Abstract
The metabolic etiology of breast cancer has been explored in the past several years using metabolomics. However, most of these studies only included non-Hispanic White individuals. To fill this gap, we performed a two-step (discovery and validation) metabolomics profiling in plasma samples from 358 breast cancer patients and 138 healthy controls. All study subjects were either Hispanics or non-Hispanic African Americans. First, a panel of 14 identified metabolites significantly differed between breast cancer cases and healthy controls in both the discovery and validation sets. Most of these identified metabolites were lipids. In the pathway analysis, citrate cycle (TCA cycle), arginine and proline metabolism, and linoleic acid metabolism pathways were observed, and they significantly differed between breast cancer cases and healthy controls in both sets. From those 14 metabolites, we selected 9 non-correlated metabolites to generate a metabolic risk score. Increased metabolites risk score was associated with a 1.87 and 1.63-fold increased risk of breast cancer in the discovery and validation sets, respectively (Odds ratio (OR) =1.87, 95% Confidence interval (CI): 1.50, 2.32; OR=1.63, 95%CI: 1.36, 1.95). In summary, our study identified metabolic profiles and pathways that significantly diffed between breast cancer cases and healthy controls in Hispanic or non-Hispanic African American women. The results from our study might provide new insights on the metabolic etiology of breast cancer.
Keywords: metabolomics, breast cancer, racial minority
Introduction
Breast cancer is the most common form of cancer and the second most common cause of cancer death in women worldwide. The expected number of new breast cancer cases is predicted to increase in the next two decades [1]. Especially, the cancer incidence among Hispanic and African American populations continues to grow, mainly estrogen receptor positive (ER+) breast cancer, despite the mortality rate decline in some populations [2]. Although racial disparities in breast cancer are well-established, the contributing factors largely remain to be determined. A better understanding of the differences in the biological characteristics of breast cancers among racial groups could provide important insights into observed racial differences.
The investigation of metabolites may help solve the mystery of breast cancer racial disparity. Metabolites are the end products of intercellular pathways and are sensitive to host and environmental stimuli, thus, metabolite profiles may reflect changes in phenotype and function and serve as indicators of the overall physiological alterations in chronic disease [3]. Some of those stimuli are racial specific, then the corresponding metabolic profiles may be racial specific too. Thus, studying racial difference in metabolic profiles may shed light on the racial difference in host and environmental exposure. Furthermore, evidence has shown that metabolic profiles are at least partially genetically determined [4].
In breast cancer, metabolomics profiling in circulation (e.g. serum and plasma) has been successfully applied in identifying cancer risk factors [5–7] as well as prognostic factors [8–10] and early detection biomarkers [11,12,7,13]. Unfortunately, racial minorities, including Hispanics and non-Hispanic African Americans are rarely included in those studies. Such omission leads to a missed opportunity to explore the role of metabolomics in racial disparity of breast cancer.
In the current study, we conducted a two-step (discovery and validation) metabolomics analysis in plasma samples from 358 breast cancer patients and 138 healthy controls. All study subjects were either Hispanics or non-Hispanic African Americans. Our goal was to identify metabolic profiles that could distinguish between breast cancer cases and controls.
Materials and Methods
Study population
Breast cancer patients were recruited from The University of Texas M. D. Anderson Cancer Center (Houston, TX) from patients with newly diagnosed (defined by the presence of malignant breast epithelial cells) and histologically confirmed (by microscopic analysis and molecular subtype) breast cancer. The blood samples were drawn prior to any cancer treatment. Specifically, the breast cancer patients included in the discovery set were selected from an existing breast cancer early detection study from 2003 to 2012 [14]. Written informed consent was obtained from each study subject. A total of 134 cases were included in the discovery set. They were selected according to the availability of plasma samples and clinical characteristics at baseline, as well as race and ethnicity (Hispanics and non-Hispanic African Americans). Self-reported ethnic background was used to define the race and ethnicity. The validation set included 224 breast cancer patients selected from an ongoing breast cancer case control study that has been recruiting participants since 2012. The same selection criteria were used. In addition to the cases, each study also recruited control subjects. Controls for the discovery study were identified from healthy women who participated in breast cancer screening at MD Anderson Cancer Center and were found to be negative for the disease. Controls for the validation study were identified largely from female residents of Harris County using random digit dialing. In the current study, the discovery set had 57 Hispanic or non-Hispanic African American female controls selected from the existing breast cancer early detection study, and the validation set had 81 Hispanic or non-Hispanic African American female controls selected from the ongoing breast cancer case control study. The selection criteria included the availability of plasma samples and epidemiological characteristics at baseline, as well as race and ethnicity (Hispanics and non-Hispanic African Americans). The study protocol was approved by the Institutional Review Board at M D Anderson Cancer Center.
Metabolomics Analysis
Metabolomics profiling was conducted on plasma samples at Metabolon Inc (Durham, NC) using ultra high performance liquid chromatography/mass spectroscopy and gas chromatography/mass spectroscopy. The protocol was described previously [7]. Forty batches of case‐control pairs and quality control (QC) replicate samples (N=16) were analyzed. Raw data was extracted, peak‐identified and QC processed on the assay platform as previously described [15,16]. After excluding metabolites that had ≥30% missing values, 561 identified compounds remained for analysis. The missing values were treated as the result of low signal intensity. They were replaced by half of the minimum positive values detected in the data. Based on existing literature, metabolites were categorized as eight mutually exclusive chemical classes (amino acids, carbohydrates, cofactors and vitamins, energy metabolites, lipids, nucleotides, peptides and xenobiotics). The median and interquartile range of the coefficient of variation (CV) across the metabolites was 10% (4–20%), similar to previous reports analyzed by Metabolon [17].
Statistical Analysis
Any missing values were assumed to be below the limits of detection, and these values were imputed with the compound minimum (minimum value imputation). Basic statistical analysis of log-transformed data was conducted using MetaboAnalyst 3.0 (http://www.metaboanalyst.ca/). Non-parametric Wilcoxon rank tests comparing metabolites differences between cases and controls were used in metabolomics data analyses and performed independently for the discovery and validation sets. To control the multiple comparison, we set the false discovery rate (FDR) at 0.05 [18]. To assess the association with breast cancer risk, logistic regression analysis was used, adjusting for age, obesity status and race differences. The assumptions of logistic regression, including the independence of each observation, collinearity among the independent variables, and linearity of independent variables and log odds, were assessed. Using significant metabolites in both discovery and validation sets, we applied partial least squares – discriminant analysis (PLS-DA) to assess the difference between the case and control groups. Metabolic pathway analysis was conducted to identify the enriched metabolite sets and significant metabolic pathways that could differentiate breast cancer cases and controls. The latest version of The Human Metabolome Database (HMDB) 3.0 was used in metabolic pathway analysis [19]. In addition, pathway topology analysis was used to estimate the impact of a certain metabolite or a group of metabolites in a certain metabolic pathway, and relative-betweeness centrality test was used to estimate the impact [20]. To further assess whether associations were independent, we evaluated the pairwise correlations between all metabolites significantly associated with breast cancer risk in both discovery and validation sets among the controls. Metabolites whose pairwise correlations greater than 0.5 were considered highly correlated and to have possible redundancy. The selection of 0.5 as the cutoff point is based on our previously published studies [21,22]. Using 9 non-redundant metabolites, we generated a metabolic risk score. For each metabolite, using the median level in the controls as the cutoff point, we stratified the study subjects into high and low level categories. Next, based on the relationship between metabolite levels and breast cancer risk, we scored the study subjects as either high or low breast cancer risk (0 or 1) and summed scores across 9 metabolites to generate a metabolic risk score (range: 0–9). Then, we generated a metabolic risk score based on the number of high level of those 9 metabolites. Logistic regression analysis was applied to assess the relationship between the metabolic risk score and breast cancer risk, adjusted for co-variates above. All analyses were performed with STATA software version 10.1 (STATA, College Station, TX).
Results
Characteristics of the study populations
The discovery set included 134 breast cancer patients and 57 healthy controls, and the validation set included 224 breast cancer patients and 81 healthy controls. Characteristics of case and control groups were described in Table 1. All study subjects are either Hispanic or non-Hispanic African American women. Specifically, 77 (57.46%) cases and 38 (66.67%) controls were Hispanics in the discovery set, and 133 (58.48%) cases and 24 (29.63%) controls were Hispanics in the validation set. In the discovery set, the mean ages were 51 and 51 for the cases and controls. And in the validation set, the mean ages were 51 and 58 for the cases and controls, subsequently. In the discovery set, 71 (52.99%) cases and 25 (43.96%) controls were obese. Similarly, in the validation set, 133 (59.64%) cases and 45 (55.56%) controls were obese.
Table 1.
Selected demographic variables by case-control status by study
| Discovery | Validation | |||||
|---|---|---|---|---|---|---|
| Case | Control | P-value* | Case | Control | P-value* | |
| (N=134) | (N=57) | (N=224) | (N=81) | |||
| Age, mean(SD) | 51.1(12.0) | 51.1(6.7) | 0.990 | 50.6(10.7) | 58.4(14.8) | <0.001 |
| BMI, mean(SD) | 31.1(6.2) | 30.4(7.0) | 0.499 | 32.8(7.7) | 33.4(8.3) | 0.599 |
| BMI category N(%) | ||||||
| normal(<25) | 18(13.43) | 17(29.82) | 28(12.11) | 10(12.35) | ||
| overweight(25–<3 0) | 45(33.58) | 15(26.32) | 63(28.25) | 26(32.10) | ||
| obese(30+) | 71(52.99) | 25(43.86) | 0.027 | 133(59.64) | 45(55.56) | 0.790 |
| Race | ||||||
| African Americans | 57(42.54) | 19(33.33) | 93(41.52) | 57(70.37) | ||
| Hispanic | 77(57.46) | 38(66.67) | 0.234 | 131(58.48) | 24(29.63) | <0.001 |
| Family history of breast cancer | ||||||
| 0 | 113(84.33) | 50(87.72) | 195(87.05) | 70(86.42) | ||
| 1 | 21(15.67) | 7(12.28) | 0.544 | 29(12.95) | 11(13.58) | 0.885 |
| ER | ||||||
| positive | 100(74.63) | 157(70.09) | ||||
| negative | 34(25.37) | 67(29.92) | ||||
| PR | ||||||
| positive | 77(57.46) | 134(59.82) | ||||
| negative | 57(42.54) | 90(40.18) | ||||
| HER2 | ||||||
| positive | 30(22.38) | 35(15.63) | ||||
| negative | 104(77.61) | 189(84.37) | ||||
| Stage | ||||||
| I | 44(32.83) | 72(32.14) | ||||
| II | 44(32.83) | 90(40.18) | ||||
| III & IV | 46(34.33) | 62(27.68) | ||||
Student T test was used to calculate the P value for continuous variables; and Chi square was used to calculate the P value for categorical variables.
Metabolites differed between breast cancer cases and controls
In the discovery set, we identified a total of 31 metabolites whose levels significantly differed between breast cancer cases and controls, after adjusting age, BMI category, race/ethnicity, and multiple comparison (Table 2). Among them, 9 were significantly higher in the cases than controls, and 22 were significantly lower in the cases than controls. The top three metabolites higher in the cases than controls were thioproline, inosine, and 3-hydroxyoctanoate. Higher levels of those three metabolites were associated with 3.90, 2.83, and 1.69-fold increased odds of breast cancer. On the other hand, the top three metabolites lower in the cases than controls were cysteines-sulfate, sphingomyelin, and threonylphenylalanine. Higher levels of those three metabolites were associated with 76, 70, and 56% decreased odds of breast cancer. In the validation set, 14 of those 31 metabolites were confirmed. Among them, 7 were higher in the cases than controls, and 7 were lower in the cases than controls. In the validation set, the top 3 metabolites whose levels were most significantly increased in breast cancer cases than controls were linoleate (18:2n6) (OR=1.98, 95%CI: 1.46, 2.70), dihomo-linoleate (OR=1.82, 95%CI: 1.34, 2.47), and 10-nonadecenoate (19:1n9) (OR=1.76, 95%CI: 1.31, 2.36). On the other hand, the top 3 metabolites in the validation set whose levels were most significantly decreased in breast cancer cases than controls were bilirubin (OR=0.48, 95%CI: 0.34, 0.69), glucuronate (OR=0.56, 95%CI: 0.35, 0.90), and 1-(1-enyl-stearoyl)-2-dihomo-linolenoyl-GPE (OR=0.65, 95%CI: 0.46, 0.92). In terms of metabolic pathways, 10 metabolites were lipids (including 6 fatty acids), 1 amino acid, 1 xenobiotics, 1 cofactors and vitamins, and 1 carbohydrate. Using those 14 metabolites, we generated the 3-D plot to differentiate the breast cancer cases and controls. We found that the cases and controls were separated with three components. Component 1, 2 and 3 could account for 30, 16.2, and 14.4% of the difference between the cases and controls. We also investigated whether the significant metabolites differed between African American and Hispanic women among the controls. However, none of them differed.
Table 2.
Serum metabolites associated with breast cancer risk
| Discovery | Validation | |||||
|---|---|---|---|---|---|---|
| Metabolites | Pathways | Sub-Pathways | OR(95%CI)* | P-value | OR(95%CI)* | P-value |
| thioproline | Xenobiotics | Chemical | 3.90 (2.34,6.51) | <0.001 | 1.42 (0.95, 2.13) | 0.087 |
| inosine | Nucleotide | Purine Metabolism, (Hypo)Xanthine/Inosine | 3.83 (2.49, 5.90) | <0.001 | 0.73 (0.55, 0.96) | 0.025 |
| 3-hydroxyoctanoate | Lipid | Fatty Acid, Monohydroxy | 1.69 (1.16, 2.45) | 0.006 | 1.62 (1.20, 2.18) | 0.002 |
| 3-hydroxybutyrate (BHBA) | Lipid | Ketone Bodies | 1.67 (1.18,2.35) | 0.004 | 1.62 (1.17, 2.23) | 0.004 |
| stearate (18:0) | Lipid | Long Chain Fatty Acid | 1.52 (1.10, 2.09) | 0.011 | 1.66 (1.24, 2.22) | <0.001 |
| 3-hydroxydecanoate | Lipid | Fatty Acid, Monohydroxy | 1.48 (1.04, 2.10) | 0.028 | 1.67 (1.22, 2.27) | 0.001 |
| linoleate (18:2n6) | Lipid | Polyunsaturated Fatty Acid (n3 and n6) | 1.42 (1.04, 1.95) | 0.028 | 1.98 (1.46, 2.70) | <.0001 |
| 10-nonadecenoate (19:1n9) | Lipid | Long Chain Fatty Acid | 1.38 (1.01,1.89) | 0.049 | 1.76 (1.31, 2.36) | <0.001 |
| dihomo-linoleate | Lipid | Polyunsaturated Fatty Acid (n3 and n6) | 1.35 (1.01, 1.86) | 0.044 | 1.82 (1.34, 2.47) | <0.001 |
| fumarate | Energy | TCA Cycle | 0.69 (0.50, 0.95) | 0.025 | 0.73 (0.54, 1.00) | 0.052 |
| 3-methylxanthine | Xenobiotics | Xanthine Metabolism | 0.67 (0.48, 0.94) | 0.021 | 0.68 (0.51, 0.90) | 0.008 |
| bilirubin | Cofactors and Vitamins | Hemoglobin and Porphyrin Metabolism | 0.66 (0.47, 0.94) | 0.019 | 0.48 (0.34, 0.69) | <0.001 |
| 1-palmitoyl-GPA (16:0) | Lipid | Lysophospholipid | 0.65 (0.46, 0.92) | 0.014 | 0.74 (0.55, 0.99) | 0.049 |
| isobutyrylcarnitine | Amino Acid | Leucine, Isoleucine and Valine Metabolism | 0.64 (0.45, 0.90) | 0.011 | 0.84 (0.62, 1.13) | 0.246 |
| 1-palmitoyl-2-linoleoyl-GPC | Lipid | Phosphatidylcholine (PC) | 0.63 (0.42, 0.94) | 0.025 | 1.01 (0.74, 1.39) | 0.951 |
| N-acetylleucine | Amino Acid | Leucine, Isoleucine and Valine Metabolism | 0.62 (0.42, 0.91) | 0.015 | 0.86 (0.64, 1.15) | 0.308 |
| 1-(1-enyl-oleoyl)-2-docosahexaenoyl-GPE | Lipid | Plasmalogen | 0.60 (0.40, 0.90) | 0.014 | 0.71 (0.50, 1.03) | 0.070 |
| glucuronate | Carbohydrate | Aminosugar Metabolism | 0.59 (0.43, 0.83) | 0.002 | 0.56 (0.35, 0.90) | 0.016 |
| uridine | Nucleotide | Pyrimidine Metabolism, Uracil containing | 0.58 (0.41, 0.82) | 0.002 | 1.47 (1.11, 1.95) | 0.007 |
| 3-hy droxy-3-methy lglutarate | Lipid | Mevalonate Metabolism | 0.58 (0.40, 0.83) | 0.003 | 0.78(0.56, 1.07) | 0.118 |
| N-acetylvaline | Amino Acid | Leucine, Isoleucine and Valine Metabolism | 0.57 (0.39, 0.83) | 0.003 | 0.88 (0.66, 1.18) | 0.409 |
| 1-(1-enyl-stearoyl)-2-dihomo-linolenoyl-GPE | Lipid | Plasmalogen | 0.55 (0.38, 0.80) | 0.002 | 0.65 (0.46, 0.92) | 0.014 |
| urea | Amino Acid | Urea cycle; Arginine and Proline Metabolism | 0.54 (0.37, 0.79) | 0.002 | 0.74 (0.55, 0.99) | 0.040 |
| retinol (Vitamin A) | Cofactors and Vitamins | Vitamin A Metabolism | 0.54 (0.37, 0.78) | 0.001 | 0.78 (0.59, 1.04) | 0.089 |
| trans-4-hydroxyproline | Amino Acid | Urea cycle; Arginine and Proline Metabolism | 0.51 (0.36, 0.74) | <0.001 | 0.79 (0.60, 1.04) | 0.097 |
| perfluorooctanesulfonic acid (PFOS) | Xenobiotics | Chemical | 0.47 (0.31, 0.71) | <0.001 | 0.99 (0.69, 1.42) | 0.949 |
| 1-linoleoy1-GPA(18:2) | Lipid | Lysophospholipid | 0.46 (0.32, 0.68) | <0.001 | 0.81 (0.61, 1.07) | 0.144 |
| beta-citrylglutamate | Amino Acid | Glutamate Metabolism | 0.45 (0.29, 0.69) | <0.001 | 1.70 (1.23, 2.36) | 0.001 |
| threonylphenylalanine | Peptide | Dipeptide | 0.44 (0.29, 0.67) | <.001 | 1.64 (1.24, 2.17) | <0.001 |
| sphingomyelin | Lipid | Sphingolipid Metabolism | 0.30 (0.19, 0.48) | <0.001 | 0.68 (0.51, 0.90) | 0.008 |
| cysteine s-sulfate | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism | 0.24 (0.14, 0.40) | <0.001 | 3.25 (2.14, 4.95) | <0.001 |
Logistic regression was used to calculate the ORs and P value with FDR<0.05 was used. Age, obesity status and race were adjusted.
Metabolic pathways differed between breast cancer cases and controls
Next, metabolic pathway analysis was conducted to identify the enriched metabolic pathways that differed between breast cancer cases and controls (Table 3). In the discovery set, we identified 12 metabolic pathways with P value less than 0.10. The top 3 most significant pathways were Glycerophospholipid metabolism (P<0.001), Sphingolipid metabolism (P<0.001), and Citrate cycle (TCA cycle) (P<0.001). Using P values of 0.10 and 0.05 as the cutoff points, 6 and 3 of the 12 identified metabolic pathways in the discovery set were confirmed in the validation set. After further considered the impact factor (the significance of each individual metabolite in a relative metabolic pathway), the most significant and consistent metabolic pathways were Linoleic acid metabolism, Arginine and proline metabolism, and Citrate cycle (TCA cycle). Particularly, the significant metabolites involved in linoleic acid metabolism had the highest impact factor, 0.656 in both sets, suggesting the identified significant metabolites involved in linoleic acid metabolism pathway are significantly involved in breast cancer development.
Table 3.
Metabolic pathways associated with breast cancer risk
| Discovery | Validation | |||
|---|---|---|---|---|
| Metabolic pathway | P value | Impact factor | P value | Impact factor |
| Glycerophospholipid metabolism | 2.17E-05 | 0.314 | 0.071 | 0.205 |
| Sphingolipid metabolism | 2.12E-04 | 0.283 | Not observed | |
| Citrate cycle (TCA cycle) | 9.65E-04 | 0.236 | 0.012 | 0.090 |
| Butanoate metabolism | 0.013 | 0.108 | 0.076 | 0.040 |
| D-Arginine and D-ornithine metabolism | 0.014 | 0.500 | Not observed | |
| Alanine, aspartate and glutamate metabolism | 0.017 | 0.003 | 0.119 | 0.003 |
| Ascorbate and aldarate metabolism | 0.019 | 0.049 | 0.310 | 0.033 |
| Arginine and proline metabolism | 0.031 | 0.096 | 0.011 | 0.120 |
| Pyruvate metabolism | 0.037 | 0.320 | Not observed | |
| Linoleic acid metabolism | 0.046 | 0.656 | 0.005 | 0.656 |
| Nicotinate and nicotinamide metabolism | 0.081 | 0.017 | 0.095 | 0.038 |
| Caffeine metabolism | 0.084 | 0.045 | 0.413 | 0.015 |
Metabolites risk score differed between breast cancer cases and controls
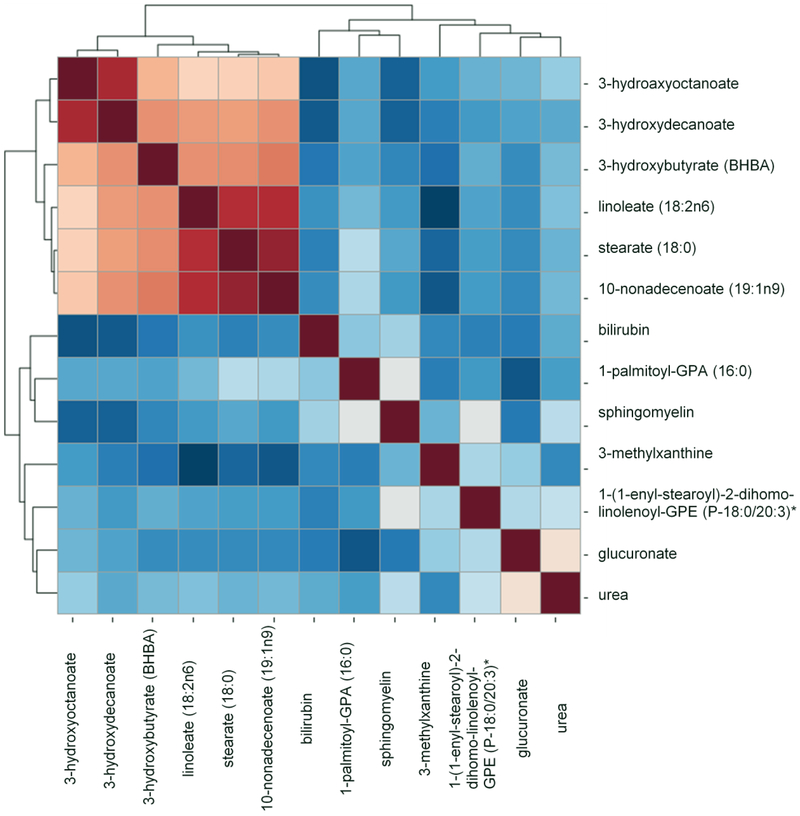
To further analyze the relationships among those 14 identified significant metabolites, we performed the pairwise correlation analysis among the controls (Figure 1). Using correlation coefficient (γ) of 0.5 as the cutoff point, 9 metabolites were found to have median to low correlation (γ<0.50) with any of the other metabolites. This includes glucuronate, stearate (18:0), 3-hydroxyoctanoate, 1-palmitoyl-GPA (16:0), 1-(1-enyl-stearoyl)-2-dihomo-linolenoyl-GPE, sphingomyelin, bilirubin, urea, and 3-methylxanthine. Using those 9 metabolites, we generated a metabolic risk score. The method used to generate the risk score was detailed in previous Statistical Analysis section. Using the risk score as a continuous variable, we found that increased risk score was 1.87-fold and 1.63-fold more likely observed in breast cancer cases than controls after adjusting for age, race, and BMI category in the discovery and validation sets, respectively (adjusted OR=1.87,95%CI=1.50–2.32; adjusted OR=1.63,95%CI=1.36–1.95) (Table 3). Next, we stratified the study subjects into 3 strata based the risk score among the control subjects: low (0 to 4), medium (5–6) and high (at least 7). When compared with study subjects with low risk scores, medium risk scores were more likely observed in the breast cancer cases than controls in both discovery and validation sets (adjusted OR=8.68, 95%CI=3.31–22.75; and adjusted OR=4.12, 95%CI=2.06–8.22). The odd was further elevated when we compared high to low risk score group. Those with high risk score were associated with 18.44-fold and 6.96-fold increased odds of breast cancer in the discovery and validation sets, respectively (adjusted OR=18.44, 95%CI=6.75–50.37; adjusted OR=6.96, 95%CI=3.27–14.81).
Figure 1.
Pair-wise correlation analysis to assess the redundancy among significant metabolites.
Discussion
In the current two-stage study of metabolomics profiling in plasma samples from 358 breast cancer patients and 138 healthy controls, we identified metabolic signatures and pathways significantly associated with breast cancer risk. Intriguingly, all seven metabolites, namely 3-hydroxyoctanoate 3-hydroxybutyrate (BHBA), stearate (18:0), 3-hydroxydecanoate, linoleate (18:2n6), 10-nonadecenoate (19:1n9), and dihomo-linoleate, whose levels were increased in breast cancer cases than controls, were lipids. The finding is consistent with our previous study in non-Hispanic Whites and African Americans that showed the levels of all 7 metabolites were higher in breast cancer cases than the controls [7]. The agreement between two studies suggested that lipids may play a key role in breast carcinogenesis.
Among the 7 lipids mentioned above, 6 were fatty acids, including 2 monohydroxy fatty acids, 2 long chain fatty acids, and two polyunsaturated fatty acids. Elevated levels of monohydroxy and long chain fatty acids may indicate altered fatty acid β-oxidation in patients with breast cancer [7]. Fatty acid β-oxidation has been reported to support functional mitochondria and is essential for the accelerated proliferation of cancer cells [23,24]. Interestingly, in a recent study, Wang et al reported that fatty Acid β-Oxidation plays a critical role in breast cancer stem cell self-renewal and chemoresistance [25]. Therefore, altered fatty acids β-oxidation may promote the aggressive tumorigenic phenotypes and breast carcinogenesis [26]. Similar observations were also seen in several other breast cancer case control analyses [27,28]. Furthermore, another significant metabolite, ketone body 3-hydroxybutyrate (BHBA), which is the end product of ketogenesis and downstream of fatty acid β-oxidation, was also found associated with breast cancer risk in our study. The higher BHBA level in the cases is another sign of altered fatty acid β-oxidation in breast cancer subjects. Consistent with our previous study [7], the essential fatty acids linoleate (18:2n6) and dihomo-linolenate (20:3n3 or n6) were significantly higher in the cases than controls. Linolenic acid has been suggested to promote the tumorigenic phenotype of breast cancer [29]. In a genome-wide association study, genetic variants in genes involved in the regulation of linolenic acid metabolism were associated with breast cancer risk [30].
Another finding of interest is that all 5 significant amino acids in the discovery set were associated with decreased risk of breast cancer. Although only urea remained statistically significant in the validation set, the impact on breast cancer risk is still evident. In fact, this observation is consistent with the notion that amino acids play important roles in carcinogenesis. For example, Maddocks and colleagues found that restriction of dietary Serine and Glycine could reduce tumor growth in xenograft and allograft models [31]. In circulation, amino acids have been reported to be reduced in patients of a variety of cancers [32–34]. The reduced levels of amino acids may reflect the increased amino acid demand by tumors and therefore amino acid malnutrition seen in tumors. In the current study, our observation on urea is consistent with a previous report that showed plasma urea level was significantly lower in breast cancer cases than healthy women [35]. However, the biological relevance of urea in breast carcinogenesis is still unclear.
Our study is first to report the association between plasma bilirubin and breast cancer risk. Endogenous antioxidant bilirubin has shown to inhibit cancer development [36]. Bilirubin was associated with reduced breast cancer risk in our study, which was consistent with its anti-carcinogenic property [37,38]. Serum bilirubin has been reported to associated with reduced lung cancer risk in two prospective studies [39,40] and reduced cancer mortality in a population-based set [41]. However, how bilirubin modulates breast carcinogenesis remains largely unknown.
In the pathway analysis, 3 metabolic pathways were significantly associated with higher risk of breast cancer risk in both sets, including Citrate cycle (TCA cycle), Arginine and proline metabolism, and Linoleic acid metabolism. Even though fumarate was the only significant metabolite directly involved in the TCA cycle, several other significant metabolites were involved in biological pathways relevant to the TCA cycle, including fatty acid β- oxidation, amino acid metabolism and ketone bodies [42]. Dysfunction of mitochondrial based metabolic pathways, including TCA cycle and oxidative phosphorylation, has been frequently observed in breast cancer [43–47,42]. Arginine and proline metabolism pathway plays an important role in cancer by impacting various regulatory targets, influencing apoptosis, autophagy, and regulating redox homeostasis [48]. In breast cancer, altered proline and arginine metabolism has also been linked to metastasis formation [49]. It’s not a surprise that Linoleic acid metabolism was significantly associated with breast cancer risk. As mentioned above, the omega-3 polyunsaturated fatty acids linoleate (18:2n6) and dihomo-linolenate (20:3n3 or n6) have been reported to upregulate breast cancer growth, enhance tumor progression, and play an important role in breast tumorigenesis [50].
Recently, two breast cancer metabolomics studies have been published using samples from the Prostate, Lung, Colorectal and Ovarian (PLCO) cohort [5,6]. A few diet and BMI related metabolites were found associated with the risk of breast cancer. Although both studies identified lipids as the top significant metabolites, they were not overlapped. More than 90% of the study participants included in their studies were postmenopausal non-Hispanic Whites aged 55–74 years old. While our study only included Hispanics and non-Hispanic African Americans with mean age less than 55 years old. It is possible that the difference in population demographics may contribute to the difference in significant metabolites. As one example, 3-hydroxyoctanoate was one of the significant metabolites identified in our study. While in their studies, 2-hydroxyoctanoate, which was fried food–associated, was found associated with estrogen receptor positive (ER+) breast cancer.
In summary, we identified a panel of plasma metabolites associated with breast cancer risk among African American and Hispanic women, which may provide new insights in breast tumorigenesis in different populations. The major limitations of this study are that the plasma samples were collected after diagnosis, and matched tissue samples were not available. However, validity of using plasma as the surrogate tissue has been reported [51]. Due to the nature of case control study, we cannot infer the causal relationship between metabolites and breast cancer risk. Other limitations may include the lack of dietary data and single metabolite measurement, etc. Thus, we need to be very cautious to interpret our results. Even though our study had biggest sample size among similar studies to compare the blood metabolic signatures between breast cancer cases and controls, validation of our results in larger and prospective populations in the future are warranted.
Finally, it seems that metabolite profiles didn’t differ between African American and Hispanic women among the controls. However, the number of controls in two races (n=138 in total) are very too small, so it is very likely that we don’t have enough power to detect the difference. In our previous metabolomics study in African American (n=24) and Caucasian (n=36) female controls, metabolite profiles were found significantly differed between them [7]. Clearly, whether metabolic profiles differ by the race/ethnicity and whether the potential difference may play a role in the association between plasma metabolites and breast cancer risk are still unclear. Future large multi-ethnic studies are needed.
Table 4.
Metabolite risk score associated with breast cancer risk
| Discovery | Validation | |||
|---|---|---|---|---|
| Continuous variable | 1.87 (1.50–2.32) | <0.001 | 1.63 (1.36–1.95) | <0.001 |
| Categorical variable | ||||
| Low (0–4) | Reference | Reference | ||
| Medium (5–6) | 8.68 (3.31–22.75) | <0.001 | 4.12 (2.06–8.22) | <0.001 |
| High (7–8) | 18.44 (6.75–50.37) | <0.001 | 6.96 (3.27–14.81) | <0.001 |
Logistic regression was used to calculate the ORs and P value. Age, obesity status and race were adjusted.
Acknowledgements
The study was supported by U01 CA179655 from NCI/NIH.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures performed in this study were approved by the Institutional Review Board at M D Anderson Cancer Center and in accordance with the ethical standards of 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all participants.
References
- 1.Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68 (1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Yedjou CG, Tchounwou PB, Payton M, Miele L, Fonseca DD, Lowe L, Alo RA (2017) Assessing the Racial and Ethnic Disparities in Breast Cancer Mortality in the United States. Int J Environ Res Public Health 14 (5). doi: 10.3390/ijerph14050486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claudino WM, Goncalves PH, di Leo A, Philip PA, Sarkar FH (2012) Metabolomics in cancer: a bench-to-bedside intersection. Crit Rev Oncol Hematol 84 (1):1–7. doi: 10.1016/j.critrevonc.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 4.Yu B, Zheng Y, Alexander D, Morrison AC, Coresh J, Boerwinkle E (2014) Genetic Determinants Influencing Human Serum Metabolome among African Americans. Plos Genet 10 (3). doi:ARTNe1004212 10.1371/journal.pgen.1004212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Playdon MC, Ziegler RG, Sampson JN, Stolzenberg-Solomon R, Thompson HJ, Irwin ML, Mayne ST, Hoover RN, Moore SC (2017) Nutritional metabolomics and breast cancer risk in a prospective study. Am J Clin Nutr 106 (2):637–649. doi: 10.3945/ajcn.116.150912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore SC, Playdon MC, Sampson JN, Hoover RN, Trabert B, Matthews CE, Ziegler RG (2018) A Metabolomics Analysis of Body Mass Index and Postmenopausal Breast Cancer Risk. J Natl Cancer Inst 110 (6):588–597. doi: 10.1093/jnci/djx244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen J, Yan L, Liu S, Ambrosone CB, Zhao H (2013) Plasma Metabolomic Profiles in Breast Cancer Patients and Healthy Controls: By Race and Tumor Receptor Subtypes. Translational Oncology 6 (6):757–765. doi: 10.1593/tlo.13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart CD, Vignoli A, Tenori L, Uy GL, To TV, Adebamowo C, Hossain SM, Biganzoli L, Risi E, Love RR, Luchinat C, Di Leo A (2017) Serum Metabolomic Profiles Identify ER-Positive Early Breast Cancer Patients at Increased Risk of Disease Recurrence in a Multicenter Population. Clin Cancer Res 23 (6):1422–1431. doi: 10.1158/1078-0432.Ccr-16-1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenori L, Oakman C, Morris PG, Gralka E, Turner N, Cappadona S, Fornier M, Hudis C, Norton L, Luchinat C, Di Leo A (2015) Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol Oncol 9 (1):128–139. doi: 10.1016/j.molonc.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, Bernini P, Zafarana E, Saccenti E, Fornier M, Morris PG, Biganzoli L, Luchinat C, Bertini I, Di Leo A (2011) Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol 22 (6):1295–1301. doi: 10.1093/annonc/mdq606 [DOI] [PubMed] [Google Scholar]
- 11.Asiago VM, Alverado LZ, Shanaiah N, Gowda NG, Owusu-Sarfo K, Ballas R, Raftery D (2010) Early Detection of Recurrent Breast Cancer Using Metabolite Profiling. Cancer Res 70. doi: 10.1158/0008-5472.Sabcs10-P3-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang AH, Sun H, Qiu S, Wang XJ (2013) Metabolomics in noninvasive breast cancer. Clin Chim Acta 424:3–7. doi: 10.1016/j.cca.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Hadi NI, Jamal Q, Iqbal A, Shaikh F, Somroo S, Musharraf SG (2017) Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci Rep-Uk 7. doi:ARTN 1715 10.1038/s41598-017-01924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodadadi-Jamayran A, Akgol-Oksuz B, Afanasyeva Y, Heguy A, Thompson M, Ray K, Giro-Perafita A, Sanchez I, Wu X, Tripathy D, Zeleniuch-Jacquotte A, Tsirigos A, Esteva FJ (2018) Prognostic role of elevated mir-24–3p in breast cancer and its association with the metastatic process. Oncotarget 9 (16):12868–12878. doi: 10.18632/oncotarget.24403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81 (16):6656–6667. doi: 10.1021/ac901536h [DOI] [PubMed] [Google Scholar]
- 16.DeHaven CD, Evans AM, Dai HP, Lawton KA (2010) Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminformatics 2. doi:Artn 9 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampson JN, Boca SM, Shu XO, Stolzenberg-Solomon RZ, Matthews CE, Hsing AW, Tan YT, Ji BT, Chow WH, Cai QY, Liu DK, Yang G, Xiang YB, Zheng W, Sinha R, Cross AJ, Moore SC (2013) Metabolomics in Epidemiology: Sources of Variability in Metabolite Measurements and Implications. Cancer Epidem Biomar 22 (4):631–640. doi: 10.1158/1055-9965.Epi-12-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chong EY, Huang Y, Wu H, Ghasemzadeh N, Uppal K, Quyyumi AA, Jones DP, Yu T (2015) Local false discovery rate estimation using feature reliability in LC/MS metabolomics data. Sci Rep 5:17221. doi: 10.1038/srep17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A (2013) HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res 41 (Database issue):D801–807. doi: 10.1093/nar/gks1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong J, Xia JG (2018) MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34 (24):4313–4314. doi: 10.1093/bioinformatics/bty528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore SC, Matthews CE, Sampson JN, Stolzenberg-Solomon RZ, Zheng W, Cai Q, Tan YT, Chow WH, Ji BT, Liu DK, Xiao Q, Boca SM, Leitzmann MF, Yang G, Xiang YB, Sinha R, Shu XO, Cross AJ (2014) Human metabolic correlates of body mass index. Metabolomics 10 (2):259–269. doi: 10.1007/s11306-013-0574-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, Shen J, Djukovic D, Daniel-MacDougall C, Gu H, Wu X, Chow WH (2016) Metabolomics-identified metabolites associated with body mass index and prospective weight gain among Mexican American women. Obes Sci Pract 2 (3):309–317. doi: 10.1002/osp4.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Enriquez S, Hernandez-Esquivel L, Marin-Hernandez A, El Hafidi M, Gallardo-Perez JC, Hernandez-Resendiz I, Rodriguez-Zavala JS, Pacheco-Velazquez SC, Moreno-Sanchez R (2015) Mitochondrial free fatty acid beta-oxidation supports oxidative phosphorylation and proliferation in cancer cells. Int J Biochem Cell B 65:209–221. doi: 10.1016/j.biocel.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 24.Nieman KM, Romero IL, Van Houten B, Lengyel E (2013) Adipose tissue and adipocytes support tumorigenesis and metastasis. Bba-Mol Cell Biol L 1831 (10):1533–1541. doi: 10.1016/j.bbalip.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, Somlo G, Jandial R, Ann D, Hanash S, Jove R, Yu H (2018) JAK/STAT3-Regulated Fatty Acid beta-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab 27 (1):136–150 e135. doi: 10.1016/j.cmet.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinlaw WB, Baures PW, Lupien LE, Davis WL, Kuemmerle NB (2016) Fatty Acids and Breast Cancer: Make Them on Site or Have Them Delivered. J Cell Physiol 231 (10):2128–2141. doi: 10.1002/jcp.25332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui M, Wang QL, Chen G (2016) Serum metabolomics analysis reveals changes in signaling lipids in breast cancer patients. Biomed Chromatogr 30 (1):42–47. doi: 10.1002/bmc.3556 [DOI] [PubMed] [Google Scholar]
- 28.O’Flaherty JT, Wooten RE, Samuel MP, Thomas MJ, Levine EA, Case LD, Akman SA, Edwards IJ (2013) Fatty Acid Metabolites in Rapidly Proliferating Breast Cancer. Plos One 8 (5). doi:ARTN e63076 10.1371/journal.pone.0063076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhou L, Shi W, Song N, Yu K, Gu Y (2012) A mechanism underlying the effects of polyunsaturated fatty acids on breast cancer. Int J Mol Med 30 (3):487–494. doi: 10.3892/ijmm.2012.1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Humphreys K, Heikkinen T, Aittomaki K, Blomqvist C, Pharoah PD, Dunning AM, Ahmed S, Hooning MJ, Martens JW, van den Ouweland AM, Alfredsson L, Palotie A, Peltonen-Palotie L, Irwanto A, Low HQ, Teoh GH, Thalamuthu A, Easton DF, Nevanlinna H, Liu J, Czene K, Hall P (2011) A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat 126 (3):717–727. doi: 10.1007/s10549-010-1172-9 [DOI] [PubMed] [Google Scholar]
- 31.Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, Blagih J, Vincent DF, Campbell KJ, Ceteci F, Sansom OJ, Blyth K, Vousden KH (2017) Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544 (7650):372–376. doi: 10.1038/nature22056 [DOI] [PubMed] [Google Scholar]
- 32.Heber D, Byerly LO, Chlebowski RT (1985) Metabolic Abnormalities in the Cancer Patient. Cancer 55 (1):225–229. doi:Doi 10.1002/1097–0142(19850101)55:1+<225::Aid-Cncr2820551304>3.0.Co;2–7 [DOI] [PubMed] [Google Scholar]
- 33.Bi X, Henry CJ (2017) Plasma-free amino acid profiles are predictors of cancer and diabetes development. Nutr Diabetes 7 (3):e249. doi: 10.1038/nutd.2016.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, Saruki N, Bando E, Kimura H, Imamura F, Moriyama M, Ikeda I, Chiba A, Oshita F, Imaizumi A, Yamamoto H, Miyano H, Horimoto K, Tochikubo O, Mitsushima T, Yamakado M, Okamoto N (2011) Plasma free amino acid profiling of five types of cancer patients and its application for early detection. Plos One 6 (9):e24143. doi: 10.1371/journal.pone.0024143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quevedo-Coli S, Crespi C, Benito E, Palou A, Roca P (1997) Alterations in circulating fatty acids and the compartmentation of selected metabolites in women with breast cancer. Biochem Mol Biol Int 41 (1):1–10 [DOI] [PubMed] [Google Scholar]
- 36.Kuhn T, Sookthai D, Graf ME, Schubel R, Freisling H, Johnson T, Katzke V, Kaaks R (2017) Albumin, bilirubin, uric acid and cancer risk: results from a prospective population-based study. Br J Cancer 117 (10):1572–1579. doi: 10.1038/bjc.2017.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XA, Meng QH, Ye YQ, Hildebrandt MAT, Gu J, Wu XF (2015) Prognostic significance of pretreatment serum levels of albumin, LDH and total bilirubin in patients with non-metastatic breast cancer. Carcinogenesis 36 (2):243–248. doi: 10.1093/carcin/bgu247 [DOI] [PubMed] [Google Scholar]
- 38.Srivastava P, Hira SK, Srivastava DN, Gupta U, Sen P, Singh RA, Manna PP (2017) Protease-Responsive Targeted Delivery of Doxorubicin from Bilirubin-BSA-Capped Mesoporous Silica Nanoparticles against Colon Cancer. Acs Biomater Sci Eng 3 (12):3376–3385. doi: 10.1021/acsbiomaterials.7b00635 [DOI] [PubMed] [Google Scholar]
- 39.Horsfall LJ, Rait G, Walters K, Swallow DM, Pereira SP, Nazareth I, Petersen I (2011) Serum Bilirubin and Risk of Respiratory Disease and Death. Jama-J Am Med Assoc 305 (7):691–697. doi:DOI 10.1001/jama.2011.124 [DOI] [PubMed] [Google Scholar]
- 40.Wen CP, Zhang FM, Liang D, Wen C, Gu J, Skinner H, Chow WH, Ye YQ, Pu X, Hildebrandt MAT, Huang MS, Chen CH, Hsiung CA, Tsai MK, Tsao CK, Lippman SM, Wu XF (2015) The Ability of Bilirubin in Identifying Smokers with Higher Risk of Lung Cancer: A Large Cohort Study in Conjunction with Global Metabolomic Profiling. Clin Cancer Res 21 (1):193–200. doi: 10.1158/1078-0432.Ccr-14-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Temme EHM, Zhang JJ, Schouten EG, Kesteloot H (2001) Serum bilirubin and 10-year mortality risk in a Belgian population. Cancer Cause Control 12 (10):887–894. doi:Doi 10.1023/A:1013794407325 [DOI] [PubMed] [Google Scholar]
- 42.Yadava N, Schneider SS, Jerry DJ, Kim C (2013) Impaired Mitochondrial Metabolism and Mammary Carcinogenesis. J Mammary Gland Biol 18 (1):75–87. doi: 10.1007/s10911-012-9271-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn CS, Metallo CM (2015) Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab 3. doi:ARTN 1 10.1186/s40170-015-0128-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grzybowska-Szatkowska L, Rzymowska J, Slaska B (2016) Oxidative phosphorylation genes in breast cancer cells. Int J Mol Med 38:S78–S78 [Google Scholar]
- 45.Marini C, Bianchi G, Buschiazzo A, Ravera S, Martella R, Bottoni G, Petretto A, Emionite L, Monteverde E, Capitanio S, Inglese E, Fabbi M, Bongioanni F, Garaboldi L, Bruzzi P, Orengo AM, Raffaghello L, Sambuceti G (2016) Divergent targets of glycolysis and oxidative phosphorylation result in additive effects of metformin and starvation in colon and breast cancer. Sci Rep-Uk 6. doi:ARTN 19569 10.1038/srep19569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meric-Bernstam F, Evans K, Zheng XF, Su XP, Yuca E, Scott S, Akcakanat A, Ueno N, Lim B, Litton J, Valero V, Symmans F, Hortobagyi G, Perou C, Tripathy D, Draetta G, Marszalek J, Gonzalez-Angulo AM, Moulder S (2017) Oxidative phosphorylation as a target in triple negative breast cancer therapy. Cancer Res 77. doi: 10.1158/1538-7445.Am2017-4970 [DOI] [Google Scholar]
- 47.Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P (2008) Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Tr 110 (3):439–452. doi: 10.1007/s10549-007-9738-x [DOI] [PubMed] [Google Scholar]
- 48.Phang JM, Liu W, Hancock CN, Fischer JW (2015) Proline metabolism and cancer: emerging links to glutamine and collagen. Curr Opin Clin Nutr 18 (1):71–77. doi: 10.1097/Mco.0000000000000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elia I, Broekaert D, Christen S, Boon R, Radaelli E, Orth MF, Verfaillie C, Grunewald TGP, Fendt SM (2017) Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat Commun 8. doi:ARTN 15267 10.1038/ncomms15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouradian M, Kikawa KD, Dranka BP, Komas SM, Kalyanaraman B, Pardini RS (2015) Docosahexaenoic Acid Attenuates Breast Cancer Cell Metabolism and the Warburg Phenotype by Targeting Bioenergetic Function. Mol Carcinogen 54 (9):810–820. doi: 10.1002/mc.22151 [DOI] [PubMed] [Google Scholar]
- 51.Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE (2010) Metabolic signatures of exercise in human plasma. Sci Transl Med 2 (33):33ra37. doi: 10.1126/scitranslmed.3001006 [DOI] [PMC free article] [PubMed] [Google Scholar]