Summary
YUC flavin monooxygenases catalyze the rate-limiting step of auxin biosynthesis. Here we report the vacuolar targeting and degradation of GFP-YUC1. GFP-YUC1 fusion expressed in Arabidopsis protoplasts or transgenic plants was primarily localized in vacuoles. Surprisingly, we established that GFP-YUC1, a soluble protein, was sorted to vacuoles through the ESCRT pathway, which has long been recognized for sorting and targeting integral membrane proteins. We further show that GFP-YUC1 was ubiquitinated and in this form GFP-YUC1 was targeted for degradation, a process that was also stimulated by elevated auxin levels. Our findings revealed a molecular mechanism of GFP-YUC1 degradation and demonstrate that the ESCRT pathway can recognize both soluble and integral membrane proteins as cargoes.
Auxin is required for many aspects of plant growth and development. In plants, auxin is synthesized from the amino acid tryptophan through the TAA/YUC two-step pathway (Zhao 2018). The YUC family of flavin monooxygenases catalyzes the rate-limiting step of auxin biosynthesis, by converting indole-3-pyruvate (IPA) to indole-3-acetic acid (IAA), the primary natural auxin. It has been shown that the expression level of YUC genes directly impacts local auxin concentrations. Disruption of YUC gene expression results in defects in all major developmental processes, including embryogenesis, root development, floral organ formation, and vascular patterning (Cheng et al. 2006; Chen et al. 2014). An increase in YUC gene expression leads to dramatic auxin overproduction phenotypes, such as elongated hypocotyls and hyponastic cotyledons (Zhao et al. 2001). Previous studies on YUC genes mainly focused on the transcriptional regulation. The post-translational regulation of YUC genes is largely unknown.
Degradation in vacuoles is usually considered as the final fate for many plant proteins. Ubiquitination is generally considered as the initiation signal for protein degradation. Ubiquitinated proteins, such as plasma membrane (PM)-localized receptors and transporters, are internalized into vesicles and are directed to the Trans-Golgi Network (TGN) that is also called the early endosome (EE). Some receptors and transporters are recycled back to the PM, whereas others are sorted to the intraluminal vesicles (ILVs) located inside the pre-vacuolar compartment (PVC) /multivesicular body (MVB), and are targeted to vacuoles for degradation upon fusion between the PVC/MVB and the vacuole.
The sorting of cargoes into ILVs inside PVC/MVB is regulated by the ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT (ESCRT) system (Gao et al. 2017). Membrane-localized auxin efflux carrier PIN2 (Spitzer et al. 2009) and Brassinosteroid receptor BRI1 (Martins et al. 2015) have been reported to be PVC cargo proteins, and are sorted for vacuolar degradation by the ESCRT system. Although the ESCRT pathway has long been considered specifically targeting integral membrane proteins, recent studies have demonstrated that non-integral membrane proteins, such as the Arabidopsis ABA receptor PYR1-LIKE 4 (PYL4), also employ ESCRT machinery components FYVE1 and VPS23A for endosomal sorting (Belda-Palazon et al. 2016; Yu et al. 2016).
Intracellular locations of auxin biosynthesis have not been fully defined. Bioinformatic analysis and transgenic approaches have shown that TAAs are cytosol localized, whereas YUCs are either in the cytosol or ER (Poulet and Kriechbaumer 2017). According to the SignalP-4.1 prediction and TMHMM analysis, YUC1 apparently lacks a transmembrane domain and does not contain a signal peptide, suggesting that YUC1 is likely a cytosolic protein (Figure S1A, B).
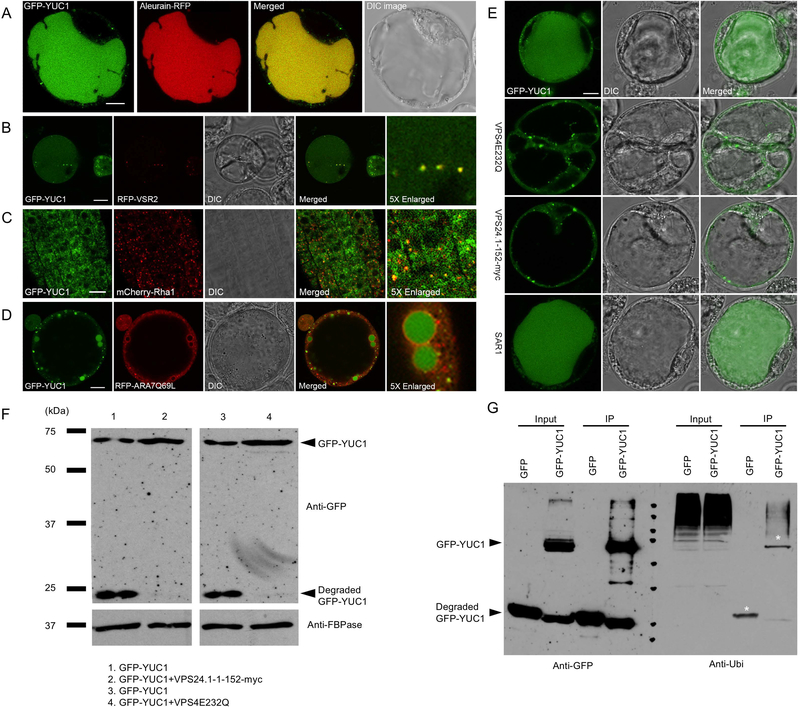
To further investigate YUC1 subcellular localization, we expressed GFP-YUC1 and YUC1-RFP fusion proteins in Arabidopsis protoplasts. Surprisingly, 12 h after transformation, the fluorescence signals were predominantly detected in the vacuole lumen of the majorities of protoplasts, while the cytosolic GFP signal was very weak. Moreover, some discrete fluorescence spots could be detected in the cytosol (Figure 1A, S2A). The GFP-YUC1 was co-localized with the vacuole marker Aleurain-RFP (Figure 1A). These findings suggested that, after translation, the GFP-YUC1 and YUC1-RFP fusions were quickly transported to vacuole for degradation (Figure 1A, S2A).
Figure 1. ESCRT-mediated vacuolar targeting and sorting of YUC1 flavin monooxygenase.
(A) Co-localization of the GFP-YUC1 fusion with the vacuole marker Aleurain-RFP in Arabidopsis protoplasts. Scale bar = 10 μm. (B) Co-localization of the GFP-YUC1 with the PVC marker RFP-VSR2 in Arabidopsis protoplasts. Scale bar = 10 μm. (C) Co-localization of freshly produced GFP-YUC1 with the PVC marker mCherry-Rha1 in Arabidopsis plants. The GFP-YUC1 fusion was placed under the control of an inducible promoter XVE. 7 days old F1 seedlings were treated with 10μM estradiol for 26 hours before confocal microscope imaging. Scale bar = 10 μm. (D) GFP-YUC1 was trapped within the lumen of the enlarged PVCs induced by overexpressing mRFP–ARA7Q69L in Arabidopsis protoplasts. mRFP–ARA7Q69L is the mutant version of ARA7 which localized in PVC and functions in vacuolar trafficking. Scale bar = 10 μm. (E) Overexpression of the dominant negative mutant VPS4E232Q and the VPS24.1–1-152-myc altered the vacuolar targeting and sorting of GFP-YUC1 and enlarged the PVCs in Arabidopsis protoplasts. Both VPS4 and VPS24 are necessary for the ESCRT system regulated cargoes sorting and degradation. Overexpression of the mutant version of VPS4 or VPS24 blocked the vacuolar sorting of GFP-YUC1. Vacuole targeting and sorting of GFP-YUC1 was unaltered by overexpression of the ER-Golgi trafficking mutant, SAR1. Scale bar = 10 μm. (F) Effects of ESCRT dominant mutants VPS4E232Q and VPS24.1–1-152-myc on the degradation of GFP-YUC1. Proteins were extracted from Arabidopsis protoplasts expressing GFP-YUC1, GFP-YUC1+VPS4E232Q, GFP-YUC1+VPS24.1–1-152-myc. The GFP-YUC1 fusion and its degradation products were detected using a GFP antibody. As shown in line 2 and 4, the GFP-YUC1 degradation was greatly decreased compared with line 1 and 3, when VPS4E232Q or VPS24.1–1-152-myc is co-expressed with GFP-YUC1. (G) Immunoprecipitation (IP) assay of GFP-YUC1. Arabidopsis protoplasts expressing GFP or GFP-YUC1 were subjected to protein extraction and IP with GFP-trap. Input protein and IP products were detected using GFP antibody (left) and Ubiquitin antibody (right). Mono-ubiquitination, poly-monoubiquitination, and poly-ubiquitination of GFP-YUC1 were detected.
We also tested the subcellular localization of Arabidopsis TAA1, which provides a substrate for YUC1. Here, both GFP-TAA1 and TAA1-RFP localized to the cytosol, similar to the single GFP localization pattern, with no spot-like signal being detected (Figure S2B, C). To test whether the GFP-YUC1 fusion is functional, we expressed the fusion in Arabidopsis, using the CaMV 35S promoter. These transgenic plants exhibited typical auxin overproduction phenotypes, including elongated hypocotyls, reduced growth of main roots, and hyponastic cotyledons (Figure S2D).
To explore the subcellular localization of the observed punctate GFP-YUC1 signal (Figure 1A), GFP-YUC1 was co-expressed with the punctate organelle markers, Man1-RFP (Golgi) and RFP-VSR2 (PVC), respectively. As shown in Figure 1B and Figure S3A, the GFP-YUC1 punctate signals were co-localized with the PVC marker RFP-VSR2, but not with the Golgi marker Man1-RFP in Arabidopsis protoplasts. We next constructed β-estradiol-inducible XVE::GFP-YUC1 Arabidopsis lines (Figure S3B) that were crossed to the Arabidopsis transgenic organelle marker lines: mCherry-SYP32 (Golgi), mCherry-Rha1 (PVC), VHAa1-mRFP (TGN), and mCherry-VAMP711 (tonoplast). In F1 generation plants, the induced GFP-YUC1 signals were separated from Golgi and TGN markers, but largely overlapped with the PVC marker (Figure 1C, S3C).
In plant cells, Wortmannin treatments have been reported to enlarge PVC and to inhibit the PVC-vacuole trafficking. As shown in Figure S3D, the number of GFP dots in the XVE::GFP-YUC1 root cells was decreased and the sizes were enlarged after an 18 h Wortmannin treatment. To further test the subcellular localization of GFP-YUC1, we co-expressed GFP-YUC1 with the RFP-ARA7Q69L, which is a constitutively-active version of ARA7, a Rab small GTPase that is localized in PVC and functions in vacuolar trafficking.
Expression of the RFP-ARA7Q69L in the early phases (6–8 h) resulted in formation of enlarged ring-like PVC in Arabidopsis protoplasts (Jia et al. 2013). As shown in Figure 1D, the GFP-YUC1 was clearly trapped within the ring-like PVC lumen and no GFP signal was detected in vacuoles. Therefore, our results support the hypothesis that GFP-YUC1 is being sorted to the vacuole for degradation through a PVC/MVB-mediated sorting route.
VPS4/SKD1 is an Arabidopsis AAA ATPase that functions as an ESCRT-III-associated protein that localizes to the endosomal membrane such as PVC. VPS4E232Q is a dominant-negative mutant version, which has lost the ATPase activity of Vps4 (Cai et al. 2014). Vps24 is part of the ESCRT-III core complex. Overexpression of VPS4E232Q or VPS24.1–1-152-myc (dominant-negative mutant of Vps24) (Cai et al. 2014) caused a reduction in ILV numbers, with an enlargement of PVC and disruption of ESCRT-dependent cargo sorting to the vacuole in Arabidopsis protoplasts.
To test whether vacuole targeting of GFP-YUC1 is mediated through the ESCRT complex, VPS4E232Q and VPS24.1–152-myc were co-expressed with GFP-YUC1 in Arabidopsis protoplasts, respectively. As shown in Figure 1E, GFP-YUC1 localization was restricted to the cytosol, and the punctate GFP signals were enlarged when VPS4E232Q or VPS24.1–1-152myc was co-expressed. The effect of VPS24.1–1-152myc on the GFP-YUC1 localization was analyzed 12 and 16 h after transient expression in protoplast.
After 12 h, some 70% of GFP-YUC1 expressing cells displayed strong vacuole-localized GFP signal, and after 16 h, the ratio of vacuole-localized cells increased to almost 80% (Figure S4). However, when VPS24–1-152myc was co-expressed with GFP-YUC1, the ratio of vacuole-localized cells decreased from 69.1% to 1.0%, and cytosol-localized cells increased from 30.96% to 99.0%. After 16 h, the ratio of vacuole-localized cells decreased from 79.3% to 6.9% and the ratio of cytosol-localized cells increased from 20.7% to 93.2% when VPS24.1–152-myc was co-expressed.
Arabidopsis Sar1 GTPase regulates the biogenesis of the COPII vesicle, which mediates ER-to-Golgi transport. Overexpression of the mutant version, SAR1(H74L) blocks the ER export and causes retention of the ER-derived vacuolar proteins, like aleurain, in the ER (Zeng et al. 2015). When GFP-YUC1 and the mutant SAR1(H74L) were co-expressed into Arabidopsis protoplasts, vacuole localization of GFP-YUC1 was not compromised (Figure 1E), indicating that GFP-YUC1 is not synthesized in the ER, consistent with its lack of any transmembrane domain and N-terminal located signal peptide in the YUC1 protein (Figure S1A, B).
To further investigate the effects of the ESCRT system on trafficking and vacuolar degradation of GFP-YUC1, we isolated proteins from protoplasts expressing GFP-YUC1 and GFP-YUC1, with or without ESCRT mutants, and conducted Western blot analysis. As shown in Figure 1F (first and third lanes), the 75-kDa band represented the GFP-YUC1 fusion protein, and the 25-kDa band represented the GFP-core, which is relatively resistant to digestion by lytic enzymes within vacuoles., When the ESCRT mutant was co-expressed, the signal of degraded YUC1-GFP was greatly reduced (Figure 1F, second and fourth lanes). Meanwhile, the full-length GFP-YUC1 fusion protein signal was stronger than those in the control samples. Taken together, these findings indicate that the vacuolar sorting and degradation of GFP-YUC1 is dependent on the ESCRT pathway.
To test the GFP-YUC1 degradation level in plants, proteins were isolated from β-estradiol-treated XVE::GFP-YUC1 plants, at different time points, followed by Western blot analysis. Degraded YUC1-GFP could be discerned 0.5 h after treatment, and the accumulation of degraded product increased, thereafter (Figure S5A). To understand how GFP-YUC1 degradation is regulated, and to test the hypothesis that overproduction of auxin by GFP-YUC1 can serve as a negative feedback regulator to promote YUC1 degradation, we next treated the GFP-YUC1 expressing protoplasts with different concentrations of IAA. As shown in Figure S5B, with higher IAA concentrations, more degraded YUC1-GFP was detected.
We next investigated whether GFP-YUC1 protein was ubiquitinated, in vivo. Proteins were extracted and purified from GFP-YUC1 expressing protoplasts, or GFP expressing lines as a negative control. GFP and GFP-YUC1 were immunoprecipitated (IP) using GFP-trap agarose beads. The input and IP production were followed by Western blot analysis, with GFP and ubiquitin antibodies (P4D1), which recognizes both mono-ubiquitination and poly-ubiquitination. As shown in Figure 1G (right, GFP-YUC1 line), a single 75-kDa band, corresponding to the likely mono-ubiquitinated GFP-YUC1 and a high-molecular-weight smear were detected, indicating that GFP-YUC1 was also poly-ubiquitinated or poly-mono-ubiquitinated, or both. Surprisingly, we also detected the ubiquitinated GFP. Our hypothesis is that the lysine residues in the GFP are responsible for this ubiquitination. However, when GFP alone was expressed in Arabidopsis protoplasts, it was cytosolic- and nuclear-localized (Figure S2D), suggesting that vacuole-targeted trafficking of GFP-YUC1 was caused by the ubiquitination on YUC1 protein, but not GFP.
Our study uncovered vacuole targeting and degradation of GFP-YUC1. The ubiquitinated GFP-YUC1 was sorted to vacuole through the ESCRT pathway. Although YUC1 is a soluble protein, cytosolic YUC1 might be recruited to the endosomal membrane by unidentified interaction partners, perhaps by membrane-localized E3 ligases after stimulation by high auxin levels.
Supplementary Material
ACKNOWLEDGEMENTS
This work is supported by NIH grant R01GM114660 (to Y.Z.), the China Scholarship Council (201606760041 to C.N.G.), and by grants from the National Natural Science Foundation of China (31671467 and 31870171 to C.G.).
REFERENCES
- Belda-Palazon B, Rodriguez L, Fernandez MA, Castillo MC, Anderson EM, Gao C, Gonzalez-Guzman M, Peirats-Llobet M, Zhao Q, De Winne N, Gevaert K, De Jaeger G, Jiang L, Leon J, Mullen RT, Rodriguez PL (2016) FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell 28: 2291–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhuang X, Gao C, Wang X, Jiang L (2014) The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol 165: 1328–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zhuang X, Shen J, Jiang L (2017) Plant ESCRT complexes: Moving beyond endosomal sorting. Trends Plant Sci 22: 986–998 [DOI] [PubMed] [Google Scholar]
- Jia T, Gao C, Cui Y, Wang J, Ding Y, Cai Y, Ueda T, Nakano A, Jiang L (2013) ARA7(Q69L) expression in transgenic Arabidopsis cells induces the formation of enlarged multivesicular bodies. J Exp Bot 64: 2817–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaitre B, Jaillais Y, Chory J, Geldner N, Vert G (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Kriechbaumer V (2017) Bioinformatics analysis of phylogeny and transcription of TAA/YUC auxin biosynthetic genes. Int J Mol Sci 18: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS (2009) The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Lou L, Tian M, Li Q, Ding Y, Cao X, Wu Y, Belda-Palazon B, Rodriguez PL, Yang S, Xie Q (2016) ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol Plant 9: 1570–1582 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong KB, Schekman R, Jiang L (2015) Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 14360–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y (2018) Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.