A functional allele of FRUITFULL MADS-box (CsFUL1) gene regulates fruit length by inhibiting the activity of CsSUP and auxin transporters CsPIN1/7 in cucumber.
Abstract
Fruit length is a prominent agricultural trait during cucumber (Cucumis sativus) domestication and diversifying selection; however, the regulatory mechanisms of fruit elongation remain elusive. We identified two alleles of the FRUITFULL (FUL)–like MADS-box gene CsFUL1 with 3393C/A Single Nucleotide Polymorphism variation among 150 cucumber lines. Whereas CsFUL1A was specifically enriched in the long-fruited East Asian type cucumbers (China and Japan), the CsFUL1C allele was randomly distributed in cucumber populations, including wild and semiwild cucumbers. CsFUL1A knockdown led to further fruit elongation in cucumber, whereas elevated expression of CsFUL1A resulted in significantly shorter fruits. No effect on fruit elongation was detected when CsFUL1C expression was modulated, suggesting that CsFUL1A is a gain-of-function allele in long-fruited cucumber that acts as a repressor during diversifying selection of East Asian cucumbers. Furthermore, CsFUL1A binds to the CArG-box in the promoter region of SUPERMAN, a regulator of cell division and expansion, to repress its expression. Additionally, CsFUL1A inhibits the expression of auxin transporters PIN-FORMED1 (PIN1) and PIN7, resulting in decreases in auxin accumulation in fruits. Together, our work identifies an agriculturally important allele and suggests a strategy for manipulating fruit length in cucumber breeding that involves modulation of CsFUL1A expression.
INTRODUCTION
During crop domestication, plants with superior agronomic traits such as high yield, robust architecture, and ease of harvest have been deliberately selected for human consumption and environmental adaptation (Doebley et al., 2006). Fruits, the major product in most horticultural crops, play essential roles in our daily diet and for seed dispersal (Seymour et al., 2013). Fruit size, which is determined by fruit length, diameter, or the length:diameter ratio, was under intensive selection during crop domestication or diversification (van der Knaap et al., 2014). Despite reports of several genes controlling fruit size (Frary et al., 2000; Xu et al., 2015; van der Knaap and Østergaard, 2018), molecular insights into fruit length regulation are limited.
Fruits can be generally classified into dry fruits and fleshy fruits, and fruit development is well characterized in model plants such as Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum; Seymour et al., 2013). The Arabidopsis fruit belongs to a type of dry fruit, named the silique, comprising a valve (seedpod walls), replum (central ridge between the valves), and valve margin where the valves detach from the replum during fruit opening (Roeder et al., 2003). A complex network of transcription factors has been identified to regulate fruit opening for seed dispersal in Arabidopsis (Dinneny et al., 2005). The basic helix-loop-helix genes INDEHISCENT (IND) and ALCATRAZ (ALC) as well as the MADS-box genes SHATTERPROOF1 (SHP1) and SHP2 control the development of the valve margin (Liljegren et al., 2000, 2004; Rajani and Sundaresan, 2001). The MADS-box gene FRUITFULL (FUL) is specifically expressed in the valve and restricts the expression of IND, ALC, and SHP1/2 in the valve margin (Gu et al., 1998; Ferrándiz et al., 2000; Liljegren et al., 2000). Siliques of the fruitfull (ful) mutant are short, bumpy, and open irregularly due to ectopic expression of the valve margin genes in the valves (Gu et al., 1998). Tomato bears fleshy berry fruits. Similar genes have been found to participate in tomato fruit development, but they display distinct functions compared with their Arabidopsis counterparts. For example, TDR4/FUL1 and MBP7/FUL2 function redundantly to regulate fruit ripening through forming heterodimers with ripening factor RIPENING INHIBITOR (RIN), and these complexes fine-tune ethylene biosynthesis and the expression of ripening-related genes in tomato (Vrebalov et al., 2002, 2009; Bemer et al., 2012; Shima et al., 2013; Fujisawa et al., 2014; Wang et al., 2014). Tomato AGAMOUS-LIKE1 (TAGL1, the putative ortholog of SHP1/2) regulates both expansion and ripening of fleshy fruits via the ethylene-dependent pathway, and expression of RIN and TAGL1 is downregulated by FUL1/2 (Vrebalov et al., 2009; Bemer et al., 2012). In addition, SUN, OVATE, LOCULE NUMBER, and FASCIATED modulate fruit shape and size in tomato (Rodríguez et al., 2011).
Cucumber (Cucumis sativus) is an important vegetable crop bearing pepo fruits (a modified berry fruit) that are harvested immaturely and consumed freshly or processed as pickles (Pan et al., 2017). Global production was 80.6 million tons in 2016, of which China accounts for 76.8% (Food and Agriculture Organization Corporate Statistical Database). The typical cucumber fruit is roughly cylindrical with tapered ends. Fruit length is a key domestication trait in cucumber (Qi et al., 2013), and it displays tremendous variation, from 5 to 60 cm in length (Sebastian et al., 2010; Yang et al., 2012). Indigenous to India, cucumber was domesticated over 3000 years ago from its wild relative C. sativus var hardwickii, which bears small and spheroid fruits (3 to 5 cm in length; Figure 1A; Sebastian et al., 2010; Yang et al., 2012). Based on molecular marker data, cucumbers could be roughly divided into several geographic groups such as Indian, Xishuangbanna, Eurasian, and East Asian (Qi et al., 2013; Wang et al., 2018). Morphologically, cucumbers in the East Asian group (North China type or Japanese Long) have characteristically long fruit (>35 cm; Figure 1A; Qi et al., 2013). Previous studies have identified at least 10 quantitative trait loci (QTLs) underlying fruit-length variation in cucumber (Bo et al., 2015; Weng et al., 2015; Pan et al., 2017); however, no fruit-length regulator except CsSUN has been cloned (Pan et al., 2017), and none of the identified quantitative-trait loci has been functionally validated.
Figure 1.
Allelic Variation of CsFUL1 in Cucumber.
(A) Representative fruits of the wild cucumber C. sativa var hardwickii (left) and the East Asian type cucumber ‘Chinese Long’ (right). Bar = 5 cm.
(B) Boxplots of distribution of fruit length in 150 cucumber lines harboring the CsFUL1A or CsFUL1C allele. Median values are shown by horizontal lines within boxes, and the range of the 25% to 75% of the data is represented by box height (P = 1.83E−17). The P-value is calculated based on two-tailed Student's t tests.
(C) The C/A SNP at 3393 in the sixth exon caused an amino acid change from Gln (Q) to Lys (K). Boxes represent exons and lines represent introns.
(D) Lys-168 (K) in the K domain of CsFUL1 and alternatively conserved Gln (Q) in that of other species (red box). Strictly conserved residues are shaded in black, and partly conserved residues are shaded in gray. At, Arabidopsis thaliana, AT5G60910; Cla, Citrullus lanatus, Cla018544; Cm, Cucumis melo, XP_008453619.1; Cs, Cucumis sativus, Csa1G039910; Nt, Nicotiana tabacum, ABF82231; Os, Oryza sativa, AGX01588; Ph, Petunia hybrida, AAF19164; Rh, Rosa hybrid cultivar, ACS74808; Sl, Solanum lycopersicum, NP_001234173 and AAP83380; Vv, Vitis vinifera, NP_001268211; Zm, Zea mays, ABW84393.
(E) and (F) Predicated three dimensional (3D) model of CsFUL1C protein (E) and the enlarged view of the helix (boxed in [E]) where residue Gln-168 (Q168) is situated (F).
Here, we identified two allelic variants of the FUL MADS-box gene CsFUL1, with the CsFUL1A allele being specifically present in the long-fruited East Asian cucumbers from China and Japan. Unlike its FUL homologs regulating fruit opening in Arabidopsis and fruit ripening in tomato (Gu et al., 1998; Bemer et al., 2012), the CsFUL1A allele is identified here as a gain-of-function allele that acts as a transcriptional repressor via reducing SUPERMAN CsSUP-mediated cell division and expansion and limitingCsPIN1/CsPIN7-mediated auxin transport during cucumber elongation.
RESULTS
A Unique SNP Allele in the K Domain of CsFUL1 Was Selected in East Asian Type Cucumber with Long Fruits
To identify genes regulating fruit elongation in cucumber, we analyzed the resequenced data of 150 cucumber lines with varying fruit length. Among these lines, two alleles with 3393C/A SNP variations of the FUL MADS-box gene (Csa1G039910, denoted hereafter as CsFUL1) were identified, in which the CsFUL1A allele was specifically present in the long-fruited East Asian cucumbers from North China and Japan, while the CsFUL1C allele was shared among wild and semiwild varieties, cultivated landraces, and other market classes (Supplemental Data Set 1). The average fruit length of CsFUL1A lines was 34.4 ± 7.3 cm (n = 32), a length that was significantly longer than that of CsFUL1C lines, with an average fruit length of 18.6 ± 8.5 cm (n = 118; P = 1.83E−17; Figure 1B), suggesting that this SNP in CsFUL1 may have been selected and fixed in Asian long cucumbers during domestication and diversification (Qi et al., 2013).
A BLAST search identified two FUL genes in the cucumber draft genome, CsFUL1 and CsFUL2 (Csa4G126480). Phylogenetic analysis showed that CsFUL1 and AtFUL were in the same clade, while CsFUL2 and AGAMOUS-like79 (AGL79) belonged to a different clade (Supplemental Figure 1; Supplemental Data Set 2). CsFUL1 encodes a typical type II MADS-box protein with four modular domains (MIKC), the N-terminal DNA binding MADS domain (M), the intervening (I) and keratin-like (K) regions, and a C-terminal region (C). Despite general conservation of the MIKC organization, CsFUL1 and its homolog in cantaloupe (Cucumis melo) contain 13 additional amino acids in the I region (Supplemental Figure 2). The 3393C/A SNP was located in the sixth exon of CsFUL1, resulting in an amino acid change from Gln (Gln-168) to Lys (Lys-168; Figure 1C). Residue Gln-168 is highly conserved in the K domain in 11 distantly related species (Figure 1D). In cucumber, Gln-168 is located in the α helix of CsFUL1. Substitution of Gln-168 by Lys-168 is predicted to form a positively charged patch with the residues Lys-169 and Lys-172 in that helix (Figure 1E), which may affect interaction of CsFUL1 with other protein partners, promoter sequences, or trans-elements.
CsFUL1 Expression Pattern
We analyzed CsFUL1 expression in 12 cucumber inbred lines with various fruit lengths, in which seven North China type cucumber lines are homozygous for the CsFUL1A allele and five South China type cucumber lines are homozygous for the CsFUL1C allele (Figures 2A and 2B). At the commercial harvest stage (10 to 15 d after anthesis [DAA]), fruit length of CsFUL1A lines varied from 25 to 44 cm, while that of CsFUL1C lines varied from 8 to 17 cm (Figure 2B). Quantitative real-time PCR (RT-qPCR) analysis showed that CsFUL1 expression was significantly lower in lines harboring the CsFUL1A allele compared with those harboring the CsFUL1C allele. Within the CsFUL1A lines, CsFUL1 expression was negatively correlated with fruit length (correlation coefficient = 0.83; Figure 2B).
Figure 2.
Expression Analyses of CsFUL1 in Cucumber.
(A) Representative fruits of 12 cucumber lines at 10 DAA. Bar = 5 cm.
(B) Negative correlation of CsFUL1A expression with fruit length. Green bars indicate fruit length, orange and pink bars represent CsFUL1 expression in lines harboring the CsFUL1A or CsFUL1C allele, respectively. Three biological replicates from different plants were performed for each expression analysis. Error bars indicate se. Values are means ± se (n = 4 fruits).
(C) to (L) In situ hybridization of CsFUL1 in lines harboring the CsFUL1A allele. (C) The shoot apex of 12-d-old plant. (D) Stage 2 floral bud. (E) and (H) Stage 3 floral bud. (F) and (I) Longitudinal section of a stage 8 female flower. (G) and (J) Cross sections of stage 9 ovaries. (K) and (L) Negative controls hybridized with the sense probe. Bars = 50 µm.
Next, we performed RT-qPCR and in situ hybridization to analyze CsFUL1 expression in lines homozygous for the CsFUL1A allele. CsFUL1 is highly expressed in the male and female flowers at anthesis, followed by flower buds and young fruits (Supplemental Figure 3A). Among fruit tissues at 10 DAA, accumulation of CsFUL1 transcripts gradually decreased from the exocarp to ventricle (Supplemental Figure 3B). mRNA in situ hybridization showed that CsFUL1 is expressed throughout the floral meristem at the early stages (Figure 2C) and then becomes restricted to the central region where the stamen and carpel primordia will initiate (Figures 2D and 2E). In the female flower, CsFUL1 is expressed in the developing ovary, the vascular tissues, and the ovule (Figures 2F and 2G). No signal was detected when hybridization was performed with the sense probe of CsFUL1 (Figure 2K). Subcellular localization analysis showed that CsFUL1 is localized to the nucleus (Supplemental Figure 3C), consistent with its role as a transcription factor. CsFUL2 displayed a similar expression pattern as CsFUL1, with high expression levels in male flowers and fruits (Supplemental Figures 4A to 4G). However, no correlation was found between the expression of CsFUL2 and fruit length variation in cucumber, neither between that of the homologs of the FUL-interacting genes cucumber HECATE3 (CsHEC3) and CsALC and fruit length variation (Supplemental Figures 4H to 4J; Ripoll et al., 2011).
Functional Divergence of CsFUL1 and FUL in Arabidopsis
To explore whether CsFUL1 is functionally similar to FUL in Arabidopsis, we first transformed both CsFUL1A and CsFUL1C alleles driven by the 35S promoter into the Arabidopsis ful-1 mutant. FUL promotes fruit opening by repressing valve margin identity genes, and the silique in the ful-1 mutant is short and fails in dehiscence due to a disrupted patterning of lignification (Gu et al., 1998). This mutant fruit phenotype of ful-1 plants is not rescued by ectopic expression of either CsFUL1A or CsFUL1C (Figure 3A to 3E), suggesting that there is functional divergence between FUL in Arabidopsis and CsFUL1 in cucumber. Instead, in 35S:CsFUL1A transgenic plants, but not in 35S:CsFUL1C plants, improper fruit opening was frequently observed at the valve, and the replum zone was reduced compared with that in ful-1 siliques (Figures 3A and 3B). Next, we transformed both 35S:CsFUL1A and 35S:CsFUL1C constructs into the wild-type Arabidopsis (Figures 3F to 3K). Overexpression of CsFUL1A led to reduced fruit length in Arabidopsis, while no difference was observed in fruit length in CsFUL1C overexpression lines (Figures 3F to 3K).
Figure 3.
Heterologous expression of CsFUL1 in Arabidopsis.
(A) to (E) Heterologous expression of CsFUL1A and CsFUL1C in ful-1 mutant Arabidopsis. (A) Representative silique images of 35S:CsFUL1A/ful-1 plants. (B) Scanning electron microscopic images of siliques in ful-1 and 35S:CsFUL1A/ful-1. Red lines indicate the replum zones. (C) RT-qPCR analysis of CsFUL1 in the 35S:CsFUL1A/ful-1 transgenic plants. (D) Representative images of fruits of 35S:CsFUL1C/ful-1 plants. (E) RT-qPCR analysis of CsFUL1 in 35S:CsFUL1C/ful-1 transgenic plants. Values are means ± se (n = 3).
(F) to (K) Overexpression of CsFUL1A and CsFUL1C in the wild-type Arabidopsis. (F) and (G) Representative images of fruits of 35S:CsFUL1A plants (F) and 35S:CsFUL1C plants (G). (H) and (I) RT-qPCR analysis of CsFUL1 in the 35S:CsFUL1A (H) and 35S:CsFUL1C (I) background. (J) and (K) Statistical analyses of fruit length in the 35S:CsFUL1A (J) and 35S:CsFUL1C lines (K). Values are means ± se (n = 30 fruits). Ler, Landsberg erecta.
Three independent samples from different plants were used for gene expression. Two-tailed Student's t tests were used to compare the transgenic lines and the corresponding controls (*P < 0.05; **P < 0.01). Bar = 1 mm in (A), (D), (F), and (G) and 100 µm in (B).
CsFUL1A Allele Negatively Regulates Fruit Length in Cucumber
To further differentiate the biological functions of CsFUL1A and CsFUL1C alleles in cucumber, we transformed the 35S:CsFUL1A and 35S:CsFUL1C constructs into the long-fruit inbred line R1407 homozygous for the A allele (Figure 4). In total, 20 and 15 stable transgenic lines were obtained for CsFUL1A and CsFUL1C, respectively. Three representative lines from each overexpression (OX) construct, A-OX-1, A-OX-4, and A-OX-29 for CsFUL1A and C-OX-26, C-OX-39, and C-OX-42 for CsFUL1C, were chosen for further characterization.
Figure 4.
Phenotypic Characterization of 35S:CsFUL1 in Long-Fruited Inbred Line R1407 Harboring the A Allele.
(A) to (D) Representative fruits of three CsFUL1A and CsFUL1C overexpression lines at 10 DAA (A, C) and at maturity (>30 DAA) (B, D). Bars = 5 cm. V/C, empty vector/control plants.
(E) and (F) CsFUL1 expression in young fruits of CsFUL1A (E) and CsFUL1C overexpression lines (F). Three independent samples from different plants were used for expression analysis. Values are means ± se.
(G) and (H) Fruit length quantification of CsFUL1A (G) and CsFUL1C overexpression lines (H). Values are means ± se (n = 4 fruits from different plants).
(I) and (J) Longitudinal sections of the fruit pericarp at maturity in CsFUL1A overexpression lines. Bars = 200 µm.
(K) Cell area in the fruit pericarp in CsFUL1A overexpression lines. Values are means ± se (n = 3 pericarps from different fruits).
Significance analysis was conducted with the two-tailed Student's t tests (*P < 0.05; **P < 0.01).
Our data showed that overexpression of CsFUL1A, but not CsFUL1C, resulted in reduced fruit length compared with control plants (Figures 4A to 4F). The short-fruit phenotype in 35S:CsFUL1A transgenic lines was discernible at anthesis (Supplemental Figure 5A), and the average fruit length at harvest (10 DAA) was 38.0 ± 1.2 cm (n = 25) for control plants and 24.0 ± 1.3 cm (n = 25) for the severe transgenic line A-OX-29 (Figure 4G). Fully mature fruits (>35 DAA) were also dramatically shorter but wider in CsFUL1A overexpression plants than in control plants (Figure 4B; Supplemental Figures 5B and 5C). Longitudinal sections of the fruit pericarp at maturity showed 16 to 30% smaller pericarp cells in 35S:CsFUL1A transgenic plants (Figures 4I to 4K), suggesting that CsFUL1A may regulate fruit length by mediating cell expansion and cell division. In the 35S:CsFUL1C transgenic plants, despite the expression of CsFUL1 being significantly increased (Figure 4F), no difference in fruit length or fruit diameter was detected between transgenic lines versus control plants (Figures 4C, 4D, and 4H; Supplemental Figures 5D to 5F).
To evaluate how downregulation of the two CsFUL1 alleles affects fruit length, a double-stranded RNA interference (RNAi) construct (CsFUL1-RNAi) containing 315 bp of the 3′ coding sequence (CDS) and an additional 121 bp of untranslated sequence of CsFUL1 were transformed into the long-fruit inbred line R1407 harboring the A allele and a short-fruit inbred line 32X harboring the C allele (Figure 5). Knockdown of CsFUL1A, but not CsFUL1C, resulted in increased fruit length compared with control plants (Figures 5A to 5D). In the CsFUL1A-RNAi lines, CsFUL1 expression was reduced by 67 to 79% (Figure 5E), without affecting the expression of the related gene CsFUL2 (Figure 5F). Compared with control plants (transformed with an empty vector), all CsFUL1A-RNAi lines produced differentially elongated fruits from 10 DAA to the fully mature stage (Figures 5A, 5B, 5I, and 5J). Fruits in the CsFUL1A-RNAi line 15 (46.0 ± 2.2 cm, n = 20) were 25% longer than control fruits at 10 DAA (36.7 ± 2.8 cm, n = 20; Figure 5I). Longitudinal sections of the fruit pericarp at maturity showed larger cells in CsFUL1A-RNAi transgenic plants (Figures 5K to 5M). However, in the CsFUL1C-RNAi plants, despite a similar degree of reduction of CsFUL1 expression (69 to 82%), fruit length was unaffected compared with controls (Figures 5C, 5D, 5G, and 5H). These data suggested that only the CsFUL1A allele, not the CsFUL1C allele, functions as a repressor of cucumber fruit elongation.
Figure 5.
CsFUL1A-RNAi Resulted in Longer Fruits in Inbred Line R1407 with the A Allele.
(A) to (D) Representative fruits at 10 DAA (A, C) and at maturity (B, D) in CsFUL1A-RNAi and CsFUL1C-RNAi lines. Bars = 5 cm. V/C, empty vector/control plants.
(E) to (G) CsFUL1 and CsFUL2 expression in young fruits of CsFUL1A-RNAi plants (E, F) and CsFUL1C-RNAi plants (G) as detected by RT-qPCR. Three independent samples from different plants were used for expression analysis. Values are means ± se. V/C, empty vector/control plants.
(H) to (J) Fruit length at 10 DAA and at maturity. Values are means ± se (n = 4 fruits). V/C, empty vector/control plants.
(K) and (L) Longitudinal sections of fruits at maturity in control (K) and CsFUL1A-RNAi (L) lines. Bars = 200 µm. V/C, empty vector/control plants.
(M) Cell area in the fruit pericarp of CsFUL1A-RNAi plants. Values are means ± se (n = 3 pericarps).
Significance analysis was conducted using two-tailed Student's t tests (*P < 0.05; **P < 0.01). V/C, empty vector/control plants.
Transcriptome Analysis Revealed CsSUP, Auxin Transporters, and Cell Division–Related Genes as Candidate Genes Regulated by CsFUL1A
To dissect how CsFUL1A regulates fruit length, transcriptome analysis was performed with ovaries 4 d before anthesis of CsFUL1A overexpression line A-OX-29 and control plants to identify putative downstream targets of CsFUL1. Differentially expressed genes (DEGs) were identified using the false discovery rate set at <0.05 and fold change > 2 as cutoffs. In total, 3374 genes were upregulated and 4586 genes were downregulated in OX-29 ovaries (Supplemental Table 1; Supplemental Data Set 3). RT-qPCR was performed with 10 DEGs related to cell division and cell expansion (Supplemental Figure 6; Supplemental Table 2) and showed the same expression pattern revealed by RNA sequencing (RNA-seq) analysis (Pearson’s correlation coefficient = 0.91; Supplemental Figure 6).
There were 270 downregulated and 215 upregulated transcription factor genes in the CsFUL1-OX samples (Supplemental Figure 7A; Supplemental Data Set 4). Notably, expression of the gene encoding the C2H2-type zinc finger transcription factor SUP (Csa3G141870) was dramatically repressed (log2 fold change = −9.32, P = 4.7E−12) in CsFUL1A-OX ovaries (Supplemental Figure 7B). RT-qPCR analysis confirmed downregulation of CsSUP expression in CsFUL1A-OX in contrast to its upregulation in CsFUL1A-RNAi samples (Figure 6A), making CsSUP a candidate target gene of CsFUL1A (Zhao et al., 2014). Second, 24 genes regulating auxin transport and signaling had differential expression (Supplemental Table 3). In particular, five PIN genes mediating polar auxin transport were significantly downregulated in CsFUL1A-OX ovaries (Supplemental Figure 7C), suggesting probable regulation of auxin transport or distribution by CsFUL1A. Third, MapMan analysis showed that most DEGs related to cell cycle and cell division were dramatically downregulated in CsFUL1A-OX ovaries (Supplemental Figure 7D). A previous transcriptome analysis examined the short fruits (line 409) and long fruits (line 408) of near-isogenic lines (NILs) both harboring the CsFUL1A allele at the same stage (Jiang et al., 2015); therefore, we analyzed these two data sets (A-OX-29 versus empty vector/control plants; 409 versus 408) together to identify common DEGs. Gene ontology (GO) term enrichment analysis (P ≤ 0.05) indicated that the top four GO terms were all related to cell division and cell expansion, including microtubule motor activity, microtubule binding, kinesin complex, and microtubule-based movement (Supplemental Figure 7E). Specifically, 24 microtubule-related genes were downregulated in the common DEGs (Supplemental Table 4). Such downregulation of microtubule and kinesin genes was consistent with the short fruit in CsFUL1A overexpression lines (Figure 4) and line 409 (Jiang et al., 2015), suggesting that CsFUL1A may inhibit fruit elongation, at least in part, by repressing the transcription of microtubule- and kinesin-related genes in cucumber.
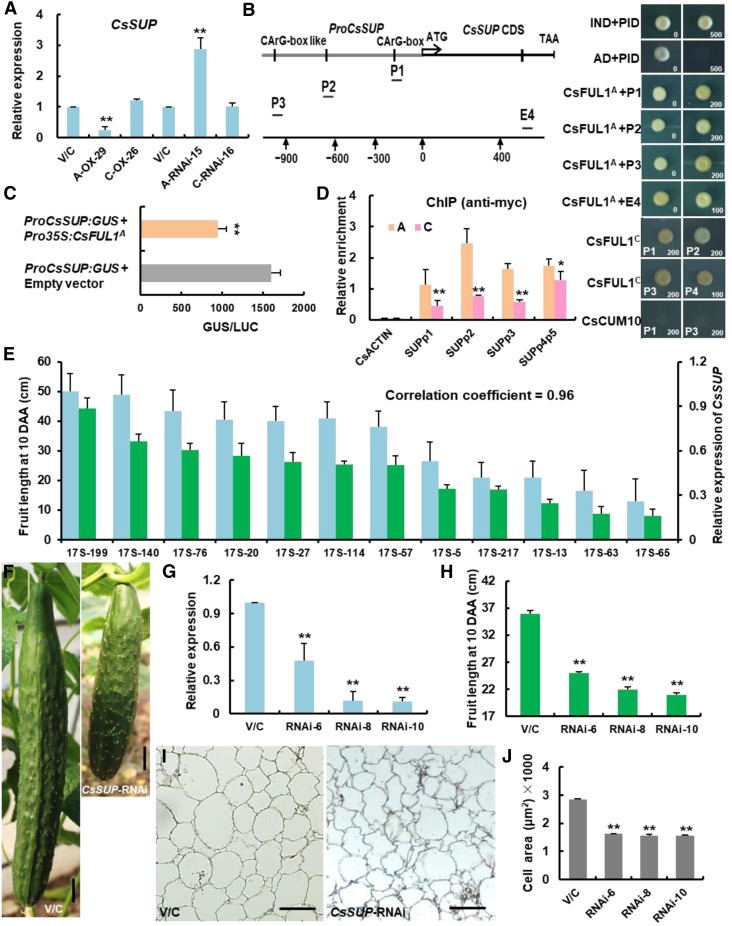
Figure 6.
CsFUL1A Inhibited Expression of CsSUP in Cucumber.
(A) CsSUP expression in ovaries 4 d before anthesis from A-OX-29, C-OX-26, A-RNAi-15, and A-RNAi-16 plants. Three independent samples from different plants were used for expression analysis. Values are means ± se. V/C, empty vector/control plants.
(B) CsFUL1A and CsFUL1C bound to CArG-box containing fragments of CsSUP in a Y1H assay, while the type II MADS-box protein CsCUM10 did not. Interaction of IND-AD with the PID-E box as a positive control and the empty AD and PID-E box as a negative control. AD, activation domain; E, exon; P, promoter.
(C) GUS activities after transient coexpression of ProCsSUP:GUS and Pro35S:CsFUL1A. Four independent transfection experiments were performed. Values are means ± se.
(D) ChIP-PCR showing the in vivo binding of CsFUL1A and CsFUL1C to the CsSUP promoter. The asterisk indicates a significant difference between the CsFUL1A allele and CsFUL1C allele. Three biological replicates were performed. Values are means ± se.
(E) Positive correlation of CsSUP expression with fruit length. Green and orange bars indicate fruit length and CsSUP expression, respectively. Three independent samples were used for expression analysis. Values are means ± se (n = 4 fruits from different transgenic lines).
(F) Representative images of fruit in control and CsSUP-RNAi lines. Bars = 2 cm.
(G) CsSUP expression was significantly reduced in different CsSUP-RNAi lines. Three independent samples from different plants were used for expression analysis. Values are means ± se. V/C, empty vector/control plants.
(H) Fruit length at 10 DAA decreased in CsSUP-RNAi lines. Values are means ± se (n = 4 fruits from different transgenic lines). V/C, empty vector/control plants.
(I) Transverse sections of the fruit pericarp at maturity. Bars = 200 µm. V/C, empty vector/control plants.
(J) Cell area in the fruit pericarp in CsSUP-RNAi lines. Values are means ± se (n = 3 pericarps from different fruits). V/C, empty vector/control plants.
Significance analysis was conducted with the two-tailed Student's t tests (*P < 0.05; **P < 0.01).
CsFUL1A Directly Represses the Expression of CsSUP in Cucumber
MADS-box transcription factors can bind to the CArG-box of target genes (Smaczniak et al., 2012a). CsSUP has three CArG-boxes in the promoter region and one CArG-box in the coding region, including one A type CArG-box located at −200 bp from the transcription start site that is well known for MADS-box protein binding (Riechmann et al., 1996). A yeast one-hybrid (Y1H) assay showed that both CsFUL1A and CsFUL1C bound to all four different CArG-containing CsSUP fragments, while another type II MADS-box protein Cucumber MADS box gene 10 (CsCUM10) could not bind to these fragments (Figure 6B). A β-glucuronidase (GUS) transactivation assay was performed to validate the interaction of CsFUL1A with CsSUP. When Pro35S:CsFUL1A was cotransformed with ProCsSUP:GUS into Nicotiana benthamiana leaves, the intensity of GUS signals was significantly reduced compared with the empty vector (Figure 6C), suggesting binding of the CsSUP promoter and negative regulation of CsSUP transcription by CsFUL1A. To confirm the binding of CsFUL1 to CsSUP in vivo, a chromatin immunoprecipitation (ChIP)–PCR assay was performed using CsFUL1A-MYC and CsFUL1C-MYC transgenic R1407 cucumber harboring the CsFUL1A allele. Our data showed that the presence of CsFUL1A or CsFUL1C substantially enhanced the PCR-based detection of the CsSUP promoter after immunoprecipitation, and the enrichment was much higher in CsFUL1A than in CsFUL1C (Figure 6D). Indeed, CsSUP showed consistently opposite expression patterns of CsFUL1 in NILs 409 and 408 (Supplemental Figure 8A), as well as in ethyl methane sulfonate–generated short-fruit mutants M85 and M573 harboring the CsFUL1A allele (Supplemental Figures 8B to 8J). In the 12 inbred cucumber lines with various fruit lengths, CsSUP expression was positively associated with fruit length (Figure 6E), implying that CsSUP may function as a positive regulator of fruit elongation in cucumber.
CsSUP-RNAi plants were then generated with cucumber inbred line R1461, and three lines (RNAi-6, RNAi-8, and RNAi-10) showed a 52 to 89% reduction in CsSUP expression compared with control plants (Figures 6F and 6G). As expected, corresponding CsSUP-RNAi fruits were obviously shorter early at anthesis (Supplemental Figures 9A and 9B) and showed a 31 to 42% reduction in length at 10 DAA (Figure 6H). RNAi-10 fruits were even more dramatically shorter at maturity (Supplemental Figures 9C to 9E). Transverse sectioning of the fruit pericarp at 10 DAA revealed smaller, disorganized cells in CsSUP-RNAi fruits compared with control fruits (Figures 6I and 6J). Notably, the expression of 10 cell division– and cell expansion–related genes was downregulated in CsSUP-RNAi (S-RNAi-10) and CsFUL1A overexpression (A-OX-29) fruits (Supplemental Figure 6; Supplemental Table 2), supporting that CsFUL1 and CsSUP may function in the same pathway in regulating fruit elongation via modulating cell division and cell expansion.
CsFUL1A Altered Auxin Distribution by Directly Repressing the Auxin Transporters CsPIN1 and CsPIN7
Auxin plays important roles in regulating cell division, cell expansion, and fruit set (Boonkorkaew et al., 2008; Kumar et al., 2014; Schaller et al., 2015). We measured auxin (indole-3-acetic acid) levels in fruits at five different developmental stages before anthesis (Figures 7A and 7B). The auxin concentration in A-OX-29 fruits was significantly lower at two early stages (N1 and N2-1) compared with that in control fruits (Figure 7B). Auxin accumulation is generally mediated by polar efflux transporters, PIN proteins (Friml, 2003; Ljung et al., 2005). Of the 24 differentially expressed auxin-related genes in CsFUL1A transgenic fruits (Supplemental Figure 7C; Supplemental Table 3), CsPIN1, CsPIN7, and CsPINOID (CsPID) had much larger variations in expression in different genotypes in the RNA-seq analysis and were therefore chosen for further study. Consistent with the RNA-seq data, expression of CsPIN1, CsPIN7, and CsPID was almost abolished in the CsFUL1A-OX fruits and dramatically upregulated in the CsFUL1A-RNAi samples as detected by RT-qPCR, while no changes were observed in the CsFUL1C transgenic plants (Figure 7C). A Y1H assay showed that CsFUL1A and CsFUL1C bound to the CArG-box in the promoter of CsPIN1 and CsPIN7, but not to that of CsPID (Figure 7D; Supplemental Figures 10A and 10B).
Figure 7.
CsFUL1A Downregulated CsPIN1 and CsPIN7 Transcription.
(A) Young fruits in CsFUL1A-OX and control lines at green bud (N1, N2-1), green-yellow bud (N2-2), yellow bud (N3), and flowering (N4) stages.
(B) The auxin concentration in CsFUL1A (dark gray) and control (light gray) fruits. Three samples from different plants were used for auxin measurement. Values are means ± se. FW, fresh weight, IAA, indole-3-acetic acid.
(C) Expression of auxin transport genes (CsPIN1, CsPIN7, and CsPID) in young fruits of transgenic A-OX-29, C-OX-5, A-RNAi-15, and C-RNAi-16 plants. Three biological replicates were performed for gene expression. Values are means ± se.
(D) Y1H showed that CsFUL1A and CsFUL1C bound to different CArG-box–containing motifs of CsPIN1 and CsPIN7, but not to those of CsPID. CsCUM10 was used as the negative control; +, positive interaction; –, no interaction. AD, activation domain.
(E) Quantification of GUS activities in N. benthamiana leaves after transient expression of Pro35S:CsFUL1A and Pro35S:CsFUL1C with ProCsPIN1:GUS and ProCsPIN7:GUS. Four independent transfection experiments were performed for each combination. Values are means ± se.
(F) ChIP-PCR showing the in vivo binding of CsFUL1A and CsFUL1C to the CsPIN1 and CsPIN7 promoters. The asterisk indicates significant difference between the CsFUL1A and CsFUL1C alleles. Three independent experiments were performed. Values are means ± se.
Significance analysis was conducted with two-tailed Student's t tests (*P < 0.05; **P < 0.01).
A GUS transactivation assay showed significantly weaker GUS signals upon cotransformation of Pro35S:CsFUL1A with ProCsPIN1:GUS or ProCsPIN7:GUS and mild reduction upon cotransformation of Pro35S:CsFUL1C with ProCsPIN1:GUS or ProCsPIN7:GUS compared with the vector control (Figure 7E). To confirm the in vivo binding of CsFUL1A to CsPIN1 and CsPIN7 promoters, a ChIP-PCR assay was performed, and our data showed that the presence of CsFUL1A or CsFUL1C significantly enriched the PCR-based detection of the CsPIN1 and CsPIN7 promoters after immunoprecipitation (Figure 7F), supporting the direct binding of both CsFUL1A and CsFUL1C proteins to CsPIN1 and CsPIN7, while the binding activity was relatively reduced for the CsFUL1C protein.
CsFUL1 Interacts with Other MADS-Box Proteins and an Auxin Response Factor
MADS-box transcription factors form higher order protein complexes that regulate multiple developmental processes such as flower patterning and fruit ripening (Smaczniak et al., 2012b). The SEPALATA (SEP) class MADS-box protein RIN interacts with the MADS-box proteins SlFUL1/2 and TAGL1 in the form of a SlFUL1/2-RIN-TAGL1 tetramer to regulate fruit ripening in tomato (Fujisawa et al., 2014). In Arabidopsis, FUL interacts with auxin response factors (ARFs) ARF6 and ARF8 that directly activate expression of a microRNA miR172-encoding gene to promote fruit valve growth (Ripoll et al., 2015). To identify potential protein interactors of CsFUL1, a yeast-two hybrid (Y2H) assay was performed with 10 homologs of known interacting partners from other species (Shima et al., 2013; Ripoll et al., 2015; Zhang et al., 2017). Our data showed that CsFUL1A interacts with seven proteins in yeast, including five MADS-box proteins (CsSEP2, CsSEP3, CsSEP4, CsAGL10, CsAGL20) and two auxin signaling proteins (CsARF12, CsARF13; Figures 8A and 8B). Notably, CsFUL1 did not form a homodimer or heterodimer between the two alleles. No interactions were detected between CsFUL1A and CsFUL2, nor between CsFUL1A and CsHEC3 and CsALC (Figures 8A and 8B; Robles and Pelaz, 2005).
Figure 8.
Protein Interactions Detected with Y2H and BiFC.
(A) Summary of interactions performed in this study. + indicates positive interaction; * represents confirmed by BiFC; − indicates no interaction; and NA indicates nonapplicable. BD and AD represented fusion of a given protein with the DNA binding and activation domain of GAL4, respectively. Gene fusion with vector pGBKT7 or pGADT7 was used as negative controls.
(B) Y2H assays showed CsFUL1A interacts with seven proteins including CsAGL20, while CsFUL1C does not interact with CsAGL20. No interactions were detected between CsFUL1A and CsFUL1C, CsFUL2, CsALC, or CsHEC3. No homodimer or heterodimer formed between CsFUL1A and CsFUL1C.
(C) Interaction of CsFUL1A with CsSEP2, CsSEP4, CsAGL20, and CsARF12 by BiFC. IND-YFPC and SPT-YFPN was used as the positive control. Columns from left to right are fluorescence, differential interference contrast, and merged channels, respectively. YFPC and YFPN represent fusion of a given protein with the C terminus and N terminus of YFP in frame, respectively. Bars = 50 µm.
Next, a bimolecular fluorescence complementation (BiFC) assay was performed in Nicotiana tabacum leaves. SPATULA (AtSPT) and AtIND were used as the positive controls (Girin et al., 2011). CsFUL1A did interact with CsSEP2, CsSEP4, CsAGL20, and CsARF12, but not with CsSEP3, CsAGL10, or CsARF13 in the BiFC assay (Figure 8C). Interestingly, CsFUL1C failed to interact with CsAGL20 in a Y2H assay (Figures 8A and 8B), suggesting the functional importance of SNP 3393C/A in protein–protein interactions of CsFUL1. A GUS transactivation assay showed that either CsFUL1A or CsAGL20 significantly suppressed CsFUL1 transcription, which was further downregulated by coexpression of CsAGL20 and CsFUL1A (Figure 9A). Cotransformation of Pro35S:CsFUL1A and Pro35S:CsAGL20 also inhibited CsPIN7 transcription (Figure 9B). However, single or combined expression of CsFUL1C with CsAGL20 did not affect the transcription of CsFUL1 and CsPIN7 (Figures 9A and 9B). In situ hybridization showed that CsAGL20 displayed an overlapping expression pattern with CsFUL1, with enriched signals in the developing carpel primordia and ovary (Figures 2H to 2J), indicating that CsFUL1A and CsFUL1C have different functions in fruit length regulation, partially due to differential interaction with CsAGL20 in cucumber.
Figure 9.
GUS/LUC Assay and the Working Model of CsFUL1 Regulating Fruit Length in Cucumber.
(A) and (B) Quantification of GUS activities in N. benthamiana leaves after transient expression of Pro35S:CsFUL1A, Pro35S:CsFUL1C, and Pro35S:CsAGL20 with ProCsFUL1:GUS (A) or ProCsPIN7:GUS (B). Four independent transfection experiments were performed for each combination. Values are means ± se. Significance analysis was conducted with the two-tailed Student's t tests (*P < 0.05; **P < 0.01).
(C) CsFUL1A controls fruit elongation via two regulatory pathways. Upon binding of CsFUL1A with CsAGL20 and/or other as-yet unidentified proteins (gray square) to form an active polymer complex, (1) CsFUL1A binds to the CArG-box to inhibit CsSUP expression and thus regulates cell division and cell expansion; and (2) CsFUL1A downregulates the expression of CsPIN1 and CsPIN7, resulting in decreases in auxin accumulation in the fruit. The active polymer may feedback on CsFUL1A to maintain its expression at appropriate levels and thus maintain proper levels of auxin accumulation for fruit elongation. Black arrows, positive regulation; black boxes, CArG-box; crosses, inhibit of transcription; lines, promoter region; red arrows, transcription start.
(D) Evolutionary diagram of the generation of the CsFUL1A allele in the East Asian long-fruited cucumbers during domestication and diversification.
DISCUSSION
Cucumber with fleshy fruit is a major vegetable crop worldwide (Weng et al., 2015). Wild cucumber produces tiny fruits, whereas modern varieties bear fruits with large variations in length across the world (Qi et al., 2013). How fruit length is programmed in cucumber during diversification remains an intriguing biological question of agronomic importance.
CsFUL1A Acts as a Key Regulator of Fruit Elongation in Cucumber
In this study, we found that the CsFUL1A allele is specifically present in East Asian cucumbers with long fruits (Figure 1B; Supplemental Data Set 1). We further showed that CsFUL1A negatively regulates fruit elongation under a certain expression window in cucumber. Heterologous and overexpression of CsFUL1A led to reduced fruit length in Arabidopsis and cucumber, respectively (Figures 3and 4), whereas long fruits grew even longer after downregulation of CsFUL1A expression (Figure 5). Consistently, expression of CsFUL1A in NIL 409 and M85 (with reduced fruit length) was higher than that in NIL 408 and M573, respectively (Supplemental Figure 8; Jiang et al., 2015). Moreover, we found that CsFUL1A is able to bind to CsSUP (a positive regulator of fruit length) and represses its expression (Figure 6). Previous studies showed that SUP regulates cell proliferation at the boundary during floral organ development in Arabidopsis (Bowman et al., 1992), mediates cell expansion and cell elongation in Petunia hybrida (Kater et al., 2000; Nakagawa et al., 2004), and participates in cell growth and proliferation through modulating auxin signaling in N. tabacum (Nibau et al., 2011). Considering that CsSUP-RNAi and CsFUL1A overexpression resulted in shorter fruits with significantly smaller cell size than control fruits (Figures 4J and 6I) and that cell division and cell cycle–related genes showed similar downregulated expression in these transgenic plants, we propose that CsFUL1A regulates fruit length, at least in part, through inhibiting the CsSUP-mediated cell division and cell expansion in cucumber (Figure 9C).
In addition, CsFUL1A overexpression significantly reduced auxin accumulation at an early stage of ovary development (Figure 7A). Auxin has been shown to play essential roles in multiple aspects of plant organogenesis, growth, and development (Zhao, 2010). Auxin accumulation, mediated by the auxin efflux carrier PIN proteins (Schaller et al., 2015), dramatically increases in the fertilized ovule of tomato and Arabidopsis, which stimulates auxin signaling and subsequent cell division and expansion to promote fruit growth (de Jong et al., 2009; Seymour et al., 2013). Consistently, expression analyses and biochemical data revealed that CsFUL1A can bind to the promoters of CsPIN1 and CsPIN7 and downregulate their expression (Figure 7). Therefore, another regulatory pathway through which CsFUL1A regulates fruit elongation may be to reduce auxin accumulation in developing fruits via repressing the transcription of the auxin polar transporters CsPIN1 and CsPIN7 (Figure 9A). There is also the possibility that the CsSUP pathway and the CsPIN-mediated auxin pathway crosstalk at a certain point and act in a combinatorial fashion to modulate fruit length in cucumber.
Moreover, previous studies showed that MADS-box proteins often function in the formation of protein complexes, and the K domain plays an essential role in mediating protein–protein interactions (Piwarzyk et al., 2007; Kaufmann et al., 2009). Substitution of CsFUL1C by CsFUL1A changes the residue Gln-168 to Lys-168 in the K domain of CsFUL1 (Figure 1). CsAGL20 can interact with CsFUL1A, but not with CsFUL1C (Figure 8), and coexpression of CsAGL20 and CsFUL1A further represses the expression of CsFUL1 and CsPIN7 (Figures 9A and 9B). Considering that CsFUL1 expression is relatively low in long-fruited lines harboring the CsFUL1A allele (Figure 2B) and that both too much and too little auxin have an inhibitory effect on cell division and expansion (Vanneste and Friml, 2009; Suzaki et al., 2012), CsFUL1A may interact with CsAGL20 and/or other as-yet unidentified proteins to form an active polymer complex that downregulates the expression of CsFUL1, as well as its downstream targets such as CsSUP, CsPIN1, and CsPIN7, so as to fine-tune CsFUL1A expression and auxin accumulation at appropriate levels for fruit elongation in cucumber (Figure 9C).
CsFUL1A Is a Gain-of-Function Allele during Selection for Long Fruits in Cucumber
During crop domestication, favorable alleles of domestication genes have been selected and fixed for added value in human use (Doebley et al., 2006). Although many mutations identified in domestication genes are shown to be complete or partial loss-of-function alleles, resulting from cis-regulatory element changes, amino acid changes, or premature transcriptional termination (Østerberg et al., 2017), gain-of-function alleles can be generated occasionally in a defined population during the domestication and breeding processes (Shi and Lai, 2015). In cucumber, fruit length and bitterness and leaf size are the major domestication traits (Qi et al., 2013). The wild cucumber relative C. sativus var hardwickii bears short and round fruits that are only 3 to 5 cm in diameter (Sebastian et al., 2010; Yang et al., 2012). The semiwild cucumber C. sativa var xishuangbannanensis was found in the Xishuangbanna geographic group that bears melon-like fruits with a low length:diameter ratio and a relatively large fruit size. The fruit length of cultivated cucumber exhibits large variation in length worldwide (Sebastian et al., 2010; Yang et al., 2012). Particularly, the East Asian long-fruited cucumbers bear long fruits (generally >35 cm in length) and are widely cultivated in North China and Japan (Figure 9D). Fruit length is an important agronomic trait in relation to many genes or quantitative trait loci including CsSUN and Fruit Size genes (FS1.1, FS1.2, FS3.1, FS3.2, FS3.3, FS4.1, FS5.1, FS6.1, FS6.2, and FS7.1), although their specific functions remain unclear (Bo et al., 2015; Pan et al., 2017).
Here, we found that CsFUL1A is specifically present in East Asian cucumbers with long fruits, while the CsFUL1C allele prevails in wild and semiwild varieties, landraces, or other regional lines (Figure 1B; Supplemental Data Set 1). We further showed that CsFUL1A, but not CsFUL1C, acts as a repressor of fruit elongation in cucumber (Figures 3 and 5), suggesting that CsFUL1A is a gain-of-function allele during cucumber domestication. In East Asia, long-fruited (30 to 40 cm) cucumber was preferred; thus, positive regulators of fruit length may have been pyramided for long fruits. However, over-elongated fruits break more frequently during harvest or storage, appear misshapen, and are unfavorable on the market. The CsFUL1A allele thus arose as a repressor of fruit length. To avoid over-repression, CsFUL1A expression was fine-tuned to maintain relatively low levels in the Asian long-fruited cucumber, likely via negative feedback of the polymer complex forming between CsFUL1A and CsAGL20 (Figure 9). Thus, CsFUL1A can serve as a target allele for manipulation of fruit length by modulating CsFUL1A at the proper expression window during cucumber breeding (Figure 9D). CsFUL1C overexpression or RNAi caused no change in fruit length, neither in transcription levels of CsSUP nor CsPIN1/7 (Figures 4, Figure 7 and 9), suggesting that the CsFUL1C allele is either nonfunctional in regulating fruit length or insufficient to confer visible phenotypes, likely due to functional redundancy with other unknown genes. Even though most germplasms harboring the CsFUL1C allele bear short fruit, some landraces with the CsFUL1C allele produce long fruits. The elongation mechanism of long-fruit landraces with the CsFUL1C allele awaits further investigation.
Functional Diversification of FUL in Different Species with Distinct Fruit Types
MADS-box transcription factors play fundamental roles in regulating flower patterning and fruit ripening (Smaczniak et al., 2012b), many of which, such as APETALA1, APETALA3, and PISTILLATA, act as bifunctional transcription factors (activator and repressor; Kaufmann et al., 2010; Wuest et al., 2012). In Arabidopsis, the MADS-box gene FUL mediates valve elongation and fruit opening by restricting the expression of IND, ALC, and SHP1/2 in the valve margin (Gu et al., 1998; Ferrándiz et al., 2000; Liljegren et al., 2000). Loss of function of FUL leads to short fruits and dehiscence failure due to ectopic adoption of the valve margin cell fate (Gu et al., 1998). In tomato, which has a fleshy berry fruit, TDR4/FUL1 and MBP7/FUL2 were found to interact with RIN to regulate fruit ripening via promoting the expression of ethylene biosynthesis genes including ACC SYNTHASE2 and ACC SYNTHASE4 (Vrebalov et al., 2002; Bemer et al., 2012; Shima et al., 2013; Fujisawa et al., 2014). MBP7/FUL2 was also shown to regulate fruit shape by mediating cell division and expansion in the pericarp (Wang et al., 2014). Here, in cucumber bearing fleshy pepo fruits, we found that CsFUL1A acts as a key repressor of fruit elongation in East Asian long-fruited cucumbers by inhibiting the transcription of CsSUP, CsPIN1, and CsPIN7 (Figure 4 to Figure 7; Figure 9). Ectopic expression of CsFUL1 failed to rescue the ful-1 mutant phenotype in Arabidopsis (Figure 3). Even though both FUL and CsFUL1A act as transcriptional repressors, the biological outcome may vary: the ful-1 mutant produces short siliques, while CsFUL1A knockdown plants produce long fruits (Figure 5; Gu et al., 1998), probably due to their regulation of different downstream targets. Thus, FUL homologs implement diverse regulatory functions during fruit development (fruit opening, ripening, or elongation), depending on fruit types and the evolutionary context (Pabón-Mora et al., 2014).
METHODS
Plant Materials and Growth Conditions
Lines 408 and 409 are NILs characterized previously (Jiang et al., 2015). M85 and M573 are ethyl methane sulfonate mutants derived from inbred ‘6457’ and ‘ZND649,’ respectively. Long-fruit inbred lines R1407 and R1461 and short-fruit inbred line 32X were used for genetic transformation. Inbred lines 17s-199, 17s-140, 17s-76, 17s-20, 17s-27, 17s-114, 17s-57, 17s-5, 17s-217, 17s-13, 17s-63, and 17s-65 were selected from over 200 varieties with different fruit length and used for expression analyses. Seeds were germinated in Petri dishes at 28°C in the dark overnight and grown under a 16-h day (350 µmol photons m−2 s−1, GXZ-500D-LED, JIANGNAN Instrument; 25°C)/8-h night (18°C) cycle in a growth chamber. At the two-true-leaf stage, seedlings were transferred to a standard glass greenhouse at China Agricultural University (Beijing). Water and fertilization management as well as pest control were performed according to standard practices.
Fruit length of 150 cucumber lines was measured at the mature fruit stage. Field data were collected across 3 years at the University of Wisconsin Hancock Agriculture Research Station and averaged.
For Arabidopsis (Arabidopsis thaliana), the ecotype Landsberg erecta (Ler) was used as the wild type and the ful-1 mutant from Marty Yanofsky’s lab at University of California, San Diego, was described previously (Gu et al., 1998). Seeds were germinated on Murashige-Skoog medium at 4°C for 3 d and then transferred to growth chambers at 22°C with a 16-h light (250 µmol photons m−2 s−1, GXZ-500D-LED, JIANGNAN Instrument)/8-h dark cycle. Seedlings were transplanted to the soil 7 to 10 d after germination.
CsFUL1 Cloning, Boxplot, and Phylogenetic Analysis
Total RNA was extracted from female flower buds of cucumber line 1407 using an RNA extraction kit (Huayueyang Biotechnology), and cDNA was synthesized using TUREscript H− Reverse Transcriptase (Aidlab). The CDS of CsFUL1 was amplified using gene-specific primers (Supplemental Table 5). The gene structure of CsFUL1 was analyzed using the online software GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). The CsFUL1-predicated three-dimensional structure was generated with PyMOL Molecular Graphics System, Version 1.8 (Schrodinger). The boxplot with the swarm layout was plotted using the R boxplot and beeswarm function from the beeswarm package (Wang et al., 2016). The cucumber sequences CsFUL1 and CsFUL2 were blasted in National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify homologous protein and CDS sequences in dicot and monocot plants. The protein alignment was conducted using MUSCLE 3.6 (http://www.drive5.com/muscle/), and the alignment was manually adjusted using GeneDoc 3.2. The CDS matrix was performed by PAL2NAL (http://www.bork.embl.de/pal2nal/) to construct the phylogenetic tree, which was based on the protein matrix. The maximum likelihood phylogenetic analysis was generated in RaxML-HPC2 BlackBox on the Cyberinfrastructure for Phylogenetic Research Science (CIPRES) Gateway (https://www.phylo.org), the bootstrap was performed 1000 replications, and other parameters were set as default.
Quantitative Real-Time RT-PCR
The leaf, tendril, flower bud, flower, and fruit at 4 d before anthesis; fruit at anthesis and 3 DAA; exocarp, mesocarp, endocarp, and ventricle from the central part of fruits at 10 DAA of cucumber line R1407; and the inflorescence of Arabidopsis were frozen in liquid nitrogen and stored at −80°C for total RNA extraction and cDNA synthesis. RT-PCR was performed on an ABI PRISM 7500 real-time PCR system (Applied Biosystems), with cucumber Ubiquitin (Csa000874) as the internal reference. Three biological and three technical replicates were included for each gene. Primers are listed in Supplemental Table 5.
RNA In Situ Hybridization
The shoot apex of 12-d-old seedlings, flower buds, and young fruits at various stages were fixed in 3.7% formol-acetic-alcohol and stored at 4°C until use. Probes of CsFUL1 and CsAGL20 were designed according to unique gene fragments. Sense and antisense probes were synthesized using SP6 and T7 RNA polymerase, respectively. Sample fixation, sectioning, and in situ hybridization were performed as described previously (Zhang et al., 2013). Primers for probe synthesis are listed in Supplemental Table 5.
Subcellular Localization
The full-length CDS of CsFUL1 was cloned into the pEZS-NL vector to generate green fluorescent protein (GFP)-CsFUL1 fusion protein. The empty pEZS-NL vector was used as a positive control. Constructs were bombarded into onion epidermal cells using a PDS-1000/He particle delivery system (Bio-Rad) as described previously (Varagona et al., 1992). After incubation in the dark at 22°C for 24 h, fluorescence signals were imaged using an BX 51 microscope (Olympus).
Plant Transformation
CsFUL1 and CsSUP RNAi constructs were generated using two gene-specific fragments that were inversely inserted into the pFGC1008 vector after amplification. The full-length CDS of CsFUL1 was inserted into the pCAMBIA1305.1 vector to generate the Pro35S:CsFUL1 overexpression vector. All constructs were transformed into Agrobacterium by electroporation. Cucumber transformation was performed as described previously (Ding et al., 2015). The transgenic plants were verified by PCR using vector- and gene-specific primers. Positive plants transformed with the empty vector were control plants (V/C). At least three representative transgenic lines and five plants per line were used for further analysis.
Transformation with the wild-type (Ler) and ful-1 mutant plants followed the floral dip method (Clough and Bent, 1998). Transgenic lines were screened on the Murashige-Skoog medium with 25 mg L−1 hygromycin. Primers are listed in Supplemental Table 5.
Histology
Mature fruit tissues were fixed, embedded, sectioned 8 μm thick, and dewaxed as described by Zhao et al. (2016a). Images were taken under a D72 light microscope (Olympus). Cell size was calculated as the 30 selected areas in the fruit pericarp under the microscope divided by cell number. Three fields were observed for each sample, and at least six biological samples for each line were included.
Transcriptome Analysis
The RNA-seq library was prepared following the manufacturer’s instructions as described previously (Jiang et al., 2015). RNA sequencing was performed on an Illumina HiSeq 2000 platform to generate 100-bp paired-end reads. Bioinformatic analysis was performed as described by Zhao et al. (2016a). GO term enrichment analysis and MapMan category enrichment were performed using the R package topGO and JAVA software MapMan, respectively (Alexa et al., 2006). The enriched GO terms or categories with a P-value < 0.05 were identified as significant.
Auxin Measurements
To examine auxin accumulation, ∼100 mg of young fruit tissue was harvested at different stages. Sample treatment and hormone measurement were performed using an ELISA (Sun et al., 2016).
Y1H Assay
The CDS of CsFUL1 was cloned into the pGADT7 vector (Clontech) using the EcoRI and BamHI sites. To obtain sense and antisense oligonucleotides, two antiparallel oligonucleotides containing a triplicate of the CArG-box variant from the promoter of CsPIN1, CsPIN7, CsPID, and CsSUP plus three flanking nucleotides on both sides were synthesized and ligated into the pAbAi vector (Clontech). A mutated version of CArG-box was used as the negative control. The linearized plasmids were transformed into the yeast Y1H Gold strain following the manufacturer’s instructions. Next, pGADT7-CsFUL1 was transformed into YIH Gold containing the linear plasmid and selected by aureobasidin A on synthetic dropout medium/−Leu (Clontech). Primers for oligonucleotide synthesis are listed in Supplemental Table 5.
GUS/LUC Assay
The transient GUS/luciferase (LUC) assay was performed as described previously (Zhao et al., 2016b). The 2-kb promoter sequence of CsPIN1, CsPIN7, CsSUP, and CsFUL1 was separately fused into pCAMBIA 1381 and introduced into Agrobacterium tumefaciens strain GV3101 together with pSuper1300 containing Pro35S:CsFUL1 and/or Pro35S:AGL20. The resulting bacterial fluid was injected into Nicotiana benthamiana leaves. Three days later, the GUS activity was measured using the methyl umbelliferyl glucuronide (Sigma-Aldrich). LUC was used as an internal control and quantified using luciferin (Promega). The GUS:LUC ratio was used to analyze activities of CsFUL1 binding to gene promoters. Primers for oligonucleotide synthesis are listed in Supplemental Table 5.
Chromatin Immunoprecipitation
ChIP-PCR was performed as described previously (Gendrel et al., 2005), with some modifications. Approximately 1 g of floral and leaf tissues from control and transgenic cucumber lines was harvested and fixed in 30 mL of 1% formaldehyde and cross-linked for 15 min under vacuum infiltration, terminated by the addition of Gly. Nuclei were isolated and lysed, and chromatin was sonicated to an average size of 300 to 500 bp. The sonicated chromatin served as input and was stored at −20°C until use. Immunoprecipitation reactions were performed using anti-Myc antibody (anti-Myc tag antibody, lot no. GR310953-4, Abcam). The complex of chromatin-antibody was captured with protein A beads (Abcam), and DNA purified using a QIAquick PCR purification kit (QIAGEN) served as a template (∼1 ng) for RT-qPCR. No antibody was used as the negative control. The enrichment of DNA sequence segments in gene promoters was chosen to perform RT-qPCR. Two biological repeats and three technical replicates were performed for each sequence segment. CsACTIN was used as the internal gene control. The primer pairs used in ChIP-PCR are listed in Supplemental Table 5.
Y2H Assay
Full-length coding sequences of HANABA TARANU (HAN), CsFUL1, CsSEP2, CsSEP3, CsAGL10, CsAGL20, CsARF12, and CsARF13 were obtained using gene-specific primers and cloned into pGADT7 (bait vector) and pGBKT7 (prey vector; Supplemental Table 5). All constructs were verified by sequencing and then transformed into yeast strain AH109. Y2H assays were conducted and analyzed as described previously (Zhang et al., 2013).
BiFC Assay
Full-length coding sequences of SPT, IND, CsFUL1, CsSEP2, CsSEP3, CsSEP4, CsAGL10, CsAGL20, CsARF12, and CsARF13 without stop codons were amplified and ligated into pSPYNE-35S and pSPYCE-35S vectors containing the N- or C-terminal half of yellow fluorescent protein (YFP). Constructs were checked by sequencing and then transformed into A. tumefaciens strain GV3101. Two plasmids were cotransformed into the abaxial side of 4- to 6-week-old N. benthamiana leaves to detect specific interactions as described previously (Zhang et al., 2013). After 48 h co-infiltration, N. benthamiana leaves were observed using an LSM 510-Meta confocal laser microscope (Zeiss). YFP signals were imaged under 488 nm excitation wavelength. The primers are listed in Supplemental Table 5.
Scanning Electron Microscopy
Stage 17 siliques of ful-1 and CsFUL1A-OX/ful-1 plants were fixed overnight at 4°C in 3.7% formol-acetic-alcohol, dehydrated through an ethanol series, and critical point dried. Samples were coated with gold and palladium and imaged using a scanning electron microscope (S-4700, Hitachi) with an accelerating voltage of 2 kV.
Statistical Analysis
Two group comparisons between control and experiment groups were performed using two-tailed Student’s t tests by Excel 2010. Means were considered significantly different based on a threshold value corresponding to P < 0.05 and P < 0.01 as indicated by * and **, respectively. Detailed results of tests are shown in Supplemental Data Set 5.
Accession Numbers
Sequence data were deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE103592. The accession numbers for genes in this study are as follow: CsFUL1 (Csa1G039910), CsFUL2 (Csa4G126480), CsSUP (Csa3G012410), CsPIN1 (Csa1G042820), CsPIN7 (Csa4G430820), CsHEC3 (Csa6G483450), CsALC (Csa010227), CsSEP2 (Csa4G126990), CsSEP3 (Csa6G095270), CsSEP4 (Csa6G367080), CsAGL20 (Csa6G076720), CsAGL10 (Csa1G051580), CsARF12 (Csa6G141390), CsARF13 (Csa6G445210), CsPID (CsaaG537400), CsUBI (Csa000874), ACTIN2 (AT3G18780), and AtHAN (AT3G50870).
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of predicted amino acid sequences of CsFUL1 and its homologs in different species.
Supplemental Figure 2. Protein alignment of CsFUL1, CsFUL2, and their homologs in different species.
Supplemental Figure 3. Expression analyses and subcellular localization.
Supplemental Figure 4. Expression analysis of CsFUL2, CsHEC3, and CsALC.
Supplemental Figure 5. Phenotypic quantification of fruit length and fruit diameter in 35S:CsFUL1A and 35S:CsFUL1C lines in cucumber.
Supplemental Figure 6. RT-qPCR analyses of cell division– and cell expansion–related genes in CsFUL1A overexpression (A-OX-29) and CsSUP-RNAi (S-RNAi-10) fruits.
Supplemental Figure 7. Transcription profiling of 35S:CsFUL1A transgenic plants.
Supplemental Figure 8. Expression patterns of CsFUL1 and CsSUP in genetic lines with different fruit length.
Supplemental Figure 9. Expression analyses and phenotypic characterization of the CsSUP-RNAi lines in cucumber.
Supplemental Figure 10. Y1H assay.
Supplemental Table 1. Summary of transcriptome sequencing data.
Supplemental Table 2. Genes used for qPCR verification in Supplemental Figure 6.
Supplemental Table 3. Auxin-related genes differentially expressed in CsFULA-OX versus control.
Supplemental Table 4. List of microtubule related genes that were differentially expressed in CsFULA-OX versus control and in lines 409 versus 408.
Supplemental Table 5. List of primers used in this study.
Supplemental Data Set 1. Allelic distribution of CsFUL in 150 cucumbers lines with fruit-length data.
Supplemental Data Set 2. Predicted amino acid sequences of CsFUL1 and its homologs in different species.
Supplemental Data Set 3. List of genes differentially expressed between CsFULA-OX and control samples.
Supplemental Data Set 4. Transcription factors differentially expressed in CsFULA-OX versus control.
Supplemental Data Set 5. Results of Student’s t tests of quantitative data.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
ACKNOWLEDGMENTS
We thank Martin Yanofsky for sharing the ful-1 seeds and to Elliot M. Meyerowitz, Michael J. Scanlon, Martin Yanofsky, Adrienne Roeder, Jinsheng Lai, Yan Guo, Ying Fu, Chuanqing Sun, and Xuexian Li for critical reading and comments on the article. This study was supported by the National Key Research and Development Program (2016YFD0101007), the National Natural Science Foundation of China (grants 31572132 and 31772315), and 111 Project (B17043). Work in Y.W.’s lab was supported by the Agriculture and Food Research Initiative Competitive Grants Program from the U.S. Department of Agriculture’s National Institute of Food and Agriculture (grants 2015-51181-24285 and 2017-67013-26195 to Y.W.).
AUTHOR CONTRIBUTIONS
J.Z., L.J., and X.Z. designed experiments. J.Z., L.J., G.C., Y.P., Y.H., W.Z., Y.Z., L.D., and S.Y. performed the experiments. J.Z., L.J., G.C., Y.L., R.L., Y.P., Y.W., L.Y., C.S., T.W., and X.L. analyzed the data. J.Z., L.J., G.C., Y.W., and X.Z. wrote the article. All authors read and approved the final version.
References
- Alexa A., Rahnenführer J., Lengauer T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607. [DOI] [PubMed] [Google Scholar]
- Bemer M., Karlova R., Ballester A.R., Tikunov Y.M., Bovy A.G., Wolters-Arts M., Rossetto Pde.B., Angenent G.C., de Maagd R.A. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo K., Ma Z., Chen J., Weng Y. (2015). Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 128: 25–39. [DOI] [PubMed] [Google Scholar]
- Boonkorkaew P., Hikosaka S., Sugiyama N. (2008). Effect of pollination on cell division, cell enlargement, and endogenous hormones in fruit development in a gynoecious cucumber. Sci. Hortic. (Amsterdam) 116: 1–7. [Google Scholar]
- Bowman J.L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- de Jong M., Mariani C., Vriezen W.H. (2009). The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 60: 1523–1532. [DOI] [PubMed] [Google Scholar]
- Ding L., Yan S., Jiang L., Zhao W., Ning K., Zhao J., Liu X., Zhang J., Wang Q., Zhang X. (2015). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet. 11: e1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Weigel D., Yanofsky M.F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132: 4687–4696. [DOI] [PubMed] [Google Scholar]
- Doebley J.F., Gaut B.S., Smith B.D. (2006). The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Liljegren S.J., Yanofsky M.F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438. [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T.C., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Tanksley S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. [DOI] [PubMed] [Google Scholar]
- Friml J. (2003). Auxin transport - Shaping the plant. Curr. Opin. Plant Biol. 6: 7–12. [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Nakagawa H., Kitagawa M., Kimbara J., Nakano T., Kasumi T., Ito Y. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Girin T., Paicu T., Stephenson P., Fuentes S., Körner E., O’Brien M., Sorefan K., Wood T.A., Balanzá V., Ferrándiz C., Smyth D.R., Østergaard L. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23: 3641–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q., Ferrándiz C., Yanofsky M.F., Martienssen R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517. [DOI] [PubMed] [Google Scholar]
- Jiang L., Yan S., Yang W., Li Y., Xia M., Chen Z., Wang Q., Yan L., Song X., Liu R., Zhang X. (2015). Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.). Sci. Rep. 5: 8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater M.M., Franken J., van Aelst A., Angenent G.C. (2000). Suppression of cell expansion by ectopic expression of the Arabidopsis SUPERMAN gene in transgenic petunia and tobacco. Plant J. 23: 407–413. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kumar R., Khurana A., Sharma A.K. (2014). Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 65: 4561–4575. [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770. [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Roeder A.H., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853. [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Ferrario S., Angenent G.C., Kobayashi A., Takatsuji H. (2004). The petunia ortholog of Arabidopsis SUPERMAN plays a distinct role in floral organ morphogenesis. Plant Cell 16: 920–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C., Di Stilio V.S., Wu H.M., Cheung A.Y. (2011). Arabidopsis and Tobacco superman regulate hormone signalling and mediate cell proliferation and differentiation. J. Exp. Bot. 62: 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østerberg J.T., et al. (2017). Accelerating the domestication of new crops: Feasibility and approaches. Trends Plant Sci. 22: 373–384. [DOI] [PubMed] [Google Scholar]
- Pabón-Mora N., Wong G.K., Ambrose B.A. (2014). Evolution of fruit development genes in flowering plants. Front. Plant Sci. 5: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Liang X., Gao M., Liu H., Meng H., Weng Y., Cheng Z. (2017). Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 130: 573–586. [DOI] [PubMed] [Google Scholar]
- Piwarzyk E., Yang Y., Jack T. (2007). Conserved C-terminal motifs of the Arabidopsis proteins APETALA3 and PISTILLATA are dispensable for floral organ identity function. Plant Physiol. 145: 1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45: 1510–1515. [DOI] [PubMed] [Google Scholar]
- Rajani S., Sundaresan V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11: 1914–1922. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll J.J., Roeder A.H., Ditta G.S., Yanofsky M.F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138: 5167–5176. [DOI] [PubMed] [Google Scholar]
- Ripoll J., Bailey L.J., Mai Q.A., Wu S.L., Hon C.T., Chapman E.J., Ditta G.S., Estelle M., Yanofsky M.F. (2015). microRNA regulation of fruit growth. Nat. Plants 1: 15036. [DOI] [PubMed] [Google Scholar]
- Robles P., Pelaz S. (2005). Flower and fruit development in Arabidopsis thaliana. Int. J. Dev. Biol. 49: 633–643. [DOI] [PubMed] [Google Scholar]
- Rodríguez G.R., Muños S., Anderson C., Sim S.C., Michel A., Causse M., Gardener B.B., Francis D., van der Knaap E. (2011). Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A.H., Ferrándiz C., Yanofsky M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13: 1630–1635. [DOI] [PubMed] [Google Scholar]
- Schaller G.E., Bishopp A., Kieber J.J. (2015). The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian P., Schaefer H., Telford I.R., Renner S.S. (2010). Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. USA 107: 14269–14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour G.B., Østergaard L., Chapman N.H., Knapp S., Martin C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64: 219–241. [DOI] [PubMed] [Google Scholar]
- Shi J., Lai J. (2015). Patterns of genomic changes with crop domestication and breeding. Curr. Opin. Plant Biol. 24: 47–53. [DOI] [PubMed] [Google Scholar]
- Shima Y., Kitagawa M., Fujisawa M., Nakano T., Kato H., Kimbara J., Kasumi T., Ito Y. (2013). Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82: 427–438. [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012b). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G., Angenent G.C., Kaufmann K. (2012a). Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Sun C., Li Y., Zhao W., Song X., Lu M., Li X., Li X., Liu R., Yan L., Zhang X. (2016). Integration of hormonal and nutritional cues orchestrates progressive corolla opening. Plant Physiol. 171: 1209–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006. [DOI] [PubMed] [Google Scholar]
- van der Knaap E., Østergaard L. (2018). Shaping a fruit: Developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 79: 27–36. [DOI] [PubMed] [Google Scholar]
- van der Knaap E., Chakrabarti M., Chu Y.H., Clevenger J.P., Illa-Berenguer E., Huang Z., Keyhaninejad N., Mu Q., Sun L., Wang Y., Wu S. (2014). What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Varagona M.J., Schmidt R.J., Raikhel N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4: 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346. [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Pan I.L., Arroyo A.J., McQuinn R., Chung M., Poole M., Rose J., Seymour G., Grandillo S., Giovannoni J., Irish V.F. (2009). Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lu G., Hou Z., Luo Z., Wang T., Li H., Zhang J., Ye Z. (2014). Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 65: 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Liu S., Ferjani A., Li J., Yan J., Yang X., Qin F. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48: 1233–1241. [DOI] [PubMed] [Google Scholar]
- Wang X., Bao K., Reddy U.K., Bai Y., Hammar S.A., Jiao C., Wehner T.C., Ramírez-Madera A.O., Weng Y., Grumet R., Fei Z. (2018). The USDA cucumber (Cucumis sativus L.) collection: Genetic diversity, population structure, genome-wide association studies, and core collection development. Hortic. Res. 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Colle M., Wang Y., Yang L., Rubinstein M., Sherman A., Ophir R., Grumet R. (2015). QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 128: 1747–1763. [DOI] [PubMed] [Google Scholar]
- Wuest S.E., O’Maoileidigh D.S., Rae L., Kwasniewska K., Raganelli A., Hanczaryk K., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 109: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Yang L., Koo D.H., Li Y., Zhang X., Luan F., Havey M.J., Jiang J., Weng Y. (2012). Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 71: 895–906. [DOI] [PubMed] [Google Scholar]
- Zhang S., Lu S., Yi S., Han H., Liu L., Zhang J., Bao M., Liu G. (2017). Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia. Planta 245: 439–457. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhou Y., Ding L., Wu Z., Liu R., Meyerowitz E.M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu M., Jiang L., Ding L., Yan S.S., Zhang J., Dong Z., Ren H., Zhang X. (2014). Cucumber SUPERMAN has conserved function in stamen and fruit development and a distinct role in floral patterning. PLoS One 9: e86192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li Y., Ding L., Yan S., Liu M., Jiang L., Zhao W., Wang Q., Yan L., Liu R., Zhang X. (2016a). Phloem transcriptome signatures underpin the physiological differentiation of the pedicel, stalk and fruit of cucumber (Cucumis sativus L.). Plant Cell Physiol. 57: 19–34. [DOI] [PubMed] [Google Scholar]
- Zhao S., Zhang M.L., Ma T.L., Wang Y. (2016b). Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28: 3005–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]