Abstract
Skeletal regenerative medicine aims to repair or regenerate skeletal tissues using pharmacotherapies, cell-based treatments, and/or surgical interventions. The field is guided by biological principles active during development, wound healing, aging and carcinogenesis. Skeletal development and tissue maintenance in adults represent highly intricate biological processes that require continuous adjustments in the expression of cell type specific genes that generate, remodel and repair the skeletal extracellular matrix. Errors in these processes can facilitate musculoskeletal disease, including cancers, or injury. The fundamental molecular mechanisms by which cell type specific patterns in gene expression are established and retained during successive mitotic divisions require epigenetic control, which we review here. We focus on epigenetic regulatory proteins that control the mammalian epigenome at the level of chromatin with emphasis on proteins that are amenable to drug intervention to mitigate skeletal tissue degeneration (e.g., osteoarthritis and osteoporosis). We highlight recent findings on a number of druggable epigenetic regulators, including DNA methyltransferases (e.g., DNMT1, DNMT3A, DNMT3B) and hydroxylases (e.g., TET1, TET2, TET3), histone methyltransferases (e.g., EZH1, EZH2, DOT1L), as well as histone deacetylases (e.g., HDAC3, HDAC4, HDAC7), and histone acetyl readers (e.g., BRD4) in relation to the development of bone or cartilage regenerative drug therapies. We also review how histone mutations lead to epigenomic catastrophe and cause musculoskeletal tumors. The combined body of molecular and genetic studies focusing on epigenetic regulators indicates that these proteins are critical for normal skeletogenesis and viable candidate drug targets for short-term local pharmacological strategies to mitigate musculoskeletal tissue degeneration.
Keywords: Bone, cartilage, skeletal development, osteoarthritis, osteoporosis, chromatin, oncohistones
Introduction
Musculoskeletal tissue degeneration is a principal reason for surgical and non-surgical orthopedic procedures that aim to promote repair and regeneration, reduce pain, and increase mobility in injured and/or aging patients. Pre-surgical options for orthopedic regenerative medicine include pharmacological or cell-based therapies that emerged from a thorough understanding of the molecular pathways that control inflammation, as well as musculoskeletal development and degeneration. Regenerative processes must ultimately modulate the activity of biochemical and molecular pathways that operate in cells residing within or surrounding the joint (e.g., cartilage, synovium, bone, ligament, tendon, muscle, blood vessels and nerves). Many non-surgical therapies have traditionally relied on cell surface-related pathways or on compounds that pass easily through the cell membrane, but our rapidly expanding knowledge of epigenetic mechanisms that operate within the nucleus now permit consideration of a fundamentally new class of pharmaceutical drug therapies to promote local repair or regeneration of joint tissues.
Epigenetics (beyond genetics) is a field of study that examines molecular events altering chromosomes, gene expression and heritable phenotypes without affecting the DNA sequence. Epigenetic changes are a hallmark of aging and senescent cells (1) and underlie many diseases including cancer. The origins of epigenetic theory are credited to Conrad Waddington who postulated in 1942 that environmental stimuli (including nutrition, injury and inflammation) irreversibly alter cell fate and phenotypes (2). At the time, gene structures were not known and DNA was not yet proven to be the transmitter of heritable information. The use of the term epigenetics has been debated and evolved over the last 75 years as more was learned about nucleic acids (DNA, RNA), chromatin, and genomes. Traditionally, the term referred to chemical modifications of DNA (i.e., CpG methylation) or post-translational modifications (PTMs) of histone proteins that package DNA into chromatin (3–5). Yet, in a broader sense, post-transcriptional processes by which mRNAs levels and protein synthesis are controlled during musculoskeletal development and homeostasis also involve non-chromatin related epigenetic mechanisms, including the modulation of mRNA translation and degradation by microRNAs (miRNAs), as well as long non-coding RNAs (lncRNAs) and other non-coding RNA species (6, 7). MiRNA-based mechanisms are considered for RNA therapeutics, but delivery methods for small nucleic acids and off-target effects remain logistic hurdles for clinical implementation (8). This review focuses on epigenetic mechanisms that control musculoskeletal-related gene expression through effects on chromatin structure, because our current appreciation of chromatin-related epigenetic mechanisms permits realistic adaptation of epigenetic drugs for short-term local applications in orthopedic regenerative medicine (9).
The development, maintenance, repair and functional performance of musculoskeletal tissues require intricate intracellular chemical reactions involving enzymes, transcription factors, microRNAs and epigenetic regulators (3, 4, 10). Classical cell signaling pathways are initiated by the interactions of protein ligands (i.e., growth factors or morphogens) with their cognate receptors at the cell surface. This signal is ultimately transduced via enzymes (i.e., kinases, which are proteins that phosphorylate tyrosine, serine or threonine residues) to transcription factors in the nucleus (Fig 1). In some cases, transcription factors bind to regions of open chromatin and recruit epigenomic factors and other proteins to control gene expression and chromatin, a protein/DNA structure that condenses the virtually infinite linear dimension of DNA within the limited three-dimensional space of the nucleus. In other cases, pioneering transcription factors act as selective scaffolding proteins to recruit additional proteins (i.e., epigenetic regulators) required for the remodeling of chromatin. This transcriptional process by which tissue-specific genes are selectively activated or silenced by chromatin remodeling is established during development and may need to be re-established during regeneration through epigenetic mechanisms. Activation of gene expression during skeletal development or reprogramming requires remodeling of chromatin from highly condensed heterochromatin to less dense euchromatin (Fig 2). The term chromatin was originally used in the context of dyes that bind the essential substance (chromatin) of chromosomes (11). Genetically active good (‘eu’) chromatin stains poorly, while genetically inactive other (‘hetero’) chromatin stains strongly in light microscopy (11). These terms also correlate with dispersed (light) or dense (dark) chromatin in transmission electron microscopy. Euchromatin and heterochromatin are equivalent to the current architectural concepts ‘open’ versus ‘closed’ chromatin, respectively. The conversion of open to closed chromatin, and vice versa, can be achieved by changing DNA methylation, histone PTMs or both (3, 4). Genes that are open and accessible to RNA polymerase II (RNAP II) can be transcribed into mature translatable mRNAs (Fig 1). In the next section the enzymes that chemically modify other proteins post-translationally to open or close chromatin and consequently activate or suppress gene expression in all cells are reviewed. We also discuss particular chromatin modifying enzymes and epigenomic mechanisms that are known to have a role in skeletal development and disease.
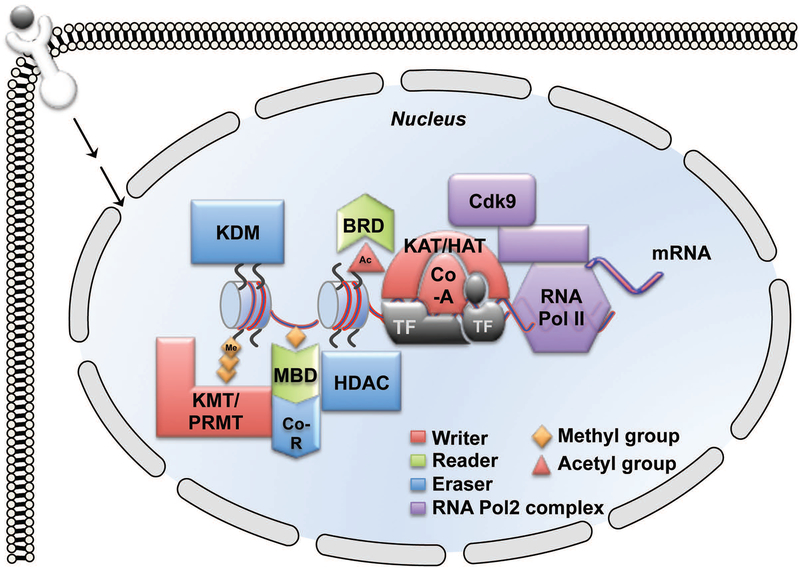
Figure 1: Transcriptional Regulation of Gene Expression.
Gene transcription by RNA polymerase II (RNAP II) is directed by combinatorial binding of transcription factors (TF) to accessible DNA sequences. The TF recruit chromatin-modifying enzymes, including co-activators (Co-A), co-repressors (Co-R) and histone modifying enzymes such as HAT/KAT, HDAC, KMT and KDM. Chemical modifications (e.g., Ac or Me) of histone tails and cytosines recruit reader proteins (e.g., BRD and MBD) that facilitate the opening and closing of the chromatin.
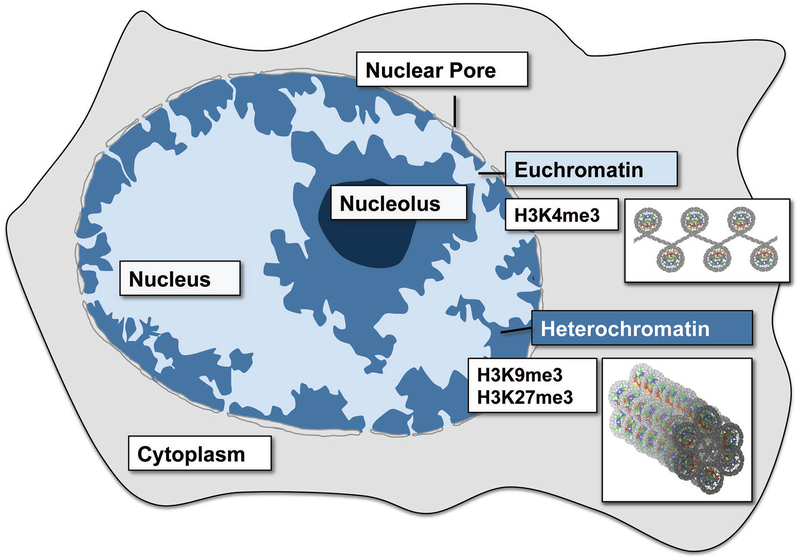
Figure 2: Nuclear organization of chromatin.
Transcriptionally silent and compacted (‘closed’) heterochromatin is marked by H3K9me3 and H3K27me3 modifications and typically resides in the nuclear periphery. In contrast, transcriptionally active and remodeled chromatin (‘open’) typically resides in the center of the nucleus.
The Basics of Epigenetics
DNA in each human diploid cell, which encodes the instructions for development and function of every tissue type, is 3 billion base pairs or 2 meters long. This genomic information is efficiently organized into the narrow confines of the nucleus (~6 to 10 microns in diameter) by histone protein complexes. Histones compact DNA into higher order structures (e.g., chromatin, Fig 3) that permit read-out of only those genes needed for the function of a specialized cell type or tissue at a particular time. For example, resting chondrocytes will express cartilage-related extracellular matrix proteins, while genes specific for bone, fat or muscle cell types are silenced.
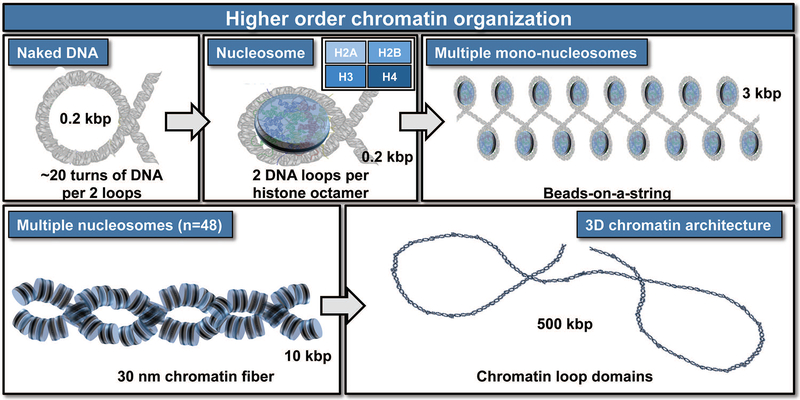
Figure 3: Epigenetics and chromatin organization.
Epigenetic mechanisms control the accessibility of genes embedded in chromatin by altering the spatial organization of nucleosomes into higher structures including bead-on-a-string structures, 30 nm chromatin fiber and higher order chromatin loops. The nucleosome is the basic unit of organization and consists of eight histone proteins (i.e., the histone octamer which contains two each of H2A, H2B, H3, and H4) to lock two full loops of DNA (~166 bp) into place. Together with a short amount of linker DNA that extends towards the next nucleosome and another histone protein type (linker histone H1), each histone octamer organizes about 200 bp of DNA. Multiple nucleosomes together form a “beads on a string” structure that can be further folded into higher order structures in a very dynamic process that permits accessibility of DNA for transcription to proceed.
The chromatin landscape refers not only to the DNA and histone proteins, but also to chemical modifications of both DNA and histones. These chemical changes are the signals for transcription initiation, elongation and termination, but they also have additional roles in processes such as DNA repair and replication. Histones are arranged within the nucleosome such that their N-terminal ‘tails’ extend away from the nucleosome (Fig 4) so epigenetic regulatory enzymes can generate dozens of post-translational modifications (PTMs) on select amino acids, including lysines (K), arginines (R) and serines (S) (12). Serines can be phosphorylated by kinases and dephosphorylated by phosphatases. For example, phosphorylation of serine at position 10 in histone 3 (H3S10) usually precedes gene transcription and is a mark of active transcription during interphase, though it appears more broadly during mitosis. Lysines can be modified in more ways, including by acetylation (Ac), crotonylation, methylation (Me), sumoylation, and mono-ubiquitination (Ub). Of these, the functional significance of lysine acetylation and methylation are currently best understood. Histone acetylation is generally associated with gene activation, but histone methylation can be either an active or repressive mark depending on the exact position of the lysine within the histone protein. For example, H3K9 and H3K27 can be acetylated to form active marks H3K9ac or H3K27ac, or they can be methylated one to three times (me1, me2, me3) to form ultimately the repressive marks H3K9me3 and H3K27me3. Similarly, the H4K20me2/3 mark, which is still less understood, is typically associated with transcriptional repression. However, the H3K4me2/3 and H3K79me3 marks indicate a transcriptionally active or poised locus.
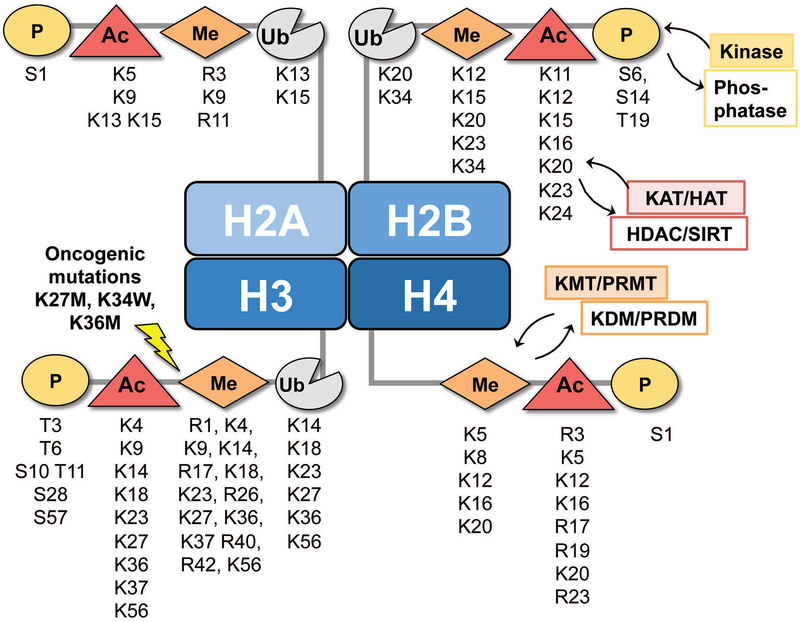
Figure 4: Post-translational modifications in histone proteins.
Post-translational modifications in nucleosomes are distributed over the N-terminal tails of the four core histone proteins H2A, H2B, H3 and H4. Each histone has multiple residues that can be covalently and reversibly modified by phosphorylation (P), acetylation (Ac), methylation (Me) or ubiquitination (Ub) using specific kinases, phosphatases, acetyl transferases (KAT/HATs), deacetylases (HDAC/SIRTs), histone methyl transferases (KMT/PRMTs) or demethylases (KDM/PRDMs). There are more than 300 distinct epigenetic regulators that generate, recognize or remove histone post-translational modifications.
Chemical modification of DNA by methylation, which typically silences a gene, occurs by covalent addition of methyl groups to the 5-carbon of the cytosine ring (5-mC), although methylation at N6 of adenine can also occur in mammals (13). Cytosines are selectively methylated in somatic cells, depending on the cell type and the environment (14). Typically, 5-mC occurs adjacent to a guanine to form a symmetrical dinucleotide (CpG, p representing the phosphate between the C and G) (Fig 5). Clusters of CpGs present near promoters of genes represent CpG islands and methylation of the majority (but not necessarily all) of CpGs in such islands typically silences gene expression.
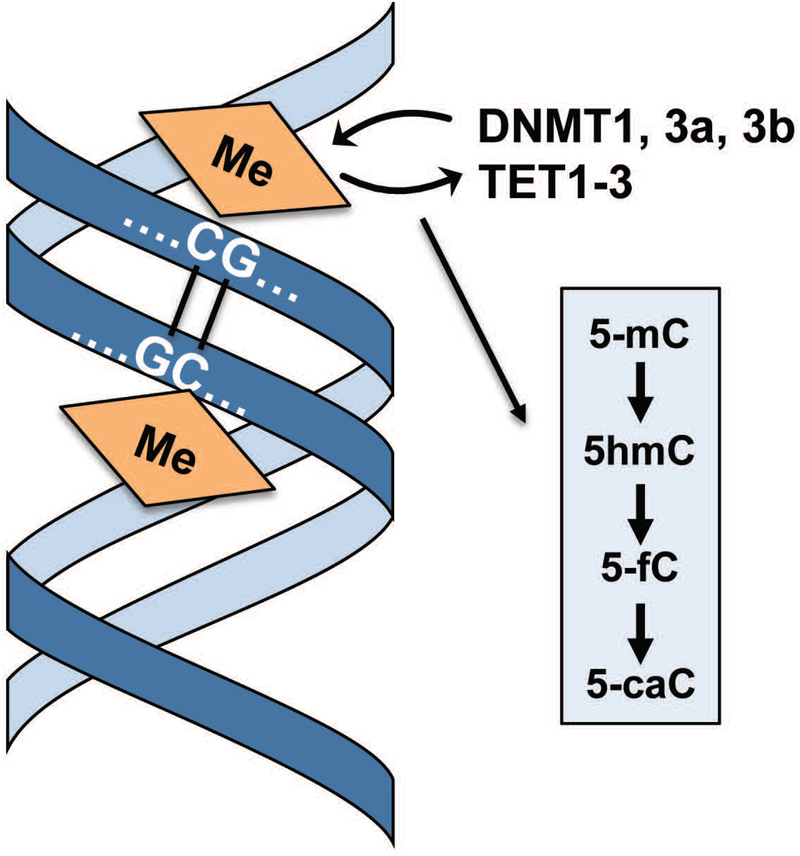
Figure 5: Covalent modifications of DNA.
Reversible methylation of DNA through maternal and patterning imprinting, as well as re-organization of methylation during embryogenesis and post-natal development, represents a canonical mechanism for epigenetic control. Active methylation (5-mC) is established by DNA methyl transferases (DNMTs) and so-called TET proteins that mediate DNA demethylation and generate several stable intermediates that generate a DNA methylome code (i.e., 5-mC, 5-hmC, 5-fC and 5-caC).
Control of skeletal development and repair by CpG methylation
CpG methylation is mediated by three distinct DNA methyl-transferases (DNMT1, DNMT3A and DNMT3B) that each covalently methylate the 5 position of cytosine (5mC) (4, 15). These enzymes support the formation of heterochromatin by DNA methyl binding proteins that recruit erasers of activating histone marks to silence genes. DNMTs actively maintain methylation status during DNA replication, essentially by copying methylation marks on the newly replicated strand from the parent strand. Otherwise, 5mC marks are passively lost if DNA replication proceeds without C methylation. DNMT1 maintains the DNA methylation marks during cell division, while DNMT3A and DNMT3B catalyze de novo DNA methylation during development. Methylated DNA is recognized by methyl-binding proteins (MBDs), which recruit large proteins complexes that include histone deacetylases and methyltransferases to suppress gene expression.
DNMT3A and DNMT3B exert similar functions in stem cells and embryos but are expressed in distinct cell types in later development. Mice lacking DNMT3A are normal at birth, but mice deficient in DNMT3B die during embryogenesis at times (E14.5–16.5) when skeletal elements are developing (16). Xu and colleagues demonstrated that DNMT3B expression in COL2-expressing chondrocytes is essential for proper lengthening and mineralization of both axial and appendicular bones (17). Chondrocyte proliferation and survival were not affected by DNMT3B deletion, but chondrocyte hypertrophy, matrix mineralization, and BMP2/SMAD1 signaling were suppressed. Thus, DNMT3B is required for chondrocyte maturation and endochondral bone formation though direct activities in chondrocytes.
Many developmental programs are reactivated in response to injury. Wang and colleagues showed that DMNT3B is expressed in early stages of fracture repair and declines at later stages of repair (18). DMNT3B expression in chondrocytes is necessary for proper callus formation and coupling to angiogenesis and ossification of the stabilized fracture due to hypomethylation and suppression of CXCL12 and osteopontin. DNMT3B, but not DNMT3A, is expressed in healthy articular cartilage but expression decreases with age and following injury (19). This decline corresponds with altered DNA methylation signatures in osteoarthritis patients. Postnatal deletion of Dnmt3b in articular chondrocytes altered the epigenomic stability of chondrocytes and accelerated the appearance of osteoarthritic pathologies while modifying metabolic processes in chondrocytes (19). These results demonstrate that DNA methylation is important for preserving articular chondrocyte function and cartilage maintenance.
Methyl marks can be removed from cytosines by passive or active mechanisms. Passive demethylation occurs when newly synthesized DNA strands are not methylated during cell division. Active demethylation is a multistep process that is mediated by ten-eleven translocation proteins (TET1, TET2 and TET3) that successively convert 5-mC to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC) by hydroxylation (15) (Fig. 5). Beyond intermediary reaction products during demethylation, the four known C modifications (i.e., 5-mC, 5-hmC, 5-fC, 5-caC) may represent a ‘methyl cytosine code’ where each modification represents distinct epigenetic marks (15). The TET proteins have distinct biological roles in musculoskeletal development. Recent studies have shown that therapeutic modulation of TET activity (e.g., using natural micronutrients like sulforaphane) represents a viable strategy for bone regeneration to mitigate bone loss (20, 21). Other studies have shown that TET enzymes are important for cartilage development and homeostasis, and that alterations in the activity of distinct TET isoforms can predispose to osteoarthritis (22–24). Our understanding of how the enzymatic activities and gene targets of distinct TET enzymes support skeletogenesis has therapeutic implications for clinical joint disorders, because drugs that modulate the activities of these enzymes may have utility in pharmacotherapy.
The phenotypes of mice with null mutations in TET1, TET2 and/or TET3 revealed that any of these proteins can functionally compensate for the loss of another during skeletogenesis (25–27); however, at least one TET protein is required for formation of live animals with a normal skeletal body plan. While single knock-outs do not result in overt skeletal defects, TET1 and TET2 double-knockout mice have developmental defects during mid-gestation (26). Interestingly, a small fraction of TET1/TET2 double knock-out mice can survive spuriously and reproduce, even though 5hmC and 5mC status is altered at imprinted loci (26), indicating that TET3 alone can support skeletal development. Triple-knockout of TET1, TET2 and TET3 depletes 5hmC in embryonic stem cells and prevents embryonic development (27), demonstrating that at least one TET protein is required for normal fetal development. Roles for TET proteins in bone formation are anticipated based on the importance of 5-hmC during osteoblast differentiation (20, 21) and knock-down studies revealing that the three TET proteins have distinct functional roles. TET1 and TET2 are required, while TET3 may attenuate osteoblast differentiation at specific stages of differentiation.
Studies by the Bhutani laboratory examined the relationship between 5-hmC and TET1 during both normal cartilage development (24) and post-natal cartilage degeneration observed in osteoarthritis (22, 23). TET1 is necessary for in vitro differentiation and its levels functionally correlate with 5-hmC levels in the regulatory regions of cartilage-related genes during chondrogenesis (24). Levels of 5hmC also increase in patients with end-stage osteoarthritis (21). Subsequent genome-wide sequencing of 5-hmC-enriched DNA revealed a very large number (>70,000) of regions with differences in hydroxymethylation in OA chondrocytes (e.g., MMP3, LRP5, GDF5, and COL11A1) and that presence of 5hmC correlates with activated, but not repressed, genes as expected (22). The relative importance of TET1, 2 and 3 in contributing to these 5hmC changes in OA remains to be determined.
Taken together, the studies discussed here clearly indicate that TET proteins are necessary for active regulation of the DNA methylome, have important functional roles during skeletal development, as well as each have both redundant and unique biological activities.
Control of skeletal development and repair by Histone Writers, Erasers and Readers
Understanding the mechanisms of how cells place (write), remove (erase) and interpret (read) the PTMs that are placed histones and DNA in response to environmental stimuli is important for the development and application of epigenetic therapies. Histone post-translational modifications are produced and erased by enzymes that exist in large, multi-protein complexes and are recruited to DNA sequences and genomic regions by transcription factors (Fig 1 and Fig 4). Thus, cell- or tissue-specific gene expression is dictated by the combinations of the transcription factors that bind to a gene regulatory regions and their ability to form a platform for activator and repressor complexes. Acetyl groups are transferred from acetyl co-A to free amino-groups of lysine residues in histones as well as non-histone proteins (including transcription factors) by histone/lysine acetyltransferases (HAT/KAT). There are approximately 22 HATs/KATs in humans and they are classified by function and cellular location (28). In human cells, histone de-acetylation is catalyzed by 18 histone lysine deacetylases (HDAC/KDAC). The acetylated lysine is a docking site for proteins containing bromodomains or YEATS domains. Bromodomain-containing proteins (BRD1 to BRD9) recruit RNA polymerase II complexes, which are required for transcription of most eukaryotic genes. As discussed below, many HATs, HDACs and BRDs contribute to bone formation and osteoarthritis.
Histone Acetylation in Bone and Cartilage:
It is well established that KAT/HATs, particularly p300 and CBP, are active in chondrocytes and regulate the expression of genes that control OA pathogenesis by acetylating histones or other factors (29–32). However, overall HAT activity is not drastically different in OA synovial tissues as compared to normal tissue (33). In contrast, HDAC activity is reduced in diseased tissues as compared to normal tissues, indicating an important function role of HDACs in OA pathogenesis. As reviewed previously (34, 35), inhibition of individual HDACs (e.g, HDAC1, HDAC3, HDAC7) by loss-of-function mutations causes delays in skeletal development and loss of bone mass. Specifically, HDAC/SIRT suppression causes abnormalities in physiological development such as craniofacial dysmorphisms, short stature, and bone frailty that are associated with several human diseases or conditions (34, 35). In contrast, activation of SIRTs may protect the skeleton from aging and immobilization-related bone loss. Of interest is the growing amount of evidence indicating that HDAC-induced alterations in gene expression may contribute to OA pathogenesis (36). Even though genetic (permanent) or chemical inhibition (reversible) of HDACs during early skeletogenesis has detrimental effects on the endochondral skeleton, HDAC inhibitors may protect permanent and adult articular cartilage from aging related OA by decreasing production of catabolic cartilage matrix genes in the diseased joint. Activation of other HDACs, particularly SIRT1, with roles in homeostasis could also have chondroprotective benefits in OA (36). While overall HDAC activity is reduced in synovial tissues of OA patients (33), tissues may respond to the disease by reducing HDAC activity to support cartilage tissue repair.
Acetylated lysines in histones H3 and H4 are recognized by “Bromodomain (BRD) and Extra-Terminal Domain” (BET) proteins. BRD proteins associate directly with CDK9, which phosphorylates RNAP II to promote transcription (Fig 1). Among these proteins, BRD4 appears to regulate bone homeostasis by direct effects on differentiation of both osteoclasts and osteoblasts (37). However, treatment of estrogen-deficient mice with a BRD4 inhibitor (JQ1) show a net positive effect on bone accrual, suggesting that BET proteins like BRD4 may represent are novel drug targets for bone degenerative disorders (e.g., osteoporosis). Mechanistically, BRD4 supports bone formation by interacting with osteoblast-lineage specific promoters and enhancers to control bone-related gene expression (38). Recent studies indicate that BRD4 also has a role in cartilage homeostasis and degeneration (39). BRD4 levels positively correlate with disease-state in articular cartilage of patients with osteoarthritis and activation of the inflammation related NF-κB signaling pathway. The BRD4 antagonist JQ1 reduces inflammatory and catabolic pathways in chondrocytes, and attenuates cartilage destruction in mice with destabilized joints (i.e., due to anterior cruciate ligament transection). Similar results were with the CDK9 inhibitor flavopiridol (40). Hence, local administration of inhibitors that target the BRD4/CDK9 complex in the joint may provide a pharmacological avenue to slow expression of early response and inflammatory genes in chondrocytes and mitigate progression of osteoarthritis.
Histone Methylation by DOT1L and EZH2 in Bone and Cartilage:
Like acetylation, the methylation of lysines and arginines serves as a mark that recruits specific regulatory proteins to either promote or inhibit transcription. Up to three methyl groups can be added to amino groups of lysine or arginine residues in H3 and H4 histones by lysine methyltransferases (KMTs) (28) or protein arginine methyltransferases (PRMTs). There are at least 28 KMTs, several of which have been studied in cartilage, including DOT1L (KMT4), EZH2 (KMT6), and SET2 (KMT3A), and nine PRMTs. Lysine methyl marks are removed from histones by at least 18 lysine demethylases (KDMs) that include JMJD (KDM2A-4D, KDM6B), JARID (KDM5A-D), UTX (KDM6A) and UTY (KDM6C) proteins (28), but only one arginine demethylase has been discover thus far (JMJD6) and it has dual lysine/arginine demethylase activity.
DOT1L (Disruptor of telomeric silencing 1-like) is a histone methyl transferase that adds methyl groups to histone 3 at lysine 79 (H3K79) and is essential for cartilage health and development. DOT1L was identified in a genome-wide association study (GWAS) as a gene that may contribute to height (41) and hip OA (42–44). DOT1L is expressed in growth plate cartilage, with particularly high levels found in prehypertrophic and hypertrophic chondrocytes. Cartilage specific deletion of DOT1L in mice disrupted the proliferative and prehypertrophic areas of the growth plate and retarded growth (45). DOT1L was also detected in articular cartilage from normal and OA tissues. However, H3K79 methylation was reduced in damaged areas of OA joints (42, 45). These results suggest that DOT1L activity is important for maintaining cartilage health. Reducing DOT1L expression in chondrogenic cell lines suppressed COL2a, COL10a1 and aggrecan gene expression, but increased osteogenic genes COL1a1 and osteocalcin (42). A DOT1L inhibitor, EPZ-5676, produced similar changes in gene expression and triggered OA in vivo (45). DOT1L negatively regulates TCF-1, a transcription factor that mediates canonical Wnt signaling, and suppression of DOTL1L in chondrocytes may increase canonical Wnt (42). Accordingly DOT1L suppression reduced H3K79 methylation while increasing H3K9 acetylation and H3K4 trimethylation (45). DOT1L interacts with and suppresses a class III HDAC, SIRT1, to increase binding of SIRT1-controlled co-activators to Wnt target genes. Thus, DOT1L is essential for maintaining chondrocyte maturation and cartilage health. DOT1L inhibitors are currently in trials for certain leukemias, but it appears that care should include monitoring of bone growth and joint health in patients receiving drugs that inhibit DOT1L activity.
The Polycomb repressive complex 2 (PRC2) with its catalytic subunit ‘enhancer of zeste homolog 2’ (EZH2) mediates H3K27 trimethylation (H3K27me3), regulates chromatin compaction, and represses gene expression during early stages of embryogenesis (46–48). Complete knockout of EZH2 in mice produced non-viable embryos with abnormal accumulation of mesodermal cells (48). EZH2 also has key roles in skeletal development as conditional loss of EZH2, affects skeletal patterning (49, 50), as well as skeletal growth (51–53) and bone formation (54–56). For example, conditional loss of EZH2 in neural crest–derived tissues (using the Wnt1-Cre driver) causes craniofacial defects (49). Loss of EZH2 in mouse mesenchymal stem cells (using the Prrx1-Cre driver) results in a neo-natal phenotype of shorter limbs and vertebrae due to premature maturation of the physis, as well as accelerated closure of cranial sutures reflecting effects on intra-membranous bone formation (51, 56). The phenotype of these mice is reminiscent of hypomorphic mutations in EZH2 that cause Weaver syndrome (57). These mice also have distinct skeletal phenotypes during post-natal endochondral bone formation (51, 56). Loss of EZH2 in osteoblasts (using the SP7/Osx1-Cre driver) causes a transient low bone mass phenotype that recedes by the time mice reach skeletal maturity (54). Studies on the EZH2 null mutation in chondrocytes (using the Col2a1-Cre driver) revealed that EZH2 deficiency reduces H3K27me3 levels, yet remarkably had minimal impact on skeletal development or maturation of articular cartilage (58). Nevertheless, EZH2 target gene analysis in either osteoblasts or chondrocytes revealed that EZH2 enhances expression of osteogenic markers in chondrocytes, including BMP2 and BMP2 responsive transcription factors SP7 and extracellular matrix proteins (e.g., IBSP and BGLAP)(58), as well as a large number other targets that have been identified in MSCs(59). However, EZH1 partially compensates for loss of EZH2 during skeletal development. Dual inactivation of EZH1 and EZH2 in chondrocytes delays skeletal growth (52). The most dramatic skeletal phenotypic changes were observed in mice with a conditional null mutation in EED, a scaffolding protein that is required for PRC2 complexes that contain either EZH1 or EZH2 as active enzymatic subunits the sites (53). Loss of EED caused kyphosis, reduced body size and increased chondrocytic hypertrophy (53). From a biochemical perspective, EZH1 and EZH2 are interchangeable catalytic subunits of the PRC2 complex. Yet, EZH1 is more potent than EZH2 in compacting chromatin independent of histone methylation, even though EZH1 regulates only a subset of EZH2 target genes (46). EZH1 appears to be expressed at different times of skeletal development. EZH2 is most abundant in proliferating uncommitted mesenchymal stem cells, while EZH1 is expressed most prominently in post-proliferative lineage-committed cells (52–54). Hence, EZH1 and EZH2 are expressed in different spatio-temporal patterns and may have distinct mechanistic activities during mesenchymal differentiation and skeletal development.
Possible uses and limitations of epigenetic drug therapies for musculoskeletal disorders.
From a therapeutic perspective, EZH2 inhibition using pharmacological drugs (e.g., GSK126, UNC1999, EPZ005687) has been considered for delaying progression of osteoarthritis (58, 60, 61), or to promote bone anabolic effects (55). EZH2 inhibition mitigates bone loss in a mouse model for estrogen deficiency (55), but also inhibits osteoclastogenesis (62). Thus, EZH2 inhibitors may have bone anabolic activity and anti-bone resorptive activity. Furthermore, enhanced osteogenic differentiation of human MSCs using EZH2 inhibitors (51, 61, 63) or molecular pathways drugs that indirectly impinge on EZH2 (64, 65) may have practical implications for bone tissue engineering.
One limitation of possible epigenetic pharmacotherapies is that epigenetic regulators may act broadly in multiple cell types. Studies with epigenetic inhibitors in cancer patients have revealed a remarkable short-term tolerance for these drugs in patients, but instead there is concern for unpredictable long-term side effects of systemic use (66). Epigenetic inhibitors used for cancer treatment typically target proliferating cells, but skeletal cells are normally quiescent. Their biological effects can be sustained beyond the initial dosing, but are not necessarily irreversible. Hence, these drugs could specifically be considered for short term and/or localized pharmacological treatments, cell therapies, or combination therapies where epigenetic drugs may prime a more targeted biological mechanisms. For example, anti-steroidal therapies appear to be more effective in the presence of an epigenetic drug (66) and delivery of epigenetic drugs can be restricted to the microenvironment of the joint or fracture callus to promote tissue healing. These considerations are encouraging and warrant further investigation of the specific role of epigenetic regulators and their inhibitors in skeletal development, degeneration and regeneration.
Epigenetics and Orthopedic Oncology
Histone mutations and orthopedic cancers:
Genetic mutations in genes (H3F3A and H3F3B) that encode variants of histone 3 (H3.3) are found at such high frequency in chondroblastomas and giant cell tumors of bone (GCT) that they are now molecular markers of these cancers (67). In the seminal 2013 report describing histone mutations in musculoskeletal tumors, point mutations in H3F3B that change the lysine at position 36 to methionine (K36M) were found in 95% of chondroblastomas and mutations in H3F3A that change glycine 34 to tryptophan (G34W) or leucine (G34L) were detected in 92% of GCTs (68). Introduction of the K36M mutation into mesenchymal cells was sufficient to generate an undifferentiated sarcoma in vivo in a subcutaneous injection model (69). Human chondrocytes engineered to express H3.3K36M formed more colonies in vivo, were resistant to apoptotic stimuli, failed to respond to Bmp2, and expressed more cancer-associated genes (70). Exogenous expression of a mutant H3.3 K36M protein into cells caused a global reduction in di- and tri- histone methylation (71, 72). The mutated H3.3K36M protein drastically altered the epigenome by sequestering and inhibiting the activity of H3K36 histone methyltransferases (e.g., MMSET, NSD2, SETD2) (69, 70, 73). The consequent reduction in global H3K36 methylation resulted a gain in H3K27 methylation and a redistribution of polycomb repressive complexes, leading to many changes in gene expression that blocked chondrocytes from terminal differentiation in vitro and promoted self-renewal (69).
Recurrent mutations in histone genes that alter H3 and nucleosome structure have now been found in several cancers besides chondroblastomas (i.e., H3.3 K27M in gliomas (71, 72) and H3.3 K34W in GCT (68)). There may also be a low frequency of histone mutations and “oncohistones” (74) in other tumors. For example, H3.3 mutations were detected in a small fraction of osteosarcomas (75) and K36M/I mutations in H3.1 were found in a pediatric undifferentiated mesenchymal sarcoma (69). All mutations are believed to cause analogous havoc on the chromatin landscape as K36M does in chondrogenic cells. However, there appears to be tissue specificity with the mutations that is not understood yet but may be related to histone writers, readers or erasers that are expressed at discrete stages of lineage differentiation.
DNA methylation and orthopedic cancers:
Alterations in DNA methylation patterns and hypermethylation of CpG islands in genes encoding tumor suppressors are common alterations in cancer genomes. However, global hypomethylation of other DNA regions likely also contributes to tumor progression by destabilizing the genome and promoting oncogene expression. This paradigm was recently demonstrated elegantly in Ewing sarcomas carrying the chromosomal translocation that produces the onco-protein EWS-FLI (76). Enhancers driving expression of the EWS-FLI1 fusion protein were consistently hypomethylated but a characteristic pattern of CpG island hypermethylation also emerged. Significant variability in global DNA methylation patterns was observed between and within Ewing tumors consistent with a spectrum of disease phenotypes.
Chondrosarcomas may also be driven by epigenomic changes. More that half of chondrosarcomas and the majority of benign enchondromas harbor gain-of-function mutations in isocitrate dehydrogenase (IDH)1/2 (77, 78). During the TCA cycle, IDH1/2 convert alpha-ketoglutarate to D-2-hydroxyglutarate, an oncometabolite that inhibits TET enzymes and histone and DNA demethylases and thus indirectly alters the epigenome of chondrosarcomas (79). DNA and histone methylation changes are observed in chondrosarcomas but do not correlate on a broad scale with IDH1/2 mutations (80). These examples highlight the importance of studying epigenomic aspects of musculoskeletal cancers, even those that are characterized by recurrent genetic mutations, because they could predict responders and non-responders to therapies.
Summary and Future Directions
This review focused on epigenetic mechanisms and enzymes that control skeletal development, cancer, and aged or injury-related tissue degeneration to clarify how these proteins are part of a new frontier of druggable targets that can be considered for musculoskeletal regenerative therapies. Among the targets currently under consideration for strategies related to either osteoarthritis or osteoporosis are DNA methyl transferases (e.g., DNMT3A, DNMT3B) and DNA methyl hydroxylases (e.g., TET1, TET2, TET3), histone deacetylases (e.g., HDAC3, HDAC4, HDAC7) and histone acetyl readers (e.g., BRD4), as well as a number of histone methyl transferases (e.g., DOTL1, EZH1, EZH2). However, beyond this dozen of epigenetic regulators, there are more than 300 such proteins and hence the number of currently characterized epigenetic proteins in bone and cartilage tissues is only a fraction of the realm of potential targets. At present, there is a paucity of knowledge on epigenetic mechanisms in other tissues (e.g., tendon, ligament and synovium) relevant to orthopedics and musculoskeletal regenerative medicine. Furthermore, in-depth knowledge is lacking for how epigenetic mechanisms in mesenchymal cells can be leveraged for stem cell therapies or skeletal and connective tissue engineering approaches, especially at the single cell level. For example, when and how epigenetic interventions can be safely and effectively delivered in vitro or in vivo must be determined. It is anticipated that in the years ahead the number of epigenetic studies in musculoskeletal biology will rapidly expand with a concomitant increase in knowledge of epigenomes of tissues as well as in single cells within tissues.
Acknowledgments:
This work was supported by grants from the National Institutes of Health (R01 AR68103 to JJW and AR049069 to AvW) and Regenerative Medicine Minnesota (JJW).
Footnotes
Disclosures: JJW is a member of the Board of Directors of the Orthopedic Research Society, receives royalties from Springer Nature publishing and is a consultant for Terumo BCT Inc. AJvW is a member of the Executive Board of the Bone Marrow Adiposity Society.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41(1):10–3. [DOI] [PubMed] [Google Scholar]
- 3.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–54. [DOI] [PubMed] [Google Scholar]
- 5.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8(4):212–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18(2):109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. [DOI] [PubMed] [Google Scholar]
- 9.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V The functions of animal microRNAs. Nature. 2004;431(7006):350–5. [DOI] [PubMed] [Google Scholar]
- 11.Passarge E Emil Heitz and the concept of heterochromatin: longitudinal chromosome differentiation was recognized fifty years ago. Am J Hum Genet. 1979;31(2):106–15. [PMC free article] [PubMed] [Google Scholar]
- 12.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18(9):517–34. [DOI] [PubMed] [Google Scholar]
- 16.Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133(6):1183–92. [DOI] [PubMed] [Google Scholar]
- 17.Xu T, Wang C, Shen J, Tong P, O’Keefe R. Ablation of Dnmt3b in chondrocytes suppresses cell maturation during embryonic development. J Cell Biochem. 2018;119(7):5852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Abu-Amer Y, O’Keefe RJ, Shen J. Loss of Dnmt3b in Chondrocytes Leads to Delayed Endochondral Ossification and Fracture Repair. J Bone Miner Res. 2018;33(2):283–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Wang C, Li D, Xu T, Myers J, Ashton JM, et al. DNA methyltransferase 3b regulates articular cartilage homeostasis by altering metabolism. JCI Insight. 2017;2(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaler R, Spitzer S, Karlic H, Klaushofer K, Varga F. DMSO is a strong inducer of DNA hydroxymethylation in pre-osteoblastic MC3T3-E1 cells. Epigenetics. 2012;7(6):635–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaler R, Maurizi A, Roschger P, Sturmlechner I, Khani F, Spitzer S, et al. Anabolic and Antiresorptive Modulation of Bone Homeostasis by the Epigenetic Modulator Sulforaphane, a Naturally Occurring Isothiocyanate. J Biol Chem. 2016;291(13):6754–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SE, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A global increase in 5-hydroxymethylcytosine levels marks osteoarthritic chondrocytes. Arthritis Rheumatol. 2014;66(1):90–100. [DOI] [PubMed] [Google Scholar]
- 23.Taylor SE, Li YH, Wong WH, Bhutani N. Genome-wide mapping of DNA hydroxymethylation in osteoarthritic chondrocytes. Arthritis Rheumatol. 2015;67(8):2129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor SE, Li YH, Smeriglio P, Rath M, Wong WH, Bhutani N. Stable 5-Hydroxymethylcytosine (5hmC) Acquisition Marks Gene Activation During Chondrogenic Differentiation. J Bone Miner Res. 2016;31(3):524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29(1):102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–6. [DOI] [PubMed] [Google Scholar]
- 29.Iioka T, Furukawa K, Yamaguchi A, Shindo H, Yamashita S, Tsukazaki T. P300/CBP acts as a coactivator to cartilage homeoprotein-1 (Cart1), paired-like homeoprotein, through acetylation of the conserved lysine residue adjacent to the homeodomain. J Bone Miner Res. 2003;18(8):1419–29. [DOI] [PubMed] [Google Scholar]
- 30.Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, et al. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280(42):35203–8. [DOI] [PubMed] [Google Scholar]
- 31.Aoyama T, Okamoto T, Fukiage K, Otsuka S, Furu M, Ito K, et al. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J Biol Chem. 2010;285(39):29842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otero M, Peng H, Hachem KE, Culley KL, Wondimu EB, Quinn J, et al. ELF3 modulates type II collagen gene (COL2A1) transcription in chondrocytes by inhibiting SOX9-CBP/p300-driven histone acetyltransferase activity. Connect Tissue Res. 2017;58(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber LC, Brock M, Hemmatazad H, Giger OT, Moritz F, Trenkmann M, et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum. 2007;56(4):1087–93. [DOI] [PubMed] [Google Scholar]
- 34.Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME, Westendorf JJ. Histone Deacetylases in Bone Development and Skeletal Disorders. Physiol Rev. 2015;95(4):1359–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in skeletal development and bone mass maintenance. Gene. 2011;474(1–2):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpio LR, Westendorf JJ. Histone Deacetylases in Cartilage Homeostasis and Osteoarthritis. Curr Rheumatol Rep. 2016;18(8):52. [DOI] [PubMed] [Google Scholar]
- 37.Baud’huin M, Lamoureux F, Jacques C, Rodriguez Calleja L, Quillard T, Charrier C, et al. Inhibition of BET proteins and epigenetic signaling as a potential treatment for osteoporosis. Bone. 2017;94:10–21. [DOI] [PubMed] [Google Scholar]
- 38.Najafova Z, Tirado-Magallanes R, Subramaniam M, Hossan T, Schmidt G, Nagarajan S, et al. BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic Acids Res. 2017;45(1):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, Zhu L, Zhang T, Lu H, Wang C, Xue B, et al. BRD4 has dual effects on the HMGB1 and NF-kappaB signalling pathways and is a potential therapeutic target for osteoarthritis. Biochim Biophys Acta Mol Basis Dis. 2017;1863(12):3001–15. [DOI] [PubMed] [Google Scholar]
- 40.Hu Z, Yik JH, Cissell DD, Michelier PV, Athanasiou KA, Haudenschild DR. Inhibition of CDK9 prevents mechanical injury-induced inflammation, apoptosis and matrix degradation in cartilage explants. Eur Cell Mater. 2016;30:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FM, Doherty SA, Hart DJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci U S A. 2012;109(21):8218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castano-Betancourt MC, Evans DS, Ramos YF, Boer CG, Metrustry S, Liu Y, et al. Novel Genetic Variants for Cartilage Thickness and Hip Osteoarthritis. PLoS Genet. 2016;12(10):e1006260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evangelou E, Valdes AM, Castano-Betancourt MC, Doherty M, Doherty S, Esko T, et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann Rheum Dis. 2013;72(7):1264–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monteagudo S, Cornelis FMF, Aznar-Lopez C, Yibmantasiri P, Guns LA, Carmeliet P, et al. DOT1L safeguards cartilage homeostasis and protects against osteoarthritis. Nat Commun. 2017;8:15889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. [DOI] [PubMed] [Google Scholar]
- 48.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarz D, Varum S, Zemke M, Scholer A, Baggiolini A, Draganova K, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141(4):867–77. [DOI] [PubMed] [Google Scholar]
- 50.Wyngaarden LA, Delgado-Olguin P, Su IH, Bruneau BG, Hopyan S. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development. 2011;138(17):3759–67. [DOI] [PubMed] [Google Scholar]
- 51.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, et al. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J Biol Chem. 2015;290(46):27604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lui JC, Garrison P, Nguyen Q, Ad M, Keembiyehetty C, Chen W, et al. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat Commun. 2016;7:13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirzamohammadi F, Papaioannou G, Inloes JB, Rankin EB, Xie H, Schipani E, et al. Polycomb repressive complex 2 regulates skeletal growth by suppressing Wnt and TGF-beta signalling. Nat Commun. 2016;7:12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudakovic A, Camilleri ET, Paradise CR, Samsonraj RM, Gluscevic M, Paggi CA, et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J Biol Chem. 2018;293(33):12894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O’Toole TM, et al. Enhancer of Zeste Homolog 2 Inhibition Stimulates Bone Formation and Mitigates Bone Loss Caused by Ovariectomy in Skeletally Mature Mice. J Biol Chem. 2016;291(47):24594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemming S, Cakouros D, Codrington J, Vandyke K, Arthur A, Zannettino A, et al. EZH2 deletion in early mesenchyme compromises postnatal bone microarchitecture and structural integrity and accelerates remodeling. FASEB J. 2017;31(3):1011–27. [DOI] [PubMed] [Google Scholar]
- 57.Cohen AS, Yap DB, Lewis ME, Chijiwa C, Ramos-Arroyo MA, Tkachenko N, et al. Weaver Syndrome-Associated EZH2 Protein Variants Show Impaired Histone Methyltransferase Function In Vitro. Hum Mutat. 2016;37(3):301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camilleri ET, Dudakovic A, Riester SM, Galeano-Garces C, Paradise CR, Bradley EW, et al. Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development. J Biol Chem. 2018;293(49):19001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemming S, Cakouros D, Vandyke K, Davis MJ, Zannettino AC, Gronthos S. Identification of Novel EZH2 Targets Regulating Osteogenic Differentiation in Mesenchymal Stem Cells. Stem Cells Dev. 2016;25(12):909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Wu Y, Wu Y, Wang Y, Sun L, Li F. The inhibition of EZH2 ameliorates osteoarthritis development through the Wnt/beta-catenin pathway. Sci Rep. 2016;6:29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jing H, Liao L, An Y, Su X, Liu S, Shuai Y, et al. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Mol Ther. 2016;24(2):217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang C, Qiao Y, Mun SH, Lee MJ, Murata K, Bae S, et al. Cutting Edge: EZH2 Promotes Osteoclastogenesis by Epigenetic Silencing of the Negative Regulator IRF8. J Immunol. 2016;196(11):4452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32(3):802–15. [DOI] [PubMed] [Google Scholar]
- 64.Samsonraj RM, Dudakovic A, Manzar B, Sen B, Dietz AB, Cool SM, et al. Osteogenic Stimulation of Human Adipose-Derived Mesenchymal Stem Cells Using a Fungal Metabolite That Suppresses the Polycomb Group Protein EZH2. Stem Cells Transl Med. 2018;7(2):197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen Y, et al. MiR-101 Targets the EZH2/Wnt/beta-Catenin the Pathway to Promote the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. Sci Rep. 2016;6:36988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25(3):403–18. [DOI] [PubMed] [Google Scholar]
- 67.Cleven AH, Hocker S, Briaire-de Bruijn I, Szuhai K, Cleton-Jansen AM, Bovee JV. Mutation Analysis of H3F3A and H3F3B as a Diagnostic Tool for Giant Cell Tumor of Bone and Chondroblastoma. Am J Surg Pathol. 2015;39(11):1576–83. [DOI] [PubMed] [Google Scholar]
- 68.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45(12):1479–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352(6287):844–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science. 2016;352(6291):1344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan KM, Han J, Fang D, Gan H, Zhang Z. A lesson learned from the H3.3K27M mutation found in pediatric glioma: a new approach to the study of the function of histone modifications in vivo? Cell Cycle. 2013;12(16):2546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang S, Zheng X, Lu C, Li GM, Allis CD, Li H. Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 2016;30(14):1611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohammad F, Helin K. Oncohistones: drivers of pediatric cancers. Genes Dev. 2017;31(23–24):2313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koelsche C, Schrimpf D, Tharun L, Roth E, Sturm D, Jones DTW, et al. Histone 3.3 hotspot mutations in conventional osteosarcomas: a comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin Sarcoma Res. 2017;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schonegger A, Schuster M, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23(3):386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–43. [DOI] [PubMed] [Google Scholar]
- 78.Damato S, Alorjani M, Bonar F, McCarthy SW, Cannon SR, O’Donnell P, et al. IDH1 mutations are not found in cartilaginous tumours other than central and periosteal chondrosarcomas and enchondromas. Histopathology. 2012;60(2):363–5. [DOI] [PubMed] [Google Scholar]
- 79.L MG, Boulay K, Topisirovic I, Huot ME, Mallette FA. Oncogenic Activities of IDH1/2 Mutations: From Epigenetics to Cellular Signaling. Trends Cell Biol. 2017;27(10):738–52. [DOI] [PubMed] [Google Scholar]
- 80.Cleven AHG, Suijker J, Agrogiannis G, Briaire-de Bruijn IH, Frizzell N, Hoekstra AS, et al. IDH1 or −2 mutations do not predict outcome and do not cause loss of 5-hydroxymethylcytosine or altered histone modifications in central chondrosarcomas. Clin Sarcoma Res. 2017;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]