADPH was used as a reductant, whereas no measurable activity was detected when using NADH. The F5H activity in the presence of NADPH in either cb5d or atr2-1 was significantly lower than that in the wild type, and the reductions in F5H activity were similar in these two types of mutant lines (Figure 12C; Supplemental Figure 8). These data indicate that both CB5D and ATR2 are required for the F5H-catalyzed hydroxylation reaction.
Figure 12.
Enzymatic Activity Assay of C4H and F5H in Planta.
Microsomes were prepared from 2-week-old seedlings of different genotypes, and P450 protein concentration was monitored by immunoblot analysis using anti-C4H or anti-F5H antibodies, as shown in Figure 7.
(A) and (B) C4H activity in the Col-0 wild type, cb5d-1, cb5d-2, atr2-1, and ref3-3 mutant lines in the presence of NADPH (A) or NADH (B) as the reductant.
(C) F5H activity in the Col-0 wild type, cb5d-1, cb5d-2, atr2-1, and fah1-2 mutant lines in the presence of NADPH as the reductant. No measurable activity was detected when using NADH as the reducing power.
The data represent means ± sd of three biological replicates. Three independent batches of experiments were conducted with the same results. Different letters indicate significant differences (P < 0.05; one-way ANOVA, Tukey’s HSD test) of different plant genotypes.
DISCUSSION
Cytochrome P450 enzymes play central roles in many metabolic processes. Notably, the monooxygenases are not self-sufficient enzymes; they require redox partner(s) as cofactors to deliver two electrons for the cleavage of an oxygen molecule (Hannemann et al., 2007). Typically, the endosomal P450s rely on the diflavin reductase CPR to transfer reducing power from NADPH (or NADH) via its flavin adenine dinucleotide and flavin mononucleotide domains to the prosthetic heme group of P450 (Urban et al., 1997; Jensen and Møller, 2010). Indeed, the Arabidopsis NADPH-dependent cytochrome P450 oxidoreductase ATR2 was demonstrated to functionally associate with C4H (Urban et al., 1997) and to be involved in lignin biosynthesis based on the coexpression of its gene with monolignol biosynthetic genes and the detectable effect of the disruption of ATR2 on lignin accumulation (Sundin et al., 2014). This defined role of ATR2 was corroborated in this study by the observation that disrupting ATR2 resulted in an up to ∼20% reduction in total lignin content and substantially reduced levels of both G- and S-lignin subunits in the cell walls of mutant plants (Figure 11). A more marked impairment of lignin biosynthesis was observed in atr2-1 in this study than what was reported previously (Sundin et al., 2014). This discrepancy is likely due to the different plant growth conditions used in the studies (i.e., we used a long-day photoperiod for plant growth). Intriguingly, besides ATR2, our biochemical and genetic analyses explicitly demonstrated that the ER-resident cytochrome b5 family member D acts as an additional indispensable electron shuttle component that contributes to lignin biosynthesis, especially S-lignin formation.
Although due to the nature of affinity purification and LC-MS-based proteomics of membrane proteins, we could not ascertain whether the copurified proteins were in direct physical interactions or in spatial proximity with the baits, our study revealed several electron transfer proteins that coeluted with monolignol P450 enzymes (Table 1). This discovery coincides with the previous demonstration that mammalian and human microsomal P450s physically interact with their redox partners, CPR and CB5, via electrostatic interactions at the basic, positively charged proximal surfaces of P450s (Schenkman and Jansson, 1999; Im and Waskell, 2011; Kandel and Lampe, 2014; Scott et al., 2016). The interaction of P450 with its electron carrier was proposed to form a functional 1:1 complex (Miwa and Lu, 1984). However, the concentration of P450 redox partners was estimated to be 10 to 100 times lower than that of total P450s (Watanabe et al., 1993). Since there are multiple P450s in excess of their redox partners in the same membrane, it is impossible for the redox proteins to interact with all P450s at the same time. This suggests that either the P450s are organized in complexes to facilitate interactions with the limited number of electron carriers or the monomeric P450s and their electron carriers interact stochastically and dynamically. The detection of a common set of electron transport components in our affinity purification experiment using the three monolignol biosynthetic P450s as baits further suggests the possibility that these P450s are spatially organized with their redox partners on the ER membrane. The individual monolignol biosynthetic P450 in the complex is expected to associate with different sets of electron carriers. The copurification of CB5s in addition to CPRs along with monolignol biosynthetic P450 enzymes points to their potential involvement in monolignol biosynthesis. This notion was confirmed by our subsequent genetic and biochemical analyses. The disruption of CB5D resulted in a drastic and specific reduction in S-lignin and sinapoyl ester synthesis but not of G-lignin deposition (Figure 5).
This phenomenon is in sharp contrast to the consequences of disturbing ATR2 on lignin content and composition, in which the accumulation of both G- and S-lignin monomers and ultimately total lignin was compromised (Figure 11). This observation suggests that ATR2, a known electron donor protein for P450s, likely functionally associates more broadly with the C4H, C3′H, and F5H catalytic reactions, thus influencing both G- and S-lignin synthesis, whereas CB5D appears to function more specifically in the F5H-catalyzed reaction conferring S-lignin formation in planta. This notion was further confirmed by the detection of P450 enzymatic activity in mutant plants. Elimination of CB5D depleted F5H activity but did not affect C4H activity (Figure 12). In addition, metabolic profiling of aqueous and methanolic extracts indicated that CB5D is required for the CYP84A4-catalyzed formation of α-pyrones but not for the F3′H (CYP75B)-catalyzed accumulation of flavonols (Supplemental Figure 3). CYP84A4 is a paralog of F5H in Arabidopsis, whereas F3′H is evolutionarily more closely related to the CYP84A subfamily than to the others (Weng et al., 2008, 2012). Taken together, these findings demonstrate that CB5D is an obligate electron donor protein that is specifically required for reactions catalyzed by F5H and its closest subfamily member, CYP84A, in planta.
Notably, a defect in ATR2 not only impaired the activity of C4H but also that of F5H, suggesting that ATR2 is biochemically necessary for F5H-catalyzed reactions as well. Mechanistically, CB5 is an electron shuttle component that augments P450 reactions via two mechanisms: the direct transfer of two required electrons from NADH-CBR to the P450 enzyme or the transfer of the second electron to the oxyferrous P450 from either CPR or CBR (Supplemental Figure 1; Porter, 2002). Interestingly, although a defect in Arabidopsis CBR1, the sole cytochrome b5 reductase in the ER-localized electron transfer chain, substantially affects fatty acid desaturation (Wayne et al., 2013), our study revealed that its deficiency does not dramatically impair lignin content or G- or S-lignin composition in mature cbr1 plants (Figure 10). Consistent with the genetic evidence, when NADH was used as the reductant to measure the enzymatic activity of F5H in Arabidopsis microsomes, its activity was barely detected (Figure 12; Supplemental Figure 8). These results hint at the possibility that the NADH-CBR-CB5 electron transfer chain plays little or no role in lignin biosynthesis in Arabidopsis; the effect of CB5D on S-lignin synthesis should be more functionally associated with the NADPH-CPR electron transport system, and both CPR and CB5 are indispensable for S-lignin formation. Indeed, the disruption of CB5D or ATR2 resulted in S-lignin depletion (Figures 5 and 11). The expression of ATR2 in the cb5d mutant background or, conversely, the expression of CB5D in the atr2-1 background did not restore the defective S-lignin formation in the corresponding mutant lines (Supplemental Figures 5 and 6). These data further suggest that both CB5D and ATR2 are indispensable and nonredundant components in S-lignin synthesis. It is likely that these proteins functionally partner together in the same electron transfer system, specifically to augment F5H activity. It should be noted that the effect of ATR2 on S-lignin formation might also be achieved through its augmentation of C4H- and/or C3′H-catalyzed reactions, which generate the biosynthetic intermediates for S-lignin monomer synthesis. However, the in vitro assay showed that disrupting ATR2 or CB5D strongly suppressed F5H catalytic activity, suggesting that both ATR2 and CB5D are directly associated with F5H-catalyzed S-lignin formation.
On the other hand, simultaneously disrupting both ATR2 and CB5D resulted in an obvious additive suppression of S-lignin synthesis. In cb5d atr2-1 double mutant lines, the S-monomers exhibited further ∼15 and ∼27% reductions in levels compared with the levels in the cb5d and atr2-1 single mutants, respectively (Figure 11). These findings suggest that CB5D and ATR2, in addition to functionally partnering together and playing prominent roles in S-lignin synthesis, may each also functionally associate with other homologous members of electron shuttle components, e.g., ATR2 with other CB5 family members or CB5D with ATR1. Such multiple functional associations might play minor and overlapping roles in augmenting F5H-catalzyed S-monomer formation.
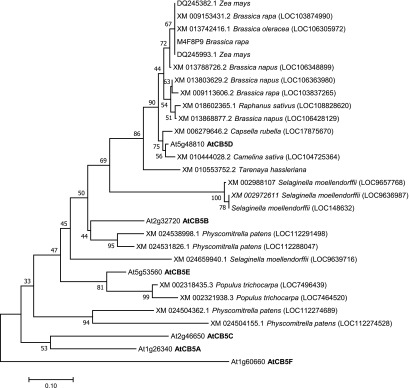
While the F5H-catalzyed S-lignin formation requires both CB5 and CPR redox partners, the G-lignin synthesis and the related C4H (and/or C3′H) reactions do not necessarily rely on CB5 components in planta, although CB5D appeared to coelute with C4H and C3′H in our affinity purification experiments. Perhaps these proteins coeluted due to their close proximity on the ER membrane. Although the exact molecular mechanism for the specific functional association of CB5D with F5H (and CYP84A4) in planta remains unclear, this may reflect its preferred binding to F5H and CYP84A4 compared with other P450s. The disparity of the functional requirements of the redox systems of F5H and C4H (as well as other CPR-dependent P450s) suggests that the most recently evolved CYP84A family members and their catalyzed reactions structurally or mechanistically differ from those of C4H and other P450 enzymes. F5H emerged with the occurrence of S-lignin in all angiosperms and lycophytes in the Selaginella genus via parallel evolutionary mechanisms (Weng et al., 2008, 2010). Phylogenetic analysis of CB5D homologs across land plant species revealed that, although the least divergent homologs of CB5D are widespread within the plant kingdom (www.pantherdb.org), the closer sequences that share over 60% identity with CB5D at the amino acid level and show node support values of >50% in the phylogenetic tree by bootstrap analysis (1000 replicates) are overrepresented only within a few lineages of angiosperms, particularly members of the Brassicaceae family as well as its sister family, the Cleomaceae (Figure 13; Supplemental File 1). Additionally, two homologous sequences (∼84% amino acid sequence similarity with CB5D) were found in maize (Figure 13), suggesting that the evolution and constraint of CB5D and its related electron shuttle system might have helped plants adapt to particular environmental niches. Consistent with the lack of S-lignin in most gymnosperms, no homologous sequence with reasonable similarity was found in gymnosperm species such as loblolly pine (Pinus taeda) and ginkgo (Ginkgo biloba), but, interestingly, three putative CB5 sequences from the lycophyte spike moss (Selaginella moellendorffii) clustered as a unique clade and are distantly connected to the CB5D homologs from angiosperms, shedding light on their potential evolutionary relationship (Figure 13). These data offer a hint that CB5D might have coevolved with the emergence of the F5H enzyme in both early and modern vascular plants with high levels of S-lignin synthesis, and that after its emergence its function as the redox partner to F5H might be further specified within a few lineages of land plants.
Figure 13.
Phylogenetic Relationship of Arabidopsis Cytochrome b5 and Related Putative Homologs (of AtCB5D) across Land Plants.
Protein sequences of 6 Arabidopsis CB5 homologs and 29 putative orthologs from different species that were obtained using the AtCB5D sequence as a query via BLAST searches of nucleotide sequences in the National Center for Biotechnology Information, and/or retrieved from the PANTHER classification system database (www.pantherdb.org), were used in the analysis. The sequences were aligned with ClustalW integrated in the MEGA v.7.0 program. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 3.19379392 is shown. The percentages (>50%) of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to construct the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method, and the units represent the number of amino acid substitutions per site.
An early study also demonstrated that disrupting a single petunia (Petunia hybrida) cb5 locus resulted in discolored flower petals and a decrease in flavonoid 3′,5′-hydroxylase activity (de Vetten et al., 1999). However, in this study, knocking down CB5D in Arabidopsis did not significantly affect flavonol accumulation (Supplemental Figure 3). Arabidopsis has several CB5 family members (Hwang et al., 2004; Maggio et al., 2007). It would be interesting to further explore the functional specification of these CB5 members as redox partners for P450 enzymes involved in different types of metabolism and to dissect how this specificity in electron donors-accepters is established at the structural and electrochemical levels. It would also be intriguing to determine the biological importance of the different electron shuttle components adopted by plants to synthesize the recently evolved angiosperm S-lignin in light of the merely one type of electron donor protein required for G-lignin synthesis.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutant lines cb5c, cb5d, cb5e, atr-2, cbr-1, fah1, and ref3-3 used in this study were obtained from the ABRC. All T-DNA insertion mutant lines were initially screened and validated for their homozygosity via antibiotic selection and genotyping using genomic DNA extracted from the corresponding seedlings. The primers used for genotyping are listed in the Supplemental Table. The seeds were sterilized with 70% (v/v) ethanol and plated on 0.5× Murashige and Skoog (Sigma-Aldrich) medium containing 0.7% agar and 1% sucrose. After sowing, the seeds were incubated at 4°C for 2 d and transferred to a FLEX series growth chamber (BioChambers) equipped with combined T5HO fluorescent lamps (Sylvania FP54/841/HO/ECO, Pentron 4100K 54W, 20,906 Hg) and halogen lamps (Topaz 40 W, 130 V A19, Rough Service Incandescent bulb) with a light intensity of 9000 lumens/m2 at 22°C under a 16-/8-h light/dark cycle for 2 weeks. The seedlings were transferred to soil (Sungro) and grown under the same conditions until mature. Wild tobacco (Nicotiana benthamiana) plants were grown for 4 to 6 weeks at the same growth conditions and used for transient expression studies.
To generate cb5d-1 atr2-1 and cb5d-2 atr2-1 double mutants, the cb5d-1 or cb5d-2 homozygous line was crossed to the atr2-1 homozygous line to obtain F1 seeds. Double mutants were obtained by genotyping the F2 population of cross progeny using the primers listed in the Supplemental Table to obtain individual plants homozygous for both mutations.
Affinity Purification of the P450 Complex and LC-MS Analysis
Proteomics analyses of monolignol P450 complexes were conducted as previously reported (Gou et al., 2018). Briefly, C4H, C3′H, and F5H genomic fragments without the stop codon were subcloned into a C4H promoter-driven pMDCpC4H-HPB vector derived from the pMDC32-HPB vector (Qi and Katagiri, 2009). The C4H and F5H constructs were transformed into the ref3-3 and fah1-2 backgrounds, respectively, while C3′H was transferred into the Col-0 wild type to obtain transgenic plants. As a control, pC4H-HPB empty vector was transformed into the Col-0 wild type. Fifteen-gram samples of stem tissue of the transgenic plants were collected after 5 weeks of growth in soil for protein extraction. Proteins were extracted and the protein complex was purified as described (Gou et al., 2018). The proteins were subjected to direct on-bead trypsin digestion or 10% SDS-PAGE, and the corresponding gels were sliced and mixed for trypsin digestion. The digested peptide mixture was analyzed by automated microcapillary liquid chromatography-tandem mass spectrometry (ThermoFinnigan). Full mass (MS) spectra were recorded on the peptides over a 400 to 2000 m/z range, followed by top five tandem MS scans (MS/MS) in the ion trap sequentially generated in a data-dependent manner on the first, second, third, fourth, and fifth most intense ions selected from the MS spectrum (at 35% collision energy). Charge state-dependent screening was turned on, and peptides with a charge state of +2 or higher were analyzed. MS/MS spectra were extracted from the RAW file with Readw.exe (http://sourceforge.net/projects/sashimi). The MS/MS data were searched with GPM X!Tandem with an expectation value of 0.01 or Inspect with a P value of 0.05 against a custom database downloaded from UniProt of the Arabidopsis proteome with added sequences for common contaminants (e.g., keratins). Fixed Cys carbamidomethylation, optional Met oxidation, and Thr, Ser, and Tyr phosphorylation were applied during the search.
Subcellular Localization Imaging and BiFC Assay
The coding regions of CB5D, ATR2, C4H, C3′H, and F5H were amplified from Col-0 wild-type cDNAs by RT-PCR using primers listed in the Supplemental Table and subcloned into the pDONR207 vector. The resulting clones were confirmed by sequencing. To generate an expression cassette for chimeric proteins with GFP, the sequence-confirmed cDNAs were subcloned into the pGWB405 vector, resulting in the p35S:CB5D:GFP, p35S:C4H:GFP, p35S:C3′H:GFP, and p35S:F5H:GFP constructs. The constructs were transformed into Agrobacterium strain GV3101 and transiently expressed in wild tobacco leaves via agroinfiltration. Fluorescence images were captured with a Leica TCS SP5 laser-scanning confocal microscope with excitation at 488 nm and an emission wavelength of 493 to 560 for GFP signals at 3 d post infiltration.
For the BiFC assay, each gene was subcloned into Gateway cloning destination vectors pDEST-GW VYNE and pDEST-GW VYCE or pDEST-GW VYCE(R) vectors to generate BiFC constructs (Gehl et al., 2009; Zhang et al., 2013). To establish negative control sets in the BiFC assay, the cDNA encoding the C-terminal transmembrane domain of Calnexin1 protein (Liu et al., 2017) and the gene encoding CYP79B2 were amplified from Col-0 wild-type cDNA using the primers listed in the Supplemental Table and subcloned into the BiFC vectors. All constructs were agroinfiltrated into 4- to 6-week-old wild tobacco leaves using working suspensions prepared by mixing a 1:1:1 ratio of three Agrobacterium strains carrying the YFPN fusion, the YFPC fusion, and the gene-silencing inhibitor pBA-HcPro, respectively. Fluorescence images were captured with a Leica TCS SP5 laser-scanning confocal microscope at 4 d post infiltration with excitation at 514 nm and emission wavelengths of 520 to 535 for YFP signals and 630 to 720 nm for chloroplast autofluorescence signals. Overlaid images of the fluorescence and bright field are shown.
Co-IP-Immunoblot Analyses
Wild tobacco leaves transiently coexpressing F5H-Myc and HA-CB5D, or F5H-Myc/HA-CB5D with the BiFC empty vector (as the control), were collected at 4 d post infiltration. Total proteins were extracted from homogenized wild tobacco leaves in buffer containing 50 mM Tris-HCl (pH 7.5), 2 mM EDTA, 150 mM NaCl, 10% glycerol, 0.2% 2-mercaptoethanol, 1% Triton X-100, 2% polyvinylpolypyrrolidone, and 1× complete protease inhibitor cocktail. EZviewRed Anti-HA Affinity Gel Beads (E6779; Sigma-Aldrich) were used for immunoprecipitation. The affinity beads were washed six times with washing buffer (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.25% nonyl phenoxypolyethoxylethanol-40, and 1× protease inhibitor cocktail). The immunoprecipitated proteins on the affinity beads were eluted by boiling with 2× protein loading buffer and subjected to 10% SDS-PAGE. Immunoblot analysis was performed according to the ECL Western Blotting procedure (GE Healthcare) using commercial anti-HA antibody (HA.11 clone 16B12; Covance) at 1:2000 dilution and anti-Myc antibody (c-Myc [9E10]: sc-40; Santa Cruz Biotechnology) at 1:1000 dilution.
RNA Extraction and qRT-PCR Analysis
To analyze CB5D expression in different Arabidopsis tissues, roots and leaves were collected from ∼1-month-old Col-0 wild-type plants grown in a growth chamber with the conditions described above; stems and flowers were sampled from ∼40-d-old plants, while siliques were obtained from ∼50-d-old plants. The tissues from five individual plants were pooled, representing one biological replicate. For seedlings (used in all qRT-PCR analyses), 2-week-old seedlings grown on 0.5× Murashige and Skoog medium were collected. Approximately 12 seedlings were pooled as one biological replicate for RNA preparation. RNA was extracted with TRIzol reagent (Thermo Fisher Scientific) and cleaned with an RNA Cleanup kit (Qiagen). Before use, the RNA was further treated with DNase I (Sigma-Aldrich) to remove contaminating DNA. The qRT-PCR analysis was performed as described previously (Gou et al., 2017). The analysis was conducted with three biological replicates. Each biological replicate was performed with three to four technical repeats (detailed in the figure legends for different experiments). The data were calculated using the delta-delta-cycle threshold method using UBIQUITIN10 as the housekeeping reference gene. The data from three to four technical repeats were averaged as a single value for each biological replicate. Statistical analysis was performed using two-tailed Student’s t tests based on the biological replicate data.
Quantification of Soluble Phenolics
Soluble phenolics were quantified using the Agilent 1100 HPLC system as previously reported (Gou et al., 2018). Briefly, 0.5 g (fresh weight) of tissue from 2-week-old seedlings or 4-week-old leaves from approximately five plants were grouped as one biological replicate, ground into a powder in liquid N2, and extracted overnight at 4°C with 2 mL of 80% (v/v) methanol containing 80 µM chrysin as the internal standard. Three or more biological replicates were conducted per experiment. Twenty-five microliters of extract was injected into an Agilent 1100 HPLC system with a gradient of solvent B (0.1% acetic acid in acetonitrile) in solvent A (0.1% acetic acid in water) as follows: 5% B (at 2 min); 50% B (at 30 min); 100% B (at 32 min); 100% B (at 34 min); and 5% B (at 35 min), at a linear flow rate of 1 mL/min in a reverse-phase C18 column [Luna C18 (2), 5 mm; Phenomenex]. The wavelengths used for detection were 280, 254, 310, 330, and 510 nm, and diode array detector signal was recorded from 200 to 800 nm for peak characterization. To quantify the contents of soluble phenolics, the UV light absorption area of a particular peak from each sample was normalized to that of the internal standard (chrysin) and calibrated using a standard curve of sinapic acid (for sinapoyl malate and sinapoyl glucose). Statistical analysis was performed using one-way ANOVA (Tukey’s Honestly Significant Difference [HSD] test) based on the data from three or more biological replicates (Supplemental File 2).
Flavonoid quercetin was extracted from mature seeds . Briefly, 1.5 mg of seed tissue (as one biological replicate) was thoroughly ground in a 2-mL Wheaton grinder with 0.4 mL of acetonitrile:water (75:25) at 4°C. Eighty micromolar chrysin was introduced at the beginning of the extraction as an internal standard. MilliQ water (0.2 mL) was then added, and the extraction was completed by sonication for 30 min. After centrifugation at 13,000 rpm for 15 min, 400 μL of the supernatant was treated with 2 n HCl at 100°C for 1 h. The supernatant was concentrated under N2 gas and extracted twice in an equal volume of water-saturated ethyl acetate. The solvent extracts were combined and dried under an N2 gas stream, and the pellet was dissolved in 100 μL of 80% methanol. Twenty microliters of extract was injected into an Agilent 1100 HPLC system with a gradient described above for the analysis. Peaks were identified against authentic standards and based on their UV light absorption spectra. Relative abundances were calculated based on the peak area of UV light absorbance. Statistical analysis was performed using two-tailed Student’s t tests based on the data from four biological replicates.
α-Pyrone extraction was performed as previously reported (Weng et al., 2012). Briefly, the primary inflorescence stems from three to four 6-week-old plants were grouped as a biological replicate and ground into a fine powder. Approximately 0.1 g of powder was combined with 4 volumes of water (approximately 400 µL). The sample was extracted at 65°C for 2 h. The supernatant was collected after centrifugation at 12,000g for 30 min. Ten microliters of supernatant was injected for LC-UV-MS analysis with a Dionex Ultimate 3000 ultra-high performance liquid chromatography (UHPLC) system attached to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific). Four biological replicates were conducted. The product was resolved chromatographically through a UHPLC C18 column (Luna, 150 mm × 2.1 mm, 1.6 µm; Phenomenex) at room temperature with a flow rate of 0.5 mL/min. The elution solvents were water with 0.1% acetic acid (A) and acetonitrile with 0.1% acetic acid (B). The elution process was set as follows: 0 to 2 min, 2% B; 2 to 17 min, 2 to 10% B; 17 to 19 min, 10 to 99% B; 19 to 20 min, 99% B; 20 to 21 min, 99 to 2% B. A full mass scan in negative mode was performed in the range of m/z 100 to 2000. The mass spectrometer parameters were set as follows: sheath gas flow rate 40 units (negative mode); auxiliary gas unit flow rate 10; capillary temperature at 350°C; auxiliary gas heater temperature at 250°C; spray voltage 3.5 kV; and S lens RF level at 60. Peaks were identified based on UV light spectra and mass spectra of arabidopyrones reported previously (Weng et al., 2012). Relative abundances were based on the peak area of UV light absorbance at 317 nm. Statistical analysis was performed using two-tailed Student’s t tests based on the data from four biological replicates.
Histochemical Analysis
To histochemically monitor lignin content, the first basal nodes (1 cm above the ground) of 7-week-old stems of Arabidopsis plants were fresh-frozen, cut into cross sections at 50 or 80 μm thickness with a cryomicrotome (Leica CM1950), and stained with phloroglucinol-HCl (Wiesner) and Mӓule reagents as described (Pradhan Mitra and Loqué, 2014).
Cell Wall Preparation and Lignin Analysis
To measure total lignin content and monomeric lignin composition, extractive-free cell walls were prepared from 14-week-old fully mature stems with three biological replicates; each replicate included at least six plants. The total lignin content was measured using the acetyl bromide method, and monomeric composition was determined using the thioacidolysis method and quantified with gas chromatography-flame ionization detection as previously described (Zhang et al., 2012). Statistical analysis was performed using one-way ANOVA (Tukey’s HSD test) or two-tailed Student’s t tests based on the data from three biological replicates.
Complementation Test Using CB5D, CB5D Mutant Variants, and ATR2
The CB5D genomic fragment (including its native promoter, coding region, and 3′ untranslated region) was amplified from Arabidopsis genomic DNA using the primers PCR8LB-CB5DFLG-F and PCR8LB-CB5DFLG-R (Supplemental Table) and cloned into the pCR/GW/TOPO entry vector using the ligation-independent cloning method (Jeong et al., 2012). The fragment was then subcloned into binary vector pGWB501 (Nakagawa et al., 2007) by LR reaction (Invitrogen) to generate the pCB5D:CB5D expression construct. Fragments of the CB5D and ATR2 coding regions were also subcloned from the pDONR207 entry constructs into the pMDCpC4H vector to generate the pC4H:CB5D and pC4H:ATR2 constructs, respectively, by LR reaction. Both the pCB5D:CB5D and pC4H:CB5D constructs were transformed into Arabidopsis cb5d-1 and cb5d-2 plants using the floral dip method (Clough and Bent, 1998). pC4H:CB5D was also transformed into atr2-1 and pC4H:ATR2 was transformed into cb5d-1 plants. Site-directed mutagenesis of CB5D was performed following the manufacturer’s instructions (QuikChange II Site-Directed Mutagenesis Kit; Agilent) using the CB5D cDNA entry clone (in pDONR207) as the template and primers listed in the Supplemental Table, which generated the entry clones for CB5DΔ1 (with substitution of His-40 to Ala), CB5DΔ2 (His-64 to Ala), and CB5DΔ1 Δ2 (substitution of both His residues). These mutant fragments were then subcloned into pMDCpC4H vector to generate the pC4H:CB5DΔ1, pC4H:CB5DΔ1, and pC4H:CB5DΔ1Δ2 constructs. All constructs were transformed into homozygous cb5d-1 to generate the corresponding transgenic plants.
Plant Microsome Preparation, Immunoblot Analysis, and Enzymatic Activity Assays
Approximately 1 g of approximately 2-week-old Arabidopsis seedlings grown on 0.5× Murashige and Skoog plates was collected as one biological replicate for microsomal protein extraction. Seedlings were ground into a fine powder in liquid nitrogen with a mortar and pestle and resuspended in the following extraction buffers: for the C4H assay, 12 mL of 100 mM potassium phosphate buffer (pH 7.5) containing 0.4 M Suc and 5 mM 2-mercaptoethanol; for the F5H assay, 10 mL of 50 mM Tris-HCl (pH 7.4) with 2 mM EDTA and 0.6 M sorbitol, followed by rotation at 4°C for 1 h. The homogenate was centrifuged twice at 10,000g for 15 min and passed through two layers of Miracloth (Calbiochem) to remove the floating debris after the first centrifugation. The supernatant was centrifuged at 100,000g for 1 h, and the pellet was resuspended in 100 μL of reaction buffer (100 mM potassium phosphate buffer [pH 7.0] for the C4H assay or 100 mM HEPES [pH 7.4] with 1 mM EDTA for the F5H assay). The protein concentration was measured using a Quick Start Bradford Protein Assay kit (Bio-Rad).
Five micrograms of microsomal proteins per sample was used for immunoblotting, which was performed according to the enhanced chemiluminescence immunoblotting procedure using Super Signal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). The polypeptide polyclonal antibodies used in immunoblots for C4H, C3′H, and F5H were developed by Pacific Immunology as previously described (Gou et al., 2018).
For the enzyme activity assay, 100 μL of reaction mixture in the corresponding reaction buffer with 40 µg of microsomes, 0.2 mM substrate (cinnamic acid for C4H or coniferaldehyde for F5H), and 1 mM NADPH or NADH were incubated at 30°C for 30 min (for the C4H assay) or 90 min (for the F5H assay). The reaction was stopped and partitioned twice with 100 μL of water-saturated ethyl acetate. The combined organic phase was evaporated under a vacuum and dissolved in 50 μL of 80% methanol. Ten microliters of the sample was injected for LC-UV-MS analysis with a Dionex Ultimate 3000 UHPLC system attached to a Thermo Scientific Q-Exactive Plus mass spectrometer. The product was resolved chromatographically through a UHPLC C18 column (Luna, 150 mm × 2.1 mm, 1.6 µm; Phenomenex) at room temperature with a flow rate of 0.2 mL/min. The elution solvents were water with 0.1% acetic acid (A) and acetonitrile with 0.1% acetic acid (B). The elution process was set as follows: 0 to 20 min, 5 to 50% B; 20 to 22 min, 50 to 90% B; 22 to 24 min, 99% B; 24 to 25 min, 99 to 5% B. MS scan was set in the range of m/z 100 to 1500. The mass spectrometer parameters were as follows: sheath gas flow rate 35 units (positive mode) or 40 units (negative mode); auxiliary gas unit flow rate 10; capillary temperature at 350°C; auxiliary gas heater temperature at 250°C; spray voltage 3.5 kV; and S lens RF level at 60. The product coumaric acid was characterized against an authentic standard, and 5-hydroxyconiferaldehyde was determined by comparing the UV light and mass spectra with those reported previously (Osakabe et al., 1999).
To calculate C4H and F5H activity, the molar amount of detected substrate (ns) and product (np) was calculated based on UV light absorbance using standard curves for cinnamic acid, p-coumaric acid, or coniferaldehyde; the coniferaldehyde standard curve was also used to calculate the level of 5-hydroxy coniferaldehyde product. Catalytic activity (α) was calculated using the equation α = ni * np/(ns + np)/t/m, where ni represents the initial amount of substrate, t represents the reaction time, and m represents the amount of total microsomal protein. Every reaction in each batch of experiments was repeated three times with biological replicates, and at least three batches of experiments were conducted. Statistical analysis was performed using one-way ANOVA (Tukey’s HSD test) based on the data from three replicates.
Phylogenetic Analysis
The AtCB5D sequence was used to search the nucleotide collection in the National Center for Biotechnology Information using the BLASTn algorithm with default settings. The obtained homologous sequences together with the documented least divergent orthologs in the PANTHER classification system database (http://www.pantherdb.org/) were subjected to multiple sequence alignment, which was performed with ClustalW. An unrooted phylogenetic tree was initially constructed by the neighbor-joining method with 1000 bootstrap replicates using MEGA v.7.0 software with default parameters (Kumar et al., 2016). The initial tree was carefully inspected, and the sequences in the CB5D clade showing node support values less than 50% in the replicate trees in the bootstrap test (1000 replicates) were eliminated. Eventually, the remaining sequences together with other Arabidopsis CB5 family members as well as the homologous sequences from distant land plants were aligned again using the ClustalW program and subjected to phylogenetic analysis with the MEGA v.7.0 program using the neighbor-joining method with default settings (Kumar et al., 2016).
Protein Homology Modeling
The CB5D amino acid sequence was used as a query to search the Protein Data Bank (PDB) database, which resulted in a hit on the human CB5B crystal structure in the database (PDB ID: 3NER). The homology model was then built using the Swiss model online service program (https://swissmodel.expasy.org/interactive#structure) using 3NER as a template. The resulting PDB was examined, and the figures were processed using the PyMol Molecular Graphics System (version 2.0; Schrödinger).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AF332415.1 (AtCB5A, At1g26340), BT000085.1 (AtCB5B, At2g32720), BT025600.1 (AtCB5C, At2g46650), AY063073.1 (AtCB5D, At5g48810), AY114045.1 (AtCB5E, At5g53560), NM_104749.3 (AtCB5F, At1g60660), NM_001342001 (ATR2, AT4G30210), NM_128601 (C4H, AT2G30490), and U38416 (F5H, AT4G36220).
The proteomics data are available in the EMBL-EBI PRIDE archive (http://www.ebi.ac.uk/pride) under accession number PXD009033.
Supplemental Data
Supplemental Figure 1. Electron transfer chains involving cytochrome b5.
Supplemental Figure 2. Characterization of the cb5d and atr2-1 single mutants and the cb5d atr2-1 double mutant.
Supplemental Figure 3. Levels of ɑ-pyrone, flavonoids, and sinapoyl esters in cb5d and atr2.
Supplemental Figure 4. Sinapoyl malate content in t1 complementation lines of CB5D.
Supplemental Figure 5. Characterization of ATR2 overexpression lines in the cb5d-1 background.
Supplemental Figure 6. Characterization of CB5D overexpression lines in the atr2-1 background.
Supplemental Figure 7. Characterization of the C4H reaction product using UHPLC-Mass spectrometry.
Supplemental Figure 8. Characterization of the F5H enzymatic reaction product using UHPLC-mass spectrometry.
Supplemental Table. Primers used in this study.
Supplemental File 1. Alignment of CB5D homologous sequences.
Supplemental File 2. ANOVA table.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Antonius Koller and Dwight Martin from the Stony Brook University Proteomics Center for LC-MS analysis of protein complexes. This work was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences through the Physical Biosciences program of the Chemical Sciences, Geosciences, and Biosciences Division (DE-SC0012704 to C.-J.L.). The research used confocal microscope of the Center for Functional Nanomaterials, a U.S. DOE Office of Science Facility, at Brookhaven National Laboratory under the same contract number. X.Y. and X.R. were partially supported by a scholarship from the China Scholarship Council.
AUTHOR CONTRIBUTIONS
C.-J.L. and M.G. designed the experiments. M.G., X.Y., Y.Z., X.R., and Y.S. conducted experiments. C.-J.L. and M.G. analyzed and interpreted data and wrote the article. All authors edited the article.
Footnotes
Articles can be viewed without a subscription.
References
- Backes W.L., Kelley R.W. (2003). Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol. Ther. 98: 221–233. [DOI] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cowley A.B., Altuve A., Kuchment O., Terzyan S., Zhang X., Rivera M., Benson D.R. (2002). Toward engineering the stability and hemin-binding properties of microsomal cytochromes b5 into rat outer mitochondrial membrane cytochrome b5: Examining the influence of residues 25 and 71. Biochemistry 41: 11566–11581. [DOI] [PubMed] [Google Scholar]
- Dailey H.A., Strittmatter P. (1979). Modification and identification of cytochrome b5 carboxyl groups involved in protein-protein interaction with cytochrome b5 reductase. J. Biol. Chem. 254: 5388–5396. [PubMed] [Google Scholar]
- de Vetten N., ter Horst J., van Schaik H.P., de Boer A., Mol J., Koes R. (1999). A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 96: 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Chen F., Guo D., Parvathi K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57: 1069–1084. [DOI] [PubMed] [Google Scholar]
- Enoch H.G., Strittmatter P. (1979). Cytochrome b5 reduction by NADPH-cytochrome P-450 reductase. J. Biol. Chem. 254: 8976–8981. [PubMed] [Google Scholar]
- Fraser C.M., Chapple C. (2011). The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M., Mizutani M., Tanaka Y., Kusumi T., Ohta D. (1999). Microsomal electron transfer in higher plants: Cloning and heterologous expression of NADH-cytochrome b5 reductase from Arabidopsis. Plant Physiol. 119: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.R., Hänsch R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Gou M., Hou G., Yang H., Zhang X., Cai Y., Kai G., Liu C.J. (2017). The myb107 transcription factor positively regulates suberin biosynthesis. Plant Physiol. 173: 1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Ran X., Martin D.W., Liu C.J. (2018). The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat. Plants 4: 299–310. [DOI] [PubMed] [Google Scholar]
- Hannemann F., Bichet A., Ewen K.M., Bernhardt R. (2007). Cytochrome P450 systems: Biological variations of electron transport chains. Biochim. Biophys. Acta 1770: 330–344. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.T., Pelitire S.M., Henderson M.P., Andrews D.W., Dyer J.M., Mullen R.T. (2004). Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell 16: 3002–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S.C., Waskell L. (2011). The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch. Biochem. Biophys. 507: 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson I., Schenkman J.B. (1977). Studies on three microsomal electron transfer enzyme systems: Specificity of electron flow pathways. Arch. Biochem. Biophys. 178: 89–107. [DOI] [PubMed] [Google Scholar]
- Jensen K., Møller B.L. (2010). Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71: 132–141. [DOI] [PubMed] [Google Scholar]
- Jeong J.Y., Yim H.S., Ryu J.Y., Lee H.S., Lee J.H., Seen D.S., Kang S.G. (2012). One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78: 5440–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen K., Rasmussen A.V., Morant M., Nielsen A.H., Bjarnholt N., Zagrobelny M., Bak S., Møller B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291. [DOI] [PubMed] [Google Scholar]
- Kandel S.E., Lampe J.N. (2014). Role of protein-protein interactions in cytochrome P450-mediated drug metabolism and toxicity. Chem. Res. Toxicol. 27: 1474–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Wallis J.G., Skidmore C., Browse J. (2006). A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J. 48: 920–932. [DOI] [PubMed] [Google Scholar]
- Kumar R., Tran L.S., Neelakandan A.K., Nguyen H.T. (2012). Higher plant cytochrome b5 polypeptides modulate fatty acid desaturation. PLoS One 7: e31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C., Monties B., Rolando C. (1985). Thioacidolysis of lignin: Comparison with acidolysis. J. Wood Chem. Technol. 5: 277–292. [Google Scholar]
- Liu C.J. (2010). Biosynthesis of hydroxycinnamate conjugates: Implications for sustainable biomass and biofuel production. Biofuels 1: 745–761. [Google Scholar]
- Liu C.J. (2012). Deciphering the enigma of lignification: Precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 5: 304–317. [DOI] [PubMed] [Google Scholar]
- Liu D.Y., Smith P.M., Barton D.A., Day D.A., Overall R.L. (2017). Characterisation of Arabidopsis calnexin 1 and calnexin 2 in the endoplasmic reticulum and at plasmodesmata. Protoplasma 254: 125–136. [DOI] [PubMed] [Google Scholar]
- Maggio C., Barbante A., Ferro F., Frigerio L., Pedrazzini E. (2007). Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: A comparative study using different isoforms from rabbit and Arabidopsis. J. Exp. Bot. 58: 1365–1379. [DOI] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Kondo M., Shimada T., Nishimura M., Hara-Nishimura I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130: 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Cusumano J.C., Somerville C., Chapple C.C. (1996). Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc. Natl. Acad. Sci. USA 93: 6869–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Shirley A.M., Cusumano J.C., Bell-Lelong D.A., Chapple C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 6619–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa G.T., Lu A.Y. (1984). The association of cytochrome P-450 and NADPH-cytochrome P-450 reductase in phospholipid membranes. Arch. Biochem. Biophys. 234: 161–166. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Napier J.A., Smith M.A., Stobart A.K., Shewry P.R. (1995). Isolation of a cDNA encoding a cytochrome b5 specifically expressed in developing tobacco seeds. Planta 197: 200–202. [DOI] [PubMed] [Google Scholar]
- Noshiro M., Harada N., Omura T. (1979). Immunochemical study on the participation of cytochrome b5 in drug oxidation reactions of mouse liver microsomes. Biochem. Biophys. Res. Commun. 91: 207–213. [DOI] [PubMed] [Google Scholar]
- Osakabe K., Tsao C.C., Li L., Popko J.L., Umezawa T., Carraway D.T., Smeltzer R.H., Joshi C.P., Chiang V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96: 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshino N., Imai Y., Sato R. (1971). A function of cytochrome b5 in fatty acid desaturation by rat liver microsomes. J. Biochem. 69: 155–167. [DOI] [PubMed] [Google Scholar]
- Porter T.D. (2002). The roles of cytochrome b5 in cytochrome P450 reactions. J. Biochem. Mol. Toxicol. 16: 311–316. [DOI] [PubMed] [Google Scholar]
- Pradhan Mitra P., Loqué D. (2014). Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Katagiri F. (2009). Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 57: 932–944. [DOI] [PubMed] [Google Scholar]
- Rahier A., Smith M., Taton M. (1997). The role of cytochrome b5 in 4alpha-methyl-oxidation and C5(6) desaturation of plant sterol precursors. Biochem. Biophys. Res. Commun. 236: 434–437. [DOI] [PubMed] [Google Scholar]
- Schenkman J.B., Jansson I. (1999). Interactions between cytochrome P450 and cytochrome b5. Drug Metab. Rev. 31: 351–364. [DOI] [PubMed] [Google Scholar]
- Schenkman J.B., Jansson I. (2003). The many roles of cytochrome b5. Pharmacol. Ther. 97: 139–152. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782. [DOI] [PubMed] [Google Scholar]
- Scott E.E., et al. (2016). The role of protein-protein and protein-membrane interactions on P450 function. Drug Metab. Dispos. 44: 576–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Cross A.R., Jones O.T., Griffiths W.T., Stymne S., Stobart K. (1990). Electron-transport components of the 1-acyl-2-oleoyl-sn-glycero-3-phosphocholine delta 12-desaturase (delta 12-desaturase) in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons. Biochem. J. 272: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Jonsson L., Stymne S., Stobart K. (1992). Evidence for cytochrome b5 as an electron donor in ricinoleic acid biosynthesis in microsomal preparations from developing castor bean (Ricinus communis L.). Biochem. J. 287: 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L., Vanholme R., Geerinck J., Goeminne G., Höfer R., Kim H., Ralph J., Boerjan W. (2014). Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol. 166: 1956–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P., Mignotte C., Kazmaier M., Delorme F., Pompon D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272: 19176–19186. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergéres G., Waskell L. (1995). Cytochrome b5, its functions, structure and membrane topology. Biochimie 77: 604–620. [DOI] [PubMed] [Google Scholar]
- Watanabe J., Asaka Y., Fujimoto S., Kanamura S. (1993). Densities of NADPH-ferrihemoprotein reductase and cytochrome P-450 molecules in the endoplasmic reticulum membrane of rat hepatocytes. J. Histochem. Cytochem. 41: 43–49. [DOI] [PubMed] [Google Scholar]
- Wayne L.L., Wallis J.G., Kumar R., Markham J.E., Browse J. (2013). Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell 25: 3052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Li X., Stout J., Chapple C. (2008). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105: 7887–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Akiyama T., Bonawitz N.D., Li X., Ralph J., Chapple C. (2010). Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Li Y., Mo H., Chapple C. (2012). Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337: 960–964. [DOI] [PubMed] [Google Scholar]
- Zhang K., Bhuiya M.W., Pazo J.R., Miao Y., Kim H., Ralph J., Liu C.J. (2012). An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 24: 3135–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gou M., Liu C.J. (2013). Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25: 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]