Abstract
Legionella pneumophila survives and replicates inside host cells by secreting ~300 effectors through the Dot/Icm type IVB secretion system (T4BSS). Here, we used complementary electron cryotomography (ECT) and immunofluorescence microscopy (IF) to investigate the molecular architecture and biogenesis of the Dot/Icm secretion apparatus. ECT mapped the location of the core and accessory components of the Legionella core-transmembrane subcomplex revealing a well-ordered central channel that opens into a large, windowed secretion chamber with an unusual 13-fold symmetry. IF deciphered an early-stage assembly process that begins with targeting of Dot/Icm components to the bacterial poles. Polar targeting of this T4BSS is mediated by two Dot/Icm proteins, DotU and IcmF, that interestingly are homologs of the T6SS membrane complex components TssL and TssM, suggesting the Dot/Icm T4BSS is a hybrid system. Together these results revealed that the Dot/Icm complex assembles in an “axial-to-peripheral” pattern.
Main
Type IV secretion systems (T4SSs) include conjugation machines used to transfer plasmids from donor to recipient cells and adapted conjugation systems used by pathogens to modify host cells1,2. Based on component number and similarity, T4SSs are classified into three types: IVA, IVB, or other3,4. The archetype T4ASS is the Agrobacterium tumefaciens VirB/D4 T4ASS that is used to inject bacterial genes into plant cells, thereby transforming tumors1. T4BSSs include the IncI conjugative plasmids (R64 and ColIb-P9) and the Legionella pneumophila and Coxiella burnetii Dot/Icm systems5. The Dot/Icm system is made up of ~27 components, including an outer-membrane (OM) protein (DotH), OM lipoproteins (DotC, DotD, DotK), a periplasmic protein (IcmX), inner membrane (IM) proteins (DotA, DotE, DotF, DotG, DotI, DotJ, DotM, DotP, DotU, DotV, IcmF, IcmT, and IcmV), IM-associated ATPases (DotB, DotL, DotO), and soluble cytosolic proteins (DotN, IcmQ, IcmR, IcmS, IcmW, and LvgA)6. These proteins can be found in several subcomplexes including the Legionella core-transmembrane (LCTM) subcomplex, consisting primarily of five proteins (DotC, DotD, DotF, DotG, DotH), and the Legionella type IV coupling protein (LT4CP) subcomplex, consisting of six proteins (DotL, DotM, DotN, IcmS, IcmW, LvgA)7–10. Previously, we demonstrated that the L. pneumophila Dot/Icm system is localized to both poles of the bacterial cell11 and imaged the system within intact cells by electron cryotomography (ECT)12. Here we extend our ECT observations by identifying the major periplasmic densities of the Dot/Icm T4BSS, find that DotU and IcmF are required for polar localization, and characterize a number of assembly dependencies that point to an axial-to-peripheral assembly.
Structural details of the Dot/Icm T4BSS
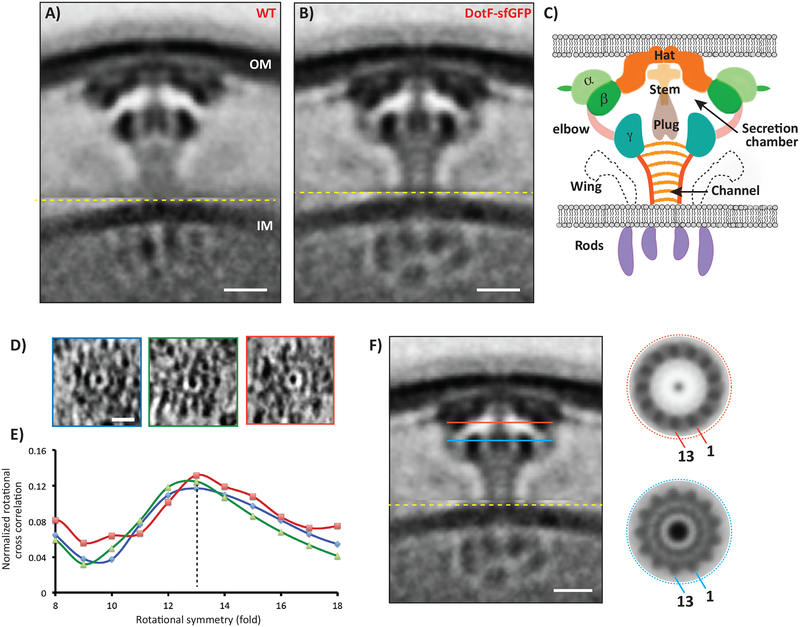
In the process of identifying the major periplasmic densities in the Dot/Icm T4BSS (Fig. 1C), we discovered that a DotF-sfGFP fusion stabilized the particles, resulting in a higher-resolution average of the complex (Fig. 1A,B, Supplementary Fig. 1). Superfolder GFP (sfGFP) was used as it is fluorescent following Sec-mediated transport into the periplasm of bacteria13. The machine containing DotF-sfGFP remained functional, as the strain grew similar to wild-type cells within host cells (Supplementary Fig. 2). This higher resolution average revealed further insights into the structure. First, the stalk is not a solid object, but rather an ~14 nm-long funnel-shaped channel with thick walls (~2 nm) and a central lumen (~4 nm) (Fig. 1B). The channel lumen is not empty, but rather contains diffuse density within and several thin striations perpendicular to the channel axis. A clear connection was seen between the beta and the gamma densities (labelled “elbow”), uncovering that the stalk channel leads into a large (~32 nm wide) “secretion chamber”. The elbows were separated by gaps circumferentially (Supplementary Movie 1). The wings were more pronounced in the DotF-sfGFP map and there was a thin additional ring of density protruding from the alpha density that likely corresponds to peptidoglycan (Fig. 1B). Distinct subunits were visible around the ring in top views of individual particles (Fig. 1D). We calculated rotational cross-correlations of slices through individual particles and found the peak was 13-fold (Fig. 1E) and clear features emerged when only 13-fold symmetry was applied (Fig. 1F). A contemporary independent analysis also observed a central channel through the apparatus leading to a structure near the outer-membrane with a similar symmetry14.
Figure 1. Overall structure of the Dot/Icm T4BSS.
Central tomographic slices through the (A) wild-type and (B) DotF-sfGFP sub-tomogram averages, showing the improved resolution of the DotF-sfGFP structure. Note that because Dot/Icm particles are flexible, all the sub-tomograms going into the average were first aligned on the outer membrane (OM)-associated densities and then on the inner membrane (IM)-associated densities, separately; the image shown is a composite of the two averages concatenated at the yellow line. Number of tomograms and particles used for the subtomogram average are listed in Supplementary Information Table 1. C) Schematic of the major densities in the structure, named for reference. D) Tomographic slices through individual particles showing top views. E) Rotational cross-correlation coefficients of the three top-view particles for symmetries from 8- to 18-fold, showing that 13-fold was the strongest in each case. Y-axis: normalized rotational cross correlation, X-axis: rotational symmetry. F) Applying 13-fold rotational symmetry at the levels of the red and blue lines in the DotF-sfGFP average produced clear structures (other symmetries failed to produce regular density patterns). For scale, the diameters of the red and blue circles are both ~32 nm. Scale bar 10 nm (A,B,F), 20 nm (D).
Dissection of the Legionella core-transmembrane subcomplex (LCTM)
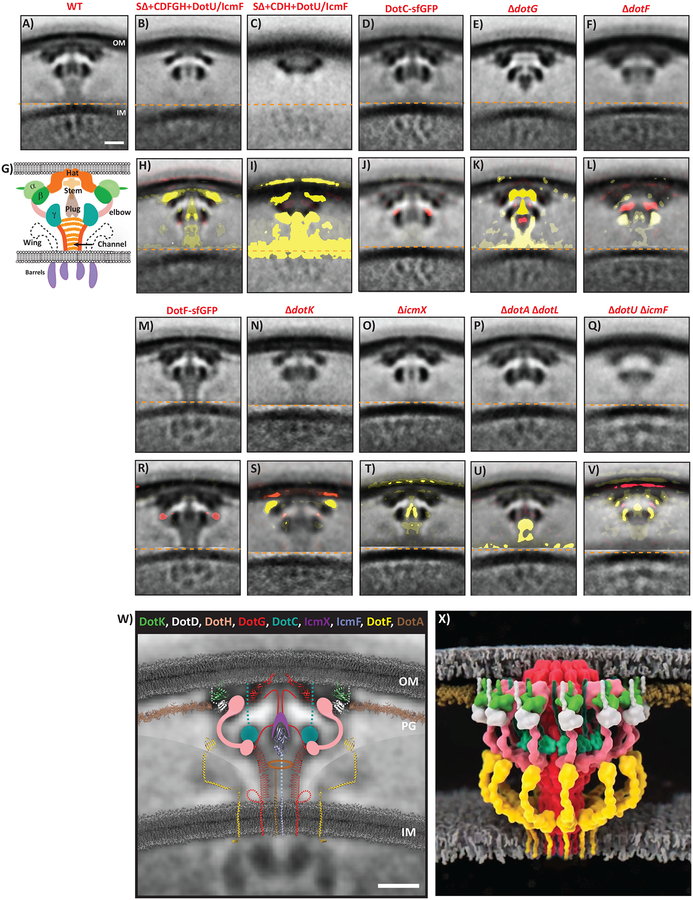
To identify the major periplasmic densities, we imaged a series of L. pneumophila mutants lacking T4BSS components or strains expressing Dot/Icm proteins fused to sfGFP. We confirmed the absence of protein expression in the deletion mutants and the presence of the sfGFP fusions by western blot analysis and demonstrated that the sfGFP fusions were functional (Supplementary Fig. 2). For strains that produced visible T4BSSs, we calculated sub-tomogram averages. The number of tomograms collected and particles used for subtomogram averaging are listed in Supplementary Table 1. In addition, we calculated difference maps between each mutant subtomogram average and the wild-type (Fig. 2). Guided by known connectivities, structural and biochemical knowledge and predictions of secondary and tertiary structures (Supplementary Fig. 3, 4), this allowed us to match component domains to densities in the overall structure (Fig. 2W).
Figure 2. Mutant structures, difference maps and architectural model of the Dot/Icm T4BSS.
Top rows (A-F, M-Q): Central slices through the sub-tomogram average structures of each strain imaged. Bottom rows (H-L, R-V): Central slices through the difference maps comparing each average to the wild-type. Yellow represents missing densities and red extra densities. Weak to strong intensities correspond to density differences from one to three standard deviations, respectively, overlaid on the mutant sub-tomogram average. Note missing densities (e.g. panels K and S), additional densities (e.g. panels J and R) or matched yellow/red pairs in an average (e.g. panel L), which likely indicate movement. Number of tomograms and particles used for each of the subtomogram average are listed in Supplementary Information Table 1. W) Based on the difference maps and evidence discussed in the text, known and predicted structures of T4BSS components are superimposed on the central slice of the DotF-sfGFP average (also see Supplementary Movie 1). The DotF-sfGFP average is generated by aligning the outer membrane (OM) and inner membrane (IM) regions separately. Note that the relative orientation of the component structures are not known – the purpose of this schematic is simply to show where in the T4BSS each component is located and how its size and shape compare to the ECT densities. Components whose structures are not known or confidently predictable are depicted as circles (e.g. DotC) or as the shape of densities seen in the sub-tomogram averages or difference maps (e.g. DotH and IcmX). Polypeptide links to the OM are shown as dotted lines. Sequences in DotG with unknown structure are shown as solid lines with speculative path. A large number of transmembrane helices are not shown for inner membrane proteins (DotA, DotE, DotL, DotM, DotP, DotU, DotV, IcmV, and IcmT). In addition, cytoplasmic components of the system are not shown. Currently, organization of the cytoplasmic complex is debated. OM = outer membrane, PG = peptidoglycan cell wall, IM = inner membrane. Lipids are shown in grey and peptidoglycan in brown. X) Three dimensional representation of the Dot/Icm complex showing windowed secretion chamber (salmon:DotH, white:DotD, green:DotK and cyan:DotC), wings (yellow:DotF) and secretion channel (red:DotG). Stoichiometry of DotF is unknown. Similar to (W), the cytoplasmic components are not shown. In this 3D representation, IcmF, IcmX and DotA are not visible. Scale bar 10 nm (A-V and W).
We began our examination of the periplasmic portion of the Dot/Icm complex by focusing on the five major proteins of the LCTM (DotCDFGH). As expected, no T4BSS particles were visible in a “super-deletion” strain (SΔ) in which all of the dot/icm genes are deleted8,12 (Supplementary Fig. 5). Expression of the five components of the LCTM subcomplex (DotCDFGH), or a smaller subcomplex of only DotCDH, in a SΔ strain expressing the proteins DotU and IcmF (represented by “SΔ(UF)”) restored a significant portion of the structure (Figs. 2B, 2C). No particles were found in the ΔdotC, ΔdotD, and ΔdotH strains (Supplementary Fig. 5), consistent with the inability to obtain biochemically isolated core complexes from these mutants15. In contrast, the DotC-sfGFP sub-tomogram average revealed additional density at the top of the gamma density (Fig. 2D, 2J). Supporting DotC being part of gamma, the gamma density was one of only two densities (with beta) seen in the SΔ(UF) + DotCDH strain (Figs. 2C, 2I). The gamma densities in individual particles in the SΔ(UF) + DotCDH strain appeared full-sized but were mobile, resulting in a smaller appearance in the average than in wild-type particles (Supplementary Fig. 6). Considering DotC is an outer-membrane lipoprotein6, 29 residues would suffice to span from the inner leaflet of the outer-membrane to the top of gamma. DotC can be responsible for only the top portion of gamma since the remaining residues are of insufficient size to represent all of gamma. As a result, we propose DotC is linked to the outer-membrane by a short linker domain and forms the top portion of gamma (Fig. 2W).
DotH was previously shown to become associated with the outer-membrane through the action of the lipoproteins DotC and DotD8. Considering that the subcomplex DotCDH exhibited beta and gamma densities, and DotC was responsible for the upper portion of gamma, we envisioned that beta and the bottom portion of gamma must consist of DotH and/or DotD. Structural predictions indicate DotH likely possesses two separate domains, similar to the outer-membrane associated T4ASS component VirB9 (Supplementary Fig. 7). The N-terminus of VirB9 is known to form part of the T4ASS inner-layer and its C-terminus binds to the periphery of the TrbI domain of VirB10 in the outer-layer16,17. Therefore, we propose that DotH forms i) the central part of beta (where a domain roughly the size of the C-terminus of VirB9 likely binds DotG), ii) the bottom of gamma (where another domain likely binds DotC), and iii) the elbow between them (Fig. 2W and Supplementary Fig. 7C).
DotD is an OM lipoprotein attached to the membrane at cysteine 1918. Following a relatively unstructured N-terminal linker, DotD contains a domain with similarity to the N0 domain of secretins, OM proteins found in T2SSs and T3SSs19. Similar to the structure of DotD, the Xanthomonas citri VirB7 protein has a similar short N-terminal linker and a N0 secretin domain, suggesting DotD may be a distant VirB7 homolog20. A crystal structure of the pKM101 T4SS OM complex showed that VirB7 lines the tops of VirB9 and VirB10 immediately adjacent to the outer-membrane16. NMR and molecular dynamics were used to dock the VirB7 N0 domain around the periphery of X. citri VirB7 (and the O-layer) (Supplementary Fig. 8)20. We suggest that 26 residues of the DotD linker line the top of DotG (the hat) and DotH (the central part of beta) and the remaining residues in the linker extend DotD from the outer-membrane to the periphery of beta, where the X. citri N0 domain has been predicted to be (Fig. 2W, Supplementary Fig. 8). Therefore, we propose DotD connects from the outer-membrane and forms the peripheral portion of the beta density.
DotG is the only protein in the T4SS superfamily with clear sequence homolog in both T4ASSs and T4BSSs6. The conserved part is the C-terminal TrbI domain (Supplementary Fig. 3), whose structure was solved in complex with the C-termini of VirB7 and VirB916. The TrbI domain forms a 14-fold-symmetric dome-shaped bowl with a central hole lined by two alpha helices per monomer thought to be embedded in the outer-membrane16. We previously speculated that DotG’s TrbI domain forms a similar dome just underneath the outer-membrane matching the Dot/Icm hat density12. Consistent with our prediction, the hat was conspicuously missing in the ΔdotG average (Fig. 2E, 2K) and the TrbI domain structure fit into the hat density (Fig. 2W). However, the hat region inexplicably persisted in a recently published ECT map of an independently derived ΔdotG mutant14, although it was absent when we re-imaged this strain (Supplementary Fig. 9).
The ΔdotG structure showed additional differences including a missing stem and stalk, a distorted gamma ring, and an altered location of the plug (Fig. 2E, 2K). The stalk is likely formed by a ~600 residue repeat region in the middle of DotG (Supplementary Fig. 3)6. We found this repeat region is predicted to fold into long β-helices (Supplementary Fig. 3) which match the apparent length, width, and curvature of the stalk/channel wall, and 13 copies of the predicted β-helix structure can form a ring with the same inner and outer dimensions as the channel (Supplementary Fig. 10). We speculate the ~100 residues between the TrbI and β-helix domains form the stem and interact with the plug (Supplementary Movie 1).
Regarding DotF, we began by comparing the densities in two reconstituted subcomplexes (SΔ(UF)+DotCDFGH versus SΔ(UF)+DotCDH). Since the latter was missing the wings, and they were also absent in a ΔdotF mutant (Fig. 2F, 2L), we propose DotF forms the wings. While faint in the averages, prominent wing densities were seen in individual particles. On closer inspection, we noticed that the positions of the wings with respect to the other major densities of the core transmembrane complex varied, explaining why they appeared faint in the averages (Supplementary Fig. 11). The DotF-sfGFP fusion generated new density at the joint of the elbow between the gamma and beta densities (Fig. 2M, 2R), implying the C-terminal domain of DotF is present at the joint. Consistent with this, several programs predicted DotF folds into a long, potentially jointed alpha-helical structure capped by a C-terminal domain similar to that of PilP/GspC in the type IV pilus/type II secretion systems (Supplementary Fig. 3). We therefore represent DotF as a flexible arm outside the stalk channel reaching to the elbow of beta/gamma (Fig. 2W).
Next, we extended our studies to identify the additional densities that were missing in the DotCDFGH reconstituted strain12 and discovered the likely positions of DotK, IcmX, DotA, and IcmF. DotK is a third outer-membrane lipoprotein5. IcmX is a soluble periplasmic protein and DotA is a polytopic inner membrane protein, although both are released into the culture media in a T4BSS-dependent manner21,22. IcmF, and its partner DotU, are homologs of the T6SS components TssL and TssM23,24, and play a critical role in targeting/assembly of the apparatus (see below).
In the ΔdotK average, all densities were present except alpha (Fig 2N,S). DotK is part of the OmpA domain family (Supplementary Fig. 3), which are bacterial peptidoglycan binding proteins. DotK’s residues 2–131 are predicted to adopt the same fold as the C-terminal peptidoglycan binding domain of Pseudomonas OprF (Supplementary Fig. 3). This structure fits the alpha density well (Fig. 2W), immediately adjacent to the thin line of density parallel to the OM, which is likely peptidoglycan. In the ΔicmX mutant, the T4BSS still assembled but a significant portion of the plug was missing (Fig. 2O,T), indicating IcmX forms part of the plug. While attempting to determine the location of the inner membrane ATPase DotL, we imaged a ΔdotA ΔdotL double mutant as ΔdotL lethality can be suppressed by inactivation of the inner membrane protein DotA9. The biggest difference observed in this strain was the absence of density in the top of the stalk channel, with diminished density in the stalk channel walls in that region (Fig. 2P,U). Since DotL does not contain a periplasmic domain9, these structural changes likely reflect the absence of DotA. Consistent with this, only the cytoplasmic densities were missing in a strain lacking all three ATPases (DotL, DotB, DotO) (Supplementary Fig. 12). Interestingly, the diameter of the upper part of the stalk roughly matched the diameter of purified DotA rings (10-nm diameter)22. As a result, we propose the periplasmic domain of DotA is located in the top of the stalk channel (Fig. 2W). Although our tomographic data provide some information on the inner membrane complex, we limited our description here to the architecture of the periplasmic portion of the complex.
Finally, DotU and IcmF are integral IM proteins that function together with only IcmF possessing a large periplasmic domain25–27. In a ΔdotU ΔicmF double mutant, the abundances of DotH, DotG and DotF are all reduced in late stationary phase, which led to the proposal that DotU/IcmF stabilize the Dot/Icm core complex25. Here we noted fewer particles at the poles with lower resolution in the ΔdotU ΔicmF mutant (Fig. 2Q), consistent with a role in stabilizing the complex and targeting it to the poles (see below). The difference map revealed an altered plug density and a decreased gamma ring diameter (Fig. 2Q, 2V). The periplasmic domain of IcmF (residues 738–972) has homology to the T6SS component TssM, whose structure has been solved28, and this domain fits nicely into the central plug density (Fig. 2W). As a result, we propose the C-terminus of IcmF forms part of the plug, consistent with the reduced diameter of the gamma ring in the absence of IcmF and explaining how it stabilizes this T4BSS.
Polar localization of the Legionella Dot/Icm T4BSS
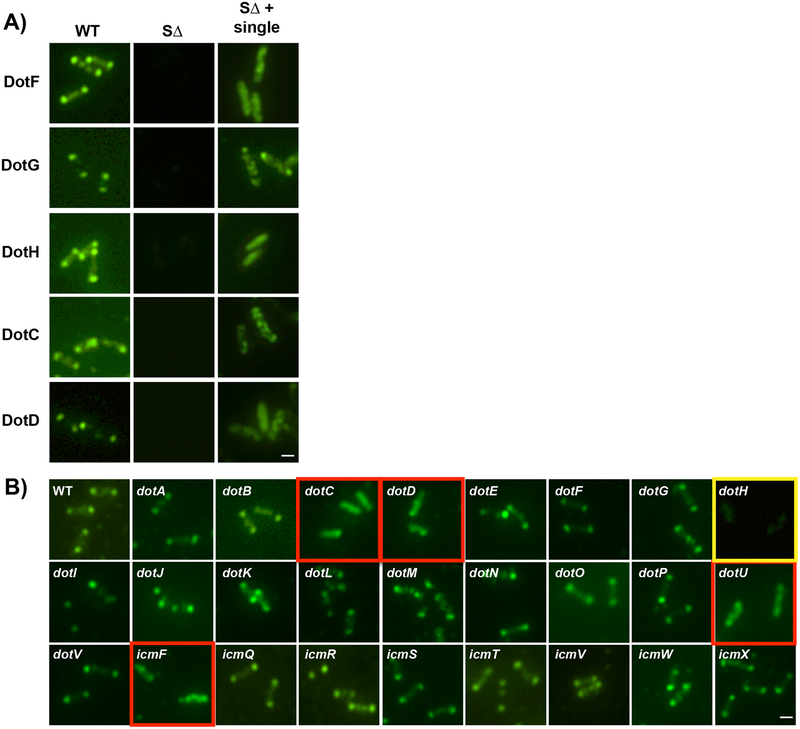
Previously we documented three components of the LCTM subcomplex, DotF, DotG, and DotH, localize to the bacterial poles of L. pneumophila11. Here we found that epitope-tagged versions of the other two proteins in the subcomplex, DotC and DotD, are functional and localize to the poles (Fig. 3A, Supplementary Fig. 13). However, each of these five proteins were found mostly at non-polar parts of the cell when expressed alone in the SΔ strain (Fig. 3A, Supplementary Fig. 14), suggesting polar localization of the core subcomplex is dependent on the presence of one or more additional Dot/Icm proteins.
Figure 3. Dot/Icm-dependent polar targeting of the Legionella core-transmembrane subcomplex.
A) Broth grown L. pneumophila cells were probed with primary antibodies (polyclonal for DotH, DotG, and DotF or HA monoclonal for DotD-HA and DotC-HA), decorated with secondary antibody conjugated with Oregon green and imaged with fluorescence microscopy. Samples assayed include the wild-type strain Lp02 (WT), a strain lacking all 27 dot/icm genes (SΔ), and the SΔ strain expressing individual core-transmembrane subcomplex components. B) DotH localization in wild-type Legionella and in individual dot/icm deletions. DotH was detected by immunofluorescence microscopy in wild-type cells (WT) and individual dot/icm mutant strains. The corresponding deletion strain is boxed in yellow and dot/icm deletions that are affected are boxed in red. Representative images are shown from three independent experiments. Scale bar 2 μm (A,B).
To identify this factor, we assayed the effect of individual dot/icm deletions on the localization of the DotH, DotG, and DotF proteins. Remarkably, while most of the dot/icm deletions did not affect the localization of the three proteins, their location was consistently perturbed by the absence of dotC, dotD, dotU or icmF (Fig. 3B and Supplementary Fig. 15–18). In addition, DotF targeting was also dependent on DotH (see below). As a result, proper localization of the LCTM subcomplex is inter-dependent between components of the LCTM subcomplex and dependent on DotU/IcmF (indicated as “UF”). Although the aberrant localization of DotH, DotG, and DotF was reminiscent of cytoplasmic staining, deconvolution microscopy revealed the proteins were localized in mostly non-polar puncta in the membrane (Supplementary Fig. 19). ECT analysis confirmed the non-polar localization of complexes in the ΔUF mutant, although they could occasionally be detected at the poles (Supplementary Fig. 5N, 20).
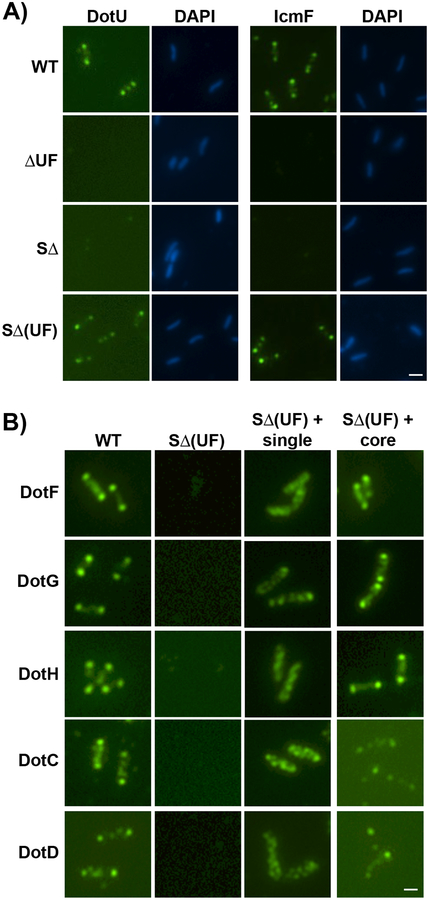
Based on these results, we hypothesized that UF may function directly as an anchor or a landmark to recruit the LCTM subcomplex to the bacterial poles. If so, then DotU and IcmF should be able to localize to the poles of the SΔ strain expressing only these two proteins (“SΔ(UF)”). As predicted, DotU and IcmF were observed at the poles in greater than 90% of such cells (Fig. 4A and Supplementary Fig. 21). To confirm the targeting properties of DotU and IcmF, we examined whether polar targeting of the LCTM subcomplex was dependent on the presence of UF (Fig. 4B). DotU and IcmF were insufficient to target the five proteins to the poles at wild-type levels when they were expressed individually in the SΔ(UF) strain (Fig. 4B, third column). However, the presence of DotU and IcmF restored polar localization when all five proteins were expressed in the SΔ(UF) strain (Fig. 4B and Supplementary Fig. 21–22).
Fig 4. DotU and IcmF localize to the bacterial poles in the absence of the Legionella T4SS.
(A) DotU and IcmF localization was assayed in the wild-type strain Lp02 (WT), ΔdotU ΔicmF mutant strain (JV1181), the super dot/icm deletion strain (SΔ, JV4044) and the SΔ strain expressing dotU and icmF from the chromosome (SΔ(UF)), JV5319). Shown are Dot staining (left) and DNA stained with DAPI (right) for each set. B) Localization of Dot proteins was assayed in the wild-type strain (WT), the SΔ strain encoding dotU and icmF (SΔ(UF)), the SΔ(UF) strain expressing individual components of the core-transmembrane subcomplex (SΔ(UF) + single), and the SΔ(UF) strain expressing all five core components (SΔ(UF) + core). Antibodies used for immunofluorescence are indicated to the left of the panels. Representative images are shown from three independent experiments. Scale bar 2 μm (A,B).
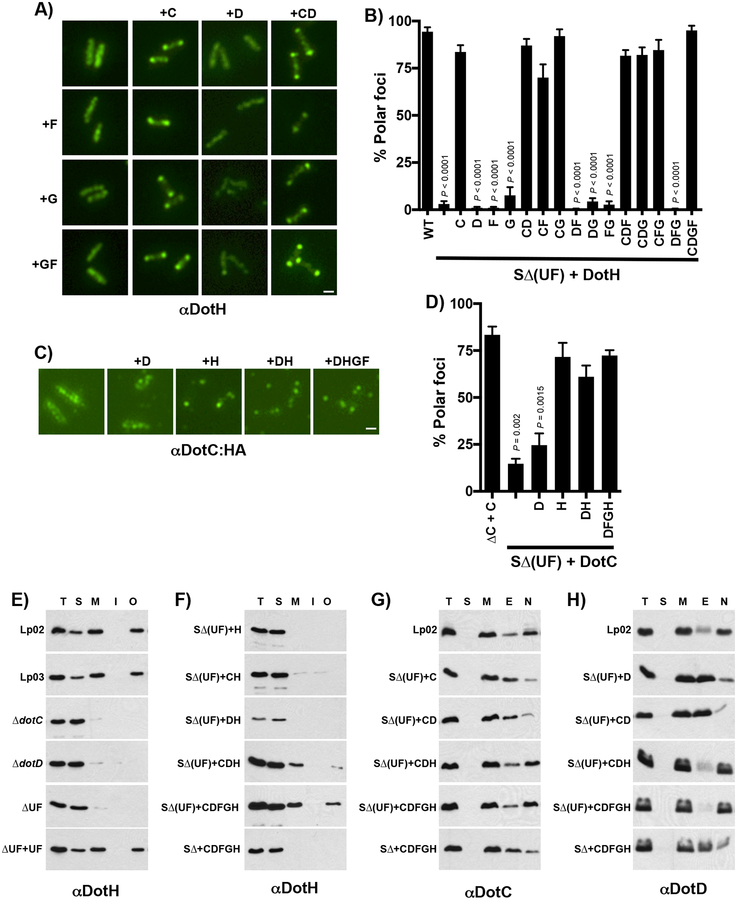
The intra-dependence between components of the LCTM subcomplex for proper polar localization by UF led us to perform a comprehensive analysis of polar targeting by expressing combinations of the LCTM subcomplex components in the SΔ(UF) strain. Polar targeting of DotH was restored in eight of the fifteen possible strain combinations (Fig. 5A,B and Supplementary Fig. 23–27). Strikingly, the presence of DotC alone was sufficient to assist UF in the proper targeting of DotH. In contrast to DotH, DotG localization was somewhat permissive as half of the cells had protein localized to the poles in the presence of only UF, but DotG was more efficiently targeted in a strain expressing all five components of the core (Supplementary Fig. 28). Similar to DotH, DotC localization dramatically increased when co-expressed with just DotH (Fig. 5C,D), whereas DotD localization depended on DotC and DotH (Supplementary Fig. 28). DotF’s localization was the most restrictive, as partial polar localization was detected in only two of the reconstituted strains, DotCDH and DotCDGH (Supplementary Fig. 28).
Figure 5. Reconstitution of the core-transmembrane subcomplex in the SΔ(UF) strain.
(A-H) Combinations of the core-transmembrane subcomplex were expressed in the super dot/icm deletion strain encoding dotU and icmF (SΔ(UF)). Representative images for DotH and DotC-HA localization are shown (A and C, respectively). Proteins expressed is indicated by labels on the left and top of each panel and the protein localized by IFM is shown below the images. The percent of cells with polar localization was determined from three independent experiments (100 cells counted from each experiment) and are shown in panels B and D. All data are representative of 3 independent biological experiments (n=3). In (B) and (D), data are presented as means ± SEM with statistical differences compared to the WT strain by unpaired two-tailed Student’s t-test. Scale bar 2 μm (A,C). (E-H) DotH association with the outer membrane require the targeting factors DotU and IcmF. (E,F) Cells were fractionated by a combination of ultracentrifugation and Triton X-100 solubility, proteins were separated by SDS-PAGE and probed in westerns using DotH specific antibodies. (E) DotH localization was determined in the wild-type strain Lp02 (WT), dotA mutant Lp03, ΔdotC (JV3743), ΔdotD (JV3572), ΔdotU ΔicmF (JV1181), and ΔdotU ΔicmF + complementing clone (JV1199). (F) DotH localization was determined in the SΔ(UF) strain expressing DotH (JV5405), DotC/DotH (JV5458), DotD/DotH (JV5459), DotC/DotD/DotH (JV5460), the core DotC/DotD/DotF/DotG/DotH (JV5443) or the core expressed in the SΔ strain without UF (JV5442). Experiments were done in triplicate and representative images are shown. (G,H) Cells were similarly fractionated and probed in westerns using DotC and DotD-specific antibodies (G and H, respectively). Interaction of the lipoproteins with DotH was determined in the following strains: wild-type Lp02 (WT), SΔ(UF) + DotC (JV5469), SΔ(UF) + DotD (JV5470), SΔ(UF) + DotC/DotD (JV5471), SΔ(UF) strain + DotC/DotD/DotH (JV5460), SΔ(UF) + core (JV5463), and the SΔ strain + core (JV5442). Fractions are indicated at the top of the panels and include total proteins (T), soluble proteins (S), membrane proteins (M), Triton X-100 extractable proteins (E), and Triton X-100 non-extractable proteins (N). Experiments were done in triplicate and representative images are shown.
To probe the role of UF in the biogenesis of the Dot/Icm complex, we examined the localization of DotC and DotH using a combination of centrifugation and detergent solubility. The majority of DotH in the wild-type strain Lp02 is membrane associated (M) and not extractable with the detergent Triton X-100, consistent with its linkage to the outer-membrane (O) (Fig. 5E and Supplementary Fig. 29)8. Membrane association of DotH was dependent on both DotC and DotD but strikingly did not occur in the ΔUF mutant (Fig. 5E). Thus, DotH outer membrane linkage (Fig. 5E) and polar localization (Fig. 3B) in a wild-type strain require the same set of four proteins (DotC, DotD, DotU and IcmF).
However, experiments using our reconstituted strains revealed only three proteins (DotC, DotU and IcmF) were sufficient to target DotH to the poles (Fig. 5A,B). Therefore, we repeated the fractionation experiments using our LCTM reconstituted strains. Interestingly, expression of DotC in the SΔ(UF) strain did not restore outer membrane association of DotH (Fig. 5F), even though it was sufficient for polar targeting of DotH (Fig. 5A,B). Instead membrane association of DotH also required DotD, although this property improved somewhat by co-expression of all five components of the LCTM (DotCDFGH). These results were specific as other combinations failed to properly link DotH to the membrane (Supplementary Fig. 29). UF did not mediate outer membrane targeting of DotC and DotD as assayed by sucrose gradients (Supplementary Fig. 30). Rather the lipoproteins became resistant to Triton X-100 extraction in the presence of UF (Fig. 5G–H), indicating their association with an outer membrane complex that is dependent on DotH. Taken together, these results demonstrate the first steps in the biogenesis pathway of the Legionella Dot/Icm complex beginning with polar targeting of DotC and DotH by DotU/IcmF prior to the stable association of DotH to the outer-membrane via the combined action of DotD (Fig. 6).
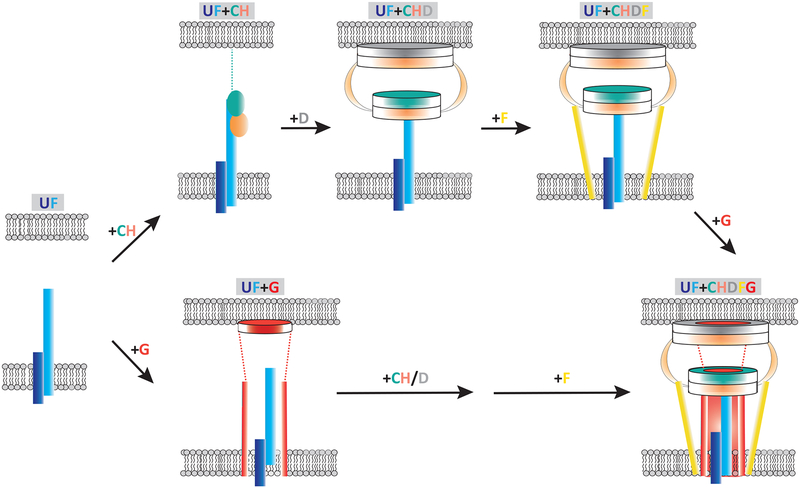
Figure 6. Polar targeting and assembly of the Legionella core-transmembrane subcomplex by DotU and IcmF.
The process begins by localization of DotU/IcmF to the poles, which can occur in the absence of any other Dot/Icm protein. This is followed by the recruitment of both DotC and DotH (pathway on the top). At this stage, the lipoprotein DotC is stably inserted in the outer membrane via its lipid domain, whereas DotH remains soluble in the periplasm, associated with DotC and the periplasmic domain of IcmF. Next, DotD arrives at the poles and assists in and/or directly mediates the outer membrane association of DotH (step 2). After formation of the DotC/DotD/DotH subcomplex, DotF and DotG are brought to the poles, thus linking the inner and outer membranes (also see Supplementary Movie 1). DotF localization is strongly dependent on the presence of DotC, DotD, and DotH but improves by the presence of DotG. DotG appears to be able to target to the poles to some level on its own but localizes more efficiently in the presence of DotC/DotD/DotH or DotC/DotD/DotH/DotF. In an alternative pathway (bottom), DotG could arrive at the poles upon arrival of UF and then subsequently DotC, DotH and DotF would join to complete the assembly process.
Summary
We used complementary approaches of ECT and immunofluorescence imaging to determine the molecular architecture of the Legionella core-transmembrane (LCTM) subcomplex and associated proteins and deciphered early-stage assembly dependencies. Our ECT analysis illuminated the periplasmic portion of the Dot/Icm T4BSS at “macromolecular” (~2–4 nm) resolution and pinpointed the locations of 9 Dot/Icm proteins (Supplementary Movie 1). The alpha, beta, and gamma rings were found to have 13-fold symmetry, which was surprising as all reported T4ASSs exhibit 14-fold symmetry16,29–31. We discovered a wide channel in the stalk of the Dot/Icm T4BSS, which immediately rationalizes how substrates are ushered from the cytoplasm to the outer membrane. This was notable in that there was no previous evidence for such a channel in other T4SSs16,29–31. Finally, we identified DotU/IcmF as the key factors that target the T4BSS to the poles.
Our ECT studies revealed conservation of several architectural elements across the T4SS family. DotG, DotH and DotD appear to be functional homologs of VirB10, VirB9 and VirB7, respectively, and form an OM complex (O-layer) present in both T4ASSs and T4BSSs12,31. DotH and DotC form a second ring below the OM complex that is likely the counterpart of the I-layer31. These two rings/layers form a conserved secretion chamber seen here to sit atop the stalk channel. This conservation of components and architecture is remarkable as only the C-terminus of DotG has detectable sequence similarity to a VirB protein.
Regarding assembly dependencies, we discovered that UF can localize to the poles on their own and serve as targeting factors for additional Dot/Icm components (Fig. 6). UF can independently recruit a DotC:DotH complex and the DotG protein. In the former case, DotH interestingly remains soluble in the periplasm at the poles until the arrival of DotD, which then facilitates the stable outer-membrane association of DotH. Following formation of the DotC/D/H complex, DotF and DotG are then incorporated into the complex. Alternatively, DotG can arrive first, followed by addition of DotC/D/H, and then DotF. A transmission electron microscopy study of purified Legionella core transmembrane complexes15 is consistent with our proposed assembly pathway.
Notably, all of these steps are dependent on DotU and IcmF, which have striking homology to two T6SS components (TssL and TssM)24. T6SSs are molecular injection machines made up of three large subcomplexes (core membrane complex, baseplate and sheath)24,32. The core membrane complex is composed of three proteins (TssL, TssM, TssJ) and plays a central role as both a docking station for cytoplasmically-expressed components and subsequently as a channel for the passage of the inner tube28. All T6SSs contain TssL/TssM proteins, whereas only the Dot/Icm T4BSSs (and not T4ASSs) have DotU/IcmF33. Intriguingly, classical T6SSs are not restricted to the bacterial poles28 and yet DotU and IcmF function as targeting factors for the Dot/Icm T4BSSs, which may reflect adaptation of these proteins to a new function and/or unique properties of Legionella cells (Supplementary Fig. 31).
Finally, our ECT analysis can be used to rationalize these dependencies by showing the periplasmic domain of IcmF forms part of a “plug” at the center of the T4BSS which touches DotG molecules and is surrounded by a ring of 13 DotC/DotH complexes. DotD was found in a second ring making contacts with one domain of DotH and with the OM, thus revealing why DotD’s localization depends on DotH and how DotD is in position to affect the outer-membrane association of DotH. DotF likely arrives last as it forms “wings” on the outside of the channel that bind a domain of DotH, thereby explaining why polar localization of DotF is strongly dependent on the presence of DotH (Supplementary Fig. 16B). Thus, assembly proceeds from the axis of the complex towards the periphery, likely reflecting the evolutionary history of a channel formed by a single primordial DotG-like protein that spanned both membranes and the periplasm and gradually accrued additional factors around its periphery for enhanced stability and function. Our ECT analyses also suggest how three additional components (DotA, DotK, and IcmX) interact with the core-transmembrane subcomplex, and show definitively that the three Dot/Icm cytoplasmic ATPases (DotL, DotO, and DotB) are not required for the assembly of the periplasmic complex. Thus, this study reveals how the periplasmic components of the Dot/Icm apparatus are recruited to the poles and incorporated into the apparatus using an axial-to-peripheral strategy dependent on a seed of the DotU/IcmF polarization factor.
Experimental Procedures
Strains and cell lines
All bacterial strains are listed in Supplementary Table 2. L. pneumophila strains were cultured in buffered AYE broth or on buffered charcoal yeast extract (CYE) plates. The media were supplemented with 100 μg/ml thymidine as needed (AYET, CYET). All media and antibiotic concentrations for E. coli and L. pneumophila were used as described previously34. The growth phase of liquid cultures was determined by the levels of motility and the optical density.
Construction of reconstituted plasmids
All used and newly constructed plasmids are listed in Supplementary Table 2. For expression in Legionella, all genes were cloned into the vector pJB908 individually or in combination. For new reconstituted plasmids, they were constructed by subcloning of open reading frames of dotD, dotC, dotF, dotG, and dotH.
Western blot analysis
Protein samples were boiled for 5 min in Laemmli sample buffer and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transfer to polyvinylidene difluoride membranes. Membranes were blocked in BLOTTO (PBS containing 5% non-fat dry milk), washed with wash buffer (PBS containing 0.05% Tween 20) and incubated for 1 hour with antibody diluted in BLOTTO. Blots were then washed with wash buffer followed by 1-hour incubation with secondary goat anti-rabbit antibody conjugated to horseradish peroxidase (Sigma) diluted 1:10,000 in BLOTTO. Blots were subsequently washed with wash buffer prior to development using an ECL detection kit (GE Healthcare).
In vitro immunofluorescence microscopy
Modified immunofluorescence microscopy (IFM) was carried out as descried previously11. In brief, five microliters of an L. pneumophila strain, grown to stationary phase culture in broth were briefly fixed in methanol, resuspended in phosphate buffered saline (PBS), and allowed to adhere to poly-L-lysine (Sigma) coated microscope slides. Lysozyme (3 mg/ml in 25 mM Tris-HCl, pH 8.0, 50 mM glucose, 10 mM EDTA) was used to permeabilize the cells, which were then washed with PBS, and incubated with various primary antibodies. After incubation, cells were washed with PBS, decorated with Oregon Green-conjugated goat anti-rabbit IgG, and stained with DAPI to detect DNA. Fluorescence anti-fade reagent was added and the immunostained cells were observed using a fluorescence microscope (Olympus, 100X objective). All images were captured and further analyzed with IPLab software (BD Bioscience).
Protein fractionations
Protein fractionation was performed by previously described method8. In brief, approximately 40 OD600 stationary cells were resuspended in 1 mL of 50 mM Tris-8, 0.5 M sucrose, 5 mM EDTA. After lysozyme treatment, 1.5 mL 50mM Tris pH 8 was added to the samples, and MgSO4 was added to 8mM final concentration and sonicated about total 5 min. Samples were centrifuged at 10,000 × g for 15 min at 4° C. Soluble and insoluble proteins were separated by centrifuging 1 mL of this fraction at 100,000 × g for 1 hour. The supernatant from the first spin was re-centrifuged at 100,000 × g for 1 hour to insure complete removal of membrane proteins, and this second supernatant was collected as the soluble protein fraction. Pelleted proteins from the first 100,000 × g spin were resuspended in ice cold 50 mM Tris pH 8 and re-centrifuged at 100,000 × g for 1 hour to remove any contaminating soluble proteins. Total membrane proteins were then resuspended in ice cold 50 mM Tris pH 8. To extract inner membrane proteins, Triton X-100 was added to a final concentration of 1% and the samples were incubated at 37° for 30 minutes. Triton X-100 insoluble proteins were removed by centrifugation of the samples at 100,000 × g for 30 minutes. The supernatants were re-centrifuged at 100,000 × g for an additional 30 minutes to ensure complete removal of Triton X-100 insoluble proteins. Triton X-100 insoluble proteins were resuspended in 1 mL 50 mM Tris pH 8, 1% Triton X-100, and re-centrifuged at 100,000 × g. Triton X-100 insoluble proteins were then resuspended in 1 mL Tris pH 8. Fractions were analyzed by SDS-PAGE followed by immunoblot analysis.
Statistical analysis
Statistical analysis was performed using Student’s t-test of GraphPad Prism 6 (Version 6.0d; GraphPad Software, La Jolla, CA, USA). Data are presented as means ± SEM. Statistical significance was declared if P<0.05.
Sample preparation for electron cryotomography
L. pneumophila Lp02 cells were grown as mentioned previously12, harvested at early stationary phase (OD600 of ~3.0), mixed with 10-nm colloidal gold beads (Sigma-Aldrich, St. Louis, MO) precoated with bovine serum albumin, and applied onto freshly glow-discharged copper R2/2 200 Quantifoil holey carbon grids (Quantifoil Micro Tools GmbH, Jena, Germany). Grids were then blotted and plunge-frozen in a liquid ethane/propane mixture35 using an FEI Vitrobot Mark IV and stored in liquid nitrogen for subsequent imaging.
Electron cryotomography, sub-tomogram averaging, and difference analysis
Tilt-series were recorded of frozen L. pneumophila Lp02 cells in an FEI Titan Krios 300 kV field emission gun transmission electron microscope (FEI Company, Hillsboro, OR) equipped with a Gatan imaging filter (Gatan, Pleasanton, CA) and a K2 Summit direct detector in counting mode (Gatan, Pleasanton, CA) using the UCSF Tomography software36 and a total dose of ~100 e/A2 per tilt-series and target defocus of ~6 μm underfocus. Energy-filtered tilt-series of images of the cells were automatically collected from 0° to −60° and then +1° to +60° at 2° intervals. Images were aligned, contrast transfer function corrected, and reconstructed using IMOD37. SIRT reconstructions were produced using TOMO3D38 and sub-tomogram averaging was performed using PEET Nicastro39. Finally, the local resolution was calculated by ResMap. As the Dot/Icm sub-tomogram average exhibited a gross two-fold symmetry around the central mid-line in the periplasm, we applied two-fold symmetry in those regions to produce the 2-D figures shown, but no symmetry was applied to the cytoplasmic densities due to their poor resolution. Sub-tomograms were aligned within masks centered either on the OM-associated densities, the IM-associated densities, or the densities between the IM and the gamma ring. The figures are composites, showing the average that was clearest in that region of the T4BSS, with lines visible at the interface to remind the viewer of this fact. Difference maps were created by mutually aligning two averages, then subtracting the densities of the mutant from the densities of the reference (usually the wildtype). Losses and increases of density were shown in yellow and red, respectively, and shaded according to significance (bright and light color for differences greater than two and one standard deviations above the mean, respectively).
Structure prediction and model building
Gene sequences were obtained from UniProt40. Signal sequences were predicted with SignalP 4.141. Transmembrane domains were predicted by Phobius42 and TMHMM Server43. Domain structures were predicted by servers Phyre244, I-TASSER45, Quark46. The cysteines attached to the OM in lipoproteins were marked as reported in the literature or predicted using SignalP. Candidate ring structures with different numbers of monomers were generated using SymmDock47 (for instance the ring of DotG N-terminal β-helices).
Supplementary Material
Acknowledgements:
We thank Dr. Ralph Isberg (Tuft’s University) for antibodies that recognize DotF and DotH, Ms. Emily Buford for technical assistance and Dr. Petra Levin (Washington University) for assistance with deconvolution microscopy. ECT data were recorded at the Beckman Institute Resource Center for Transmission Electron Microcopy at Caltech and the cryo-EM facility at Janelia Research Campus. We thank Dr. Catherine Oikonomou for creation of the domain maps and for help structuring and revising the text. We also recognize Dr. Eep Darwin for key suggestions and critical appraisal of this manuscript. This work was funded by NIH grant R01AI127401 to G.J.J. and NIH grant R01AI48052 to J.P.V.
Footnotes
Data availability: The sub-tomogram average of the Dot/Icm (DotF-sfGFP) complex that supports the findings of this study has been deposited in the Electron Microscopy Data Bank (EMDB) under the accession code: EMD-0566. All other density maps are available from the authors upon request. The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information documents.
Competing Interests: The authors declare no competing interests.
REFERENCES
- 1.Christie PJ, Whitaker N & Gonzalez-Rivera C Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843, 1578–1591, doi: 10.1016/j.bbamcr.2013.12.019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa TRD et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nature Reviews. Microbiology 13, 343–359, doi: 10.1038/nrmicro3456 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Christie PJ & Vogel JP Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends in Microbiology 8, 354–360 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guglielmini J et al. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Research 42, 5715–5727, doi: 10.1093/nar/gku194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal G, Feldman M & Zusman T The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev 29, 65–81, doi: 10.1016/j.femsre.2004.07.001 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Nagai H & Kubori T Type IVB Secretion Systems of Legionella and Other Gram-Negative Bacteria. Frontiers in Microbiology 2, 136, doi: 10.3389/fmicb.2011.00136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland MC, Nguyen TL, Tseng V & Vogel JP The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog 8, e1002910, doi: 10.1371/journal.ppat.1002910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent CD et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 62, 1278–1291 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Vincent CD, Friedman JR, Jeong KC, Sutherland MC & Vogel JP Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85, 378–391, doi: 10.1111/j.1365-2958.2012.08118.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak M-J et al. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nature Microbiology 2, 17114, doi: 10.1038/nmicrobiol.2017.114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong KC, Ghosal D, Chang Y-W, Jensen GJ & Vogel JP Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proceedings of the National Academy of Sciences 114, 8077–8082, doi: 10.1073/pnas.1621438114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosal D, Chang YW, Jeong KC, Vogel JP & Jensen GJ In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 18, 726–732, doi: 10.15252/embr.201643598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinh T & Bernhardt TG Using superfolder green fluorescent protein for periplasmic protein localization studies. J Bacteriol 193, 4984–4987, doi: 10.1128/JB.00315-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chetrit D, Hu B, Christie PJ, Roy CR & Liu J A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Micro, doi: 10.1038/s41564-018-0165-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubori T et al. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A 111, 11804–11809, doi: 10.1073/pnas.1404506111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandran V et al. Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015, doi: 10.1038/nature08588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Calzada A et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. The EMBO journal 32, 1195–1204, doi: 10.1038/emboj.2013.58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yerushalmi G, Zusman T & Segal G Additive effect on intracellular growth by Legionella pneumophila Icm/Dot proteins containing a lipobox motif. Infection and Immunity 73, 7578–7587, doi: 10.1128/IAI.73.11.7578-7587.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano N, Kubori T, Kinoshita M, Imada K & Nagai H Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS pathogens 6, e1001129, doi: 10.1371/journal.ppat.1001129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza DP et al. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS pathogens 7, e1002031, doi: 10.1371/journal.ppat.1002031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews M & Roy CR Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infection and Immunity 68, 3971–3982 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai H & Roy CR The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. The EMBO journal 20, 5962–5970, doi: 10.1093/emboj/20.21.5962 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand E et al. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. The Journal of Biological Chemistry 287, 14157–14168, doi: 10.1074/jbc.M111.338731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen VS et al. Towards a complete structural deciphering of Type VI secretion system. Curr Opin Struct Biol 49, 77–84, doi: 10.1016/j.sbi.2018.01.007 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Sexton JA, Miller JL, Yoneda A, Kehl-Fie TE & Vogel JP Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun 72, 5983–5992 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanRheenen SM, Dumenil G & Isberg RR IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun 72, 5972–5982, doi: 10.1128/IAI.72.10.5972-5982.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zusman T, Feldman M, Halperin E & Segal G Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect Immun 72, 3398–3409, doi: 10.1128/IAI.72.6.3398-3409.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand E et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560, doi: 10.1038/nature14667 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Chang Y-W, Shaffer CL, Rettberg LA, Ghosal D & Jensen GJ In Vivo Structures of the Helicobacter pylori cag Type IV Secretion System. Cell Reports 23, 673–681, doi: 10.1016/j.celrep.2018.03.085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frick-Cheng AE et al. Molecular and Structural Analysis of the Helicobacter pylori cag Type IV Secretion System Core Complex. mBio 7, e02001–02015, doi: 10.1128/mBio.02001-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low HH et al. Structure of a type IV secretion system. Nature 508, 550–553, doi: 10.1038/nature13081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basler M Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370, doi: 10.1098/rstb.2015.0021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubori T & Nagai H The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29, 22–29, doi: 10.1016/j.mib.2015.10.001 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Bardill JP, Miller JL & Vogel JP IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56, 90–103 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Tivol WF, Briegel A & Jensen GJ An improved cryogen for plunge freezing. Microsc Microanal 14, 375–379, doi: 10.1017/S1431927608080781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng SQ et al. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157, 138–147, doi: 10.1016/j.jsb.2006.06.005 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76, doi: 10.1006/jsbi.1996.0013 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Agulleiro J-I & Fernandez J-J Tomo3D 2.0--exploitation of advanced vector extensions (AVX) for 3D reconstruction. Journal of Structural Biology 189, 147–152, doi: 10.1016/j.jsb.2014.11.009 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Nicastro D et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948, doi: 10.1126/science.1128618 (2006). [DOI] [PubMed] [Google Scholar]
- 40.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Research 46, 2699, doi: 10.1093/nar/gky092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emanuelsson O, Brunak S, von Heijne G & Nielsen H Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971, doi: 10.1038/nprot.2007.131 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Käll L, Krogh A & Sonnhammer ELL A combined transmembrane topology and signal peptide prediction method. Journal of Molecular Biology 338, 1027–1036, doi: 10.1016/j.jmb.2004.03.016 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Krogh A, Larsson B, von Heijne G & Sonnhammer EL Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580, doi: 10.1006/jmbi.2000.4315 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858, doi: 10.1038/nprot.2015.053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12, 7–8, doi: 10.1038/nmeth.3213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu D & Zhang Y Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735, doi: 10.1002/prot.24065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneidman-Duhovny D, Inbar Y, Nussinov R & Wolfson HJ PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Research 33, W363–367, doi: 10.1093/nar/gki481 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.