Abstract
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease, characterized by motor neuron death in the brain and spinal cord. Mutations in the Cu/Zn superoxide dismutase (SOD1) gene account for ~20% of all familial ALS forms, corresponding to 1%–2% of all ALS cases. One of the suggested mechanisms by which mutant SOD1 (mtSOD1) exerts its toxic effects involves intracellular accumulation of abnormal mtSOD1 aggregates, which trigger endoplasmic reticulum (ER) stress and activate its adaptive signal transduction pathways, including the unfolded protein response (UPR). PERK, an eIF2α kinase, is central to the UPR and is the most rapidly activated pathway in response to ER stress. Previous reports using mtSOD1 transgenic mice indicated that genetic or pharmacological enhancement of the UPR- PERK pathway may be effective in treating ALS. We investigated the response to PERK haploinsufficiency, and the response to deficiency of its downstream effectors GADD34 and CHOP, in five distinct lines of mtSOD1 mice. We demonstrate that, in contrast to a previously published study, PERK haploinsufficiency has no effect on disease in all mtSOD1 strains examined. We also show that deficiency of GADD34, which enhances the UPR by prolonging the phosphorylation of eIF2α does not ameliorate disease in these mtSOD1 mouse strains. Finally, we demonstrate that genetic ablation of CHOP transcription factor, which is known to be pro-apoptotic, does not ameliorate disease in mtSOD1 mice. Cumulatively, our studies reveal that neither genetic inhibition of the UPR via ablation of PERK, nor genetic UPR enhancement via ablation of GADD34, is beneficial for mtSOD1-induced motor neuron disease. Therefore, the PERK pathway is not a likely target for therapeutic intervention in ALS.
Keywords: Amyotrophic lateral sclerosis, SOD1 mice, motor neurons, endoplasmic reticulum stress, unfolded protein response, PERK, GADD34, CHOP
Introduction
Amyotrophic lateral sclerosis (ALS) is a late-onset progressive neurodegenerative disease that is characterized by the selective loss of motor neurons. It leads to paralysis and ultimately death due to respiratory failure, typically within several years after onset (1–4). Most cases of ALS (90%) are without an obvious genetic component (sporadic), whereas approximately 10% are inherited in a dominant manner (familial). Since sporadic and familial forms of ALS are clinically similar, understanding the mechanisms underlying familial ALS may provide insights into both forms of the disease. Although it is unclear what causes motor neuron demise in ALS, studies conducted using spinal cord samples from sporadic ALS patients and transgenic mouse models of familial ALS detected endoplasmic reticulum (ER) stress in both forms of the disease, suggesting that the ER stress response plays an important role in ALS pathogenesis (5–10). Notably, motor neurons are thought to be particularly vulnerable to ER stress in part due to their intrinsically low expression of ER chaperones (11). Mutant superoxide dismutase SOD1 (mtSOD1) induces cell-autonomous and non-cell autonomous motor neuron death through a toxic gain of function (12–14). One of the proposed toxicities involves accumulation of intracellular SOD1 aggregates, which may trigger ER stress and activate adaptive signal transduction pathways, including the unfolded protein response (UPR). The guiding hypothesis is that the toxicity of mtSOD1 arises from its ability to inhibit ER-associated degradation machinery, which is involved in export of misfolded proteins from the ER to the ubiquitin proteasome system, via binding to the integral membrane protein Derlin-1 (15, 16).
The PERK (protein kinase RNA-activated (PKR)-like ER kinase, encoded by Eif2ak3) pathway is one of the three principal signaling branches of the UPR and is the most rapidly activated pathway triggered by the presence of mis- or unfolded proteins in the ER (17–19). It is also central to the integrated stress response (ISR), which is activated by phosphorylation of eIF2α (eukaryotic translation initiation factor 2α) by a family of protein kinases in response to cellular stresses (20). The active (phosphorylated) form of PERK directly phosphorylates eIF2α, which leads to the reduction of global protein synthesis. The resulting effect is prevention of protein overload in the ER. At the same time, there is a preferential translation of ATF4 (activating transcription factor 4), which induces the expression of cytoprotective genes whose functions are to restore proteostasis. One of these genes, Ppp1r15a, encodes GADD34 (growth arrest and DNA damage-inducible 34), which acts in a feedback loop to dephosphorylate p-eIF2α and restore general protein synthesis. Under conditions of chronic stress, however, the PERK-ATF4 axis positively regulates CHOP (C/EBP homologous protein, encoded by Ddit3), one of the key pro-apoptotic players in the UPR (21). Murine mouse models to study the UPR, including mice genetically deficient for PERK, GADD34, and CHOP, have been invaluable in understanding the contribution of the UPR during stress conditions (22).
Studies using genetic or pharmacological approaches to manipulate the UPR have revealed the potential involvement of PERK signaling in mtSOD1-induced ALS (23, 24). PERK haploinsufficiency accelerated the accumulation of misfolded SOD1 and shortened life span in G85R mice (25), and GADD34 haploinsufficiency was shown to protect G85R mice against the disease (26). Pharmacological modulation of GADD34 has been accomplished in G93A high- copy (G93A-HC) mice using several small molecule inhibitors: salubrinal, guanabenz, and Sephin1. These studies demonstrated a protective effect of blocking GADD34 activity, which results in a prolonged ER stress response (9, 27–29), with an exception of one report, where guanabenz was shown to exacerbate disease (30). In comparison, little is known regarding the role of CHOP in ALS. Although increased CHOP expression has been detected in spinal cords of sporadic ALS patients and familial ALS mouse models (5, 31), the significance of these findings to ALS pathogenesis remains uncertain.
Considering the contradictory evidence regarding the protective effects of the UPR in experimental models of ALS, the contribution of the PERK pathway to ALS needs to be better defined. Using a genetic approach in several well-characterized mtSOD1 mouse models (Table 1), we demonstrate that neither diminished, nor enhanced, UPR capacity significantly affect the disease. These results were consistent across all five mtSOD1 mouse models studied. Therefore, the PERK pathway is an unlikely therapeutic target for mtSOD1-induced ALS.
Table 1.
Lines of transgenic mice expressing human SOD1 mutation
| hSOD1 Mutation | Dismutase Activity | Disease Onset (Age) | Early-Symptomatic Disease (Symptoms, Age) | End-Stage Disease/Survival (Age) |
|---|---|---|---|---|
| G93A-high copy | Active | ~4 months | Hind limb tremors with tail suspension, ~4 months | ~6 months |
| G93A-low copy | Active | ~6.5 months | Hind limb tremors with tail suspension, ~8 months | ~10 months |
| G85R | Inactive | ~9.5 months | No obvious tremors, mild weight loss, ~10 months | ~11 months |
| G37R-42 | Active | ~4 months | Hind limb tremors with tail suspension, ~4.5 months | ~6.5 months |
| G37R-29 | Active | ~10 months | Hind limb tremors with tail suspension, ~14.5 months | ~17 months |
hSOD1=human superoxide dismutase 1. Disease onset was defined as peak body weight before a decline. Early-symptomatic disease was defined by the appearance of first clinical symptoms. End-stage disease was defined by the inability of a mouse with limb paralysis to right itself within 20 s after being placed on its side. This artificial endpoint is universally used to determine ‘survival’ reliably and humanely. Disease onset, early-symptomatic disease, and end-stage/survival time points were determined experimentally for each mouse line. For immunohistochemical and biochemical analyses, tissues were collected from mice with early-symptomatic and end-stage disease at time points as indicated.
Results
PERK haploinsufficiency does not affect disease in SOD1(G93A) high-copy transgenic mice
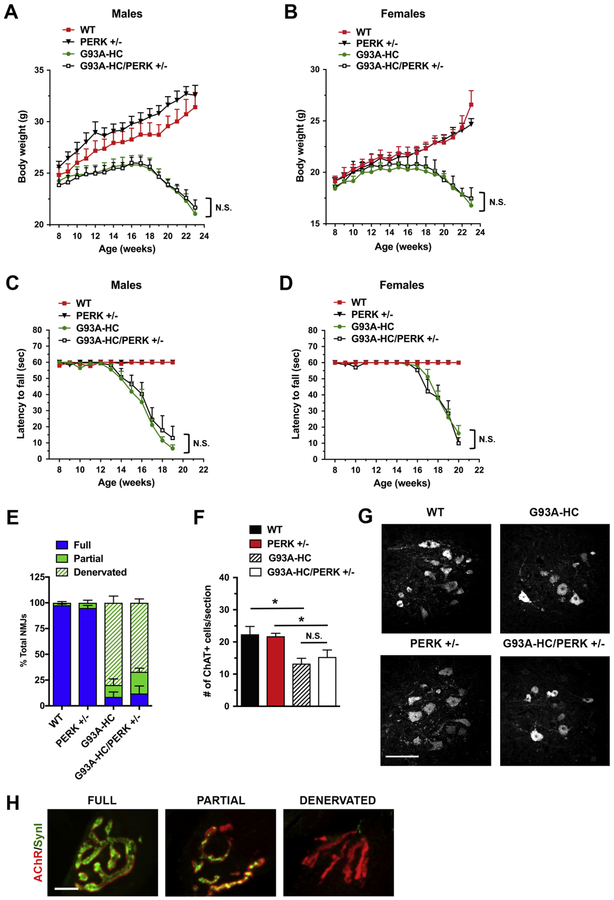
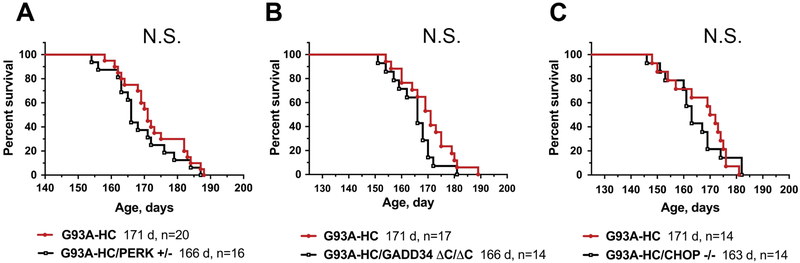
We used a genetic approach to clarify the importance of the PERK pathway in mtSOD1-induced familial ALS. To this end, we crossed G93A-HC mice onto a PERK+/‒ background. PERK+/‒ mice have no clinical phenotype and display decreased phosphorylation of eIF2α with ER stress (32). In contrast, PERK‒/‒ mice are normal at birth, but subsequently develop a rapid and progressive decline in endocrine and exocrine pancreatic functions, such that they could not be used in these studies (32). Progeny were followed for general disease progression by assessing their weight loss, muscle fatigue (using an inverted grid-hanging test), and survival. Males and females were analyzed separately. We found that both male and female G93A-HC and G93A- HC/PERK+/‒ mice displayed a nearly identical disease course, manifested by similar weight at all time points examined (Fig. 1A, B) and by comparable motor performance (Fig. 1C, D). We also found that PERK haploinsufficiency had no significant effect on the survival of G93A-HC mice (median survival: 171 d in G93A-HC versus 166 d in G93A-HC/PERK+/‒)(Fig. 4A).
Figure 1. PERK deficiency does not affect disease in G93A-HC mice.
(A, B) Body weights of male (A) and female (B) mice. No significant differences were found in G93A-HC/PERK+/‒ vs. G93A-HC mice (male or female). n = 6–7 males and n = 6–9 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (C, D) Motor fatigue measurements of male (C) and female (D) mice using inverted grid-hanging test. The ability of G93A-HC mice to cling to the wire grid declined rapidly after 13 weeks of age for males, and after 15 weeks of age for females. PERK deficiency did not change the length of time G93A-HC mice cling to the grid. n = 6–7 males and n = 6–9 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (E) Quantitation of tibialis anterior muscle innervation in early-symptomatic (15-week- old) mice. PERK deficiency had no effect on the extent of NMJ denervation in G93A-HC mice. n = 3 mice per genotype. % Fully innervated NMJs: 8.64% in G93A-HC vs. 11.95% in G93A- HC/PERK+/‒ P = 0.7264 (unpaired two-tailed t test). (F) Quantitation of ChAT-positive ventral horn motor neurons (per lumbar spinal cord section) in 15-week-old animals revealed no significant difference between G93A-HC/PERK+/‒ and G93A-HC mice. n = 3 mice per genotype. One-way ANOVA with Tukey’s post hoc test. (G) Representative images of ChAT-positive motor neurons in lumbar spinal cords of 15-week-old mice. Scale bar 100 μM. (H) Fully innervated, partially innervated, and denervated NMJs were distinguished by differences in co-localization of immunostaining for SynI (green) and AChRs (red). Scale bar 10 μM. Data are shown as mean ± SEM. *P < 0.05 N.S. = not significant.
Figure 4. PERK, GADD34, and CHOP deficiencies do not affect survival in G93A-HC mice.
No significant differences were found in the median survival times of G93A-HC mice deficient for PERK (A), GADD34 (B), and CHOP (C) compared to their respective G93A-HC littermate controls. n = number of animals of the designated genotype. Log-rank (Mantel-Cox) test: P = 0.2636 (A), P = 0.2152 (B), P = 0.2391 (C). N.S. = not significant. Mice unable to right themselves within 20 s after being placed on their sides were defined as end-stage. This artificial endpoint was used to determine ‘survival’ reliably and humanely.
We next evaluated molecular changes in tibialis anterior muscles and lumbar spinal cords of G93A-HC and G93A-HC/PERK+/‒ mice at 15 weeks of age, corresponding to an early disease stage, when the animals typically display hind limb tremor with tail suspension (graded as a clinical score of ‘1’ refer to “Methods” section for scoring system). Neuromuscular junction (NMJ) denervation is the first morphological change observed in ALS patients and mouse models (33). Because muscle denervation is associated with functional motor deficits, preservation of NMJs is commonly used as a criterion to determine a meaningful effect on target modulation. We stained tibialis anterior muscles with antibodies against synapsin I (SynI) to label nerve terminals and with α- bungarotoxin (α-BGT) to visualize acetylcholine receptors (AChRs) in muscle. Quantitation revealed fully innervated synaptic sites in WT and PERK+/‒ control mice, with nerve terminals perfectly apposed to AChRs in the postsynaptic membrane. In contrast, G93A-HC and G93A- HC/PERK+/‒ transgenics displayed significant tibialis anterior denervation, although the extent of denervation was statistically similar for both groups of mice (Fig. 1E). For motor neuron analysis, we identified motor neurons in the ventral horn of lumbar spinal cord using immunofluorescent labeling of choline acetyltransferase (ChAT). The extent of neurodegeneration was statistically similar in G93A-HC and G93A-HC/PERK+/‒ mice (~30–40% motor neuron loss compared to age-matched controls) (Fig. 1F, G). Overall, these data demonstrate that in contrast to a previously published report using G85R mice (25), PERK haploinsufficiency in G93A-HC mice affects neither their disease course, nor histopathology.
GADD34 deficiency does not ameliorate disease in SOD1(G93A) high-copy transgenic mice
GADD34 is a stress-inducible regulatory subunit of a holophosphatase complex that dephosphorylates p-eIF2α and plays a role in translational recovery (34). It has been shown that inhibition of GADD34 enhances the UPR by prolonging the phosphorylation of eIF2α (35–37). To access the impact of GADD34 deficiency on familial ALS, we bred GADD34 ΔC/ΔC mice, which express a mutation encoding a truncated GADD34 protein that lacks eIF2α phosphatase activity (36), with G93A-HC mice. The histopathology and disease progression of the offspring were then evaluated.
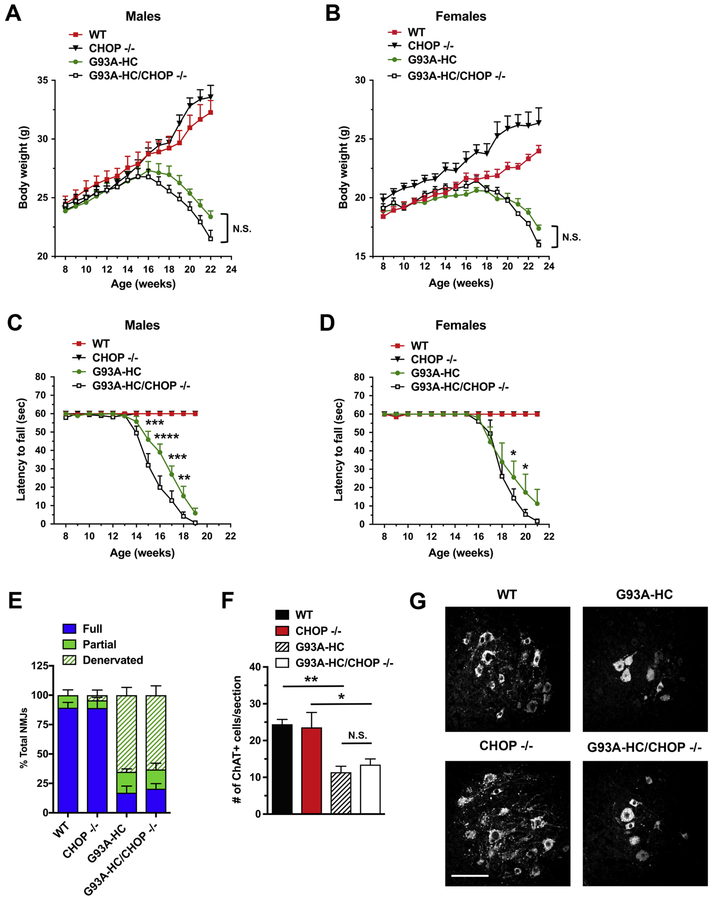
Previous genetic studies showed that G85R mice with diminished GADD34 activity (G85R/GADD34 +/ΔC) display a delay in the onset of disease and a markedly prolonged survival (26). On the contrary, our data revealed no protection against weight loss (Fig. 2A, B) or progressive motor deficits (Fig. 2C, D) in G93A-HC/GADD34 ΔC/ΔC mice compared to their G93A-HC littermates. Both male and female G93A-HC/GADD34 ΔC/ΔC mice demonstrated significantly reduced motor performance on the inverted grid-hanging test compared to their age- and sex-matched G93A-HC littermates, although motor deficits were more pronounced in females versus males (Fig. 2C, D). Importantly, no significant change in survival was observed in G93A-HC/GADD34 ΔC/ΔC mice compared to G93A-HC mice (median survival: 166 d versus 171 d) (Fig. 4B).
Figure 2. GADD34 deficiency does not ameliorate disease in G93A-HC mice.
(A, B) Body weights of male (A) and female (B) mice. No significant differences were found in G93A-HC/GADD34ΔC/ΔC vs. G93A-HC mice (male or female). n = 6–9 males and n = 6–8 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (C, D) Motor fatigue measurements of male (C) and female (D) mice using inverted grid-hanging test. Male and female G93A-HC/GADD34ΔC/ΔC mice displayed significantly diminished latency to fall, compared to their age- and sex-matched G93A-HC littermates. n = 6–9 males and n = 6–8 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (E) Quantitation of tibialis anterior muscle innervation in early-symptomatic (15-week-old) mice. G93A-HC/GADD34ΔC/ΔC mice displayed reduced NMJ innervation, although not statistically significant, compared to G93A-HC mice. n = 3 mice per genotype. % Fully innervated NMJs: 21.39% in G93A-HC vs. 1.23% in G93A-HC/GADD34ΔC/ΔC P = 0.1447 (unpaired two-tailed t test). (F) Quantitation of ChAT- positive ventral horn motor neurons (per lumbar spinal cord section) in 15-week-old animals indicated that GADD34 deficiency had no effect on motor neuron loss in G93A-HC mice. n = 3 mice per genotype. One-way ANOVA with Tukey’s post hoc test. (G) Representative images of ChAT-positive motor neurons in lumbar spinal cords of 15-week-old mice. Scale bar 100 μM. Data are shown as mean ± SEM. *P < 0.05 **P < 0.01 ***P < 0.001 N.S. = not significant.
At the histopathological level, tibialis anterior muscles and lumbar spinal cords of G93A- HC/GADD34 ΔC/ΔC and G93A-HC mice at an early diseases stage (15 weeks of age) displayed statistically similar extent of NMJ denervation (Fig. 2E) and motor neuron degeneration (Fig. 2F, G). Therefore, the loss of GADD34 activity did not exert a protective effect either at the level of the synapse (NMJ innervation), or the spinal cord (motor neuron numbers).
CHOP deficiency does not ameliorate disease in SOD1(G93A) high-copy transgenic mice
CHOP is a transcription factor downstream of the PERK-eIF2α pathway. CHOP activation has been shown to play an essential role in ER stress-induced apoptosis (21, 38, 39), although recent studies suggest that CHOP expression can be either adaptive or maladaptive, depending on cell type and disease context (40, 41). We investigated the role of CHOP in familial ALS using CHOP‒/‒ mice, which are phenotypically normal, but display an attenuated response to ER stress (39). These mice were crossed with G93A-HC mice to obtain G93A-HC/CHOP‒/‒ mice. Although we found that G93A-HC/CHOP‒/‒ mice exhibited modestly accelerated weight loss compared to their age- and sex-matched G93A-HC littermates (Fig. 3A, B), these effects were not statistically significant. Nevertheless, the G93A-HC/CHOP‒/‒ mice did display significantly reduced motor performance (on the inverted grid-hanging test) compared to their G93A-HC littermates (Fig. 3C, D). Interestingly, these motor deficiencies were more severe in males versus females. At the same time, the loss of CHOP did not affect survival in the transgenic mice (median survival: 171 d in G93A-HC versus 163 d in G93A-HC/CHOP‒/‒) (Fig. 4C).
Figure 3. CHOP deficiency does not ameliorate disease in G93A-HC mice.
(A, B) Body weights of male (A) and female (B) mice. No significant differences were found in G93A-HC/CHOP‒/‒ vs. G93A-HC mice (male or female). n = 7–9 males and n = 6–9 females per genotype. Data are shown as mean ± SEM for each time point. Two-way RM-ANOVA with Tukey’s post hoc test. (C, D) Motor fatigue measurements of male (C) and female (D) mice using inverted grid-hanging test. Male and female G93A-HC/CHOP‒/‒ mice displayed significantly diminished latency to fall, compared to their age- and sex-matched G93A-HC littermates. n = 7–9 males and n = 6–9 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (E) Quantitation of tibialis anterior muscle innervation in early-symptomatic (15- week-old) mice. CHOP deficiency had no effect on the extent of NMJ denervation in G93A-HC mice. n=3–4 mice per genotype. % Fully innervated NMJs: 17.08% in G93A-HC vs. 20.43% in G93A-HC/CHOP‒/‒ P = 0.6565 (unpaired two-tailed t test). (F) Quantitation of ChAT-positive ventral horn motor neurons (per lumbar spinal cord section) in 15-week-old animals indicated that CHOP deficiency had no effect on motor neuron loss in G93A-HC mice. n=3–4 mice per genotype. One-way ANOVA with Tukey’s post hoc test. (G) Representative images of ChAT- positive motor neurons in lumbar spinal cords of 15-week-old mice. Scale bar 100 μM. Data are shown as mean ± SEM. *P < 0.05 **P < 0.01 ***P <0.001 ****P < 0.0001 N.S. = not significant.
In parallel experiments, we examined tibialis anterior muscles and lumbar spinal cords collected from 15-week-old transgenic mice by immunohistochemistry. G93A-HC/CHOP‒/‒ and G93A-HC mice had a statistically similar extent of NMJ denervation (Fig. 3E), as well as motor neuron degeneration (Fig. 3F, G). All together, these data indicate that the loss of CHOP is not protective against disease in G93A-HC mice.
The level of SOD1(G93A) expression does not determine the response to genetic modulation of the PERK pathway
In contrast to prior genetic studies conducted in G85R mice, we did not observe modulation of disease in G93A-HC mice by genetic ablation of key molecules in the PERK pathway. A possible explanation could involve the extent of the toxic effect of the over-expressed human mtSOD1 gene. G93A-HC mice are characterized by extreme mtSOD1 overexpression (42). As a result, G93A-HC mice display an aggressive disease phenotype, with an early disease onset and a shorter life span, compared to other mtSOD1 strains (Table 1). In contrast, G85R mice used in this study express mtSOD1 at levels comparable to endogenous SOD1 expression and develop a late-onset form of disease (at ~9.5 months of age, defined by the onset of weight loss) (43). However, once initiated, the disease proceeds very rapidly and mice are completely paralyzed within 2 to 4 weeks after initial symptoms. Considering these differences in the disease course of G85R and G93A-HC mice, we next examined a variant of the G93A strain that expresses more modest levels of the human transgene, and, therefore, undergoes a prolonged disease compared to G93A-HC mice (Table 1) (44). Importantly, the average survival time of these low-copy G93A mice (G93A-LC) is similar to that in G85R mice (~10 months of age for G93A-LC versus ~11 months of age for G85R), thus enabling a more direct comparison between these two strains.
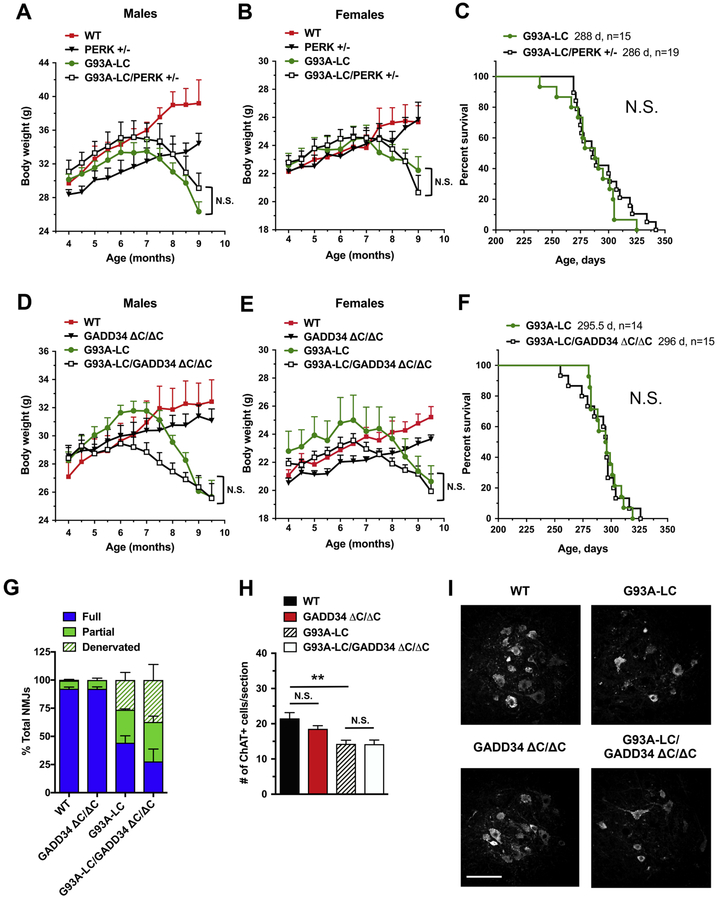
We crossed G93A-LC mice with either PERK+/‒ or GADD34 ΔC/ΔC mutant mice, and then followed the disease progression and survival in their progeny. We found that the PERK+/‒ mutation had no significant effect on weight in the G93A-LC mice, either in males or females (Fig. 5A, B). Additionally, no significant change was observed in median survival of G93A- LC/PERK+/‒ mice compared to their G93A-LC littermates (286 d versus 288 d) (Fig. 5C). The analysis of G93A-LC/GADD34 ΔC/ΔC animals revealed no protection against weight loss (Fig. 5D, E). The G93A-LC/GADD34 ΔC/ΔC males displayed modestly reduced weights throughout the disease course compared to their male G93A-LC littermates (Fig. 5D)
Figure 5. PERK and GADD34 deficiencies do not affect disease in G93A-LC mice.
(A, B, D, E) Body weights of male (A, D) and female (B, E) mice. No significant differences were found in G93A-LC/PERK+/‒ vs. G93A-LC mice (A, B) and in G93A-LC/GADD34ΔC/ΔC vs. G93A- LC mice (D, E). n = 6–10 males and n = 6–11 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (C, F) There were no significant differences in the median survival times of G93A-LC/PERK+/‒ vs. G93A-LC mice (C) and in the median survival times of G93A- LC/GADD34ΔC/ΔC vs. G93A-LC mice (F). n = number of animals of the designated genotype. Log-rank (Mantel-Cox) test: P = 0.3601 (C), P = 0.9752 (F). N.S. = not significant. (G) Quantitation of tibialis anterior muscle innervation in early-symptomatic (8-month-old) animals demonstrated that GADD34 deficiency had no significant effect on the extent of NMJ denervation in G93A-LC mice. n = 4 mice per genotype. % Fully innervated NMJs: 44.37% in G93A-LC vs. 27.83% in G93A-LC/GADD34ΔC/ΔC P = 0.2362 (unpaired two-tailed t test). (H) Quantitation of ChAT-positive ventral horn motor neurons (per lumbar spinal cord section) in 8- month-old mice indicated that GADD34 deficiency had no effect on motor neuron loss in G93A- LC mice. n = 4 mice per genotype. One-way ANOVA with Tukey’s post hoc test. (G) Representative images of ChAT-positive motor neurons in lumbar spinal cords of 8-month-old mice. Scale bar 100 μM. Data are shown as mean ± SEM. **P < 0.01 N.S. = not significant.
however, these results were not statistically significant. Moreover, G93A-LC and G93A- LC/GADD34 ΔC/ΔC mice had similar median survival times (295.5 d versus 296 d) (Fig. 5F). In agreement with our weight and survival studies, histological analysis of tibialis anterior muscles and lumbar spinal cords of early-symptomatic (8 months of age) G93A-LC and G93A- LC/GADD34 ΔC/ΔC mice revealed a comparable extent of NMJ denervation (Fig. 5G) and motor neuron degeneration (Fig. 5H, I). These data suggest that the level of the mtSOD1 transgene expression, and the associated protein toxicity, do not determine the response of mice carrying G93A mutation to genetic alterations in the PERK pathway.
Transgenic mice carrying different SOD1 mutations respond similarly to genetic ablations of PERK and GADD34
One of the hypotheses regarding the molecular mechanisms of ALS is based on mutation- driven SOD1 misfolding and subsequent deposition of its cytotoxic aggregates. More than 100 different mutations have been found in the SOD1 gene throughout all coding regions (45). It is generally thought that the different mtSOD1 proteins cause ALS by a similar mechanism, although it remains unclear how mutations in distinct SOD1 regions can lead to similar disease manifestations. We considered the possibility that if the level of mtSOD1 expression has little effect, then perhaps a specific mutation might determine the response of a mtSOD1 mouse strain to genetic alterations in the PERK pathway.
To account for this possibility and further explore the discordant results in G85R and G93A mice, we investigated two additional well-characterized mtSOD1 mouse models: G37R mice, lines 42 and 29 (46). The G37R-42 mouse line is most similar to the G93A-HC line in respect to disease onset and survival (Table 1). The other G37R mouse line (G37R-29) has dramatically prolonged disease onset (~10 months of age) and survival (~17 months of age) (Table 1). As before, we crossed G37R-42 and G37R-29 mice with either PERK+/‒ or GADD34 ΔC/ΔC mutants, and followed disease progression in their progeny. We discovered that neither PERK haploinsufficiency nor homozygous GADD34 deficiency significantly affected weight gain/loss in G37R-42 mice (male or female) throughout the disease course (Fig. 6A, B, D, E). Genetic ablations of PERK and GADD34 also had no significant effect on median survival of G37R-42 mice (192‒195.5 d in G37R-42 versus 196 d in G37R-42/PERK+/‒ versus 185 d in G37R- 42/GADD34 ΔC/ΔC) (Fig. 6C, F). In parallel experiments, we followed the disease course of G37R-29/PERK+/‒ and G37R-29/GADD34 +/ΔC mice, as well as their control G37R-29 littermates. Due to the exceedingly long life span of G37R-29 mice, we chose to follow them until 14.5 months of age (early-symptomatic disease), when the mice display a clinical score of ‘1’. We found that PERK haploinsufficiency had no effect on weight trend in G37R-29/PERK+/‒ mice (Fig. 7A). At the same time, G37R-29/GADD34 +/ΔC mice exhibited modestly reduced weight gain early in the disease course compared to G37R-29 mice, although not statistically significant (Fig. 7B). Nonetheless, these weight data suggest that GADD34 haploinsufficiency is not protective against disease. To confirm these findings at the histological level, we analyzed tibialis anterior muscles and lumbar spinal cords collected from 14.5-month-old G37R- 29/PERK+/‒ and G37R-29/GADD34 +/ΔC mice and their respective age-matched G37R-29 littermates. Although both complete NMJ denervation and motor neuron loss were observed in these two double-transgenic groups, the extent of the pathology was not significantly different from that in G37R-29 mice (Fig. 7C–H).
Figure 6. PERK and GADD34 deficiencies do not affect disease in G37R-42 mice.
(A, B, D, E) Body weights of male (A, D) and female (B, E) mice. No significant differences were found in G37R-42/PERK+/‒ vs. G37R-42 mice (A, B) and in G37R-42/GADD34ΔC/ΔC vs. G37R-42 mice (D, E). n = 6–9 males and n = 6–11 females per genotype. Two-way RM-ANOVA with Tukey’s post hoc test. (C, F) There were no significant differences in the median survival times of G37R-42/GADD34ΔC/ΔC vs. G37R-42 mice (C) and in the median survival times of G37R- 42/GADD34ΔC/ΔC vs. G37R-42 mice (F). n = number of animals of the designated genotype. Log- rank (Mantel-Cox) test: P = 0.4481 (C), P = 0.2383 (F). N.S.=not significant. Data are shown as mean ± SEM. N.S. = not significant.
Figure 7. PERK and GADD34 deficiencies do not affect disease in G37R-29 mice.
(A, B) Body weight ratios were calculated relative to baseline weight measurement (in 4 month- old mice). No significant differences were found in weight trend of G37R-29/PERK+/‒ vs. G37R- 29 mice (A) and in weight trend of G37R-29/GADD34+/ΔC vs. G37R-29 mice (B). Both male and female mice were used (n = 6–7 mice per genotype). Unpaired t test (one per time point) with Holm-Sidak correction for multiple comparisons. (C, F) Quantitation of tibialis anterior muscle innervation in early-symptomatic (14.5-month-old) mice demonstrated that PERK deficiency (C) and GADD34 deficiency (F) had no effect on the extent of NMJ denervation in G37R-29 mice. n = 4 mice per genotype. % Fully innervated NMJs: 47.49% in G37R-29 vs. 54.41% in G37R- 29/PERK+/‒ P = 0.5028 (unpaired two-tailed t test) 30.94% in G37R-29 vs. 31.91% in G37R-29/GADD34+/ΔC P = 0.8999 (unpaired two-tailed t test). (D, G) Quantitation of ChAT-positive ventral horn motor neurons (per lumbar spinal cord section) in 14.5-month-old mice indicated that PERK deficiency (D) and GADD34 deficiency (G) had no effect on motor neuron loss in G37R-29 mice. n = 4 mice per genotype. One-way ANOVA with Tukey’s post hoc test. (E, H) Representative images of ChAT-positive motor neurons in lumbar spinal cords of 14.5-month-old mice with PERK deficiency (E) and GADD34 deficiency (H). Scale bar 100 μM. Data are shown as mean ± SEM. *P < 0.05 **P < 0.01 ***P < 0.001 N.S. = not significant.
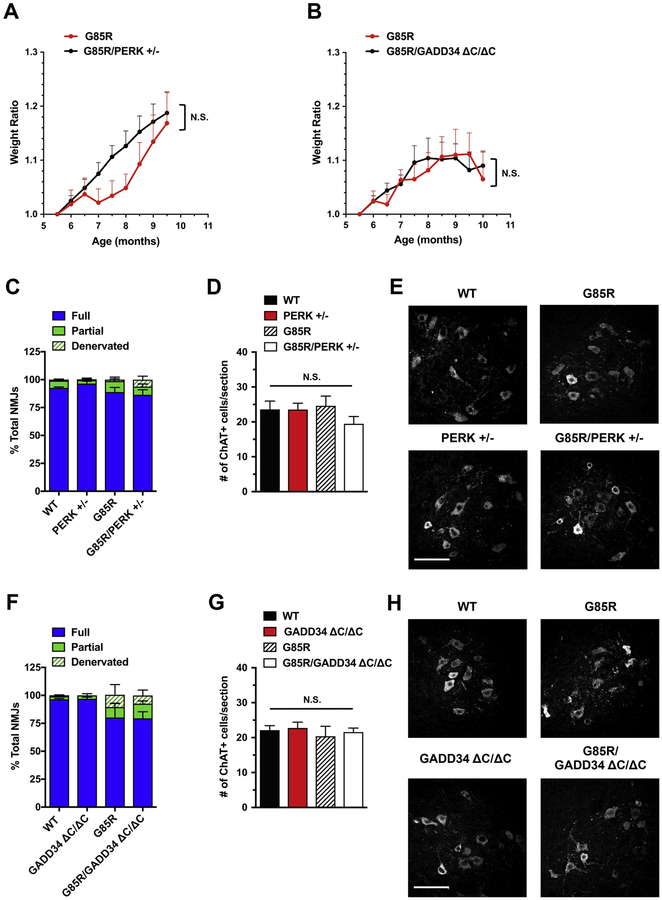
The original findings that demonstrated the potential involvement of the UPR in mtSOD1- induced ALS used the G85R mice (47). As none of the four mouse models we used in our study demonstrated a significant effect on disease after genetic inhibition or enhancement of the UPR-PERK pathway, we decided to investigate the effects of genetic PERK and GADD34 deficiencies in the G85R mouse model. Because the original G85R mouse line (47) was no longer available, we used another G85R mouse line (43). We bred these mice with PERK+/‒ and GADD34ΔC/ΔC mutants, and analyzed the resulting G85R/PERK+/‒ and G85R/GADD34ΔC/ΔC double-mutant offspring. Based on our experimental data, the G85R mouse line is distinct from other mtSOD1 lines (Table 1) in that early clinical symptoms (equivalent to a score of ‘1’ or ‘2’, denoting the severity of limb tremors) are not observed. As the first clinical symptom is limb paralysis (equivalent to a score of ‘3’), we selected time points (9.5–10 months of age) that were prior to the typical onset of limb paralysis and corresponded to the average disease onset/early disease to end our observations and collect histological samples. In contrast to a prior study (25), we did not observe an earlier disease onset in G85R/PERK+/‒ mice. Although it was previously reported that mean disease onset (defined as peak weight before a decline) of G85R/PERK+/‒ mice was at ~8.5 months of age (25), in our study G85R and G85R/PERK+/‒ animals displayed a similar trend of weight gain until 9.5 months of age, corresponding to disease onset in G85R mice and the last time point examined (Fig. 8A). We also analyzed tibialis anterior muscles and lumbar spinal cords collected from 9.5-month-old animals and did not observe accelerated neuropathological changes in G85R/PERK+/‒ mice compared to G85R mice (Fig. 8C–E). Similarly, comparisons of the weight gain trends in G85R/GADD34ΔC/ΔC and G85R mice revealed no statistically significant differences between these two groups of mice until 10 months of age, the last time point examined (Fig. 8B). The immunohistochemical analysis of NMJ innervation in tibialis anterior muscles of G85R/GADD34ΔC/ΔC and G85R mice at 10 months of age demonstrated a comparable extent of modest endplate denervation in both groups of mice (Fig. 8F), suggesting that in contrast to a prior study (26), GADD34 deficiency is not protective against disease. As expected from precipitous disease course in G85R mice, where significant motor neuron loss occurs shortly before end-stage disease (~11 months of age), motor neuron numbers in lumbar spinal cords of 10-month-old G85R/GADD34ΔC/ΔC and G85R mice were found to be normal (Fig. 8G, H). Cumulatively, these data demonstrate that all five mtSOD1 mouse strains studied show a similar response, or lack thereof, to genetic alterations in the PERK pathway.
Figure 8. PERK and GADD34 deficiencies do not affect disease in G85R mice.
(A, B) Body weight ratios were calculated relative to baseline weight measurement (in 5.5 month-old mice). No significant differences were found in weight trend of G85R/PERK+/‒ vs. G85R mice (A) and in weight trend of G85R/GADD34ΔC/ΔC vs. G85R mice (B). Both male and female mice were used (n = 6–8 mice per genotype). Unpaired t test (one per time point) with Holm-Sidak correction for multiple comparisons. (C, F) Quantitation of tibialis anterior muscle innervation demonstrated that PERK deficiency (C) did not exacerbate NMJ innervation in 9.5- month-old mice (time point corresponding to disease onset in G85R controls). n = 4 mice per genotype. % Fully innervated NMJs: 88.87% in G85R vs. 86.49% in G85R/PERK+/‒ P = 0.7121 (unpaired two-tailed t test). As expected from precipitous disease course in G85R mice, NMJ innervation in these 9.5-month-old mice was comparable to that in WT controls. (F) GADD34 deficiency had no effect on the extent of NMJ denervation in early-diseased (10-month-old) G85R mice. n = 4 mice per genotype. % Fully innervated NMJs: 80.22% in G85R vs. 79.43% in G85R/GADD34ΔC/ΔC P = 0.9568 (unpaired two-tailed t test). (D, G) Quantitation of ChAT-positive ventral horn motor neurons (per lumbar spinal cord section) indicated that PERK deficiency (D) and GADD34 deficiency (G) had no effect on motor neuron numbers in G85R mice. n = 4 mice per genotype. One-way ANOVA with Tukey’s post hoc test. As expected from the disease course in G85R mice, motor neuron numbers in these mice were normal at 9.5–10 months of age. (E, H) Representative images of ChAT-positive motor neurons in lumbar spinal cords of G85R mice with PERK deficiency (E) and GADD34 deficiency (H). Scale bar 100 μM. Data are shown as mean ± SEM. N.S.=not significant.
Biochemical analyses of the PERK pathway in spinal cords of early-symptomatic and end-stage mutant SOD1 mice
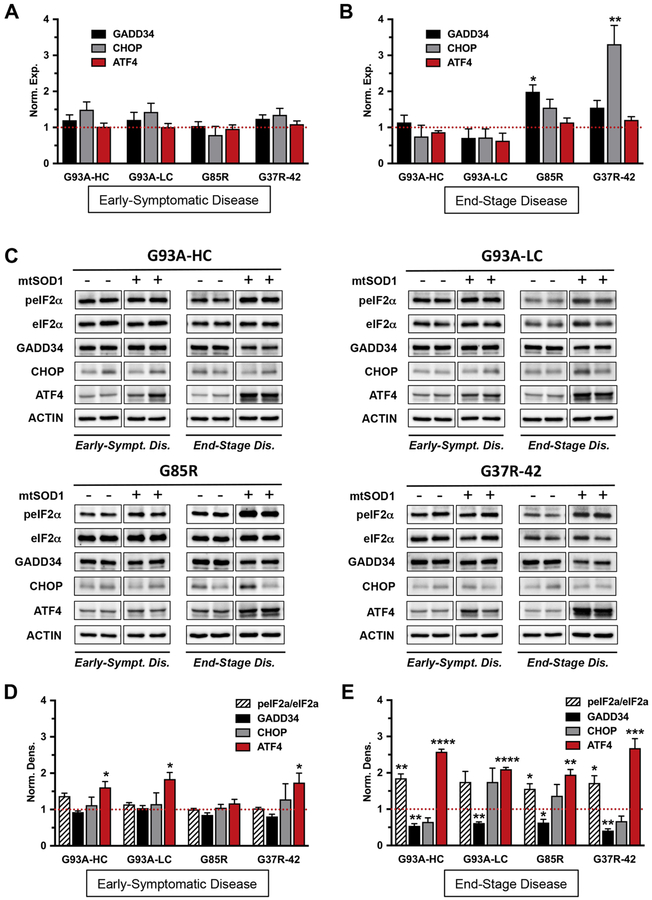
It was previously suggested that alterations to ER proteostasis play a critical role in ALS progression and represent one of the earliest pathological signatures of the disease (48). We therefore investigated molecular changes in lumbar spinal cords of early-symptomatic and paralyzed (end-stage) mtSOD1 mice (Table 1) by real-time qPCR and by western blot. We found no change in GADD34 and CHOP mRNA expression levels at an early disease stage in all five mtSOD1 mouse strains used in the study (Fig. 9A, Fig. S1A). Consistent with mRNA expression data, we also did not observe any changes in GADD34 and CHOP protein levels. Additionally, there was no evidence of increased phosphorylation of eIF2α at this early disease time point in all five mtSOD1 mouse strains examined (Fig. 9C, D and Fig. S1B, C). Interestingly, there was an increase in protein levels of the downstream UPR transcription factor ATF4 at an early disease stage (Fig. 9C, D and Fig. S1B, C), despite the apparent absence of PERK pathway activation. Together, our results indicate that PERK pathway activation is not detected early in disease (when mice have started to display clinical symptoms), at least not at the level of total RNA and protein analyses in lumbar spinal cord lysates.
Figure 9. Biochemical analyses of the PERK pathway in spinal cords of early- symptomatic and end-stage mutant SOD1 mice.
The PERK pathway activation is not detected in spinal cords of early-symptomatic mtSOD1 mice. Moreover, increased eIF2α phosphorylation in spinal cords of end-stage mtSOD1 mice does not induce downstream UPR components CHOP and GADD34. (A, B) Real-time qPCR for GADD34, CHOP and ATF4 mRNA levels in lumbar spinal cords from mtSOD1 mice with early-symptomatic disease (A) and end-stage disease (B). Data were normalized to actin reference gene and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 4 mice per group. Norm. exp. = normalized expression. (C) Representative immunoblots of p-eIF2α (phosphorylated), eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from mtSOD1 mice with early-symptomatic and end-stage disease, and their age-matched WT controls. Actin was used as a loading control. (D, E) Quantification of p-eIF2α/eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from mtSOD1 mice with early-symptomatic disease (D) and end-stage disease (E). Data were normalized to actin and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 3–4 mice per group. Norm. dens. = normalized densitometry. Unpaired t test: *P < 0.05 **P < 0.01 ***P < 0.001 ****P <0.0001.
In mice with end-stage disease, we observed an increase in GADD34 mRNA levels in the G85R mice, and in CHOP mRNA levels in the G37R-42 mice (Fig. 9B). However, there was no correlation between message and protein levels (the latter were reduced or unchanged) (Fig. 9E). Also in these mice, the enhanced phosphorylation of eIF2α was surprisingly not accompanied by an increase in GADD34 and CHOP protein levels. On the contrary, GADD34 protein levels were significantly reduced, whereas CHOP protein levels remained unchanged, relative to those in similarly aged wild-type control littermates. These results were consistent across all four mtSOD1 mouse strains analyzed (Fig. 9C, E). Additionally, we found that whereas ATF4 mRNA levels remained unaffected, the ATF4 protein levels were significantly upregulated in end-stage disease (Fig. 9B, C, E). These data demonstrate that increased phosphorylation of eIF2α in lumbar spinal cords of all end-stage mtSOD1 mouse strains examined does not result in a full UPR response.
Discussion
ALS is associated with the accumulation of abnormal intracellular protein aggregates, which can disrupt the balance between protein generation and degradation that is crucial for protein homeostasis. These alterations can trigger ER stress and ultimately contribute to neurodegeneration. Cells counteract ER stress by activating the UPR, which aims to restore proteostasis within the secretory pathway in part through regulation of genes involved in protein folding, quality control, degradation and translational repression pathways (49). PERK is the most rapidly activated UPR pathway in response to the accumulation of misfolded proteins, leading to eIF2α phosphorylation and subsequent attenuation of protein synthesis (50). The PERK/eIF2α phosphorylation axis recently emerged as a potential therapeutic target for ALS (51, 52). Therefore, we sought to conduct a comprehensive analysis of the effects of genetic PERK pathway modulation on ALS using several well-characterized mtSOD1 transgenic mouse models. We demonstrate that, in contrast to previous reports (25, 26), neither PERK haploinsufficiency nor GADD34 deficiency significantly affect survival in all mtSOD1 mouse models examined. Moreover, we show that genetic ablation of the pro-apoptotic CHOP transcription factor has no effect on survival in G93A-HC mice. Overall, our data show an absence of disease modulation by genetic alteration of the key molecules in the PERK pathway.
It was previously reported that G85R/PERK+/‒ mice display an accelerated disease course and pathology (25). In contrast, we discovered that PERK haploinsufficiency had no effect on disease in all five mtSOD1 mouse models studied, including the G85R mice. We further discovered that these divergent findings were not likely due to the level of the mtSOD1 transgene expression, as the PERK+/‒ mutation had no effect on disease progression and survival of both G93A-HC and G93A-LC mouse lines. Moreover, we found that transgenic mice carrying different SOD1 mutations displayed similarly unchanged disease and pathology in response to PERK haploinsufficiency. Taken together, our genetic data suggest that PERK signaling does not likely play a crucial role in mtSOD1-induced ALS.
With regards to G85R mice, our disparate results from what has been published (25) might reflect the use of distinct G85R transgenic lines, which were generated by different laboratories. Specifically, G85R transgenic mice used in the original UPR genetic studies (25, 26), but no longer available, were generated with a construct containing loxP sites engineered to flank the G85R mutant human SOD1 gene (47). These mice reportedly over-express the human SOD1 protein at levels approximately 50% higher than the endogenous mouse SOD1 protein, which may augment the development of ER stress in these animals. In contrast, we used transgenic mice that carry a G85R mutant human SOD1 transgene lacking the loxP sequences (43). These mice express the human SOD1 protein at levels comparable with the endogenous mouse SOD1 protein. Therefore, the lack of an effect on the mtSOD1 phenotype by the genetic manipulation of the UPR in the current study might reflect our use of a G85R mouse line that expresses more physiologically relevant levels of the mtSOD1 protein. Notably, similar discrepancies between these two G85R lines have been previously recognized (47). Our results thus caution against drawing conclusions regarding mechanistic contributors to ALS pathogenesis from a single mutant SOD1 mouse line. Additionally, it is important to mention that mutant SOD1 mouse lines display significant variability with regards to disease onset and progression (Table 1). This variability is currently unexplained, but may be an important factor that should be taken into consideration as far as translational efforts are concerned.
In agreement with our genetic studies, we did not observe PERK pathway activation at the molecular level in lumbar spinal cords of early-symptomatic mtSOD1 mice, which by that time already exhibited substantial neuropathological changes. These included significant motor neuron loss in the ventral horn of lumbar spinal cords and NMJ denervation in tibialis anterior muscles. The protein levels of p-eIF2α, as well as mRNA and protein levels of GADD34 and CHOP, were consistently unchanged in all mtSOD1 mouse strains analyzed, suggesting that PERK signaling is not involved in early disease. There is a possibility, however, that the UPR mRNA and protein expression levels were very low and thus below the sensitivity of detection using our real-time qPCR and western blot assays (in whole lumbar spinal cord lysates). It was previously suggested that adaptation to ER stress in cells chronically exposed to protein folding insults, such as those implicated in familial forms of neurodegeneration, is an intrinsic consequence of low-level activation of the UPR (53). This is in contrast to robust UPR activation in cells by acute severe stress, such as pharmacological perturbation of ER function. Along the same lines, prior evidence of ER stress activation specifically in motor neurons of mtSOD1 mice with early disease (9, 11) suggests that our analyses of whole lysates may not have detected changes that occurred exclusively in motor neurons.
Under ER stress, persistent activation of the PERK/eIF2α pathway, and the resulting global suppression of protein synthesis, play a major role in determining the cell’s fate. GADD34 (also known as PPP1R15A) is a stress-inducible regulatory subunit of the PP1/GADD34 holophosphatase complex that quickly dephosphorylates p-eIF2α to counteract PERK signaling and restore general protein synthesis. Recently, pharmacological targeting of GADD34-mediated p-eIF2α dephosphorylation has emerged as a promising strategy for modifying the course of protein misfolding diseases, including ALS (51, 52, 54). To this end, several small molecule inhibitors (salubrinal, guanabenz, and Sephin1) have been used to disrupt the PP1/GADD34 holophosphatase complex in order to enhance eIF2α phosphorylation and downstream signaling. Salubrinal attenuated disease manifestations and prolonged survival in the G93A-HC mouse model (9), while guanabenz ameliorated disease in two separate studies using G93A-HC mice (28, 29). Nevertheless, another group reported adverse effects of guanabenz in G93A-HC mice. In these studies, guanabenz treatment significantly accelerated disease onset and shortened lifespan in male mice (30). Most recently, Sephin1 prevented motor deficits and motor neuron loss in G93A-HC mice (27).
The divergent findings using small-molecule GADD34 inhibitors highlight some of the challenges of pharmacological modulation of the ER stress response in mouse models of ALS. These include issues of bioavailability, potency, specificity, or off-target effects that can be associated with using pharmacological compounds. Moreover, given that pharmacological studies were all conducted in the G93A-HC mouse model, we were interested in analyzing the effects of GADD34 deficiency in these transgenic mice by using a genetic approach. We discovered that, in direct contrast to a genetic study using G85R mice (26), as well as the pharmacological studies, GADD34 deficiency did not ameliorate disease in G93A-HC mice, and did not affect their survival. Importantly, GADD34 deficiency similarly did not alleviate disease in all other mtSOD1 mouse models studied, including the G85R model. The inconsistency of our genetic study and the pharmacological studies might suggest that the consequences of genetic GADD34 ablation during development are different from transient pharmacological inhibition of GADD34 in the adult. It is known that genetic knockout or knockdown approaches can produce phenotypes distinct from those seen with drug perturbation (55). A limitation of our G85R/GADD34ΔC/ΔC study is that we followed G85R/GADD34ΔC/ΔC mice and their G85R littermates only to 10 months of age (corresponding to early disease in control G85R mice), at which time lumbar spinal cord and tibialis anterior muscle tissues were collected for immunohistochemical analysis. At this time point, we did not detect significant differences in the extent of NMJ denervation in G85R/GADD34ΔC/ΔC mutant mice compared to their G85R littermates. Nevertheless, since none of our survival studies, which were carried out in G93A-HC, G93A-LC, and G37R-42 mouse strains, revealed a positive protective effect of the GADD34 mutation, it is possible that G85R mice are the only ones that show disease modulation by genetic GADD34 haploinsufficiency. In this case, it might be an indication that the outcomes are model-specific and possibly uninformative for human disease.
CHOP is a key transcription factor in the PERK pathway and is known to play an important role in ER stress-mediated cell death (21, 38, 39). There is also evidence suggesting that CHOP expression can be adaptive or maladaptive, depending on cell type and disease context. For instance, genetic CHOP ablation was shown to be detrimental to hippocampal neurons after seizures (40, 56), but neuroprotective in EAE/optic neuritis (57), Parkinson’s disease (58), and moderate spinal cord injury (59). Increased CHOP expression has been detected in spinal cords of sporadic ALS patients and familial ALS mouse models (5, 31). At the same time, the exact role of CHOP in ALS is not clear. Considering that in the ER stress response CHOP is a critical player that can decide cellular fate, it is important to determine whether CHOP could be a viable target for therapeutic intervention. We hypothesized that genetic CHOP deletion will be protective in G93A-HC mice. On the contrary, we discovered that CHOP deficiency did not ameliorate disease in these mice. There was also no significant difference in the survival between G93A-HC/CHOP‒/‒ and littermate G93A-HC mice, indicating that global CHOP deletion ultimately does not affect disease outcome. Therefore, the present study demonstrates that the PERK-CHOP signaling axis is unlikely to be significantly involved in mtSOD1-induced ALS.
Conclusions
In summary, we have investigated the role of the UPR-PERK pathway in mtSOD1-induced ALS using several transgenic mouse models. Our gene expression data suggest that PERK pathway might not be a prominent player in mtSOD1-induced disease. These results are in agreement with our genetic studies, where neither inhibition of the ER stress response (via ablation of PERK), nor ER stress response enhancement (via ablation of GADD34), had a significant effect on the survival of mtSOD1 mice. We also showed that genetic CHOP ablation is not protective against the disease. Therefore, targeting the PERK arm of the UPR is not likely to be an effective strategy for ALS therapy.
Materials and Methods
Animals
Mice overexpressing human SOD1 (hSOD1) were purchased from Jackson Laboratory (Bar Harbor, ME). The following mutant SOD1 strains were used: B6.Cg-Tg(SOD1-G93A)1Gur/J (stock #004435), B6.Cg-Tg(SOD1-G93A)dl1Gur/J (stock #002299), B6.Cg-Tg(SOD1- G37R)42Dpr/J (stock #008342), B6.Cg-Tg(SOD1-G37R)29Dpr/J (stock #008229), and B6.Cg- Tg(SOD1-G85R)148Dwc/J (stock #008248). These mutant SOD1 mice have been previously described (42–44, 46) and were maintained in-house as hemizygous transgenics. GADD34ΔC/ΔC and PERK+/‒ mice were kindly provided by Dr. David Ron (University of Cambridge, Cambridge, UK), and CHOP‒/‒ mice were purchased from Jackson Laboratory (stock #005530). These mice have been previously described (32, 36, 39) and were bred in-house. All mice were on the C57BL/6 background. Both males and females were used.
All mice used in this study were housed under pathogen-free conditions at controlled temperatures and relative humidity with a 12/12-hour light/dark cycle and free access to pelleted food and water. All animal experiments were conducted in compliance with The University of Chicago’s Animal Care and Use Committee guidelines.
Genotyping
Mouse genomic DNA was isolated from tail biopsies and amplified using REDExtract-N-Amp™ Tissue PCR Kit (catalog# XNAT, Sigma-Aldrich) as per manufacturer’s instructions. Genotyping PCRs were performed using the following primer sequences: human SOD1 sense (5’- CATCAGCCCTAATCCATCTGA-3’) and human SOD1 anti-sense (5’- CGCGACTAACAATCAAAGTGA-3’) primers were used in combination with positive control mouse interleukin-2 (IL-2) sense (5’-CTAGGCCACAGAATTGAAAGATCT-3’) and mouse IL-2 anti-sense (5’-GTAGGTGGAAATTCTAGCATCATCC-3’) primers. IL-2 PCR product was visualized at 324 bp and human SOD1, if present, at 236 bp. PERK sense primer (5’- CGGAGACAGTACAAGCGCAGATGA-3’) was used with either wild-type (wt) PERK anti-sense primer (5’- AAGGACCCTATCCTCCTGCTGCAC-3’) or mutant (mt) PERK anti-sense primer (5’- GCTACCGGTGGATGTGGAATGTG-3’) in separate reactions. Expected bands were 232 bp (wt), 302 bp (mt). GADD34 sense primer (5’-CCAGGAGAGAAGACCAAGGGACGTG-3’) was used in combination with wt GADD34 anti-sense primer (5’- CGAGATTGCAAGAGAGTGAACACAGC) and mt GADD34 anti-sense primer (5’- AAGCCTTCGCCATCTGCTTATCCAG-3’) in a single reaction. Expected bands were 468 bp (wt), 550 bp (mt). CHOP sense primer (5’-ATGCCCTTACCTATCGTG-3’) was used with wt CHOP anti-sense primer (5’-GCAGGGTCAAGAGTAGTG-3’) and mt CHOP anti-sense primer (5’-AACGCCAGGGTTTTCCCAGTCA-3’) in a single reaction. Expected bands were 544 bp (wt), 320 bp (mt).
hSOD1 copy number assessment
It was previously reported that the phenotype of high-copy G93A mice is dependent on the number of transgene copies in their genome (60). Because the mutant G93A transgene can sometime undergo copy number loss due to intra-locus recombination events during meiosis, it is imperative to monitor the transgene number in the breeding colony (61). To this end, we determined the SOD1(G93A) copy number using a real-time qPCR assay in all breeders and their progeny. Genomic DNA was extracted from tails with the use of an Extract-N-Amp tissue PCR kit (catalog# XNAT2R, Sigma-Aldrich). The following primers and probes (Integrated DNA Technologies Inc.) were used, as suggested by Jackson Laboratory: human SOD1 sense primer (5’-GGGAAGCTGTTGTCCCAAG-3’) human SOD1 anti-sense primer (5’-CAAGGGGAGGTAAAAGAGAGC-3’) human SOD1 FAM probe (5’- CTGCATCTGGTTCTTGCAAAACACCA-3’) mouse apolipoprotein-B (internal control) sense primer (5’-CACGTGGGCTCCAGCATT-3’) mouse apolipoprotein-B anti-sense primer (5’-TCACCAGTCATTTCTGCCTTTG-3’) mouse apolipoprotein-B Cy5 probe (5’- CCAATGGTCGGGCACTGCTCAA-3’). Extract-N-AMP PCR ReadyMix reagent (catalog #E3004, Sigma-Aldrich) was used for real-time amplification of DNA. After heating at 94°C for 3 min, the DNA was amplified by 40 cycles of 94°C for 15 s and 60°C for 1 min on a Bio-Rad CFX96 Real-Time PCR detection system. The transgene zygosity was determined by comparing ΔC(t) values of each SOD1-positive sample against standard high-copy controls, using an endogenous reference (apolipoprotein-B). We found that the copy number did not change over multiple generations and the course of experiments.
Clinical assessment and survival
Transgenic SOD1-positive mice and their littermate controls were followed for disease onset, progression, and survival as previously described in studies of familial ALS mouse models (61, 62). Weight was recorded twice a week for SOD1(G93A) high-copy mice and once a week for all other mtSOD1 strains. Onset of the disease was defined as peak weight before a decline (as a measure of denervation muscle atrophy onset). End-stage disease was defined by the inability of a mouse to right itself within 20 s after being placed on its side. This artificial endpoint is used to determine ‘survival’ reliably and humanely. In addition to weight, the neurological score of limbs was assessed at least once a week on a scale from 0 to 4 (with 0= normal, 1= hind limb tremor with tail suspension, 2= tremor with locomotion, 3= limb paralysis, and 4= mouse paralyzed and cannot right itself within 20 s after being placed on its side). The neurological assessment was necessary to identify non-ALS deaths: if a mouse dies before attaining a score of ‘2’, it is highly unlikely that the death is attributable to ALS. Additionally, to facilitate access to food and hydration, wet food pellets and gel diet were placed on the cage floor when the animals developed limb paralysis.
Behavioral test for high-copy SOD1(G93A) mice
Motor fatigue was assessed once a week, starting at 8 weeks of age, using an inverted grid- hanging test (63, 64). Individual mice were placed in the center of a wire grid, which was mounted ~80 cm above a padded surface. The grid was then gently inverted and maintained in an inverted position for 60 s. The length of time each mouse remained attached to the grid was recorded. Each mouse was tested three times with an interval of ~15 min. The average hanging time was calculated from two best attempts. Control mice routinely remained attached to the grid for the entire duration of the experiment. For mtSOD1-positive mice, the experiment was stopped once hanging time was less than 5 s, at approximately 19 to 21 weeks of age.
Histology
Mice were deeply anaesthetized with 2.5% avertin in dH2O. Avertin stock solution was prepared by dissolving 2, 2, 2-Tribromoethanol (catalog #T48402, Sigma Aldrich) in 2-methyl-2-butanol (catalog #240486, Sigma Aldrich). Upon the loss of nociceptive reflexes, animals were perfused transcardially with 0.9% NaCl, followed by cold 4% PFA in PBS. Tibialis anterior muscles were immediately dissected out. Whole muscles were post-fixed in 4% PFA for 20 min and rinsed in PBS for 5 min, followed by cryopreservation through 3 successive 5 min incubations in 5, 10, and 15% sucrose in PBS. Tissue samples were then incubated overnight in 20% sucrose in PBS, embedded in Optimal Cutting Temperature compound (OCT) and frozen on dry ice. Spinal cords were carefully dissected out, post-fixed in 4% PFA for 4 h, washed in PBS, and cryopreserved in 30% sucrose until saturation. The lumbar segments of each spinal cord were embedded in OCT and frozen on dry ice. Samples were stored in −80°C until use.
Immunohistochemistry
Longitudinal tibialis anterior muscle sections were cut at 10 μm on a cryostat, collected onto microscope slides (catalog #1358W, Globe Scientific Inc), and frozen at −80°C until use. For immunostaining, sections were allowed to thaw at room temperature (RT), permeabilized in acetone at −20°C for 10 min and washed with PBS. Sections were then incubated for 1 h at RT in a blocking solution, consisting of PBS, 5% BSA, 1% normal donkey serum, and 0.2% Triton X-100. Primary antibody staining was performed overnight at 4°C in a humidifying chamber with antibodies diluted in the blocking solution. To analyze the NMJs in tibialis anterior muscles, presynaptic nerve terminals were detected with rabbit 1:500 synapsin I antibody (catalog #ab18814, Abcam), and postsynaptic endplates (AChRs) with 1:200 Alexa 594-conjugated α- bungarotoxin (α-BGT) (catalog #B13423, Life Technologies). Sections were then rinsed 3 times with PBS, and secondary donkey-anti-rabbit IgG Alexa 488 antibody (Life Technologies) was applied for 2 h at RT. For lumbar spinal cords, transverse sections were cut at 10 μm on a cryostat, collected onto microscope slides (catalog #1358W, Globe Scientific Inc), and frozen at −80°C until use. Immunostaining was performed using an antigen retrieval technique. Briefly, sections were washed in TBS (pH 7.5), and boiled in 10mM trisodium citrate buffer (pH 6.0) for 30 min at 90°C, followed by cooling at RT for 30 min. Sections were then incubated in 10 mM glycine (in TBS with 0.25% Triton X-100) for 1h at RT to quench autofluorescence from the PFA. After several washes in TBS, blocking solution was applied for 1 h at RT, followed by overnight incubation with goat 1:200 choline acetyltransferase antibody (ChAT) (catalog #AB144P, Millipore) to visualize motor neurons. Secondary donkey-anti-goat IgG Alexa 594 antibody (Invitrogen) was applied in the blocking solution for 2 h at RT. The fluorescent-stained muscle and spinal cord tissue sections were mounted in ProLong Gold antifade reagent with DAPI (catalog # P36931, Thermo Fisher Scientific) under a glass coverslip. Tissue sections were imaged with Marianas Yokogawa type spinning disk confocal microscope using 20x Plan- Neofluar/NA 0.5 dry objective.
For muscle innervation analysis, we examined 50–100 neuromuscular junctions (NMJs) in tibialis anterior of each mouse (n=3–4 animals per genotype). Each NMJ was designated as fully innervated, partially denervated, or fully denervated. At fully innervated NMJs, nerve terminal staining completely overlapped with α-BGT staining for postsynaptic AChRs, whereas fully denervated NMJs displayed α-BGT staining only. At partially innervated NMJs, just a portion of the AChR-rich postsynaptic endplate was labeled with nerve terminal staining. Muscle innervation data (full/partial/none) were expressed as a percentage of total NMJs counted per animal. For motor neuron analysis, we defined motor neurons as cells in the ventral horn of the spinal cord that were positive for ChAT. We analyzed 4 non-consecutive lumbar spinal cord sections per mouse (n=3–4 animals per genotype). All immunohistochemistry analyses were done using mice in an early-symptomatic disease stage, as described in Table 1.
Total protein and RNA isolation
For spinal cord collection, deeply anesthetized mice were perfused with ice-cold PBS. Fresh lumbar spinal cords were then obtained by ejection of the cord from the vertebrate column (65) using a 18 g 1/2” needle (BD Biosciences) attached to a 10-mL syringe filled with PBS. Spinal cord tissues were rinsed in PBS, snap frozen in liquid nitrogen, and stored in −80°C until use.
For protein isolation, samples were homogenized in ice-cold RIPA lysis buffer (catalog #R0278, Sigma-Aldrich) supplemented with protease inhibitor cocktail (catalog #78430, Thermo Fisher Scientific), phosphatase inhibitor cocktail 2 (catalog #P2850, Sigma-Aldrich), phosphatase inhibitor cocktail 3 (catalog #P5726, Sigma-Aldrich), and 17.5 mM β-glycerophosphate (catalog #G9422, Sigma- Aldrich). After 30 min incubation on ice, protein lysates were clarified by centrifugation at 14,000 rpm for 20 min at 4°C, and stored at −80°C. Protein concentration was determined using a BCA Protein Assay Kit (catalog #23255, Thermo Fisher Scientific).
Total RNA was extracted from samples using Aurum Total RNA Fatty and Fibrous Tissue Kit (catalog #732–6870, Bio-Rad Laboratories). RNA concentration was measured with Nanodrop spectrophotometer, and RNA quality was confirmed on a model 2100 Bioanalyzer using Agilent RNA 6000 Nano Kit (catalog #5067–1511, Agilent Technologies) according to the manufacturer’s instructions. Only samples with an RNA integrity number ≥ 8 were used.
Western blot
Protein samples (30 μg) were separated by SDS-PAGE using a 4–20% gradient gel, and transferred to a nitrocellulose membrane, pore size 0.2 μm (catalog #1620112, Bio-Rad). Nonspecific binding was blocked with 4% non-fat milk in TBST for 1 h at RT. Membranes were then incubated with primary antibodies in the blocking solution at 4°C overnight. The following primary antibodies were used: rabbit 1:500 p-eIF2α (catalog #AB32157, Abcam), rabbit 1:1000 eIF2α (catalog #9722S, Cell Signaling Technology), mouse 1:500 CHOP (catalog #MA1–250, Thermo Fisher Pierce), mouse 1:250 CHOP (catalog #sc-7351, Santa Cruz), mouse 1:250 ATF4 (catalog #sc-390063, Santa Cruz), rabbit 1:2000 GADD34 (catalog #sc-825, Santa Cruz), and mouse 1:2000 β-actin (catalog #A4700, Sigma- Aldrich). Proteins were stained with species-specific HRP-conjugated secondary antibodies (GE Healthcare) and visualized with Super Signal West Pico Chemiluminescent substrate (Thermo Fisher Scientific) on ChemiDoc™ Touch machine (Bio-Rad). Bands were quantified using Image Lab software (Bio-Rad). Data were normalized to β-actin and expressed as mean fold change in relation to the control group.
Quantitative real-time PCR
RNA was cleaned of genomic DNA, and reverse-transcribed, using iScript gDNA Clear cDNA Synthesis Kit (catalog #172–5035, Bio-Rad Laboratories) according to the manufacturer’s instructions. Quantitative real-time PCR was run on a Bio-Rad CFX96 Real-Time PCR detection system using SYBR Green technology (catalog #1725271, Bio-Rad Laboratories). Relative gene expression was calculated using the ΔΔC(t) method after normalization to β-actin reference gene. Primers used (Integrated DNA Technologies Inc) are listed below:
Real-time qPCR primer sequences (Forward: Fwd Reverse: Rev).
CHOP
Fwd: 5’-CTGCCTTTCACCTTGGAGACG-3’
Rev: 5’-CTTTGGGATGTGCGTGTGACC-3’
GADD34 Exon 2
Fwd: 5’-CCCTCCAACTCTCCTTCTTCAG-3’
Rev: 5’-CAGCCTCAGCATTCCGACAA-3’
GADD34 Exons 2–3
Fwd: 5’-CCCGAGATTCCTCTAAAAGCTC-3’
Rev: 5’-CCAGACAGCAAGGAAATGG-3’
PERK
Fwd: 5’-TCTTGGTTGGGTCTGATGAAT-3’
Rev: 5’-GATGTTCTTGCTGTAGTGGGGG-3’
ATF4
Fwd: 5’-TGGATGATGGCTTGGCCAGTG-3’
Rev: 5’-GAGCTCATCTGGCATGGTTTC-3’
β-ACTIN
Fwd: 5’-GTGACGTTGACATCCGTAAAGA-3’
Rev: 5’-GCCGGACTCATCGTACTCC-3’
Statistical Analyses
Data are presented as mean ± SEM. Multiple comparisons were made using one-way ANOVA, or two-way repeated measures (RM) ANOVA, with Tukey’s post hoc test. Comparisons of two data points were made using a two-sided unpaired t test, with Holm-Sidak correction for multiple comparisons, where indicated. Comparisons of survival curves were made using log-rank (Mantel-Cox) test. A P value of <0.05 was considered significant. All statistical analyses were done using GraphPad Prism software.
Supplementary Material
(A) Real-time qPCR for GADD34, CHOP and ATF4 mRNA levels in lumbar spinal cords from early-symptomatic (14.5-month-old) G37R-29 mice. Data were normalized to actin reference gene and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 4 mice per group. Norm. exp. = normalized expression. (B) Representative immunoblots of p-eIF2α (phosphorylated), eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from 14.5-month-old G37R-29 mice and their age-matched WT controls. Actin was used as a loading control. (C) Quantification of p-eIF2α/eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from 14.5-month-old G37R-29 mice. Data were normalized to actin and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 3–4 mice per group. Norm. dens. = normalized densitometry. Unpaired t test: *P < 0.05 ***P < 0.0001.
Real-time qPCR for PERK (A), CHOP (B), and GADD34 (C) mRNA levels in lumbar spinal cords obtained from PERK, CHOP, and GADD34 mutant mice, respectively. To differentiate between wild-type and mutant transcripts, we used qPCR primer pairs which amplified a cDNA region absent in mutant mice. For instance, qPCR primers to GADD34 amplified a cDNA region that spanned from exon 2 to exon 3 (since exon 3 is deleted in GADD34 mutant mice). As expected, the results demonstrate ~50% mRNA level reduction in heterozygous mice, and no mRNA detected in homozygous mutant mice. Data were normalized to actin reference gene and expressed as mean ± SEM fold change relative to age-matched WT controls. n = 4 mice per group. Norm. exp. = normalized expression.
Highlights.
The UPR is a cytoprotective pathway activated in response to ER stress.
Genetic inhibition of UPR-PERK pathway had no effect on disease in mtSOD1 mice.
Genetic enhancement of UPR-PERK pathway did not alleviate disease in mtSOD1 mice.
Genetic ablation of pro-apoptotic CHOP did not alleviate disease in mtSOD1 mice.
Acknowledgements
We are grateful to Sharon Way for critical reading of the manuscript. We also thank Ani Solanki for technical assistance with the mice.
Funding
This work was supported by grants to BP from NIH/NINDS (R01 NS034939), Target ALS, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and to YD from NIH/NINDS (F32NS089290).
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- mtSOD1
Mutant superoxide dismutase 1
- ER
Endoplasmic reticulum
- UPR
Unfolded protein response
- PERK
Protein kinase RNA-activated (PKR)-like ER kinase
- GADD34
Growth arrest and DNA damage-inducible 34
- CHOP
C/EBP homologous protein
- ATF4
Activating transcription factor 4
- eIF2α
Eukaryotic translation initiation factor 2α
- NMJ
Neuromuscular junction
- SynI
Synapsin I
- AChR
Acetylcholine receptor
- α-BGT
α-Bungarotoxin
- ChAT
Choline acetyltransferase
- WT
Wild-type
- qPCR
Quantitative polymerase chain reaction
- RT
Room temperature
- PFA
Paraformaldehyde
- BSA
Bovine serum albumin
- PBS
Phosphate-buffered saline
- TBS
Tris-buffered saline
- OCT
Optimal cutting temperature compound
- RM-ANOVA
Repeated-measures analysis of variance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
The authors declare no conflict of interest.
Materials and Data Availability
All data are available upon reasonable request to the corresponding author.
References
- 1.Cleveland DW & Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nature reviews. Neuroscience 2(11):806–819. [DOI] [PubMed] [Google Scholar]
- 2.Gordon PH (2013) Amyotrophic Lateral Sclerosis: An update for 2013 Clinical Features, Pathophysiology, Management and Therapeutic Trials. Aging and disease 4(5):295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rezania K & Roos RP (2013) Spinal cord: motor neuron diseases. Neurologic clinics 31(1):219–239. [DOI] [PubMed] [Google Scholar]
- 4.Rothstein JD (2009) Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Annals of neurology 65 Suppl 1:S3–9. [DOI] [PubMed] [Google Scholar]
- 5.Ito Y, et al. (2009) Involvement of CHOP, an ER-stress apoptotic mediator, in both human sporadic ALS and ALS model mice. Neurobiology of disease 36(3):470–476. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi H, et al. (2006) Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proceedings of the National Academy of Sciences of the United States of America 103(15):6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagata T, et al. (2007) Increased ER stress during motor neuron degeneration in a transgenic mouse model of amyotrophic lateral sclerosis. Neurological research 29(8):767–771. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki S (2010) Endoplasmic reticulum stress in motor neurons of the spinal cord in sporadic amyotrophic lateral sclerosis. Journal of neuropathology and experimental neurology 69(4):346–355. [DOI] [PubMed] [Google Scholar]
- 9.Saxena S, Cabuy E, & Caroni P (2009) A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nature neuroscience 12(5):627–636. [DOI] [PubMed] [Google Scholar]
- 10.Walker AK & Atkin JD (2011) Stress signaling from the endoplasmic reticulum: A central player in the pathogenesis of amyotrophic lateral sclerosis. IUBMB life 63(9):754–763. [DOI] [PubMed] [Google Scholar]
- 11.Sun S, et al. (2015) Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. Proceedings of the National Academy of Sciences of the United States of America 112(50):E6993–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boillee S, Vande Velde C, & Cleveland DW (2006) ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron 52(1):39–59. [DOI] [PubMed] [Google Scholar]
- 13.Bruijn LI, et al. (1998) Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281(5384):1851–1854. [DOI] [PubMed] [Google Scholar]
- 14.Ilieva H, Polymenidou M, & Cleveland DW (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. The Journal of cell biology 187(6):761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishitoh H, et al. (2008) ALS-linked mutant SOD1 induces ER stress- and ASK1- dependent motor neuron death by targeting Derlin-1. Genes & development 22(11):1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Shibata Y, Yun C, Ron D, & Rapoport TA (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429(6994):841–847. [DOI] [PubMed] [Google Scholar]
- 17.Hetz C (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews. Molecular cell biology 13(2):89–102. [DOI] [PubMed] [Google Scholar]
- 18.Hetz C, Chevet E, & Harding HP (2013) Targeting the unfolded protein response in disease. Nature reviews. Drug discovery 12(9):703–719. [DOI] [PubMed] [Google Scholar]
- 19.Walter P & Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334(6059):1081–1086. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly N, Gorman AM, Gupta S, & Samali A (2013) The eIF2alpha kinases: their structures and functions. Cellular and molecular life sciences : CMLS 70(19):3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Guo Y, Tang J, Jiang J, & Chen Z (2014) New insights into the roles of CHOP- induced apoptosis in ER stress. Acta biochimica et biophysica Sinica 46(8):629–640. [DOI] [PubMed] [Google Scholar]
- 22.Bommiasamy H & Popko B (2011) Animal models in the study of the unfolded protein response. Methods in enzymology 491:91–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halliday M, Hughes D, & Mallucci GR (2017) Fine-tuning PERK signaling for neuroprotection. Journal of neurochemistry 142(6):812–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang C, Wang Y, Zhang H, & Han F (2017) The role of endoplasmic reticulum stress in neurodegenerative disease. Apoptosis 22(1):1–26. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Popko B, & Roos RP (2011) The unfolded protein response in familial amyotrophic lateral sclerosis. Human molecular genetics 20(5):1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Popko B, & Roos RP (2014) An enhanced integrated stress response ameliorates mutant SOD1-induced ALS. Human molecular genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das I, et al. (2015) Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348(6231):239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang HQ, et al. (2014) Guanabenz delays the onset of disease symptoms, extends lifespan, improves motor performance and attenuates motor neuron loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. Neuroscience 277:132–138. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Popko B, Tixier E, & Roos RP (2014) Guanabenz, which enhances the unfolded protein response, ameliorates mutant SOD1-induced amyotrophic lateral sclerosis. Neurobiology of disease 71:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira FG, et al. (2015) Guanabenz Treatment Accelerates Disease in a Mutant SOD1 Mouse Model of ALS. PloS one 10(8):e0135570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlug AS, et al. (2005) ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. The European journal of neuroscience 22(8):1881–1894. [DOI] [PubMed] [Google Scholar]
- 32.Harding HP, et al. (2001) Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Molecular cell 7(6):1153–1163. [DOI] [PubMed] [Google Scholar]
- 33.Vinsant S, et al. (2013) Characterization of early pathogenesis in the SOD1(G93A) mouse model of ALS: part II, results and discussion. Brain and behavior 3(4):431–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novoa I, Zeng H, Harding HP, & Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. The Journal of cell biology 153(5):1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin W, et al. (2008) Enhanced integrated stress response promotes myelinating oligodendrocyte survival in response to interferon-gamma. The American journal of pathology 173(5):1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novoa I, et al. (2003) Stress-induced gene expression requires programmed recovery from translational repression. The EMBO journal 22(5):1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsaytler P, Harding HP, Ron D, & Bertolotti A (2011) Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332(6025):91–94. [DOI] [PubMed] [Google Scholar]
- 38.Marciniak SJ, et al. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes & development 18(24):3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinszner H, et al. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes & development 12(7):982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel T, et al. (2013) CHOP regulates the p53-MDM2 axis and is required for neuronal survival after seizures. Brain : a journal of neurology 136(Pt 2):577–592. [DOI] [PubMed] [Google Scholar]
- 41.Gow A & Wrabetz L (2009) CHOP and the endoplasmic reticulum stress response in myelinating glia. Current opinion in neurobiology 19(5):505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurney ME, et al. (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264(5166):1772–1775. [DOI] [PubMed] [Google Scholar]
- 43.Bruijn LI, et al. (1997) ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18(2):327–338. [DOI] [PubMed] [Google Scholar]
- 44.Acevedo-Arozena A, et al. (2011) A comprehensive assessment of the SOD1G93A low- copy transgenic mouse, which models human amyotrophic lateral sclerosis. Disease models & mechanisms 4(5):686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philips T & Rothstein JD (2015) Rodent Models of Amyotrophic Lateral Sclerosis. Curr Protoc Pharmacol 69:5 67 61–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong PC, et al. (1995) An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14(6):1105–1116. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, et al. (2009) Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Human molecular genetics 18(9):1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozas P, Bargsted L, Martinez F, Hetz C, & Medinas DB (2017) The ER proteostasis network in ALS: Determining the differential motoneuron vulnerability. Neurosci Lett 636:9–15. [DOI] [PubMed] [Google Scholar]
- 49.Matus S, Glimcher LH, & Hetz C (2011) Protein folding stress in neurodegenerative diseases: a glimpse into the ER. Current opinion in cell biology 23(2):239–252. [DOI] [PubMed] [Google Scholar]
- 50.Harding HP, Zhang Y, Bertolotti A, Zeng H, & Ron D (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular cell 5(5):897–904. [DOI] [PubMed] [Google Scholar]
- 51.Bertolotti A (2018) Importance of the subcellular location of protein deposits in neurodegenerative diseases. Current opinion in neurobiology 51:127–133. [DOI] [PubMed] [Google Scholar]
- 52.Sundaram JR, Lee IC, & Shenolikar S (2017) Translating protein phosphatase research into treatments for neurodegenerative diseases. Biochem Soc Trans 45(1):101–112. [DOI] [PubMed] [Google Scholar]
- 53.Rutkowski DT, et al. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 4(11):e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatterjee J & Kohn M (2013) Targeting the untargetable: recent advances in the selective chemical modulation of protein phosphatase-1 activity. Curr Opin Chem Biol 17(3):361–368. [DOI] [PubMed] [Google Scholar]
- 55.Knight ZA & Shokat KM (2007) Chemical genetics: where genetics and pharmacology meet. Cell 128(3):425–430. [DOI] [PubMed] [Google Scholar]
- 56.Chen CM, Wu CT, Chiang CK, Liao BW, & Liu SH (2012) C/EBP homologous protein (CHOP) deficiency aggravates hippocampal cell apoptosis and impairs memory performance. PloS one 7(7):e40801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, et al. (2017) Neuroprotection by eIF2alpha-CHOP inhibition and XBP-1 activation in EAE/optic neuritiss. Cell death & disease 8(7):e2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva RM, et al. (2005) CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. Journal of neurochemistry 95(4):974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohri SS, et al. (2011) Attenuating the endoplasmic reticulum stress response improves functional recovery after spinal cord injury. Glia 59(10):1489–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alexander GM, et al. (2004) Effect of transgene copy number on survival in the G93A SOD1 transgenic mouse model of ALS. Brain research. Molecular brain research 130(1–2):7–15. [DOI] [PubMed] [Google Scholar]
- 61.Gurney ME (1997) The use of transgenic mouse models of amyotrophic lateral sclerosis in preclinical drug studies. Journal of the neurological sciences 152 Suppl 1:S67–73. [DOI] [PubMed] [Google Scholar]
- 62.Scott S, et al. (2008) Design, power, and interpretation of studies in the standard murine model of ALS. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases 9(1):4–15. [DOI] [PubMed] [Google Scholar]
- 63.Kaja S, et al. (2007) Severely impaired neuromuscular synaptic transmission causes muscle weakness in the Cacna1a-mutant mouse rolling Nagoya. The European journal of neuroscience 25(7):2009–2020. [DOI] [PubMed] [Google Scholar]
- 64.Perez-Garcia MJ & Burden SJ (2012) Increasing MuSK activity delays denervation and improves motor function in ALS mice. Cell reports 2(3):497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy HS, Jones C 3rd, & Caplazi P (2013) Comparison of standard laminectomy with an optimized ejection method for the removal of spinal cords from rats and mice. J Histotechnol 36(3):86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Real-time qPCR for GADD34, CHOP and ATF4 mRNA levels in lumbar spinal cords from early-symptomatic (14.5-month-old) G37R-29 mice. Data were normalized to actin reference gene and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 4 mice per group. Norm. exp. = normalized expression. (B) Representative immunoblots of p-eIF2α (phosphorylated), eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from 14.5-month-old G37R-29 mice and their age-matched WT controls. Actin was used as a loading control. (C) Quantification of p-eIF2α/eIF2α, GADD34, CHOP, and ATF4 protein levels in lumbar spinal cords from 14.5-month-old G37R-29 mice. Data were normalized to actin and expressed as mean ± SEM fold change relative to age-matched WT controls (red dashed line). n = 3–4 mice per group. Norm. dens. = normalized densitometry. Unpaired t test: *P < 0.05 ***P < 0.0001.
Real-time qPCR for PERK (A), CHOP (B), and GADD34 (C) mRNA levels in lumbar spinal cords obtained from PERK, CHOP, and GADD34 mutant mice, respectively. To differentiate between wild-type and mutant transcripts, we used qPCR primer pairs which amplified a cDNA region absent in mutant mice. For instance, qPCR primers to GADD34 amplified a cDNA region that spanned from exon 2 to exon 3 (since exon 3 is deleted in GADD34 mutant mice). As expected, the results demonstrate ~50% mRNA level reduction in heterozygous mice, and no mRNA detected in homozygous mutant mice. Data were normalized to actin reference gene and expressed as mean ± SEM fold change relative to age-matched WT controls. n = 4 mice per group. Norm. exp. = normalized expression.