Maize subsidiary cell-specific glucose transporter CST1 mediates the feedback-regulation of photosynthesis by photoassimilates at the grain-filling stage.
Abstract
It has long been recognized that stomatal movement modulates CO2 availability and as a consequence the photosynthetic rate of plants, and that this process is feedback-regulated by photoassimilates. However, the genetic components and mechanisms underlying this regulatory loop remain poorly understood, especially in monocot crop species. Here, we report the cloning and functional characterization of a maize (Zea mays) mutant named closed stomata1 (cst1). Map-based cloning of cst1 followed by confirmation with the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated protein 9 system identified the causal mutation in a Clade I Sugars Will Eventually be Exported Transporters (SWEET) family gene, which leads to the E81K mutation in the CST1 protein. CST1 encodes a functional glucose transporter expressed in subsidiary cells, and the E81K mutation strongly impairs the oligomerization and glucose transporter activity of CST1. Mutation of CST1 results in reduced stomatal opening, carbon starvation, and early senescence in leaves, suggesting that CST1 functions as a positive regulator of stomatal opening. Moreover, CST1 expression is induced by carbon starvation and suppressed by photoassimilate accumulation. Our study thus defines CST1 as a missing link in the feedback-regulation of stomatal movement and photosynthesis by photoassimilates in maize.
INTRODUCTION
Fixation of atmospheric CO2 through photosynthesis is crucial for the survival of plants, and is also pivotal for meeting the ever-increasing food, feed, and fuel demands of humans. To achieve optimal photosynthesis, leaf photosynthetic rates need to be tightly controlled according to the plant carbon status, which involves feedback regulation of photosynthesis by photoassimilates (Araya et al., 2006; Ainsworth and Bush, 2011).
Given that stomata modulate CO2 availability and consequently the rate of photosynthesis (Lawson and Blatt, 2014), it is perhaps unsurprising that various exogenous and endogenous cues, including the plant carbon status, converge at stomata to modulate photosynthesis (Farquhar and Sharkey, 1982). Several photoassimilates have been reported to regulate stomatal movement, among which Suc and Glc have been most extensively studied (Misra et al., 2015; Daloso et al., 2016). Suc modulates stomatal movement in a spatial distribution- and concentration-dependent manner. Suc was proposed to serve as an osmoticum in guard cells to facilitate stomatal opening (Talbott and Zeiger, 1996). More recently, Suc breakdown in guard cells was suggested to be an important mechanism during light-induced stomatal opening (Daloso et al., 2015). On the other hand, high concentrations of Suc in the guard cell apoplast cause reduced stomatal opening, presumably by acting as an apoplastic osmolyte (Outlaw and De Vlieghere-He, 2001; Kang et al., 2007). This hypothesis was later challenged by the finding that apoplastic Suc-stimulated stomatal closure depends on the Glc sensor hexokinase (HXK; Kelly et al., 2013). In periods of highly active photosynthesis, Suc-derived Glc overaccumulates in guard cells, leading to HXK activation and subsequently stomatal closure (Kelly et al., 2013; Lugassi et al., 2015; Li et al., 2018). The molecular events downstream of HXK in this process are currently unknown. In addition to Suc and Glc, starch (Horrer et al., 2016), triacylglycerols (McLachlan et al., 2016), trehalose (Van Houtte et al., 2013), pyruvate (Li et al., 2014; Yu and Assmann, 2014), and malate (Araújo et al., 2011; Gago et al., 2017) have been reported to regulate stomatal movement.
Prolonged perturbation of carbon homeostasis imposes adverse effects on the maintenance of the photosynthetic apparatus. In maize (Zea mays), for example, overaccumulation of carbohydrates in leaves due to ear removal (Christensen et al., 1981), prevention of pollination (Sekhon et al., 2012), or girdling (Jeannette et al., 2000) leads to reduced chlorophyll content and consequently decreased rates of photosynthesis. Similar phenomena have also been observed in maize mutants defective in phloem loading, including tie-dyed1 (Braun et al., 2006), tie-dyed2 (Slewinski et al., 2012), sucrose export defective1 (Ma et al., 2008), and sucrose transporter1 (Baker et al., 2016). However, carbohydrate deprivation/starvation induced by long-term dark or shade treatment also leads to early senescence phenotypes in plants, including chlorosis and disassembly of the photosynthetic apparatus (Hanaoka et al., 2002; Buchanan-Wollaston et al., 2005; Kim et al., 2016).
Although the role of photoassimilates as regulators of stomatal movement and photosynthesis has long been observed, the genetic components and molecular mechanisms involved remain obscure. In this study, we report the cloning and characterization of a subsidiary cell-localized Sugars Will Eventually be Exported Transporters (SWEET) family Glc transporter gene named CLOSED STOMATA1 (CST1). With physiological functions different from other reported SWEET family members, CST1 is specifically expressed in subsidiary cells and positively regulates stomatal opening and source capacity at the grain-filling stage. Our data thus define a crucial role for CST1 as a missing link in the feedback regulation of stomatal movement by photoassimilates.
RESULTS
cst1 Exhibits Reduced Stomatal Conductance, Source Capacity, and Yield
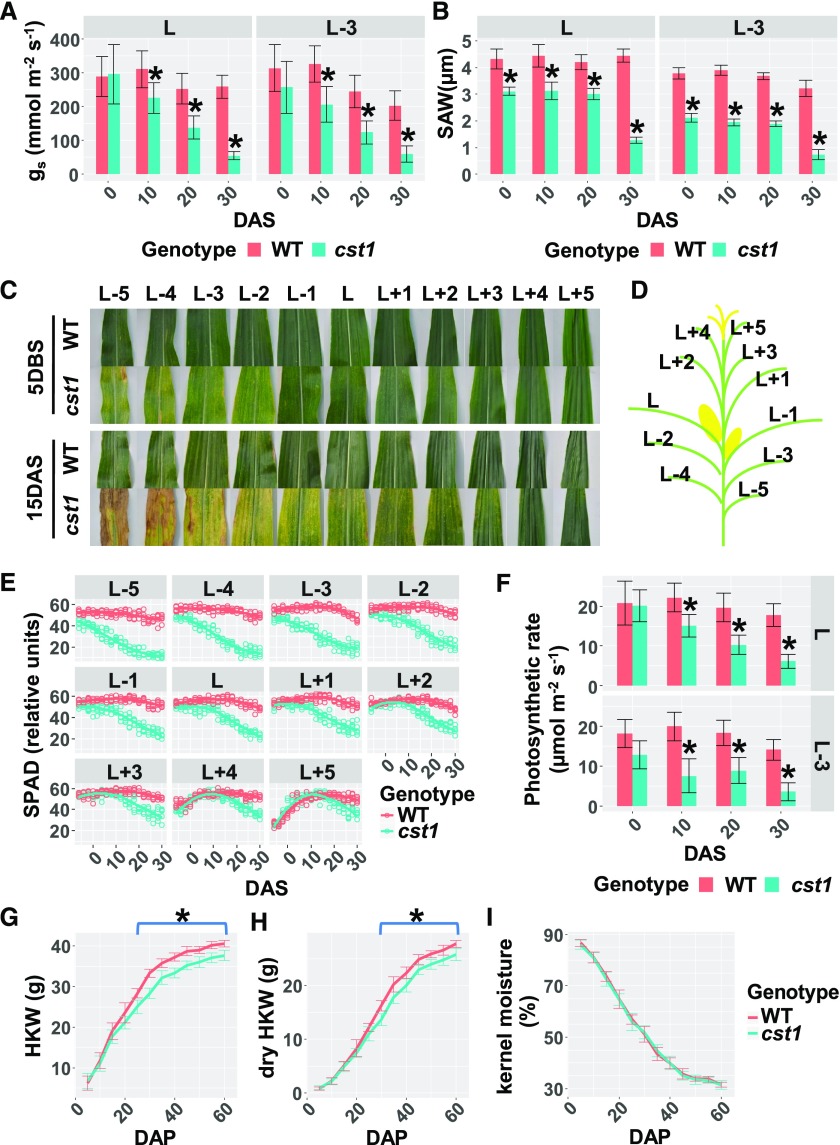
Stomata modulate the availability of CO2 to the plant and therefore exert considerable control on the photosynthetic rate. They are, however, subject to feedback regulation by plant carbon status. This regulation may be more prominent under strong source–sink interaction such as during seed/grain filling. To dissect the interplay between stomatal movement and photoassimilate dynamics, we identified a mutant in maize named closed stomata1 (cst1) from an ethyl methanesulfonate-mutagenized population created in the Zheng58 background. Stomatal conductance was significantly lower in cst1 than in wild type Zheng58 plants for leaf L and L-3 at 10, 20, and 30 d after silking (DAS; Figure 1A; Supplemental Data Set 1; leaves were named as shown in Figure 1D). In addition, the stomatal aperture width (SAW) was significantly reduced in the cst1 as compared with that in the wild type for both leaf L and L-3 from 0 DAS to 30 DAS (Figure 1B). Stomatal conductance and SAW were also measured in cst1 and wild-type plants before silking. However, no significant differences in either of the two phenotypes were observed between cst1 and wild type at 10 or 20 d before silking (DBS; Supplemental Figures 1A and 1B).
Figure 1.
The cst1 Plants Exhibit Diminished Stomatal Conductance, Reduced Source Capacity, and Lower Yield Compared With the Wild-Type (WT) Plants.
(A) and (B) The stomatal conductance (gs; A) and SAW (B) of wild-type and cst1 leaves (L and L-3) at 0, 10, 20, and 30 DAS. Leaves are labeled as shown in (D). Data are means ±sd of 10 plants for each genotype. (C) Wild-type and cst1 leaves 5 DBS and 15 DAS. Leaves are labeled as shown in (D).
(E) Total chlorophyll contents (represented as SPAD values) in wild-type and cst1 leaves from 6 DBS to 30 DAS. Five plants of each genotype were measured every 2 d. Measurements were fitted by LOESS (Local Regression) curves.
(F) Photosynthetic rates of wild-type and cst1 leaves (L and L-3) at 0, 10, 20, and 30 DAS. Shown are means ±sd (n = 10).
(G) to (I) HKW (G), dry HKW (H), and kernel moisture (I) were measured in wild-type and cst1 kernels from 5 DAP to 60 DAP. For each genotype, 9 ears (from 9 different plants) were sampled every 5 d, with 10 kernels randomly sampled from each ear. Measurements were fitted with LOESS (Local Regression) curves. Shown are means ±sd (n = 9). In (A), (B), (F), (G), and (H), asterisks indicate significant difference from wild type (p.adj < 0.05) by the Tukey's honestly significant difference test (Supplemental Data Set 1).
The cst1 plants were visually indistinguishable from the wild-type plants until 5 DBS (Supplemental Figure 2), when cst1 leaves began to exhibit senescence-like phenotypes, characterized by chlorosis in the older leaves. At 15 DAS, chlorosis became prominent in most cst1 leaves except for the youngest two leaves (Figure 1C). To study the spatio-temporal dynamics of chlorosis in cst1, total chlorophyll content represented as Soil Plant Analysis Development (SPAD) values was measured every 2 d from 6 DBS to 30 DAS in cst1 and wild-type plants. As shown in Figure 1E, the chlorophyll content in cst1 leaves peaked earlier and declined faster than in the corresponding wild-type leaves.
Consistent with the reduced stomatal opening and chlorophyll content in cst1, the photosynthetic rates of cst1 leaves were significantly lower compared with corresponding wild-type leaves (Figure 1F). As source capacity at grain filling stage is an important determinant of yield, we next investigated whether yield was reduced in cst1 plants. Fresh and dry weights of seeds were measured every five days from 5 d after pollination (DAP) to 60 DAP in cst1 and wild-type plants. The fresh and dry weights of cst1 seeds were significantly lower than wild type from 25 DAP to 60 DAP, with ∼7% reduction in both fresh hundred kernel weights (HKW) and dry HKW at 60 DAP (Figures 1G and IH). The early-senescence phenotype of cst1 was not associated with accelerated desiccation of kernels (Figure 1I). Taken together, the cst1 mutation results in reduced stomatal opening and source capacity, as well as inadequate grain filling.
Cloning of the CST1 Gene
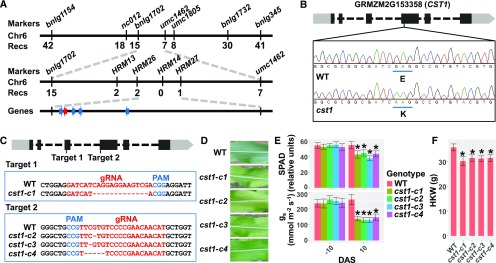
To isolate the causal gene for the observed phenotypes of cst1, we constructed an F2 mapping population by crossing cst1 with the inbred line B73. Early senescence of leaves at the grain-filling stage was used as the marker phenotype to discriminate cst1 from the wild type in field conditions. All the F1 individuals showed the wild-type phenotype, and among the 727 F2 individuals, 169 and 558 F2 progenies were found with early senescence and wild-type phenotypes, respectively, suggesting that cst1 is controlled by a single recessive locus (Chi-square=1.193, p-value=0.275). The cst1 locus was first located between the Simple Sequence Repeat markers bnlg1702 and umc1462 (3.57cM to bnlg1702 and 0.97cM to umc1462, respectively; Figure 2A). High-resolution melting (HRM) analysis of recombinants further narrowed down the location of cst1 to a 188-kb genomic region between single-nucleotide polymorphism (SNP) markers HRM26 and HRM27, which contains five genes (Figure 2A). Sanger sequencing of these five genes in cst1 and Zheng58 revealed a single nucleotide substitution of G to A on the fourth exon of GRMZM2G153358, which causes an E81K mutation in the predicted protein sequence (Figure 2B).
Figure 2.
Map-Based Cloning of cst1.
(A) Map-based cloning of cst1. Molecular markers and number of recombinants are indicated above and below the filled bars, respectively. The candidate gene (GRMZM2G153358) is indicated by a red arrow, whereas the other genes within this genomic interval are indicated by blue arrows. Chr, chromosome; Recs, recombinations.
(B) A schematic representation of the genomic structure of GRMZM2G153358. The light and dark gray filled boxes indicate untranslated region (UTR)s and coding exons, respectively. The dashed lines indicate introns. The codons that overlap with the base substitution are underlined in wild type (WT) and cst1, with corresponding amino acids labeled below the codons.
(C) The two target sites (designated Target 1 and Target 2) used for gene targeting are shown at top. At middle (for Target 1) and bottom (for Target 2) are shown the wild type and mutated sequences. gRNA, guide RNA; PAM, protospacer adjacent motif.
(D) Ear leaves of wild type, cst1-c1, cst1-c2, cst1-c3, and cst1-c4 at 5 DAS.
(E) Total chlorophyll content (SPAD) and stomatal conductance (gs) of wild type and cst1-c1, cst1-c2, cst1-c3, and cst1-c4 ear leaves at −10 and 10 DAS. Shown are mean ±sd of 10 leaves for each genotype.
(F) HKW of wild type, cst1-c1, cst1-c2, cst1-c3, and cst1-c4 at 60 DAP, when grain-filling is complete. Shown are means ±sd of 10 ears for each genotype. In (E) and (F), asterisks indicate significant difference from wild type (P < 0.05) by the Student’s t test (Supplemental Data Set 1).
To test whether the identified G-to-A substitution is the causal mutation for the cst1 phenotype, a clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated protein 9 (Cas9) vector containing two guide RNA open reading frames each targeting the third and fourth exon of GRMZM2G153358, respectively, was constructed (Supplemental Information 1). This vector was used to generate transgenic maize lines in the C01 background (Figure 2C), as Zheng58 is recalcitrant to transformation. In T1 plants, individuals with homozygous frameshift mutations but without the Cas9 transgene were identified, and they were categorized into four types based on the mutated sequences at target sites (cst1-c1, cst1-c2, cst1-c3, and cst1-c4; Figure 2C). Similar to cst1, all the knockout plants exhibited reduced stomatal conductance, early senescence, and reduced yield, although they are in a genetic background different from cst1 (Figures 2D to 2F). In sum, these results strongly suggest that the G-to-A mutation in GRMZM2G153358 is the causal mutation for the cst1 phenotypes.
cst1 Exhibits Transcriptome and Metabolome Changes Characteristic of Senescence
A time-course transcriptome analysis of cst1 and wild type at 0, 10, 20, and 30 DAS on leaf L and L-3 was conducted to obtain a global view of senescence and photosynthesis in cst1 (Supplemental Figure 3A; Supplemental Information 2;Supplemental Data Set 2). Data quality was validated by distance matrices of samples (Supplemental Figures 3B and 3C) and principal component analysis (Supplemental Figure 3D). Genes differentially expressed between cst1 and wild type in at least one of the eight comparisons (2 leaf types × 4 time points) were identified and functionally annotated (Supplemental Data Set 3).
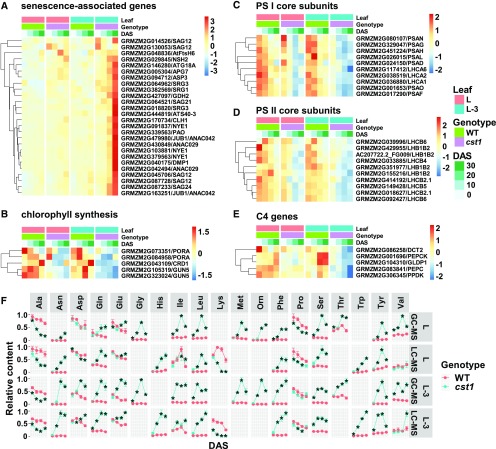
We found that the expression levels of senescence-associated genes (SAGs) were significantly higher in cst1 than in wild-type leaves (Figure 3A). These SAGs are of diverse functional groups. Several of them are putative orthologs of well-characterized senescence regulators, such as AtS40-3 (Fischer-Kilbienski et al., 2010), ANAC029 (Guo and Gan, 2006), and ANAC042 (Wu et al., 2012). They also include genes involved in degradation of chlorophyll or the light harvesting complex, including chlorophyllase encoding genes (Benedetti and Arruda, 2002), NON-YELLOWING1 (Li et al., 2017), PHEOPHORBIDE A OXYGENASE (Süssenbacher et al., 2015), SENESCENCE-RELATED GENE1 (Callard et al., 1996), and FtsH6 (Zelisko et al., 2005). Strikingly, several genes involved in nitrogen remobilization during senescence were also found, including GLUTAMATE DEHYDROGENASE2 (Diaz et al., 2008), AUTOPHAGY-RELATED7 (Doelling et al., 2002), AUTOPHAGY 18A (Zhuang et al., 2017), and SENESCENCE-ASSOCIATED GENE12 (SAG12; Otegui et al., 2005). AUTOPHAGY-RELATED7 and AUTOPHAGY 18A are autophagy-related genes, whereas SAG12 is a major Cys protease localized in senescence-associated vacuoles. SAG12 was reported to be exclusively upregulated by the natural development of senescence in Arabidopsis (Arabidopsis thaliana; Otegui et al., 2005). In contrast with SAGs, genes involved in photosynthesis were strongly downregulated in cst1, including chlorophyll biosynthetic genes (Figure 3B), photosystem I and II core subunits (Figures 3C and 3D), and C4 genes (Figure 3E).
Figure 3.
cst1 Exhibits Transcriptome and Metabolome Changes Characteristic of Senescence.
(A) to (E) Heat map representation of gene expression levels in cst1 and wild type (WT) for leaf L and L-3 at 0, 10, 20, and 30 DAS, as quantified by RNA-Seq analysis. Shown are senescence-associated genes (A), chlorophyll synthesis genes (B), PS I core subunits (C), PS II core subunits (D), and C4 genes (E). Refer to Supplemental Data Set 3 for absolute expression levels and statistical analysis.
(F) Free amino acids were measured for cst1 and wild type in leaf L and L-3 at 0, 10, 20, and 30 DAS by both gas chromatography–tandem mass spectrometry (GC-MS) and liquid chromatography–tandem mass spectrometry (LC-MS). Data are means ±sd of four biological replicates with each replicate pooled from leaf L or leaf L-3 of 10 individual plants. *Significant difference from wild type (p.adj < 0.05) by the Tukey's honestly significant difference test (Supplemental Data Set 1).
Upregulated expression of nitrogen remobilization-related genes in cst1 prompted us to investigate whether cst1 overaccumulates free amino acids. Wild-type and cst1 leaves were also subject to metabolome profiling using both liquid chromatography–tandem mass spectrometry and gas chromatography–tandem mass spectrometry (Supplemental Information 2; Supplemental Figures 4 and 5; Supplemental Data Sets 4 and 5). Nineteen amino acids were identified and quantified by at least one platform (Figure 3F), and results from the two MS platforms are highly consistent for the thirteen amino acids that were identified by both platforms (Pearson correlation coefficient = 0.8). Most amino acids (except Ala, Lys, and Pro) were significantly higher in cst1 than in wild type at 10, 20, and 30 DAS (Figure 3F). These results indicate that nitrogen remobilization, a hallmark of senescence, is more active in cst1 than in wild-type plants.
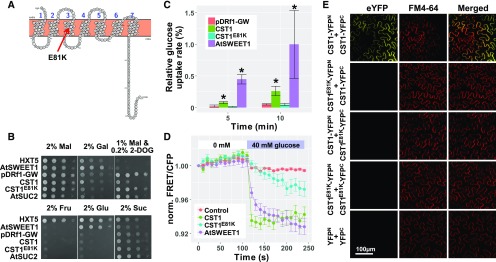
The CST1 Protein Is a Cell Membrane-Localized Glc Transporter and the E81K Mutation Substantially Impairs Its Transport Activity
Phylogenetic analysis of the CST1 protein revealed that it belongs to the Clade I subfamily of the SWEET protein family (Supplemental Figure 6; Supplemental Data Set 6). The CST1 protein was predicted by PROTTER (Omasits et al., 2014) to contain seven transmembrane domains (Figure 4A). To determine the in vivo subcellular localization of the CST1 protein, a CST1-enhanced yellow fluorescent protein (eYFP) fusion protein was transiently expressed in either maize protoplasts or Nicotiana benthamiana epidermal cells, driven by the Cauliflower mosaic virus (CaMV) 35S promoter. Ectopically expressed CST1-eYFP in both cell types colocalized with FM4-64, a lipophilic fluorescent probe that specifically stains cell membrane (Supplemental Figures 7A and 7B). Interestingly, the CST1E81K protein was also localized to the plasma membrane (Supplemental Figures 7A and 7B). Collectively, these results indicate that CST1 is a plasma membrane-localized protein and that the E81K mutation does not alter its subcellular localization.
Figure 4.
CST1 Protein Is an Active Glc Transporter, and the E81K Mutation Impairs its Transporter Activity.
(A) The CST1 protein has 7 transmembrane domains typical of SWEET family proteins, as predicted by PROTTER. The E81K mutation (indicated by the red arrow) is located in the middle portion of the third transmembrane domain.
(B) Functional analysis of CST1 activity by the yeast complementation assay. CST1 and CST1E81K were tested for complementation of the growth defect of the YSL2-1 mutant strain (only grows on maltose [Mal]) on various carbon sources. Note that CST1 and CST1E81K confer limited growth on Gal, Fru, Glc, and Suc. When tested on the toxic Glc analog 2-deoxyglucose (2-DOG), cells with CST1 and other active glucose transporters failed to grow well relative to pDRf1-GW (the empty vector) and CST1E81K.
(C) Relative Glc uptake rates of CST1, CST1E81K, and AtSWEET1 in the yeast Glc transport-deficient mutant EBY.VW4000 at 5 min and 10 min (10 mM D-glucose; 0.1μCi [14C]-D-glucose). EBY.VW4000 with the empty vector pDRf1-GW served as the negative control. Values are normalized to AtSWEET1 (100%). Data are means ±sd (n = 3). *Significantly different from the empty vector group (P < 0.05) by the Student’s t test (Supplemental Data Set 1).
(D) Normalized Glc uptake analysis using HEK293T cells expressing the Glc sensor FLIPglu600μΔ13. CST1, CST1E81K, or AtSWEET1 (positive control) were individually coexpressed with the Glc sensor. HEK293T cells transfected with the sensor vector only served as a negative control. Data are means ±seM (n = 10).
(E) CST1 and CST1E81K were tested for their oligomerization activity by a BiFC assay. CST1 and CST1E81K were fused with either YFPN or YFPC and transiently expressed in Nicotiana benthamiana epidermal cells driven by the 35S promoter. The YFPN+YFPC combination served as a negative control. The scale bar applies to all images in this panel.
SWEET family proteins were reported to catalyze facilitated diffusion of various sugars across membranes (Chen, 2014). To determine the biochemical activity and substrate specificity of the CST1 protein, CST1 and CST1E81K were tested for their complementation of the growth defect of the yeast (Saccharomyces cerevisiae) YSL2-1 mutant strain, which lacks all endogenous hexose transporters and extracellular invertase but retained the cytosolic invertase, allowing it to grow only on medium containing maltose (Chen et al., 2015). YSL2-1 cells carrying CST1 and CST1E81K exhibited limited growth on galactose, fructose, glucose, and sucrose (Figure 4B). When tested on the toxic Glc analog 2-deoxyglucose, cells with CST1 failed to grow well relative to the negative control (transformed with the empty vector), indicating that CST1 can mediate 2-deoxyglucose transport. By contrast, YSL2-1 cells with CST1E81K were able to grow at a rate similar to the negative control in the presence of 2-deoxyglucose, indicating that the E81K mutation likely impairs the Glc transport activity of CST1. To directly measure the Glc transport activity of CST1, the Glc-transport–deficient yeast strain EBY.VW4000 (Wieczorke et al., 1999) expressing CST1, CST1E81K, or AtSWEET1 was tested for [14C]-D-Glc uptake activity. When compared with EBY.VW4000 harboring the empty vector pDRf1-GW, significantly higher levels of [14C]-D-Glc uptake were detected in EBY.VW4000 expressing CST1 or AtSWEET1, but not CST1E81K (Figure 4C). To further confirm the Glc transport activity of CST1, we used a highly sensitive Glc detection system based on the mammalian HEK293T cells expressing a fluorescence resonance energy transfer (FRET) Glc sensor, FLIPglu600µΔ13V. This system was employed to identify the first member of the SWEET gene family and also used to detect the transport activity of other SWEET family members from various species (Chen et al., 2010). After Glc treatment, HEK293T cells expressing CST1 or AtSWEET1 swiftly accumulate Glc, as demonstrated by the decreased normalized FRET emission/cyan fluorescent protein (CFP) emission ratio (Figure 4D). By contrast, CST1E81K conferred a much slower and weaker response than CST1 (Figure 4D). This weak Glc transport activity of CST1E81K is undetectable in yeast, probably due to varying efficiencies in transgene expression or protein folding/sorting in yeast and mammalian cells. Taken together, these results indicate that CST1 is an active Glc transporter, and the E81K mutation significantly impairs the Glc transport activity of CST1. The nature of the E81K mutation may at least partly explain the stronger reduction of stomatal conductance and HKW in CRISPR lines than in cst1, although the varying severities of the two phenotypes in CRISPR lines and cst1 may also be caused by their different genetic backgrounds.
SWEET proteins form oligomers in order to perform their function (Xuan et al., 2013; Tao et al., 2015). Thus we tested whether CST1 oligomerizes in vivo by the bimolecular fluorescence complementation (BiFC) assay. When CST1-YFPN (CST1 fused to the N-terminal 155 amino acids of YFP) and CST1-YFPC (CST1 fused to the C-terminal 86 amino acids of YFP) were transiently coexpressed in N. benthamiana epidermal cells, the fluorescence signal of YFP was observed. The signal colocalized with that of FM4-64 (Figure 4E), indicating that CST1 oligomerized at the cell membrane. However, no fluorescence signal was detected in combinations involving CST1E81K (i.e., CST1-YFPN and CST1E81K-YFPC, CST1E81K-YFPN and CST1-YFPC, or CST1E81K-YFPN and CST1E81K-YFPC) or in the negative control (YFPN and YFPC; Figure 4C). Taken together, the E81K mutation impairs the Glc transport activity of CST1, at least in part by preventing its oligomerization.
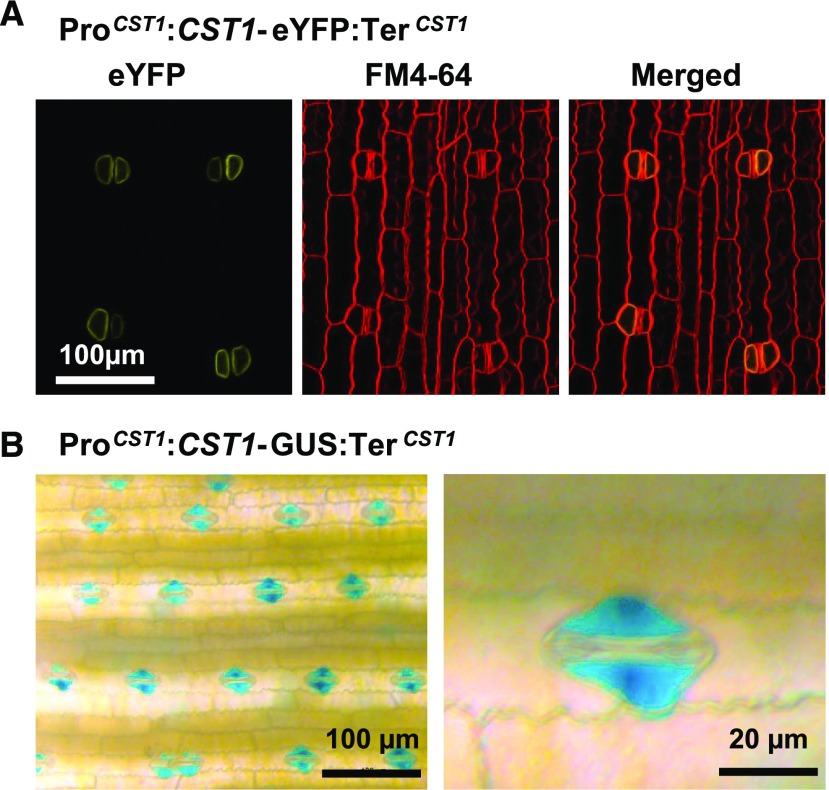
The CST1 Gene Is Specifically Expressed in Subsidiary Cells, and the cst1 Mutation Leads to Mis-Regulation of Stomatal Movement-Controlling Genes
To determine the tissue-specificity of CST1 activity, the genomic sequence of CST1 (from 2 kb upstream of the start codon to 0.5 kb downstream of the stop codon) was cloned and fused with the coding sequences (CDS) of eYFP or β-glucuronidase (GUS; with the coding sequence of each reporter gene inserted in front of the stop codon of CST1), to generate the construct ProCST1:CST1-eYFP:TerCST1 and ProCST1:CST1-GUS:TerCST1, respectively. The two constructs were used to generate stable transgenic maize plants. In the leaves of ProCST1:CST1-eYFP:TerCST1 transgenic plants, eYFP fluorescence signal was observed on the membrane of subsidiary cells at the V6 stage (with the collar of the 6th leaf visible; Figure 5A). Consistent with this, ProCST1:CST1-GUS:TerCST1 plants showed strong subsidiary cell-specific GUS staining (Figure 5B) at V6 stage. Since the GUS protein is fused to the C terminus of CST1 and CST1 has seven transmembrane domains, we conclude that the N terminus of CST1 is outside the subsidiary cells, whereas the C terminus of CST1 is inside the subsidiary cells. Localization of CST1 in subsidiary cells was also observed after flowering (Supplemental Figure 8A). Moreover, GUS staining of transgenic lines (Supplemental Figures 8B to 8D) and RT-qPCR analysis in wild-type Zheng58 maize (Supplemental Figure 8E) failed to detect CST1 expression in all tested tissues except for green leaves. CST1 was found to be slightly upregulated after flowering in both wild-type and cst1 plants, with its expression level at 30 DAS around 1.7-fold that at 0 DAS (Supplemental Figure 8F). Subsidiary cells have not been as extensively studied as guard cells have, but it is widely accepted that they assist the function of guard cells (Franks and Farquhar, 2007; Raissig et al., 2017; Apostolakos et al., 2018). The function of CST1 as a positive regulator of stomatal opening and its localization in subsidiary cells supports a role of subsidiary cells in the regulation of stomatal movement.
Figure 5.
CST1 Is Localized on the Subsidiary Cell Membrane.
(A) Transgenic maize leaf with the ProCST1:CST1-eYFP:TerCST1 transgene was stained by the membrane probe FM4-64 and observed under a confocal microscope. The scale bar applies to all images in this panel.
(B) Transgenic maize leaf with the ProCST1:CST1-GUS:TerCST1 transgene was stained for GUS activity and observed under a dissection microscope.
Stomatal movement is coordinately controlled by a number of guard cell-localized ion channels and also by regulators of these ion channels (Assmann and Jegla, 2016). Fisher's exact test revealed that stomata movement-controlling genes are significantly enriched (p-value=0.00035) in differentially expressed genes between cst1 and wild type (Supplemental Data Set 7), including pivotal stomatal movement regulators such as OPEN STOMATA1 (Imes et al., 2013), SLOW ANION CHANNEL1 (Laanemets et al., 2013), and ALUMINUM-ACTIVATED MALATE TRANSPORTER12 (Meyer et al., 2010). These results suggest that the reduced stomatal opening in cst1 may be caused by mis-regulation of stomatal movement-controlling genes.
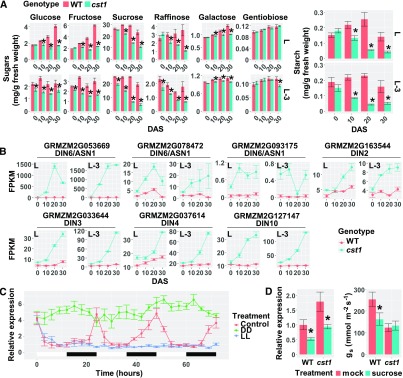
Loss-of-Function of CST1 Leads to Carbon Starvation in Leaves
The early senescence phenotype of cst1 could potentially result from either carbohydrate starvation or carbohydrate overaccumulation. Given that CST1 is a positive regulator of stomatal opening and photosynthesis, we hypothesized that cst1 leaves may accumulate less carbohydrates than wild type. To test this hypothesis, six major forms of sugar (Glc, Fru, Suc, raffinose, Gal, and gentiobiose) and starch were measured in leaves of cst1 and wild-type plants at different time points during the grain-filling period. At 10, 20, and 30 DAS, most sugars (except for gentiobiose) and starch were significantly lower in cst1 than in wild type (Figure 6A). These results were also observed in the aforementioned metabolome data (Supplemental Data Sets 4 and 5). Consistently, genes involved in Suc and starch synthesis were found to be downregulated in cst1, whereas those responsible for Suc and starch degradation were upregulated in cst1 (Supplemental Figure 9).
Figure 6.
cst1 Leaves Accumulate Less Photo-Assimilates than the Wild-Type (WT) Leaves.
(A) Sugar (left) and starch (right) contents in cst1 and wild type in L and L-3 leaves at 0, 10, 20, and 30 DAS. Data are means ±sd of four biological replicates with 10 plants in each replicate. *Significant difference from the wild type (p.adj < 0.05) by the Tukey's honestly significant difference test (Supplemental Data Set 1).
(B) Expression levels of carbon starvation marker genes in cst1 and wild type for leaf L and L-3 at 0, 10, 20, and 30 DAS, as quantified by RNA-Seq analysis (Refer to Supplemental Data Set 3 for statistical analysis). FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(C) B73 maize plants at the V4 stage (with the collar of the 4th leaf visible) were entrained in 12-h light/12-h dark cycles before they were transferred to continuous light (LL), continuous dark (DD), or kept in their original growth condition (Control). Expression levels of CST1 were quantified by RT-qPCR. White and black bars represent subjective day and night, respectively. Data are means ±sd of 3 biological replicates, with each replicate pooled from the forth leaves of 7 individual plants.
(D) Wild-type and cst1 plants were treated with either Suc or distilled water (mock) by stem infusion. The treatment started at noon of 15 DAS. At 48 h after the onset of the treatments, CST1 expression (left) and stomatal conductance (right) was quantified in wild-type and cst1 ear leaves. Data are means ±sd of 10 plants for each genotype. *Significant difference from the mock treatment (P < 0.05) by the Student’s t test (Supplemental Data Set 1).
In addition, we examined the expression of carbon starvation marker genes. ASPARAGINE SYNTHETASE (AS) has been reported to be a hallmark of carbon starvation in multiple plant species including maize, and is strongly induced by prolonged darkness or a low exogenous carbon supply (Chevalier et al., 1996; Davies et al., 1996; Downs and Somerfield, 1997; Nozawa et al., 1999; Fujiki et al., 2001). We found that AS genes are among the most highly induced genes in cst1, along with several other carbon starvation marker genes including putative orthologs of DARK INDUCIBLE genes such as DIN2, DIN3, DIN4, and DIN10 (Fujiki et al., 2001; Figure 6B). Taken together, these results suggest that the CST1 gene is a positive regulator of carbohydrate accumulation, and the cst1 mutation leads to carbon starvation in leaves.
CST1 Expression Is Feedback Regulated by Photoassimilates
We next examined whether CST1 is regulated at the transcriptional level by leaf carbon status, through perturbing the availability of carbohydrates in leaves. We tested whether there is an association between the diurnal oscillation of leaf carbon status and CST1 expression. Maize plants entrained in 12-h light/12-h dark cycles were transferred to continuous darkness (designated as DD, for prolonged dark-induced carbon starvation), continuous light (designated as LL, for continuous active photosynthesis), or kept in their original condition (control). CST1 expression is rhythmic under the control condition and peaked at the end of each night. Under the LL condition, CST1 was expressed at a constantly low level comparable with its lowest expression level in the control condition. Under the DD condition, however, CST1 was expressed at a constantly high level comparable with its highest expression level in the control condition (Figure 6C). We therefore conclude that CST1 transcription is not regulated by the circadian clock per se, but is rather negatively associated with light illumination/photosynthesis.
To tease apart the effects of light signaling and photoassimilates on CST1 expression, wild-type and cst1 plants were exogenously supplied with Suc at 15 DAS by stem infusion (Hiyane et al., 2010) starting at noon, and CST1 expression was quantified in ear leaves 48 h after the onset of the treatment. Suc was used since it is the major transport form of assimilated carbon in maize. In both wild type and cst1, the abundance of the CST1 transcript was significantly reduced by exogenous Suc supply, compared to mock-treated plants (Figure 6D, left), supporting the idea that CST1 expression is suppressed by photoassimilates. Consistent with this view and the lower levels of carbohydrates in cst1, CST1 transcript abundance is higher in cst1 than in wild type for mock-treated plants (Figure 6D, left). We subsequently investigated the role of CST1 in carbon status-regulated stomatal movement. In wild-type plants, the exogenous Suc supply significantly reduced stomatal conductance (Figure 6D, right). However, this effect was abolished in cst1, as stomatal conductance was found at a low level in both mock- and Suc-treated cst1 plants (Figure 6D, right). Therefore, CST1 is indispensable for normal stomata opening and their responsiveness to leaf carbon status.
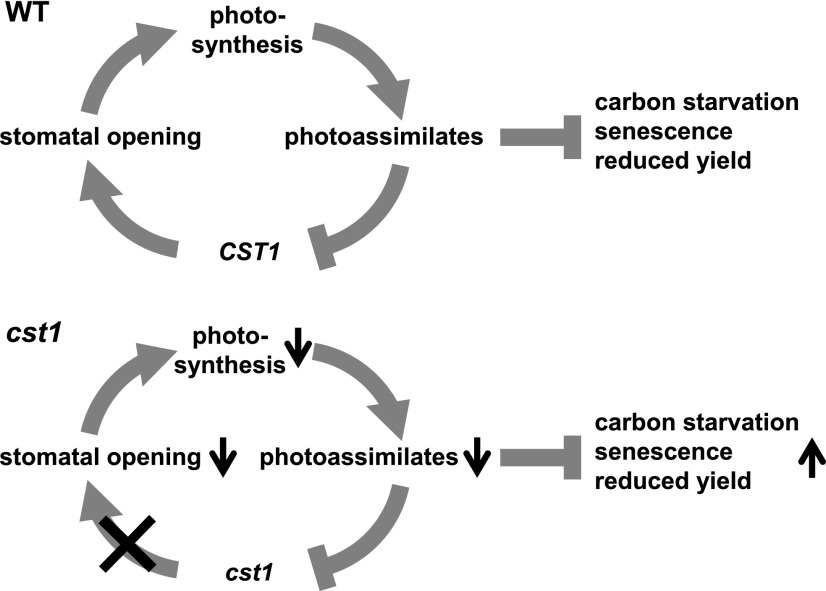
The Working Model of CST1
Based on the above results, we propose a working model of CST1 (Figure 7). In wild-type plants, the subsidiary cell-localized CST1 is an essential positive regulator of stomatal opening and photosynthesis. As CST1 is negatively regulated at the transcriptional level by photoassimilates, it possibly mediates the feedback inhibition of stomatal opening and photosynthesis by photoassimilates. In cst1 plants, loss-of-function of CST1 leads to diminished stomatal opening and photosynthesis, which in turn results in carbon starvation, early senescence in leaves, and reduced grain yield. The function of CST1 is likely specific to monocots, since 1) loss-of-function of its Arabidopsis ortholog, AtSWEET1, does not lead to any discernible aberrant phenotypes and 2) both maize and rice (Oryza sativa) harbor two orthologs of AtSWEET1 (Supplemental Figure 4), suggesting neofunctionalization of CST1 after a monocot-specific gene duplication event.
Figure 7.
Working Model for CST1.
At top, in wild-type (WT) plants, CST1 is a positive regulator of stomatal opening and photosynthesis. CST1 is also negatively regulated at the transcriptional level by photoassimilates. Thus it mediates the feedback inhibition of stomatal opening and photosynthesis by photoassimilates. At bottom, in cst1 plants, loss-of-function of CST1 leads to diminished stomatal opening and photosynthesis, which in turn results in carbon starvation, early senescence, and reduced yield.
DISCUSSION
CST1: an Example of Diverse Neofunctionalization in Clade I SWEET Family Genes
CST1 is a Clade I SWEET family gene, and is phylogenetically closest to AtSWEET1 (Supplemental Figure 6), the founding member of the SWEET gene family. Although AtSWEET1 was reported to encode a Glc transporter (Xuan et al., 2013), its physiological function is still unknown. In our studies, loss-of-function mutants of AtSWEET1 created by the CRISPR/Cas9 system do not exhibit any visible aberrant phenotypes. The other two Clade I SWEET family genes in Arabidopsis, AtSWEET2 and AtSWEET3, have tissue specificities and physiological functions distinct from CST1, although they also encode Glc transporters (Xuan et al., 2013; Chen et al., 2015). Therefore, while the SWEET family members within the same clade are highly conserved in protein sequences and substrate specificity, their subcellular localization, spatio-temporal expression pattern and physiological functions can be dramatically divergent (Eom et al., 2015). A lack of conservation in the physiological functions of Clade I SWEET family members both within and between species suggests extensive neo-functionalization following gene duplication (Schnable et al., 2011).
CST1 Controls the Grain Yield of Maize by a Distinctive Mechanism
Clade II and Clade III SWEET family genes have been reported as important genetic components contributing to grain yield in maize by different mechanisms. ZmSWEET4c is a Clade II subfamily member localized in the basal endosperm transfer layer and mediates Suc transport from maternal phloem into the seed (Sosso et al., 2015). Three paralogous Clade III subfamily genes (ZmSWEET13a, ZmSWEET13b, and ZmSWEET13c) are highly expressed in leaf vasculature and contribute to phloem loading (Bezrutczyk et al., 2018). In contrast with ZmSWEET4c, ZmSWEET13a, ZmSWEET13b, and ZmSWEET13c, CST1 encodes a subsidiary cell-specific Glc transporter, and functions to promote stomatal opening and photosynthesis.
Earlier studies demonstrated that photosynthetic capacity was closely associated with stomatal conductance, and subsequently has an impact on crop yield (Wong et al., 1979; Fischer et al., 1998). In cst1, stomatal aperture width and stomatal conductance were progressively reduced at the late developmental stage after silking, accompanied by a dramatic reduction in the photosynthetic rate and inadequate grain filling (Figures 1A, 1B, and 1F to 1H). Notably, the reduction of HKW in cst1 was around 7% (Figures 1G and 1H), which was less severe than the reduction of stomatal conductance measured in leaf L and L-3 (Figures 1A and 1B). Similar results were observed in the CRISPR lines, in which loss-of-function of CST1 reduced the stomatal conductance in the ear leaf by around 45% at 10 DAS, whereas the HKW was reduced by only around 14%. This is probably due to the fact that the leaves above the ear leaf in cst1 and the CRISPR lines lose their source capacity only at a later stage, and may produce sufficient photoassimilates to partially compensate for the reduced photosynthetic efficiency in senescent leaves, considering that leaves above the ear leaf contribute more to the grain yield than do lower leaves in maize (Subedi and Ma, 2005). It is also worth noting that maize uses C4 photosynthesis and is able to maintain normal photosynthesis at a lower intercellular CO2 level than C3 plants (Wong et al., 1979). Thus the photosynthetic capacity of maize might not be as sensitive to reduced stomatal conductance as in C3 plants.
CST1 Bridges the Gap between Stomatal Conductance and Leaf Senescence
The involvement of the stomata in the control of leaf senescence was suggested by floating leaf disc experiments using various chemical compounds that affect senescence (Thimann and Satler, 1979a, 1979b). Senescence-inhibiting compounds usually promote stomatal opening, whereas compounds that promote senescence generally induce stomatal closure. Subsequent experiments with detached leaves demonstrated that while stomatal closure accelerates senescence, stomatal opening is not directly linked to the prevention of leaf senescence (Thimann, 1985). Consistent with these observations, reduced stomata opening is associated with accelerated leaf senescence in cst1. Prolonged carbohydrate deprivation or starvation leads to early senescence in plants (Hanaoka et al., 2002; Buchanan-Wollaston et al., 2005; Kim et al., 2016), and our research demonstrated a dramatic reduction of sugar accumulation as well as upregulated sugar starvation marker genes in the cst1 mutant. Therefore, the early senescence phenotype of cst1 can be explained by its lower sugar accumulation, which may be a direct result of reduced stomatal conductance and photosynthesis. Notably, cst1 phenotypes become visible at around flowering, when the source/sink relationship changes dramatically, suggesting that the change in source/sink relationship may elevate the requirement for CST1 activity to maintain proper stomatal function. The emergence of cst1 phenotypes at flowering may also be due to weakened functional complementation from the paralogs of CST1. For example, the closest paralog of CST1, GRMZM2G039365, was found to be downregulated after flowering (Supplemental Data Set 3).
CST1 Defines the Role of Subsidiary Cells in Controlling Stomatal Movement and Source Capacity
Glc has been identified as an important signaling molecule upstream of numerous physiological processes (Sheen, 2014). Glc transporters such as CST1 are most likely components of this complex Glc signaling network. In Arabidopsis, Glc is sensed within guard cells by HXK to induce stomatal closure (Kelly et al., 2013; Lugassi et al., 2015; Li et al., 2018); thus, it is tempting to hypothesize that the function of CST1 may be mediated by HXK in maize. In contrast with HXK, CST1 functions as a positive regulator of stomatal opening. Therefore, a reasonable hypothesis is that CST1 protein may sequester Glc in subsidiary cells, thereby preventing it from entering guard cells to activate HXK signaling. It is also possible that when the Glc concentration inside subsidiary cells is higher than that of the apoplasmic space, CST1 could release Glc from subsidiary cells, leading to reduced osmotic and turgor pressure in the subsidiary cells, which in turn enables guard cells to overcome the large mechanical advantage of subsidiary cells for stomatal opening (Franks and Farquhar, 2007). In maize, potassium and chloride ions migrate from the subsidiary cells into the guard cells when the stomata open, and return to the subsidiary cells when the stomata close, suggesting a role of subsidiary cells as reservoirs for potassium and chloride ions (Raschke and Fellows, 1971). It is possible that CST1 is highly expressed at dawn to enhance Glc import into the subsidiary cells, and the Glc in turn provides energy to drive potassium and chloride ions into the guard cells from subsidiary cells, a step essential for stomatal opening. In addition, current evidence does not support a role of CST1 in stomatal closing. As the stomata of cst1 are partially closed under normal growth conditions, their insensitivity to Suc treatment cannot serve as evidence supporting the role of CST1 in stomatal closing. However, testing these hypotheses is technically challenging as it requires real-time high-resolution measurements of the levels of Glc and other ions in subsidiary cells, guard cells, and the apoplasmic space. Nevertheless, our study suggests that subsidiary cells may potentially represent a new signaling hub where exogenous and endogenous signals (besides carbon status) converge to regulate photosynthesis. It is conceivable that CST1 and possibly also other stomatal movement-controlling genes in subsidiary cells may be exploited to fine-tune the CO2 availability and photosynthetic rates of crop plants to maximize grain yield.
METHODS
Plant Materials and Growth Conditions
The cst1 mutant was isolated from an ethyl methanesulfonate mutagenized population of maize in the Zheng58 background and backcrossed to Zheng58 three times. The F2 mapping population was derived from a cross between cst1 and the maize inbred line B73. Wild-type and cst1 plants in the F2 population were phenotyped under field conditions at 20 DAS. F2 plants with cst1 phenotypes were used for gene mapping.
Time-Course Transcriptome and Metabolome Analysis of cst1 and Wild-Type Leaf Samples
Experimental procedures for time-course transcriptome and metabolome analysis are described in Supplemental Information 2.
Photosynthesis, Stomatal Conductance, and Stomatal Aperture Width Measurements
Measurements of photosynthesis were performed between 1 PM and 3 PM at 0, 10, 20, and 30 DAS using a portable photosynthesis system LI-6400 (LI-COR). Ten plants were randomly selected for each genotype. L and L-3 leaves were used to measure the photosynthetic rate and stomatal conductance (Gs). Measurements were taken at ambient CO2 (400 μL L–1), with leaf temperature at 30°C and photosynthetic photon flux density at 1800 µmol m–2 s–1. Measurements of the SAW were taken between 10 AM and 11 AM on sunny days with a temperature between 30° and 32°C. The adaxial epidermis of the leaves was carefully smeared with nail varnish from the mid-area between the central vein and the leaf edge. The thin film was peeled off the leaf surface and immediately mounted on a glass slide. The imprints were measured under an OLYMPUS AX80 (Olympus Corporation) microscope equipped with a digital camera (Diagnostic Instruments).
Development of Molecular Markers for Map-Based Cloning of cst1
Simple Sequence Repeat markers from the MaizeGDB (https://maizegdb.org/data_center/ssr) were screened to identify markers that are polymorphic between Zheng58 and B73. To develop markers for HRM analysis of recombinants in the fine-mapping of cst1, next generation sequencing reads of Zheng58 (accession numbers SRR449340, SRR449342, and SRR449343) were downloaded from the Sequence Read Archive. Reads were converted to fastq format by fastq-dump in sra-tools (Version 2.8.2), mapped to the B73 reference genome by BWA (Version 0.7.12), followed by SNP calling with the GATK pipeline (Version 3.1). Thirty high-confidence SNPs located between the simple sequence repeat markers bnlg1702 and umc1462 were selected to design primers for HRM analysis. For PCR amplification of target sequences, 1 μL 10 x LC Green (Idaho Technology Inc.) was included in 10 μL PCR volumes. The resulting PCR products were analyzed using the LightScanner system (Idaho Technology Inc.), by ramping the temperature from 55° to 95°C at 0.1°C per s. Amplicons were genotyped using the LightScanner software (Idaho Technology Inc.).
Creating Loss-of-Function Mutants of CST1 with the CRISPR-Cas9 System in Maize
Experimental procedures for the generation of loss-of-function mutants of CST1 are described in Supplemental Information 1.
Yeast Complementation Assay and [14C]-D-Glc Uptake Assay
The CDS of CST1 and CST1E81K (774 bp) were cloned using the Zero Blunt™ PCR Cloning Kit (Invitrogen) to generate pZERO-CST1 and pZERO-CST1E81K, respectively. The CST1 and CST1E81K CDS were then amplified from pZERO-CST1 and pZERO-CST1E81K using pfx polymerase (Invitrogen) with gene-specific primers CST1-attB1: ggggacaagtttgtacaaaaaagcaggcttaATGGAGGATGTGGTGAAGTTCGTCT and CST1-attB2: ggggaccactttgtacaagaaagctgggtaGACCAGGCGGTCCTCCTTGCCGCCG. The cDNA fragments were cloned into the pDONR221-f1 vector by BP clonase II (Invitrogen) and then transferred to the pDRf1-GW vector (Loqué et al., 2007) using LR clonase II (Invitrogen), resulting in the pDRf1-GW-CST1 and pDRf1-GW-CST1E81K constructs. The positive controls (pDRf1-GW-hexose transporter5 [HXT5] and pDRf1-GW-AtSWEET1) and the vector control were constructed as previously described (Chen et al., 2010). Modified yeast stain EBY.VW.4000 carrying the cytosol invertase named YSL2-1 (MATα, ura3-52, leu2-3, 112, his3-∆1, trp1-289, MAL 2-8c, hxt1-18∆::loxP, gal2∆::loxP, agt1 m2-3∆::loxP, pSUC2::pHXT7; Chen et al., 2015) was used for transformation using the LiAc method. Transformants were selected on synthetic deficient media lacking uracil (0.17% [w/v] yeast nitrogen base w/o amino acid and ammonium sulfate [Difco], 0.5% [w/v] ammonium sulfate, 2% [w/v] maltose, 1% [w/v] agar, and 0.01% [w/v] of His, Leu, and Trp). The yeast spotting assay was performed as previously described (Chen et al., 2010).
The relative Glc uptake rates of CST1, CST1E81K, and AtSWEET1 were determined by the [14C]-D-Glc uptake assay following the procedure described before (Chen et al., 2010) with minor modifications. Briefly, the yeast Glc transport deficient mutant EBY.VW4000 was transformed with the vector pDRf1-GW, pDRf1-GW-CST1, pDRf1-GW-CST1E81K, or pDRf1-GW-AtSWEET1 (see descriptions above), respectively. For each vector, three colonies were picked and cultured in Synthetic Defined medium supplemented with 2% (w/v) maltose for OD600 to reach 0.5–0.7. Cells were weighed after being harvested and washed twice with ice-cold distilled water, and then resuspended in 40-mM ice-cold potassium phosphate buffer (pH 6.0) to reach a concentration of 5% (w/v). For each colony, 330 μL cells were taken and incubated in the potassium phosphate buffer at 30°C for 5 min. To start the reaction, prewarmed equal volumes of buffer containing 10 mM Glc and 0.1μCi [14C]-D-Glc (PerkinElmer) was added into each cell suspension, and incubated at 30°C for 5 min or 10 min. At each time point, a 120 μL aliquot was withdrawn, washed twice with 10 mL of cold deionized water, and analyzed using the 1450 MicroBeta TriLuxMicroplate Scintillation and Luminescence Counter (PerkinElmer). For each vector, count per minute values were normalized to AtSWEET1 (set to 100%) after removal of background signal and correction for the efficiencies of detectors.
Glc Transport Activity Assay using FRET Glc Sensor in HEK293T Cells
The CDS of CST1 and CST1E81K were cloned into the mammalian expression vector, pcDNA3.2/V5-DEST (Invitrogen), and verified by Sanger sequencing. The Glc transport activity was analyzed as described before (Chen et al., 2010; Hou et al., 2011). Briefly, HEK293T cells were cotransfected with the plasmid carrying the Glc sensor FLIPglu600μΔ13V and a plasmid carrying CST1, CST1E81K or AtSWEET1(a positive control) using Lipofectamine2000 (Invitrogen) in 96-well plates as described before (Chen et al., 2010). A Zeiss Axio Oberve Z1/7 with Hamamatsu camera ORCA-Flash4.0 V3 Digital CMOS was used for imaging. A 60 μL Hanks Balanced Saline Salt buffer was used to replace the culture medium 30 min before imaging, and the cells were washed once every 10 min. Also, 60 μL of 80 mM Glc was added to the well to reach a final Glc concentration of 40 mM after 2 min of imaging. The filter set used for FRET emission was CFP excitation-YFP emission: 436/20–535/30. The camera exposure time was 200 ms and the interval time was 10 s. The ratio of FRET emission to CFP emission at the excitation of CFP was analyzed and normalized as described before (Hou et al., 2011). Eleven regions of interest including 10 individual cells and a region of background were chosen from one well of a 96-well plate.
BiFC Assay
The CDS of CST1 and CST1E81K were amplified using the primers BIFC-5: aactagtATGGAGGATGTGGTGAAGTTCGTC and BIFC-3: aatcgatGACCAGGCGGTCCTCCTTGCC from Zheng58 and cst1 leaf cDNA, respectively, using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The amplicons were then cloned into the pCR–BluntII-TOPO (Invitrogen) vector, and then subcloned into the binary vector pSPYNE‐35S and pSPYCE‐35S (Walter et al., 2004) at SpeI and ClaI restriction sites to generate the four vectors: pSPYNE‐35S-CST1, pSPYCE‐35S-CST1, pSPYNE‐35S-CST1E81K, and pSPYCE‐35S-CST1E81K. Various combinations of N-end and C-end constructs were then used in Agrobacterium-mediated transient transformation of Nicotiana benthamiana epidermal cells. Two days after transformation, the YFP signal was photographed using a LSM 700 camera (Zeiss). Each BiFC assay was repeated three times, and consistent results were obtained.
Quantification of Starch
Procedures for starch quantification are detailed in Supplemental Information 2.
Real-Time RT-qPCR
Total RNA was isolated from maize leaves using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The isolated RNA was treated with RNase-free DNase (Promega). First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and an oligo (dT) 18 primer. The EF1-alpha transcript was used as an internal control to normalize RNA quantity. RT-qPCR was performed with Maxima SYBR Green/ROX qPCR Master Mix (Invitrogen) using the StepOnePlus system (Applied Biosystems).
Phylogenetic Analysis of the SWEET Protein Family
Protein sequences of Arabidopsis (Arabidopsis thaliana), Oryza sativa Japonica, Zea mays, and Chlamydomonas reinhardtii were downloaded from EnsemblPlants V3.31 (http://plants.ensembl.org). SWEET family proteins in these species were identified by blastp in stand-alone BLAST (version 2.3.0) using Arabidopsis SWEET family proteins as queries. Multiple sequence alignment of SWEET proteins from the four species was performed using MEGA 7.0 (Kumar et al., 2016).
Generation of GUS and GFP Fusion Constructs and Observation of Reporter Gene Expression
For transient expression of CST1-eYFP and CST1E81K-eYFP fusion proteins in maize protoplasts and N. benthamiana epidermal cells, the CDS of CST1 and CST1E81K were amplified by the primer pair CST1EYFP-5 aggtaccATGGAGGATGTGGTGAAGTTCGTC and CST1EYFP-3 aggatccCGGACCAGGCGGTCCTCCTTGCC from Zheng58 and cst1 leaf cDNA, respectively. The amplicons were cloned into pCR–BluntII-TOPO (Invitrogen) and then subcloned into the pCAM35S::eYFP binary vector (Wang et al., 2013) at KpnI and BamHI restriction sites. To generate the ProCST1:CST1-eYFP:TerCST1 construct, the promoter and genic region of CST1 (from 2 kb upstream of the start codon to 1 bp before the stop codon) and also the terminator region of CST1 (from the stop codon to 500 bp downstream of the stop codon) were amplified from B73 genomic DNA using the Phusion High-Fidelity DNA Polymerase (New England Biolabs). These two fragments, along with a third fragment encoding eYFP, were ligated with the binary vector pCAMBIA3300 by seamless cloning with the In-Fusion Cloning Kit (Takara). The ProCST1:CST1-GUS:TerCST1 construct was generated using the same method as above. Stable transformants of ProCST1:CST1-eYFP:TerCST1 and ProCST1:CST1-GUS:TerCST1 in the C01 inbred genetic background were generated by the China National Seed Group. Histochemical GUS activity staining was performed as previously described (Wang et al., 2013). The eYFP and FM4-64 (Invitrogen, http://www.invitrogen.com) signal was observed using a Nikon C1 confocal microscope (Nikon) with the following settings: 488 nm argon laser line, 500–530 nm bandpass detection for YFP, and 560–615 nm bandpass detection for FM4-64.
Suc Feeding by Stem Infusion
Stem infusions were conducted as reported before (Hiyane et al., 2010). Each plant received 25 mL Suc at a concentration of 0.438 mM per d. The infusion was performed at the upper part of the second internode under the ear internode from noon of 15 DAS to noon of 16 DAS. A second infusion was performed at the upper part of the internode immediately under the ear internode from noon of 16 DAS to noon of 17 DAS. Sample harvesting and stomatal conductance measurements were performed at noon of 17 DAS.
Accession Numbers
Sequence data of CST1 can be found in the Maize Genetics and Genomics Database (https://maizegdb.org/) under the following accession number: GRMZM2G153358. The other accession numbers used for phylogenetic analysis can be found in Supplemental Data Set 6 and Supplemental Figure 6. RNA sequencing data are available in the Sequence Read Archive repository under Accession No. SRP148646 (https://www.ncbi.nlm.nih.gov/sra/SRP148646).
Supplemental Data
Supplemental Figure 1. Temporal dynamics of stomatal aperture width (SAW) and stomatal conductance (gs) in WT and cst1.
Supplemental Figure 2. Plant architecture and flowering time of WT and cst1 plants.
Supplemental Figure 3. Transcriptome profiling of cst1 and WT.
Supplemental Figure 4. Principal component analysis (PCA) of the untargeted metabolomic data from GC-MS and LC-MS.
Supplemental Figure 5. Multivariate statistical analysis of the untargeted metabolomic data from GC-MS and LC-MS.
Supplemental Figure 6. Phylogenetic analysis of SWEET family proteins from Zea mays, Oryza sativa, Arabidopsis thaliana, and Chlamydomonas reinhardtii.
Supplemental Figure 7. Subcellular localization of the CST1 protein.
Supplemental Figure 8. Tissue-specificity of CST1 expression.
Supplemental Figure 9. Expression levels of genes in sucrose and starch metabolism.
Supplemental Information 1. Generating loss-of-function mutants of CST1 by the CRISPR/Cas9 system.
Supplemental Information 2. Transcriptome and metabolome profiling of cst1 and WT.
Supplemental Data Set 1. Statistical tests in this study.
Supplemental Data Set 2. Summary of the RNA-Seq samples in this study.
Supplemental Data Set 3. Differentially expressed genes between cst1 and WT.
Supplemental Data Set 4. Metabolites detected by GC-MS and LC-MS.
Supplemental Data Set 5. Metabolites differentially accumulated in cst1 and WT.
Supplemental Data Set 6. Text file of the alignment used to generate the phylogenetic tree in Supplemental Figure 6.
Supplemental Data Set 7. Differentially expressed putative stomatal movement-controlling genes.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Eckhard Boles and Dr. Yanxin Zhao for the EBY.VW4000 yeast strain, Dr. Cuimin Liu for help with the [14C]-D-glucose uptake assay, and Dr. Edward S. Buckler for helpful discussions and suggestions. This work was supported by The National Key Research and Development Program of China (2016YFD0100405); The Fundamental Research Funds for the Chinese Academy of Agricultural Sciences (Y2017JC07); The Science and Technology Program of Guangdong Province, China (2016B030303007); and the Ph.D. Start-up Fund of the Natural Science Foundation of Guangdong Province, China (2014A030310489).
AUTHOR CONTRIBUTIONS
H.W., C.Z., and Z.L. supervised the project; H.W., S.Y., and L.-Q.C. designed the experiments; H.W., S.Y., H.X., W.H., H.Z., S.T., Y.-C.Y., X.L., P.L., S.L., Y.-L.R., and L.-Q.C. performed research; H.W., S.Y., A.R.F., and L.-Q.C. analyzed data and wrote the article.
References
- Ainsworth E.A., Bush D.R. (2011). Carbohydrate export from the leaf: A highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol. 155: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolakos P., Livanos P., Giannoutsou E., Panteris E., Galatis B. (2018). The intracellular and intercellular cross-talk during subsidiary cell formation in Zea mays: Existing and novel components orchestrating cell polarization and asymmetric division. Ann. Bot. 122: 679–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., et al. (2011). Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T., Noguchi K., Terashima I. (2006). Effects of carbohydrate accumulation on photosynthesis differ between sink and source leaves of Phaseolus vulgaris L. Plant Cell Physiol. 47: 644–652. [DOI] [PubMed] [Google Scholar]
- Assmann S.M., Jegla T. (2016). Guard cell sensory systems: Recent insights on stomatal responses to light, abscisic acid, and CO2. Curr. Opin. Plant Biol. 33: 157–167. [DOI] [PubMed] [Google Scholar]
- Baker R.F., Leach K.A., Boyer N.R., Swyers M.J., Benitez-Alfonso Y., Skopelitis T., Luo A., Sylvester A., Jackson D., Braun D.M. (2016). Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiol. 172: 1876–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C.E., Arruda P. (2002). Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol. 128: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrutczyk M., Hartwig T., Horschman M., Char S.N., Yang J., Yang B., Frommer W.B., Sosso D. (2018). Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 218: 594–603. [DOI] [PubMed] [Google Scholar]
- Braun D.M., Ma Y., Inada N., Muszynski M.G., Baker R.F. (2006). tie-dyed1 Regulates carbohydrate accumulation in maize leaves. Plant Physiol. 142: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.-F., Wu S.-H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585. [DOI] [PubMed] [Google Scholar]
- Callard D., Axelos M., Mazzolini L. (1996). Novel molecular markers for late phases of the growth cycle of Arabidopsis thaliana cell-suspension cultures are expressed during organ senescence. Plant Physiol. 112: 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Q. (2014). SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 201: 1150–1155. [DOI] [PubMed] [Google Scholar]
- Chen H.-Y., Huh J.-H., Yu Y.-C., Ho L.-H., Chen L.-Q., Tholl D., Frommer W.B., Guo W.-J. (2015). The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 83: 1046–1058. [DOI] [PubMed] [Google Scholar]
- Chevalier C., Bourgeois E., Just D., Raymond P. (1996). Metabolic regulation of asparagine synthetase gene expression in maize (Zea mays L.) root tips. Plant J. 9: 1–11. [DOI] [PubMed] [Google Scholar]
- Christensen L.E., Below F.E., Hageman R.H. (1981). The effects of ear removal on senescence and metabolism of maize. Plant Physiol. 68: 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daloso D.M., Antunes W.C., Pinheiro D.P., Waquim J.P., Araújo W.L., Loureiro M.E., Fernie A.R., Williams T.C. (2015). Tobacco guard cells fix CO2 by both Rubisco and PEPcase while sucrose acts as a substrate during light-induced stomatal opening. Plant Cell Environ. 38: 2353–2371. [DOI] [PubMed] [Google Scholar]
- Daloso D.M., Dos Anjos L., Fernie A.R. (2016). Roles of sucrose in guard cell regulation. New Phytol. 211: 809–818. [DOI] [PubMed] [Google Scholar]
- Davies K.M., Seelye J.F., Irving D.E., Borst W.M., Hurst P.L., King G.A. (1996). Sugar regulation of harvest-related genes in asparagus. Plant Physiol. 111: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C., Lemaître T., Christ A., Azzopardi M., Kato Y., Sato F., Morot-Gaudry J.-F., Le Dily F., Masclaux-Daubresse C. (2008). Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 147: 1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Downs C.G., Somerfield S.D. (1997). Asparagine synthetase gene expression increases as sucrose declines in broccoli after harvest. N. Z. J. Crop Hortic. Sci. 25: 191–195. [Google Scholar]
- Eom J.-S., Chen L.-Q., Sosso D., Julius B.T., Lin I.W., Qu X.-Q., Braun D.M., Frommer W.B. (2015). SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 25: 53–62. [DOI] [PubMed] [Google Scholar]
- Farquhar G.D., Sharkey T.D. (1982). Stomatal conductance and photosynthesis. Annu. Rev. Plant Physiol. 33: 317–345. [Google Scholar]
- Fischer R.A., Rees D., Sayre K.D., Lu Z.M., Condon A.G., Larque Saavedra A. (1998). Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Sci. 38: 1467–1475. [Google Scholar]
- Fischer-Kilbienski I., Miao Y., Roitsch T., Zschiesche W., Humbeck K., Krupinska K. (2010). Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 73: 379–390. [DOI] [PubMed] [Google Scholar]
- Franks P.J., Farquhar G.D. (2007). The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol. 143: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Yoshikawa Y., Sato T., Inada N., Ito M., Nishida I., Watanabe A. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111: 345–352. [DOI] [PubMed] [Google Scholar]
- Gago J., Fernie A.R., Nikoloski Z., Tohge T., Martorell S., Escalona J.M., Ribas-Carbó M., Flexas J., Medrano H. (2017). Integrative field scale phenotyping for investigating metabolic components of water stress within a vineyard. Plant Methods 13: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46: 601–612. [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyane R., Hiyane S., Tang A.C., Boyer J.S. (2010). Sucrose feeding reverses shade-induced kernel losses in maize. Ann. Bot. 106: 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrer D., Flütsch S., Pazmino D., Matthews J.S.A., Thalmann M., Nigro A., Leonhardt N., Lawson T., Santelia D. (2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26: 362–370. [DOI] [PubMed] [Google Scholar]
- Hou B.-H., Takanaga H., Grossmann G., Chen L.-Q., Qu X.-Q., Jones A.M., Lalonde S., Schweissgut O., Wiechert W., Frommer W.B. (2011). Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 6: 1818–1833. [DOI] [PubMed] [Google Scholar]
- Imes D., Mumm P., Böhm J., Al-Rasheid K.A.S., Marten I., Geiger D., Hedrich R. (2013). Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74: 372–382. [DOI] [PubMed] [Google Scholar]
- Jeannette E., Reyss A., Gregory N., Gantet P., Prioul J.L. (2000). Carbohydrate metabolism in a heat-girdled maize source leaf. Plant Cell Environ. 23: 61–69. [Google Scholar]
- Kang Y., Outlaw W.H. Jr., Andersen P.C., Fiore G.B. (2007). Guard-cell apoplastic sucrose concentration--a link between leaf photosynthesis and stomatal aperture size in the apoplastic phloem loader Vicia faba L. Plant Cell Environ. 30: 551–558. [DOI] [PubMed] [Google Scholar]
- Kelly G., Moshelion M., David-Schwartz R., Halperin O., Wallach R., Attia Z., Belausov E., Granot D. (2013). Hexokinase mediates stomatal closure. Plant J. 75: 977–988. [DOI] [PubMed] [Google Scholar]
- Kim J., Woo H.R., Nam H.G. (2016). Toward systems understanding of leaf senescence: An integrated multi-omics perspective on leaf senescence research. Mol. Plant 9: 813–825. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanemets K., et al. (2013). Mutations in the SLAC1 anion channel slow stomatal opening and severely reduce K+ uptake channel activity via enhanced cytosolic [Ca2+] and increased Ca2+ sensitivity of K+ uptake channels. New Phytol. 197: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T., Blatt M.R. (2014). Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164: 1556–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-L., Wang M., Ma X.-Y., Zhang W. (2014). NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol. Plant 7: 1508–1521. [DOI] [PubMed] [Google Scholar]
- Li Y., Xu S., Wang Z., He L., Xu K., Wang G. (2018). Glucose triggers stomatal closure mediated by basal signaling through HXK1 and PYR/RCAR receptors in Arabidopsis. J. Exp. Bot. 69: 1471–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wu S., Chen J., Wang X., Gao J., Ren G., Kuai B. (2017). NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis. Plant J. 92: 650–661. [DOI] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L.L., von Wirén N., Frommer W.B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198. [DOI] [PubMed] [Google Scholar]
- Lugassi N., Kelly G., Fidel L., Yaniv Y., Attia Z., Levi A., Alchanatis V., Moshelion M., Raveh E., Carmi N., Granot D. (2015). Expression of Arabidopsis hexokinase in citrus guard cells controls stomatal aperture and reduces transpiration. Front. Plant Sci. 6: 1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Baker R.F., Magallanes-Lundback M., DellaPenna D., Braun D.M. (2008). Tie-dyed1 and sucrose export defective1 act independently to promote carbohydrate export from maize leaves. Planta 227: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan D.H., Lan J., Geilfus C.-M., Dodd A.N., Larson T., Baker A., Hõrak H., Kollist H., He Z., Graham I., Mickelbart M.V., Hetherington A.M. (2016). The breakdown of stored triacylglycerols is required during light-induced stomatal opening. Curr. Biol. 26: 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Mumm P., Imes D., Endler A., Weder B., Al-Rasheid K.A.S., Geiger D., Marten I., Martinoia E., Hedrich R. (2010). AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63: 1054–1062. [DOI] [PubMed] [Google Scholar]
- Misra B.B., Acharya B.R., Granot D., Assmann S.M., Chen S. (2015). The guard cell metabolome: Functions in stomatal movement and global food security. Front. Plant Sci. 6: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa A., Ito M., Hayashi H., Watanabe A. (1999). Dark-induced expression of genes for asparagine synthetase and cytosolic glutamine synthetase in Radish Cotyledons is dependent on the growth stage. Plant Cell Physiol. 40: 942–948. [Google Scholar]
- Omasits U., Ahrens C.H., Müller S., Wollscheid B. (2014). Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30: 884–886. [DOI] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.-S., Martínez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41: 831–844. [DOI] [PubMed] [Google Scholar]
- Outlaw W.H. Jr., De Vlieghere-He X. (2001). Transpiration rate. An important factor controlling the sucrose content of the guard cell apoplast of broad bean. Plant Physiol. 126: 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Matos J.L., Anleu Gil M.X., Kornfeld A., Bettadapur A., Abrash E., Allison H.R., Badgley G., Vogel J.P., Berry J.A., Bergmann D.C. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Raschke K., Fellows M.P. (1971). Stomatal movement in Zea mays: Shuttle of potassium and chloride between guard cells and subsidiary cells. Planta 101: 296–316. [DOI] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Childs K.L., Santoro N., Foster C.E., Buell C.R., de Leon N., Kaeppler S.M. (2012). Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 159: 1730–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2014). Master regulators in plant glucose signaling networks. J. Plant Biol. 57: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T.L., Baker R.F., Stubert A., Braun D.M. (2012). Tie-dyed2 encodes a callose synthase that functions in vein development and affects symplastic trafficking within the phloem of maize leaves. Plant Physiol. 160: 1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47: 1489–1493. [DOI] [PubMed] [Google Scholar]
- Subedi K.D., Ma B.L. (2005). Ear position, leaf area, and contribution of individual leaves to grain yield in conventional and leafy maize hybrids. Crop Sci. 45: 2246–2257. [Google Scholar]
- Süssenbacher I., Hörtensteiner S., Kräutler B. (2015). A dioxobilin-type fluorescent chlorophyll catabolite as a transient early intermediate of the dioxobilin-branch of chlorophyll breakdown in Arabidopsis thaliana. Angew. Chem. Int. Ed. Engl. 54: 13777–13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbott L.D., Zeiger E. (1996). Central roles for potassium and sucrose in guard-cell osmoregulation. Plant Physiol. 111: 1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Cheung L.S., Li S., Eom J.-S., Chen L.-Q., Xu Y., Perry K., Frommer W.B., Feng L. (2015). Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527: 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K.V. (1985). The senescence of detached leaves of tropaeolum. Plant Physiol. 79: 1107–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K.V., Satler S. (1979a). Relation between senescence and stomatal opening: Senescence in darkness. Proc. Natl. Acad. Sci. USA 76: 2770–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann K.V., Satler S.O. (1979b). Relation between leaf senescence and stomatal closure: Senescence in light. Proc. Natl. Acad. Sci. USA 76: 2295–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houtte H., Vandesteene L., López-Galvis L., Lemmens L., Kissel E., Carpentier S., Feil R., Avonce N., Beeckman T., Lunn J.E., Van Dijck P. (2013). Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol. 161: 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang H., Lu Y., Jiang T., Berg H., Li C., Xia Y. (2013). The Arabidopsis U-box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J. 74: 511–523. [DOI] [PubMed] [Google Scholar]
- Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C.P., Boles E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464: 123–128. [DOI] [PubMed] [Google Scholar]
- Wong S.C., Cowan I.R., Farquhar G.D. (1979). Stomatal conductance correlates with photosynthetic capacity. Nature 282: 424–426. [Google Scholar]
- Wu A., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Y.H., Hu Y.B., Chen L.-Q., Sosso D., Ducat D.C., Hou B.-H., Frommer W.B. (2013). Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 110: E3685–E3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Assmann S.M. (2014). Metabolite transporter regulation of ABA function and guard cell response. Mol. Plant 7: 1505–1507. [DOI] [PubMed] [Google Scholar]
- Zelisko A., García-Lorenzo M., Jackowski G., Jansson S., Funk C. (2005). AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 102: 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Chung K.P., Cui Y., Lin W., Gao C., Kang B.-H., Jiang L. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E426–E435. [DOI] [PMC free article] [PubMed] [Google Scholar]