Abstract
The centriole is an ancient microtubule-based organelle with a conserved nine-fold symmetry. Centrioles form the core of centrosomes, which organize the interphase microtubule cytoskeleton of most animal cells and form the poles of the mitotic spindle. Centrioles can also be modified to form basal bodies, which template the formation of cilia and play central roles in cellular signaling, fluid movement and locomotion. In this review we discuss developments in our understanding of the biogenesis of centrioles and cilia and the regulatory controls that govern their structure and number. We also discuss how defects in these processes contribute to a spectrum of human diseases and how new technologies have expanded our understanding of centriole and cilium biology, revealing exciting avenues for future exploration.
Introduction
Centrioles are evolutionarily conserved microtubule-based structures that have diverse functions in controlling cell polarity, proliferation, division, motility and signaling. Centrioles recruit a surrounding pericentriolar material (PCM) to form centrosomes, which serve as the major microtubule organizing center in most animal cells (Figure 1A). Centrosomes nucleate the formation of the microtubule cytoskeleton in interphase cells and form the poles of the bipolar microtubule spindle during mitosis. In quiescent cells, a fully mature centriole can dock at the plasma membrane and act as a basal body that anchors a cilium. The cilium is comprised of axonemal microtubules that elongate from the distal end of the basal body and a ciliary membrane that surrounds the axoneme. Phylogenetic studies indicate that centrioles were present in the last eukaryotic common ancestor, but were lost in some branches of the tree of life, such as some yeasts and higher plants (1). The presence of centrioles specifically correlates with the presence of cilia and not centrosomes, suggesting that the ancestral role of centrioles was to direct formation of cilia (2).
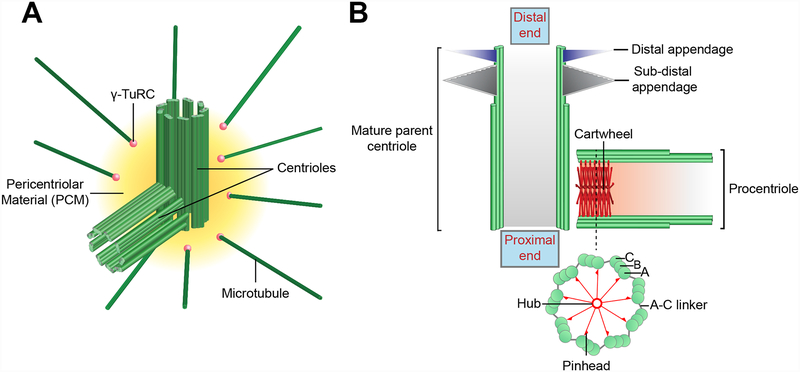
Figure 1. Centriole and centrosome structure.
A) Architecture of the mammalian centrosome. The centrosome is comprised of a pair of orthogonally oriented centrioles surrounded by a proteinaceous Pericentriolar Material (PCM). The PCM contains proteins required for microtubule nucleation and anchoring, such as the γ-Tubulin Ring Complex (γTuRC) (pink spheres). B) Schematic illustration of a mature parent centriole and associated procentriole. Centrioles are cylindrical strictures comprised of nine triplet microtubules, each of which contains a complete A-tubule and an incomplete B and C-tubule. The cartwheel is present in the proximal lumen of the procentriole and is formed by a central hub from which nine spokes emanate. Each spoke terminates in a pinhead structure that binds to the A-tubule of the microtubule triplet. The A-tubule of one triplet is linked to the C-tubule of the adjacent triplet via an A-C linker. Mature parent centrioles are decorated at their distal end with ninefold symmetric distal and sub-distal appendages.
Recent work has begun to elucidate the molecular framework that underlies centriole and cilium assembly, as well as how dysregulation in these organelles contribute to human disease. In this review, we explain recent advances in the centrosome and cilia field, with a focus on vertebrate centrosomes and cilia, but referring to other systems when necessary. We begin by discussing centriole architecture and the centriole duplication cycle. We then deal with how centrosome defects contribute to human disease before discussing how cilia are assembled and disassembled in a cell cycle-dependent manner. We briefly explain the roles of cilia in cell signaling and how cilium dysfunction contributes to disease. Finally, we end by reviewing new technologies for studying centrosomes and cilia and highlight some important open questions and future avenues for exploration.
Centriole structure
Centrioles are cylindrical in shape and are comprised of a conserved nine-fold-symmetrical array of microtubules that form their walls (3). While the rotational symmetry of centrioles is invariant across life, centrioles can vary in size and diameter in different organisms and cell types. In mammalian cells, centrioles are ~230 nm in diameter and ~420 nm in length (4). Most centrioles have a wall comprised of nine sets of interconnected triplet microtubule blades, although in some organisms the centriole wall is comprised of singlet or doublet microtubules (Figure 1B). Triplet microtubules contain a 13 protofilament A-tubule and 10 protofilament B- and C-tubules, with the A-tubule from one triplet connected to the C-tubule of the neighboring triplet through an A-C linker (5). A recent cryo-electron microscopy study has shown that mammalian centrioles are organized into two structurally distinct regions along the proximal-distal axis (4). The proximal domain is ~200 nm in length and shares a common core architecture with the shorter Drosophila centriole, while the distal region of the mammalian centriole has a distinct A-C linker, an incomplete C-tubule, and a narrower diameter. Given that fly centrioles are structurally similar to the proximal region of mammalian centrioles and do not generate motile cilia, it is plausible that the distinct architecture of the distal portion of the mammalian centriole provides this expanded functionality (4).
The centriole is polarized along the proximal-distal axis for distinct functions. The proximal end recruits and organizes PCM required for the centrosome to nucleate microtubes. In some vertebrate cell types, small aggregates of proteinaceous material termed centriolar satellites are also observed in the vicinity of centrosomes (6). Centriolar satellites traffic PCM components to the centrosome and act as assembly points for proteins required for cilia assembly (7). The distal end of a fully mature centriole carries nine distal appendages and a variable number of sub-distal appendages. Distal appendages are required for docking of centrioles at the plasma membrane during the process of ciliogenesis (8), while sub-distal appendages are involved in anchoring microtubules in interphase centrosomes and contribute to centriole cohesion (9). The cell cycle controls assembly of sub-distal appendages, as they are lost from centrioles during mitosis and reassembled in the following G1; the precise timing of distal appendage assembly is less well understood (Figure 2).
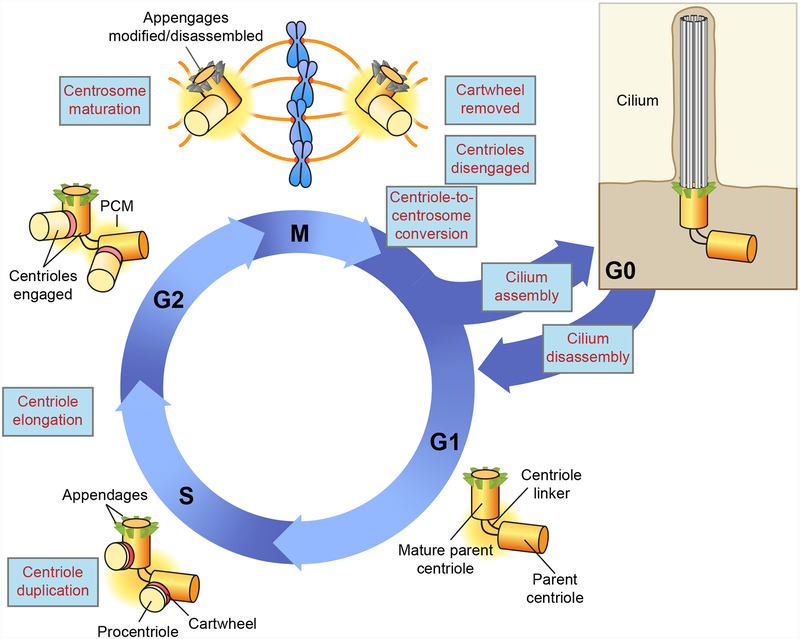
Figure 2. Regulation of centriole and cilium biogenesis during the cell cycle.
G1 cells contain two parent centrioles connected at their base by a flexible linker. At the beginning of S phase, each parent centriole assembles one new procentriole aligned orthogonal to its proximal end. This arrangement is termed ‘engagement’ and acts to prevent the reduplication of the parent centriole. The procentrioles elongate as cells progress through the cycle and in late G2, the flexible linker that holds the two parent centrioles together is dissolved to permit centrosome separation. In preparation for mitotic spindle formation, centrosome maturation occurs resulting in PCM expansion. During mitosis the cartwheel is removed from the lumen of the procentriole. At the end of mitosis, the centriole pair disengages and loses its orthogonal arrangement. This step is required to relicense the parent centriole for duplication in the next cell cycle. At the same time the procentriole is converted into a parent centriole. This ‘centriole-to-centrosome’ conversion allows the procentriole to recruit PCM material and acquire competence for duplication. The distal and sub-distal appendages are transiently modified/disassembled in mitosis. In G1 appendages form on the mature parent centriole that was created one and half cell cycle earlier. In quiescent cells, the mature parent centriole can migrate to the plasma membrane and initiate the formation of the axoneme of a cilium. Cell cycle re-entry is accompanied by the disassembly of the cilium.
Centriole biogenesis
In cycling cells, centriole number is maintained through a duplication cycle that is tightly coordinated with cell cycle progression (10) (Figure 2). At the start of the cycle, cells contain two centrioles connected by a flexible linker at their base. The younger of the two centrioles was assembled in the previous cell cycle and will be referred to as a parent centriole, while the older centriole will be termed the mature parent centriole. Centriole duplication begins at the G1-S transition, when a new procentriole grows perpendicularly from a single site at the proximal end of each existing centriole. The procentriole remains engaged in this orthogonal orientation during S and G2 phases, during which time it elongates, reaching ~80% of the length of a parent centriole prior to mitotic entry. In late G2, the flexible linker that connects the proximal end of the two parent centrioles is lost, allowing them to separate and guide the formation of the mitotic spindle apparatus. During mitosis, the procentriole disengages from the parent centriole so that the two newly created daughter cells each inherit a pair of parent centrioles that are competent for duplication in the next cell cycle. Importantly, the parent centriole that was formed 1.5 cell cycles ago reaches its full length in G1 and acquires sub-distal and distal appendages that allow it to function as a basal body.
Building a new centriole
Pioneering work in C. elegans led to the identification of a highly conserved set of five core proteins that are required for the initiation of centriole duplication: PLK4 (ZYG-1 in C. elegans), CEP192 (SPD-2 in C. elegans and Spd-2 Drosophila), CPAP (also known as CENPJ, SAS-4 in C.elegans and Sas-4 in Drosophila), STIL (SAS-5 in C. elegans and Ana-2 in Drosophila) and SAS6 (3). A list of key genes and corresponding orthologs are shown in Table 1. The kinase PLK4 is the master regulator of centriole duplication and is the earliest known marker for the site of procentriole assembly (11–13). In vertebrates, PLK4 is recruited to parent centrioles in G1 by binding to the centriole receptors CEP152 (Asterless in Drosophila) and CEP192, which encircle the proximal end of the parent centriole (14–18). While PLK4’s centriole receptors are localized as a ring throughout the cell cycle, PLK4 transitions from an initial ring-like localization around the parent centriole in G1 to a single dot at the G1-S transition (13, 16, 19, 20). This transition requires binding of PLK4 to STIL (19), which activates PLK4 dimers by inducing trans auto-phosphorylation of the kinase’s activation loop (21–23). Active PLK4 then phosphorylates STIL in a conserved STAN domain to trigger binding and recruitment of SAS6. SAS6 in turn initiates the assembly of the cartwheel, which forms a structural foundation for the procentriole (19, 21, 24, 25). PLK4 also phosphorylates STIL at additional sites that are required for the loading of STIL at the site of procentriole assembly (20, 26).
Table 1:
Key gene names
| H. sapiens | D. melanogaster | C. elegans | Chlamydomonas | Loclization |
|---|---|---|---|---|
| PLK4 | Plk4 (SAK) | zyg-1 | Cartwheel | |
| SAS-6 | DSas-6 | sas-6 | BLD12 | Cartwheel |
| STIL | Ana2 | sas-5 | Cartwheel | |
| CPAP (CENPJ) | DSas-4 | sas-4 | Centriole | |
| CEP135 | DCep135 | BLD10 | Centriole | |
| CEP152 | Asl | Centriole | ||
| CEP192 | DSpd-2 | spd-2 | Centriole/PCM | |
| CEP215 (CDK5RAP2) | Cnn | spd-5 | PCM | |
| CEP295 | Ana1 | Centriole | ||
| PCNT | Plp | PCM |
The cartwheel is comprised of stack of a ring-like assemblies that occupy the proximal ~100 nm of the human procentriole (Figure 1B) (27). Each stack is comprised of a central circular hub, from which nine spokes emanate and connect to the A-tubule of the microtubule triplets in the centriole wall. Elegant in vitro reconstitution has shown that recombinant SAS6 forms a homodimer that can oligomerize into a ninefold symmetrical cartwheel structure in vitro (28–31) and this assembly is facilitated by binding to Bld10 (CEP135 in humans) (32). Growth of the cartwheel occurs through the addition of SAS6 molecules to the proximal end of the cartwheel stack, with the rate of growth set by PLK4 activity (33). While the cartwheel plays an important role in establishing the centriole’s nine-fold radial symmetry, cartwheel-independent mechanisms also contribute to symmetry, including potentially the structural constraint imposed by the A-C linker in the microtubule wall (34, 35). Furthermore, in some species the cartwheel is a stable part of the centriole structure, but in human cells the cartwheel is removed during early mitosis and absent from mature centrioles.
Once the cartwheel has assembled, CPAP, in collaboration with its binding partners CEP135, CEP120 and SPICE1, facilitates the formation and stabilization of the procentriole’s microtubule wall (36–41). Centriolar microtubules grow at a very slow rate and are exceptionally stable, in part because of extensive post-translational modification by acetylation and polyglutamylation (42–44). In addition to canonical α- and β-tubulin, the assembly of centriolar microtubules likely requires the centriole-specific tubulin isoforms δ- and ε-tubulin. δ- and ε-tubulin form a biochemical complex with two proteins required for centriole stability named TEDC1 and TEDC2 (45). Cells lacking δ- and ε-tubulin form unstable centrioles with singlet microtubules, suggesting that these tubulin isoforms may provide critical interactions required for forming or stabilizing triplet microtubules (5, 46).
Setting centriole length
While centriole length varies among species and cell types, centrioles from a given species or cell type achieve a remarkably reproducible length. In Drosophila and C. elegans, the cartwheel extends through the entire length of the centriole, and thus cartwheel height seems to control centriole length (33). In vertebrates, the centriolar microtubule triplets extend ~300nm beyond the height of the cartwheel, suggesting that other mechanisms must determine the final length of centriolar microtubules and thus that of the organelle. A central player in setting centriole length is CPAP, which associates with centriole microtubules and controls their growth and stability (47, 48). Overexpression of CPAP, or its interacting proteins CEP120 and SPICE1, leads to hyper-elongation of centriolar microtubules in mammalian cells (36–38, 40, 49). Other proteins such as POC1 (50), hPOC5 (51), Asterless (CEP152 in humans) (52) and CEP295 (Ana1 in Drosophila) (53, 54) have also been implicated in centriole length control. CP110 and CEP97 cap the distal end of the centriole and restrict centriolar microtubule growth in mammals (49, 55); as such, the removal of CP110 is necessary for formation of the ciliary axoneme (see below) (49, 56, 57).
Centriole copy number control
In proliferating cells, centrioles duplicate every cell cycle by forming one new procentriole adjacent to each existing parent centriole. Recent years have seen an explosion of interest in understanding how cells maintain centriole copy number through successive cell cycles. In the sections that follow, we discuss three conceptually distinct levels of control that are required to maintain centriole homeostasis.
Spatial control: Build locally
During canonical centriole biogenesis, centriole formation is spatially restricted to a site close to existing centrioles. This spatial control is dictated by the preferential recruitment of PLK4 to the wall of the parent centriole by its centriole receptors CEP152 and CEP192. PCM at the proximal end of the parent centriole also provides a favorable environment for centriole assembly (58). Centrioles thus catalyze their own assembly by recruiting and locally regulating key factors required for centriole duplication.
In addition to canonical centriole biogenesis, centrioles can also form in the absence of pre-existing centrioles in a process known as de novo centriole biogenesis. One example where this occurs is in mouse embryos, where cell divisions are initially acentriolar, and centrioles are created de novo at the 64 cell stage (59). Importantly, de novo centriole formation is suppressed by the presence of even a single pre-existing centriole, ensuring that canonical biogenesis takes precedence over de novo formation (60). In mammalian somatic cells, de novo formation of centrioles occurs if centrioles are experimentally depleted, but the process is error-prone, resulting in the generation of a variable number of centrioles that often have an abnormal geometry (34, 60–63). Thus, spatially restricting centriole duplication to the parent centrioles helps ensure the structural integrity and numerical control of procentriole formation.
Numerical control: Build only one
A central feature of centriole copy number control in cycling cells is that each parent centriole forms exactly one new procentriole. This regulation depends upon finely tuned levels of the centriole duplication proteins PLK4, STIL and SAS6. Overexpression of any of these three initiator proteins induces the simultaneous production of multiple procentrioles around one parent centriole (11, 64). Centriole duplication is particularly sensitive to alterations in the level of PLK4, and accordingly, PLK4 abundance is controlled by feedback regulation. PLK4 dimerizes through a cryptic polo box domain (17, 65, 66), and the dimeric kinase phosphorylates itself in trans within a phosphodegron (67–70). This creates a binding site for the SCFβ-TrCP ubiquitin ligase, which ubiquitinates and targets active PLK4 for proteasomal destruction, thus placing the stability of PLK4 under the control of its own activity (68).
The relocalization of PLK4 to a discrete locus on the wall of the parent centriole is thought to be critical for selecting a single site for procentriole assembly (13, 16, 19, 20). However, it remains unclear how PLK4 achieves this asymmetric localization and how the kinase escapes its own degradation when concentrated at this site. In one model, PLK4 is degraded en masse around the parent centriole but is stabilized at a single site through binding to its activator STIL (19). Since PLK4 can self-organize into supramolecular assemblies, it is possible that these assemblies protect PLK4 from proteolysis at the site of procentriole formation (71, 72).
Ultimately, the transition of PLK4 from an initially symmetric localization on the parent centriole to a discrete site is a symmetry-breaking reaction that bears striking similarity to that observed for budding yeast Cdc42 GTPase during bud site selection (73, 74). Mathematical modelling of symmetry breaking has shown that two cooperating feedback loops, or one non-linear feedback loop, are required to establish asymmetry from an initially symmetric state (75). A challenge therefore, for future will be to identify the feedback loops that control PLK4 localization and/or activity. Importantly, the symmetry breaking model assumes that the site of procentriole assembly is randomly selected on the parent centriole. However, it is worth noting that in algae and ciliates, where basal bodies are anchored and triplet microtubule blades can be readily distinguished, new basal body assembly occurs at a defined location on the parent (76–79). The molecular basis for this preferential assembly site remains unclear.
Temporal control: Build once per cell cycle
In addition to spatial and numerical control, centriole biogenesis must also be licensed to ensure that duplication initiates only once per cell cycle. This is achieved through a centrosome-intrinsic block to reduplication, where duplication of the parent centriole is prevented as long as the parent and procentriole remain tightly associated or “engaged” with each another (58, 80). The dissolution of this linkage following passage through mitosis is known as “disengagement” and licenses centrioles for a new round of duplication in the next cell cycle (Figure 2). The identity of the link connecting the parent and procentriole and the molecular mechanism by which centriole duplication is inhibited remains unclear. One intriguing model postulates that the cartwheel of the procentriole acts to prevent the reduplication of the parent (81), although it remains to be determined how the parent centriole would read out the proximity of the cartwheel complex in the procentriole.
Centriole disengagement requires the activity of the kinase PLK1 and the protease Separase (82, 83). While Separase is well known for cleaving the Cohesin complex to initiate sister chromatid separation at anaphase, it has also been shown to cleave the PCM protein PCNT during mitosis. PCNT cleavage is required to license centrioles for duplication in the subsequent cell cycle (84, 85). One possibility is that PCNT cleavage alters PCM structure, allowing the procentriole to separate from its parent centriole and recruit its own PCM material in G1. While Separase activation helps ensure the correct timing of centriole disengagement, PLK1 activity plays a more central role (83). High levels of active PLK1 are sufficient to promote centriole disengagement and reduplication in interphase without passage through mitosis (86, 87). One likely target of PLK1 is PCNT, with phosphorylation of PCNT by PLK1 facilitating its Separase-mediated cleavage and centriole separation (88). Additional PLK1 centriole targets required for centriole disengagement await identification.
Once disengagement has occurred, the parent centriole is competent to reduplicate in the next cell cycle. However, a disengaged procentriole needs to acquire the ability to recruit PCM before duplication can proceed, a process termed the ‘centriole-to-centrosome conversion’ (89). In Drosophila embryos, centriole-to-centrosome conversion requires phosphorylation of SAS4 by CDK1 in mitosis; this phosphorylation generates a binding site for Plk1 that in turn recruits CEP152/Asl to license the procentriole for duplication in the next cell cycle (90, 91). Centriole-to-centrosome conversion also requires a conserved set of scaffolding proteins including CEP295 (Ana1 in Drosophila) (89, 92). CEP295/Ana1 is required to stabilize the new centriole after cartwheel removal in mitosis and is responsible for recruiting factors required for centriole duplication and PCM assembly, such as CEP152 and CEP192 (93, 94). C. elegans lack a clear CEP295/Ana1 homologue, but the SAS-7 protein may play an analogous role by recruiting CEP192/SPD-2 and endowing new centrioles with competence for duplication (95).
A final aspect of temporal control is to ensure centriole duplication, like DNA replication, is coordinated with cell cycle progression. Both centriole duplication and DNA replication initiate at the G1/S transition and rely on the activation of cyclin-dependent kinases (CDKs) that drive cell cycle transitions. CDK2 is activated at the G1/S transition and is required for centriole duplication in both Xenopus (96, 97) and mammalian cells (98, 99). Nevertheless, CDK2 knockout cells have normal centriole numbers, likely because CDK1 is able to compensate for loss of CDK2 activity (100). While CDK2 activity promotes centriole duplication at the G1/S transition, CDK1 activity suppresses centriole duplication in mitosis by inhibiting the interaction of PLK4 with STIL (101). Several additional proteins with roles in DNA replication and chromosome segregation have also been proposed to play roles in centriole duplication, but indirect effects remain difficult to exclude.
Multiciliated cells: breaking the rules
Although cycling cells construct exactly two new centrioles per cell cycle, centriole number can be modified in specialized cell types. For example, multiciliated epithelial cells that coat the airways, ventricles and oviducts of vertebrates contain hundreds of motile cilia that drive extracellular fluid flow (102). In contrast to the strict control of centriole number observed in cycling cells, multiciliogenesis relies on the production of large numbers of centrioles that are converted into basal bodies and produce motile cilia. To achieve this feat, post-mitotic multiciliated cells use specialized structures called deuterosomes to rapidly amplify centriole content (103, 104). Deuterosomes are comprised of several proteins required for centriole duplication and can be nucleated by an existing centriole (105) or form spontaneously in the cytoplasm (106). Centrioles grow on the surface of deuterosomes until they reach their correct length, when they are released into the cytoplasm and dock at the plasma membrane. To promote the distinct stages of centriole biogenesis, multiciliated cells undergo biochemical changes that are similar to those that promote cell cycle transitions in proliferating cells but avoid DNA replication and commitment to mitosis (107, 108). Therefore, centriole amplification in multiciliated cells is controlled in a specialized cycle that bypasses the tight spatial and temporal controls on centriole biogenesis that operate in cycling cells.
Another example where centriole number is modulated is in the asymmetric inheritance of centrioles during fertilization. During sexual reproduction in most mammals, centrioles are eliminated from oocytes and contributed to the zygote by the sperm. The sperm-derived centriole pair then duplicates during zygotic S phase to provide the two centrosomes required for successful mitotic divisions. Centriole elimination in oocytes is thus critical to balance centriole number following fertilization. The elimination of centrioles in Drosophila oocytes is triggered by the downregulation of Polo kinase, which leads to PCM loss and subsequent centriole elimination (109, 110). Centrioles are also lost during muscle development, but in this case the mechanism and functional significance of centriole loss remain unclear (111, 112).
Building the centrosome
To form a centrosome, the parent centriole recruits a matrix of PCM comprised of several hundreds of proteins, including many that are required to nucleate or anchor microtubules (113, 114). Unlike many cellular organelles, the centrosome lacks a delimiting membrane, raising the question of how PCM assembly and size are controlled. PCM material was initially described by electron microscopy (EM) studies as an electron-dense and amorphous cloud. However, more recently, super-resolution imaging revealed that interphase PCM has an ordered structure with many proteins localizing to distinct toroidal layers that surround the proximal end of the parent centriole (115–119). In addition, two proteins, CEP152 and PCNT, form elongated filaments with one terminus located close to the centriole wall and the other extending into the PCM (117–119). Such filaments may act as a scaffold that organizes the interphase PCM. It is worth noting that in some terminally differentiated cell types such as muscle, neurons and epithelial cells, centrosomes no longer function as the dominant microtubule organizing centers, and centrioles organize very little PCM material.
Following mitotic entry, the interphase PCM rapidly increases in size to support the robust microtubule nucleation needed for mitotic spindle assembly. This PCM expansion, or centrosome maturation, is dependent on the activity of PLK1 (Polo in Drosophila). PLK1 phosphorylates multiple proteins including the PCM components PCNT, CDK5RAP2 (Cnn in Drosophila) and CEP192 (Spd-2 in Drosophila), which are thought to form an underlying mitotic PCM scaffold (120–124). Importantly, PCNT and CDK5RAP2 also directly bind and recruit γ-tubulin ring complexes (γTuRCs) that nucleate centrosome microtubules, while CEP192 recruits γTuRCs through the adapter NEDD1 (Figure 1A) (125–127).
In contrast to the ordered interphase PCM, the mitotic PCM appears to form a more disordered gel-like scaffold. Mitotic PCM assembly is best understood in Drosophila and C. elegans. In Drosophila embryos, Cnn is recruited to the parent centriole in a Spd-2-dependent manner, where it is phosphorylated by Polo to promote multimerization and scaffold assembly (115, 128). Phosphorylated Cnn fluxes outwards from the parent centriole along centrosomal microtubules (122, 129). The outward spread of Cnn separates it from the source of Polo/PLK1 activity at the parent centriole, favoring dephosphorylation and thereby limiting scaffold assembly. Whether the flux of PCM scaffolding proteins is a general pathway to control PCM size remains unclear: outward flux of Cnn is not observed in Drosophila somatic cells and in C. elegans, SPD-5 (130), the functional ortholog of Cnn, incorporates isotropically into the PCM (131).
Similar to Drosophila, the assembly of the mitotic PCM in C. elegans requires SPD-5 phosphorylation by PLK-1 (123). Like Cnn, SPD-5 can assemble into supramolecular assemblies in vitro that are enhanced by the presence of SPD-2 and PLK-1 (123). Macromolecular crowding agents drive recombinant SPD-5 to phase separate into spherical, liquid-like condensates that rapidly ‘harden’ into solid-like structures (132). The microtubule polymerase XMAP215 and the microtubule-stabilizing protein TPX2 can partition into SPD-5 condensates, where they concentrate tubulin and promote microtubule nucleation. These studies raise the question of whether the mitotic PCM forms through phase-separation of components into condensates with liquid-like properties, or alternatively, assembles from well-ordered protein-protein interactions that form a gel-like or solid phase. One possibility is the mitotic PCM starts as a liquid-like droplet around the parent centriole that then solidifies into a porous gel-like matrix (114).
Centrosomes in cell proliferation
In most mammalian cells, centrosomes nucleate the majority of the spindle microtubules during mitosis and increase the speed and efficiency of spindle assembly. However, additional microtubule-nucleation pathways also contribute to spindle formation and allow for cell division in the absence of centrosomes (133). Although centrosomes are not required for cell division per se, they are required for the continued proliferation of many mammalian cells. Cells lacking centrosomes activate a USP28-53BP1-P53 signaling axis that leads to either cell death or a cell cycle arrest (134–137). 53BP1 is a key regulator of DNA double-strand break repair that binds P53, while USP28 is a deubiquitinase that interacts with 53BP1. While 53BP1 and USP28 have both been reported to play roles in DNA damage signaling, multiple lines of evidence have shown that growth arrest in response to centrosome loss is mechanistically distinct from the DNA damage response (138). How centrosome loss is sensed by USP28 and 53BP1 remains to be determined. Since USP28, 53BP1 and P53 are also required to arrest the cell cycle following a prolonged mitosis, one attractive possibility is that centrosome loss indirectly activates a USP28-53BP1-P53-mediated ‘mitotic surveillance pathway’ by delaying mitosis (134–136).
There are likely to be tissue and organism specific differences in the function of the mitotic surveillance pathway. For instance, the mitotic surveillance pathway must be inactive in early mouse embryos, which proliferate in the absence of centrosomes until the 64-cell stage. Additionally, the mitotic surveillance is not present in flies, where centrosomes are only required for the rapid divisions of the syncytial embryo but are dispensable for cell divisions thereafter (139, 140). Future work will be required to understand how the mitotic surveillance pathway is triggered and to establish its role in normal physiology and disease.
Centrosome defects in disease
Given the central role of centrosomes in diverse cellular processes, it is unsurprising that centrosome dysfunction has been linked to several human diseases. A wealth of data has shown that centrosome aberrations are commonly observed in human tumors and are often correlated with clinical aggressiveness (10, 141). Centrosome defects in tumors take the form of either numerical or structural alterations. Numerical alterations reflect increases in the number of centrosomes (known as centrosome amplification), while structural alterations encompass alterations in the shape and size of centrosomes. While conceptually distinct, numerical and structural alterations in centrosomes frequently co-exist in human cancers. A causal link between centrosome amplification and cancer recently emerged with the demonstration that extra centrosomes can trigger and/or accelerate tumorigenesis in mice (142–144). Exactly how centrosome aberrations contribute to tumorigenesis remains to be clarified. Supernumerary centrosomes can promote genomic instability by increasing the rates of chromosome missegregation and micronucleus formation (145–148), and consistently, the tumors that form in mice with extra centrosomes show dramatically altered karyotypes (143). In addition, the presence of supernumerary centrosomes can alter the interphase microtubule cytoskeleton to increase tumor cell migration and invasion (149). Similarly, structural defects in centrosomes have also been shown to increase the extrusion of individual mitotic cells from an epithelial layer, possibly providing a route for metastasis (150). Importantly, cellular extrusion is a non-cell-autonomous process that relies on the cooperation of cells within the epithelium. Thus, centrosome aberrations could contribute to metastasis without the disseminating cells themselves harboring centrosomal alterations.
In addition to a role in tumorigenesis, there are also clear links between congenital centrosome defects and developmental disorders. Primary autosomal microcephaly (MCPH) is a rare condition in which individuals are born with a brain that is considerably smaller than normal (151). MCPH is caused by a depletion of the neural progenitor cell (NPC) pool during embryonic development, resulting in the production of fewer mature neurons. Surprisingly, more than half of the known MCPH genes encode proteins that localize to the centrosome and play important roles in centriole biogenesis. It remains to be understood why mutations in ubiquitously expressed centrosome proteins specifically impair brain development. One intriguing possibility is that centrosome defects delay mitosis and lead to pathological activation of the mitotic surveillance pathway in NPCs (10). Indeed, an increase in the length of NPC mitosis has been observed in several mouse models of centrosome-associated microcephaly, and P53 deletion is able to rescue cell death and reduced brain size in these mice (152–154). In this model, the tissue specificity could be explained if NPCs have a lower threshold for activation of the mitotic surveillance pathway compared with other cell types (155). Understanding how cells measure mitotic duration and what sets the sensitivity of this response are important questions for future exploration.
The cilium: a centriole-dependent organelle
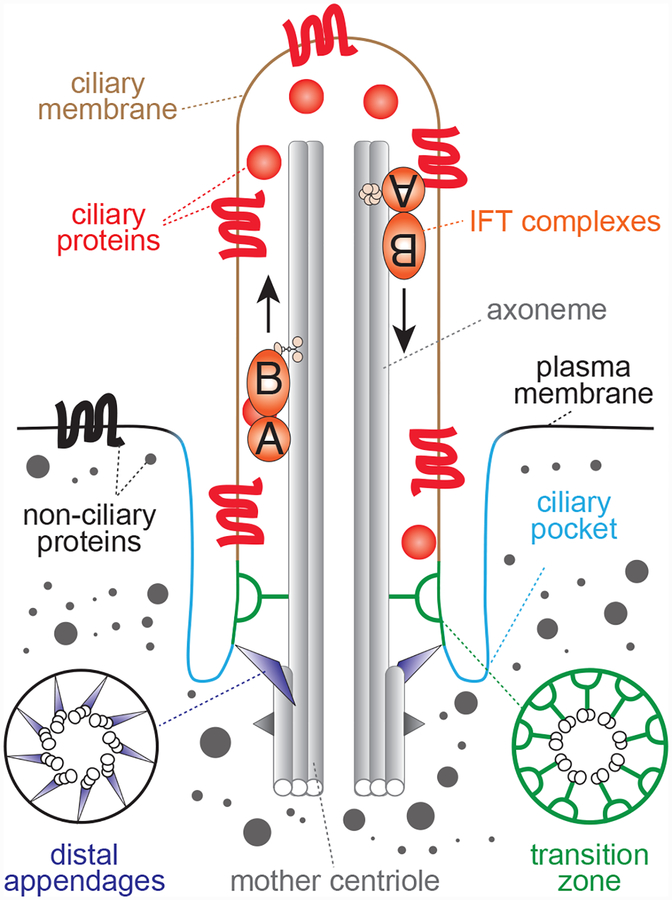
In most mammalian cells, the mature parent centriole templates the formation of a cilium that protrudes from the cell surface. Cilia are typically ~350 nm in diameter and 1–10 μm in length, and like centrosomes, lack a delimiting membrane. As a result, the ciliary lumen is continuous with the cytosol, and the ciliary membrane is likewise continuous with the plasma membrane (Figure 3). Nonetheless, cilia maintain a unique complement of biomolecules through the combined action of dedicated trafficking machineries and diffusional barriers at the cilium base (156–158). A region of particular importance for compartmentalization of the cilium is the transition zone, a proximal domain of the cilium where linkers tether the axonemal microtubules to the surrounding ciliary membrane (157, 159) (Figure 3). Additionally, the cilium often lies within a ‘ciliary pocket’ formed by invagination of the plasma membrane adjacent to the ciliary membrane (160).
Figure 3. Primary cilium structure.
Architecture of a mammalian primary cilium, highlighting key structural features. The axonemal microtubules form the core of the cilium and extend from the mature parent centriole, which is docked at the plasma membrane. This docking is mediated by the mature centriole’s distal appendages and often occurs at a site on the cell surface where the plasma membrane is invaginated. This invaginated region of the plasma membrane adjacent to the cilium is known as the ciliary pocket is a key site for exo/endocytosis of ciliary materials. Although the cilium lacks a delimiting membrane, it contains a distinct complement of soluble and membrane proteins. This compartmentalization is enabled by diffusion barriers near the base of the cilium at a region known as the transition zone (TZ). The transition zone is made up of several functional and physical modules, including MKS and NPHP proteins, which are mutated in Meckel Syndrome and Nephronophthisis, respectively. Selective trafficking of proteins to cilia across the transition zone is mediated by trafficking machineries, such as IFT-A and IFT-B, that cooperate with ciliary kinesin and dynein motors. Additionally, IFT-A and IFT-B mediate protein transport within cilia along the axonemal microtubules and are required for ciliogenesis.
Cilia were likely present in the last eukaryotic common ancestor and are found today in a diverse array of organisms ranging from single-celled protists to vertebrates (161). Many of the structural features of cilia and the genes required for their function are highly conserved. Despite these commonalities, cilia in different cell types and organisms exhibit considerable diversity in their axonemal structure, length, morphology, and function. We will focus here on non-motile primary cilia, as they are widespread in vertebrates and exhibit many of the essential features of cilia; for those interested in the specialized features and functions of motile cilia, we refer the reader to recent review articles (102, 162, 163).
The assembly of primary cilia is a tightly regulated, multi-step process that is strictly dependent on the mature parent centriole. The nine doublet microtubules of the ciliary axoneme are formed through elongation of the A and B tubules of the parent centriole (Figure 3). Additionally, the centriolar distal appendages form the interface that connects the centriole to the nascent ciliary membrane and anchor it to the cell surface when a mature cilium has formed (157, 159). These essential roles of the parent centriole have several critical consequences. First, because each cell has only one mature parent centriole, cells are limited to generating a single cilium (except in specialized cases, e.g. multiciliated cells). Second, the position within the cell of the mature parent centriole and the base of the cilium are by necessity coupled. Finally, centrioles must live dual lives, acting both as basal bodies that anchor primary cilia and as key components of centrosomal microtubule organizing centers. A dichotomy between these functions is evident in the regulation of cilia: across many species, the cilium must be disassembled prior to mitosis so that the mature centriole can help organize the mitotic spindle (164–166). Indeed, mammalian primary cilia are predominantly found on cells in the G0 or G1 phases of the cell cycle, and ciliogenesis is commonly initiated for cells in culture by withdrawal of serum growth factors (Figure 2). Thus, the cell cycle and coordinately regulated events in centriole duplication and maturation are intimately linked to the cilium assembly and disassembly programs.
Pathways for initiating ciliogenesis
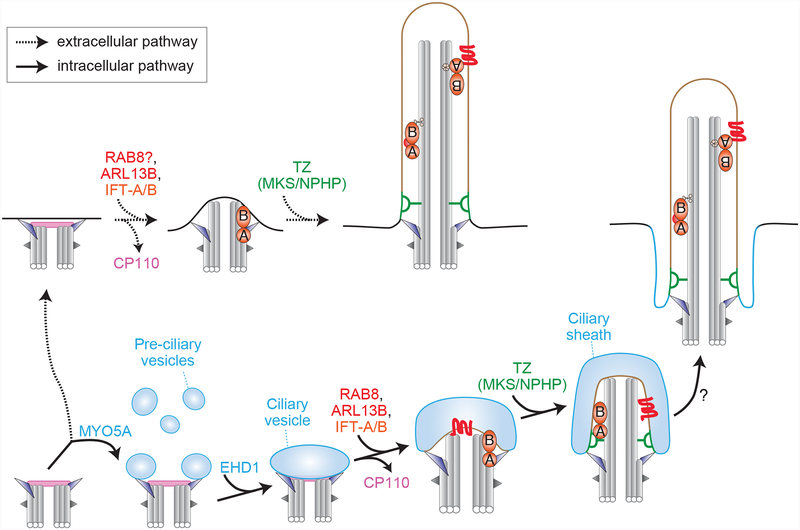
Foundational studies on ciliogenesis have revealed two principal pathways for cilia assembly in vertebrate cells (167, 168). One, a so-called ‘extracellular’ pathway, is characterized by the migration and docking of the mature parent centriole to the plasma membrane via the centriolar distal appendages (Figure 4). After centriole docking, the axonemal microtubules extend, the transition zone forms, and ciliary trafficking machineries such as intraflagellar transport (IFT) complexes A and B deliver material to the growing cilium. The second, ‘intracellular’ pathway shares many commonalities with the extracellular pathway but instead begins with the recruitment, docking and fusion of vesicles at the distal appendages of the mature parent centriole (Figure 4). The resulting ciliary vesicle is then deformed as the axonemal microtubules extend and the transition zone forms, giving rise to a nascent ciliary structure that is entirely inside the cell. Finally, fusion of the ciliary vesicle with the plasma membrane leads to external exposure of a mature primary cilium. In this pathway, the outer region of the ciliary vesicle gives rise to the ciliary pocket adjacent to the ciliary membrane.
Figure 4. Pathways for primary cilium assembly.
The mature parent centriole (bottom left) serves as the foundation for primary cilium assembly via either an ‘extracellular’ pathway (top, dashed arrows) or an intracellular pathway (bottom, solid arrows). In the intracellular pathway, key steps include i) MYO5A-dependent recruitment of pre-ciliary vesicles to the distal appendages, ii) EHD1-mediated fusion of these vesicles to form an enlarged ciliary vesicle, iii) the growth of the ciliary vesicle via the joint action of RAB8, ARL13B, and the IFT complexes, a process that occurs in conjunction with removal of the CP110 cap from the distal end of the fully mature centriole, iv) the growth of the axoneme, formation of the transition zone, and maturation of the ciliary vesicle into distinct domains corresponding to the ciliary sheath and the nascent ciliary membrane, and v) the fusion of the ciliary sheath with the ciliary membrane, which exposes the cilium to the external environment. In the extracellular pathway, a key distinction is that the mature parent centriole initially migrates to the cell surface and docks to the plasma membrane via its distal appendages. Subsequent steps appear to occur in a similar fashion as the intracellular pathway, although the precise sequence of events and molecular requirements are not fully knowns.
Recent studies have provided an increasingly detailed view of the sequence of events and proteins needed for ciliogenesis. In the intracellular pathway, cilium formation begins with the trafficking and capture of vesicles at the mature parent centriole. Vesicles are first transported to the centriole through the sequential action of dynein and Myosin-5a (169). These ‘pre-ciliary’ vesicles associate with the distal appendages and then fuse to form a larger ciliary vesicle in a manner that depends on the EHD family of membrane-tubulating proteins (169, 170). MYO5A and EHDs are among the first factors to be recruited to the mature parent centriole during ciliogenesis and localize to the sub-domain of the growing ciliary vesicle that gives rise to the ciliary pocket. Soon after MYO5A and EHDs are recruited, several other proteins that are enriched in the ciliary membrane can be detected at the ciliary vesicle, including the small GTPase ARL13B, and components of the RAB8-RAB11 GTPase cascade (169, 170). In this cascade, RAB11 recruits its effector RABIN8, which then serves as the guanine nucleotide exchange factor (GEF) that activates RAB8 (171). RAB8 and ARL13B in turn promote the growth of the ciliary membrane and the selective trafficking of ciliary proteins to the cilium (172). Following growth of the nascent cilium, the ciliary vesicle fuses with the plasma membrane to give rise to a surface-exposed cilium. At present, the proteins required for this fusion event are not known.
These initial events differ significantly in the extracellular ciliogenesis pathway. In this case, the mature parent centriole does not capture vesicles in the cytoplasm but instead migrates to the cell surface and docks to the plasma membrane. This migration is oriented toward the apical side of polarized epithelial cells and is promoted by the distal appendage protein CEP164 and by the microtubule and actin cytoskeletons (173, 174). In particular, actin is cleared from the region of the apical membrane where the centriole docks (175), perhaps explaining why actin inhibitors can promote ciliogenesis (176). After plasma membrane docking, the axoneme extends and the transition zone forms (see below for details). These latter processes appear to occur in a similar fashion for the intracellular and extracellular pathways.
What determines whether specific cell types utilize the intracellular or extracellular pathway for ciliogenesis, and does the choice of pathway dictate different molecular requirements for ciliogenesis or different functional properties for the mature cilia? The answers to these questions are not well understood, but the mode of ciliogenesis appears to be a characteristic feature of particular cell and tissue types. For example, fibroblasts and Retinal Pigment Epithelium (RPE1) cells predominantly use the intracellular pathway, while polarized epithelial cells typically use the extracellular pathway (167, 168, 177). Moreover, because the process of ciliogenesis is often linked to the ultimate position of the cilium, there may well be functional implications for the mature cilia. For example, in epithelial cells that use the extracellular pathway, the cilium is typically positioned apically, with almost the entire length of the cilium protruding from the cell (168). These cilia are therefore ideally positioned to sense extracellular fluid flow. By contrast, cells that use the intracellular ciliogenesis pathway typically maintain the basal body near the nucleus and Golgi, deep within the cell (9, 167, 177). These ‘submerged’ cilia are often associated with a pronounced ciliary pocket and may barely protrude into the extracellular environment, making them poor sensors of fluid flow (160). A recent study tested this idea, finding that conversion of ‘submerged’ cilia into ‘surfaced’ cilia promotes flow sensing but dysregulates ciliary Hedgehog signaling (9). It will be interesting to further examine how cilium positioning is regulated by factors including cell shape and contractility (173) and how this feature of cilia influences their functional properties.
A second area for future study is how the composition of the ciliary vesicle is specified during intracellular ciliogenesis. It is noteworthy that RAB8, ARL13B, and SMO-RFP are present in the ciliary vesicle soon after its formation (169, 170). The rapid enrichment of cilium-specific proteins in the ciliary vesicle highlights the need to identify the origin of the vesicles that give rise to the ciliary membrane and how their cargos are specified. Further work will also be required to understand how the association of proteins with the ciliary membrane is dynamically regulated during ciliogenesis. For example, while some early ciliary markers such as ARL13B remain at the ciliary membrane through the completion of ciliogenesis, others such as RAB8 and MYO5A are typically absent from mature cilia (169, 178). An additional feature of RAB8 and MYO5A that warrants further study is the discrepancy between the phenotypes associated with their disruption in vitro versus in vivo: knockdown or knockout of these genes in cultured cells blocks ciliogenesis at an early stage (169, 170), while mouse mutants do not exhibit overt ciliary defects (179, 180).
Cilium growth and maintenance
After the initial membrane association of the mature parent centriole, the axoneme elongates, the ciliary membrane grows, and the transition zone forms. These events begin with the removal of CP110 from the distal end of the mature parent centriole, which allows the centriolar microtubules to extend and form the axoneme (55). CP110 removal is driven by TTBK2, a kinase which binds to distal appendage protein CEP164 and phosphorylates substrates including CEP164 and the kinesin KIF2A (181–183). At a similar time as CP110 removal, the IFT machinery is recruited to the distal appendages and mediates trafficking of ciliary axonemal precursors such as tubulin in conjunction with the ciliary motors kinesin-II and dynein-2 (170). IFT proteins are organized into two large protein complexes, IFT-A and IFT-B, that have conserved roles in ciliary trafficking and are universally required for cilium assembly (Figure 3) (158, 184). IFT-B is thought to primarily associate with kinesin and mediate anterograde (base-to-tip) movement along axonemal microtubules, while IFT-A may primarily associate with dynein and mediates retrograde (tip-to-base) movement. However, recent studies have revealed additional complexities in ciliary trafficking, with IFT-A also promoting ciliary entry of some membrane proteins (172, 185). Additional information on how ciliary trafficking is mediated by IFT-A, IFT-B, and a protein complex known as the BBSome is provided in recent reviews (158, 172, 184, 186).
Shortly after IFT complex recruitment to the mature parent centriole, the transition zone that partitions the cilium from the cell body begins to form. The transition zone contains Y-shaped linkers that tether the axonemal microtubules to the ciliary membrane. The outer face of the ciliary membrane also exhibits a periodic series of particles termed the ciliary necklace (187). While the precise protein components that correspond to these structures is unknown, many proteins localize to the transition zone and are organized into physical and functional modules (159, 188, 189). Together these components are required to form a barrier that limits diffusional exchange of proteins between the cilium and the cell body, and transition zone defects impair ciliogenesis (190, 191).
After these early events in ciliogenesis, the cilium grows to a steady-state length and is maintained by the ongoing trafficking of components to and from cilia. How the length of cilia is determined after ciliogenesis and homeostatically maintained in mature cilia is an area of ongoing investigation (110, 192, 193). One area of the cilium that is likely to harbor structural elements and regulatory factors that control cilium length is the distal tip of the cilium. In particular, delivery of IFT cargos, incorporation/turnover of axonemal building blocks, and ectocytosis from the ciliary membrane are all processes occurring at the cilium tip that are likely to influence cilium length (194–198). A dynamic balance of these events is likely required for cilium homeostasis.
Mechanisms of cilium disassembly
It has long been recognized that cilia are disassembled when cells progress through the cell cycle or upon differentiation of certain cell types. A number of different models have been proposed for how this disassembly is achieved including, excision of all or some of the protruding cilium, retraction of the axoneme into the cell body, and progressive shortening of the cilium followed either by un-docking of the basal body from the plasma membrane or by endocytosis-like resorption of the final ciliary remnant. Critically, each of these models implies a different sequence of events and associated set of enzymatic activities needed for disassembly. Work in different experimental systems has generated support for several of these disparate models. For example, Chlamydomonas reinhardtii cilia appear to undergo excision at the base, promoted in part by the microtubule severing activity of katanin (166, 199), whereas chytrid fungi retract the axoneme into the cell through a ‘reeling’-type mechanism while discarding the ciliary membrane (T. Stearns, personal communication). In mammalian cells, different modes of disassembly have been observed in different experimental systems. In cultured mouse cells, a ‘decapitation’ or excision event near the tip may initiate cilium shortening and disassembly (197). During chick neurogenesis, a similar disassembly process has been observed in which the apical, cilium-containing portion of the cell undergoes actomyosin-dependent abscission. However, in this case cilium shortening and basal body dissociation from the membrane precede abscission (200). In another variation on these events, others have observed progressive shortening of cilia followed by endocytosis of the ciliary membrane remnant (201, 202). Notably, this membrane remnant can remain associated with the mature parent centriole throughout mitosis, and the daughter cell inheriting this centriole is able to more rapidly reassemble a signaling-competent cilium after mitosis (201). Thus, by controlling the timing of ciliary signaling, the mechanism of cilium disassembly may contribute to asymmetric cell fates after cell division.
Mirroring the variety of pathways for cilium disassembly, a number of distinct disassembly factors have also been identified. These can be divided into proteins that serve as mediators of disassembly (discussed here) versus proteins that regulate initiation of disassembly or that suppress aberrant cilium assembly (discussed below). Factors that directly participate in cilium disassembly include microtubule-modifying enzymes such as katanin, depolymerizing kinesins, and the HDAC6 tubulin deacetylase (56, 166, 203–206). These proteins likely contribute to axoneme disassembly, while regulators of the actin cytoskeleton such as phosphoinositide lipids, CDC42 and myosin may promote scission of the ciliary membrane (197, 200, 202, 207). These actin-associated proteins likely work in conjunction with clathrin, dynamin and RAB5 to promote endocytosis of disassembling cilia (202). Given that dynamic remodeling of the cilium is needed for its disassembly, it is not surprising that ubiquitin-proteasome system components and ciliary trafficking regulators also participate in cilium disassembly (208, 209). Specifically, the IFT complexes and dynein regulators DYNLT1, NDE1 and NDEL1 have been shown to promote cilium disassembly (210–212). Lastly, centrosomal proteins such as TCHP, an Aurora A kinase regulator, and CPAP promote cilium disassembly by poorly defined means (213, 214).
Regulation of cilium assembly and disassembly
Due to the importance of ciliary signaling and the dual role of centrioles in the formation of basal bodies and centrosomes, the assembly and disassembly of cilia are tightly regulated processes. Longstanding observations that cilia are disassembled before mitosis and re-assembled after mitotic exit or upon mitogen deprivation indicate that the cell cycle is a central regulator (Figure 2). But how do specific events in the cell cycle control the activity of cilium assembly/disassembly factors? And conversely, how does cilium assembly/disassembly regulate cell cycle progression? These are key questions for future research, but it appears that mitogens (e.g. EGF and PDGF) both suppress ciliogenesis and activate disassembly (164, 215–217). These signals appear to converge on kinases including PLK1, NEK2, and Aurora A that stimulate depolymerizing kinesins, HDAC6, and other effectors (203, 206, 218, 219). Notably, these pathways appear to have conserved roles in cilium disassembly, with C. reinhardtii orthologs of AURKA and NEK2 also regulating cilium disassembly (220, 221). Furthermore, in both C. reinhardtii and mammalian cells, inhibition of cilium disassembly leads to a block in cell cycle progression. This block is specifically due to cilium maintenance, as it can be bypassed by disrupting ciliogenesis genes (166, 206, 210, 211, 213). Elucidating how cilium disassembly exerts this checkpoint-like regulation of the cell cycle will be a key area for future study, as the mechanism appears to be distinct from that of other cell cycle checkpoints (210, 211, 213).
One promising strategy to understand regulation of cilium assembly/disassembly is to examine ciliated versus non-ciliated tissues and cell types. For example, while most cells in the body are ciliated, it has long been recognized that some cell types, such as cells of the immune lineage, lack cilia. Recently, Bangs et al. examined cilia during mouse embryonic development and found that all epiblast cells are ciliated at E8.0 (except cells in mitosis) (222). In contrast, the extra-embryonic cells of the visceral endoderm and trophectoderm lack cilia, in part due to activity of the Aurora A and HDAC6 disassembly factors. Given that the ciliated epiblast cells give rise to all cell types found in the adult, certain non-ciliated cell lineages must selectively lose the ability to ciliate during development, although the underlying mechanisms are not known. Interestingly, in the case of non-ciliated immune cells, a latent capacity for cilium assembly is suggested by the ability of some cell lines derived from B cells and T cells to form cilia at low rates in culture (223). Moreover, primary T cells can successfully carry out some key initial steps in ciliogenesis. For example, in activated T cells the fully mature centriole migrates to the plasma membrane and undergoes CEP164-dependent docking at the immune synapse, although an axoneme is not extended (224, 225). Parallels between the immune synapse and cilium are further supported by findings that ciliary proteins such as Unc119b, Arl13b, and IFT complexes localize to the immune synapse and modulate T cell signaling (226–228). Unraveling these similarities and differences in detail will be an exciting area for further research.
Cilia in physiology: signaling, ciliopathies, and cancer
The importance of understanding cilium assembly and disassembly is underscored by the vital roles of cilia in signaling (for a detailed discussion of how cilia enable signaling see the following (229–234)). It is clear that cilia contribute to a wide range of signaling processes that control embryonic development, tissue homeostasis and sensory signaling. Specifically, cilia have been shown to be essential for Hedgehog (Hh) pathway signaling, left-right symmetry breaking, phototransduction, and olfaction; there is also mounting evidence that cilia modulate the PDGF, mTor, Notch, TGF-β, and Wnt pathways. Similarly, a host of signaling receptors and effectors localize to cilia, including components of the Hh pathway, PDGF receptor alpha, energy-sensing kinases LKB1 and AMPK, multiple adenylyl cyclase isoforms, the polycystin-2 ion channel, and GPCRs such as SSTR3, D1R, 5HT6, MC4R, olfactory receptors, and rhodopsin (229, 231–236). This role for cilia in signaling is widely conserved, with many examples of cilium-dependent signaling seen in diverse organisms.
Understanding how cilia regulate signaling remains a central challenge in the field. Current obstacles include the fact that the functional outputs of some putative ciliary signaling pathways are not well characterized, and in other cases, cilia appear to modulate but not be strictly required for signaling to occur. However, even in the case of vertebrate Hh signaling, in which cilia are absolutely required for transcriptional output and all core pathway components localize to cilia, the precise role of cilia is not yet known (229). A particularly perplexing feature is that Drosophila Hh signaling does not depend on cilia, thereby indicating that the same basic set of signaling components can require cilia for signal transduction in one species but not in another (237). How then do cilia enable signaling? One possibility is that cilia promote signaling by confining signaling components in a small compartment that may have unique features with respect to second messenger content, membrane lipid composition, and ratio of surface area to volume (230, 234, 238, 239). Alternatively, some ciliary components such as the IFT machinery may directly participate in signal transduction (240).
The many contexts in which cilia promote signaling are illustrated by a group of pediatric disorders caused by inherited ciliary defects. These diseases are collectively known as ciliopathies and include Joubert Syndrome, Bardet-Biedl Syndrome, Meckel-Gruber Syndrome, Short Rib Thoracic Dysplasia, Polycystic Kidney Disease, Retinitis Pigmentosa, and Nephronophthisis (190, 191). In brief, ciliopathies are characterized by intellectual disability, retinal degeneration, anosmia, kidney cysts, skeletal and craniofacial malformations, obesity, and congenital heart defects (191, 241). Ciliopathy gene products include ciliary motors and trafficking complexes, transition zone components, and a host of other proteins needed for the assembly and function of cilia (190, 191). The many characteristic symptoms of ciliopathies reflect the diverse tissues and signaling pathways regulated by cilia. In some cases, particular symptoms can be ascribed to specific signaling pathways, such as polydactyly and Hh signaling and retinal degeneration and phototransduction by rhodopsin (229, 242). However, the molecular basis of other symptoms awaits further characterization. Additionally, while the developmental roles of ciliary signaling are highlighted by ciliopathies, cilia also play important but incompletely characterized roles in adult tissue homeostasis (243).
In addition to roles in development, several lines of evidence have linked cilia to cancer. First, aberrant ciliary signaling, for example in the Hh pathway, can drive tumorigenesis in basal cell carcinoma and medulloblastoma (244–246). Second, the finding that cilium disassembly regulates cell cycle progression suggests that dysregulated cilium assembly/disassembly may contribute to uncontrolled cell growth in cancer. In particular, loss of cilia may bypass a brake on the cell cycle and promote tumorigenesis. Consistent with this possibility, many tumors lack cilia despite arising from ciliated tissues (247–251). Moreover, a recent study observed progressive loss of cilia as breast cancer cells became more aggressive, and restoring ciliogenesis to these cells by inhibiting a depolymerizing kinesin reduced tumor cell proliferation (206). Further study of cilium loss during tumorigenesis may therefore provide opportunities not only to understand how cilium biogenesis is regulated but also to evaluate the potential therapeutic benefit of targeting pathways controlling cilium assembly or disassembly.
New tools for studying centrosomes and cilia
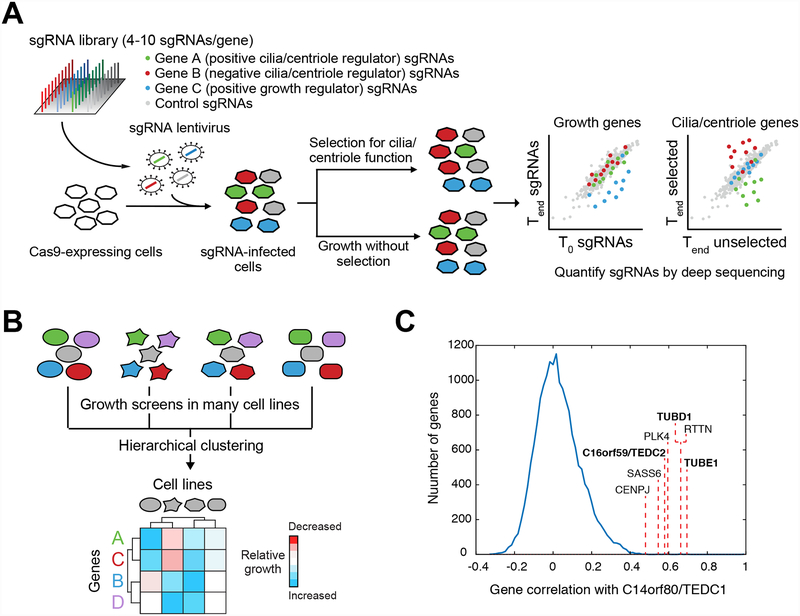
Several new technologies have recently emerged as powerful tools to study cilia and centrioles, led by prominent developments in the areas of functional genomics and proteomics. For example, the recent development of high-throughput screening using CRISPR-based gene disruption has made it possible to conduct genome-wide screens with unprecedented precision and sensitivity. A key success of initial CRISPR-based screens was identification of genes that affect growth of cultured cells (252, 253). To do so, a pool of single guide RNAs (sgRNAs) is introduced in bulk into a large number of cells (Figure 5A). After a defined period of growth, essential genes are identified by the depletion of sgRNAs targeting these genes from the pool. Similarly, by applying a specific stress or perturbation, genes can be identified that participate in a biological process of interest. However, an initial challenge in applying these approaches to study cilia and centrioles was the need to identify conditions in which ciliary or centriolar functions specifically modulate growth (or otherwise confer an isolatable phenotype suitable for pooled screening). In the case of centriole biology, a key breakthrough was the elucidation of the mitotic surveillance pathway that stops proliferation in centriole-deficient cells (134–136). Here functional screening for cells that escape growth arrest following centriole loss provided insight into a cellular process that had previously been poorly characterized.
Figure 5. Application of functional screening to study cilia and centrioles.
A) Overview of pooled functional screening using CRISPR/Cas9. A pool of sgRNAs is introduced into Cas9-expressing cells by lentiviral transduction. Transduced cells can then be grown under conditions that select for a functional property of centrioles or cilia or in the absence of such selection (note that the cells in question may need to engineered such that centrioles/cilia control a selectable phenotype). Deep sequencing is then used to analyze the composition of sgRNAs present at the outset of the experiment (T0 – e.g. the sgRNA library used to make lentiviral particles), in the unselected pool at the end of the experiment (Tend unselected), and in the selected pool at the end of the experiments (Tend selected). If sgRNAs targeting a particular gene are consistently depleted (or enriched) in the final selected sample relative to the final unselected sample, then the gene in question regulates centriole or cilium function. Similarly, changes in sgRNA abundance between the T0 sample and the final unselected sample reveal genes that affect cell growth. B) Schematic illustration of using growth phenotype screens conducted in different cell lines (indicated by cells of different shape) to identify genes with shared function. Hierarchical clustering of growth phenotypes across all cell lines can be used to group and identify genes having a shared function. C) Several centriolar genes, including members of the TED complex (bold labels), exhibit highly correlated patterns of growth phenotypes to that of C14orf80/TEDC1 across 436 cell lines in the Achilles dataset (Avana public 18Q2). The growth phenotypes for knockout of C14orf80/TEDC1 were compared to those for all other genes in the dataset, yielding the plotted distribution of correlation coefficients. Correlation values between TEDC1 and other TED complex components are indicated.
To study ciliary signaling, a mouse fibroblast cell line was engineered in which cilium-dependent Hh signaling drives expression of a reporter gene that confers resistance to the antibiotic blasticidin (45). In this fashion, genes were identified that affect ciliary Hh signaling through their modulation of blasticidin resistance. Known ciliary proteins and ciliopathy genes were identified with high precision and sensitivity, while previously uncharacterized hits revealed new insights into cilia and ciliary disorders. Moreover, because the NIH-3T3 cell line used is deficient in the mitotic surveillance pathway, several centriolar genes were among the hits, as expected given the essential role of the basal body in ciliogenesis. Importantly, a similar screen used a GFP-based Hh reporter and fluorescence-activated cell sorting to isolate hits, illustrating the variety of means by which screens can be tailored to investigate specific pathways or processes (254). Furthermore, by conducting screens under different conditions, it was possible to shift the focus of hit genes identified to specific functional categories, such as positive versus negative regulators of Hh signaling (254) or genes acting at a particular step in Hh signal transduction (45).
In contrast to these targeted screens, it is also possible to systematically probe diverse cellular processes through untargeted, growth-based screens. In particular, because such screens have now been carried out in over 400 cell lines that encompass diverse genetic mutations and epigenetic states (255), it is commonly observed that a given gene’s inactivation has variable effects on growth across cell lines (256–258). These context-dependent phenotypes are tightly linked to the gene’s molecular function and thus can be viewed as a gene-specific functional signature. Systematic comparison of these signatures reveals genes with shared functions and can thus be used to define the functions of uncharacterized genes (45, 256–258) (Figure 5B). For example, in the Achilles Project collection of over 400 CRISPR growth screens (255), many genes required for centriole duplication, such as PLK4, SASS6, STIL, RTTN, and CENPJ, exhibit highly correlated patterns of growth phenotypes. Unsupervised hierarchical clustering of this data revealed several clusters of functionally related centriole genes, including CEP97 and CCP110; CEP120, CEP44, HYLS1 and POC5; and CEP135 and SPICE1 ((45) and our unpublished observations). Indeed, nearly all centriolar genes can be identified by analyzing CRISPR-based screens that were not focused on centriole biology, and functionally relevant sub-groups can be defined. Further illustrating the value of this data, two uncharacterized hit genes from the Hh signaling screen, TEDC1/C14orf80 and TEDC2/C16orf59, exhibit growth phenotype patterns that are highly correlated to each other as well as to those for genes encoding δ-tubulin, ε-tubulin, and several other centriolar proteins (Figure 5C) (45, 257). This finding suggests a shared function for these genes, and indeed TEDC1, TEDC2, δ-tubulin, and ε-tubulin form a protein complex required for centriole stability (45, 257).
In contrast to centriolar genes, most cilia-associated genes do not exhibit a distinct phenotypic signature, likely because their knockout had little effect on cell proliferation under the growth conditions and cell lines examined. Furthermore, this type of approach may miss proteins that have additional roles outside of cilium/centriole function that lead to distinct phenotypic signatures. Going forward, we anticipate that both highly targeted screens and large-scale growth datasets will provide complementary means to investigate the biology of cilia and centrioles.
In addition to these functional genomic approaches, new proteomic technologies have also been applied to cilia and centrioles. Shotgun proteomics of partially purified cilia and centrioles has provided important insights into the composition of these organelles (259–265). However, cilia and centrioles are not fully membrane-enclosed, making it difficult to biochemically isolate them while ensuring that that their contents remain stably associated (with the notable exception of organisms that can be induced to release their cilia intact, such as Chlamydomonas reinhardtii). This challenge is further compounded by the small size of cilia and centrioles and their low copy number per cell. Recently, proximity labeling has emerged as a proteomic approach that can overcome some of these obstacles. Proximity labeling takes advantage of enzymes that generate radical forms of biotin-containing compounds that, due to their high reactivity and short half-life, covalently react with and label nearby proteins (266, 267). Following in situ labeling, biotinylated proteins can be purified and analyzed by mass spectrometry. At present, the two enzymes most commonly used to generate biotinyl radicals are BirA-R118G (known as BioID) and variants of soybean ascorbate peroxidase named APEX or APEX2 (for further discussion, see (266, 267)).
One of the first applications of proximity labeling to cilia and centrioles was reported by Gupta et al. (268). In this study, BioID fusions were analyzed for a host of proteins that localize to the cilium-centriole interface, yielding an extensive proximity-based protein network containing known and novel components. In parallel, Mick et al. (235) and Kohli et al. (207) used APEX labeling to define a ciliary proteome. In both cases, the APEX enzyme was fused to cilia-targeted proteins that localize throughout the ciliary membrane. The labeling reactions therefore led to biotinylation of a range of known and novel ciliary proteins. These studies also investigated changes in ciliary proteome composition in mutant cells deficient in the IFT-B subunit IFT27 (235) and in response to stimulation of cilium disassembly (207). These examples illustrate how ciliary APEX labeling may be used to investigate how the ciliary proteome changes during dynamic cellular processes or in specific disease states. With ongoing improvements to proximity labeling methodology, ciliary and centriolar proteomics is likely to be a powerful complement to the functional screening approaches described above.
Finally, we note that advances in light and electron microscopy are also providing important new insights into the biology of cilia and flagella. While a full discussion of such approaches is beyond the scope of this review, we note that super-resolution fluorescence microcopy methods are providing increasingly detailed molecular maps of ciliary and centriolar structures (118, 169, 170, 269–271). For example, 3D-STORM imaging has been used to generate a map of proteins that form the distal appendages and transition zone, identifying for the first time distinct functions and localizations for distal appendage blade versus distal appendage matrix proteins (269, 270). Additionally, EM approaches are revealing the in situ organization of cilia (272–275) and the elaborate structure of the centriole (4, 276). Given the nanometer scale of many key ciliary and centriolar structures, the continued application of these technologies is likely to be an important complement to genomic and proteomic approaches.
Perspective
The past several years has seen tremendous advances in our understanding of cilium and centriole biology. The key steps in the biogenesis of cilia and centrioles have been defined, and many of the important proteins have been identified. Since the molecular players are now largely known, a key challenge for the future will be to define molecular mechanisms and to better understand the roles of cilia and centrioles in normal physiology and disease. Some critical questions that remain to be addressed include:
Can cells ‘sense’ the presence of a cilium or centrioles, and if so, what is the underlying mechanism?
How is centriole biogenesis restricted to a single new procentriole per parent centriole in each cell cycle?
Given differences in how cilia are assembled and disassembled in different cell types or organisms, what aspects of these processes are invariant and which exhibit plasticity?
How is centriole and cilium function regulated through transcriptional, translational, and post-translational means and in different tissues and cell types?
What are the physiological consequences of dysregulated cilium disassembly?
How do centriole and ciliary defects lead to the phenotypes observed in microcephaly and ciliopathies?
Can insights into centriole and cilium biogenesis be leveraged for therapeutic benefit?
With the advent of new technologies and a growing interesting in the biology of cilia and centrioles, we anticipate exciting new findings as answers to these questions emerge.
Acknowledgements
We thank our lab members for helpful discussions and apologize to colleagues whose work could not be cited due to space limitations. D.K.B. is a member of the Yale Cancer Center and is supported by the Alfred P. Sloan Foundation, the Charles H. Hood Foundation, and the National Institutes of Health (R00HD082280). A.J.H is supported by the National Institutes of Health (R01GM114119) and an American Cancer Society Scholar Grant (129742-RSG-16-156-01-CCG).
Terms and definitions
- Pericentriolar material
The electron dense material that surrounds the centrioles and makes up part of the centrosome
- Centrosome
A cellular structure consisting of a pair of centrioles embedded in pericentriolar material; often forms the major microtubule-organizing center of the cell
- Basal body
A mature centriole that docks at the plasma membrane to nucleate the formation of a cilium
- Axoneme
A ring of nine doublet microtubules and associated proteins that form the cilium core; can also contain a central microtubule pair
- Centriole satellites
Electron dense cytoplasmic granules occurring around the centrosome
- Distal appendages
Structures that radiate from the distal end of a fully mature parent centriole and mediate membrane docking during ciliogenesis
- Sub-distal appendages
Structures projecting from the sub-distal end of a fully mature parent centriole that anchor the minus-ends of microtubules in interphase cells
- Parent centriole
A centriole that is able to duplicate but is not fully mature and lacks appendages. Sometimes known as a daughter centriole
- Mature parent centriole
A mature centriole that is able to duplicate and is decorated with appendages that enable ciliogenesis. Sometimes known as a mother centriole
- Procentriole
A newly formed centriole that is not competent for duplication
- Cartwheel
A scaffolding structure comprised of a hub and nine-radially arranged spokes located at the proximal end of the procentriole
- Deuterosome
A protein structure formed in the cytoplasm of multiciliated cells that templates the formation of multiple procentrioles
- Mitotic surveillance pathway
A USP28-53BP1-P53 signaling pathway that prevents the proliferation of unfit cells that undergo centrosome loss and/or delay in mitosis
- Transition zone
A domain at the base of the cilium that links the axoneme to the ciliary membrane and that controls protein entry and exit from cilia
- Ciliary pocket
An invaginated plasma membrane domain found adjacent to some cilia that may participate in membrane trafficking to and from the cilium
- Ciliary vesicle
A vesicle associated with the mature parent centriole during ciliogenesis that is the precursor to the ciliary membrane
- Ciliopathy
A disease that is part of a group of human developmental disorders that are caused by cilium dysfunction
References
- 1.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194: 165–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickett-Heaps JD. 1971. The autonomy of the centriole: fact or fallacy? Cytobios 3: 205–14 [Google Scholar]
- 3.Gonczy P 2012. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 13: 425–35 [DOI] [PubMed] [Google Scholar]
- 4.Greenan GA, Keszthelyi B, Vale RD, Agard DA. 2018. Insights into centriole biogenesis and evolution revealed by cryoTomography of doublet and triplet centrioles. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JT, Stearns T. 2018. The ABCs of Centriole Architecture: The Form and Function of Triplet Microtubules. Cold Spring Harb Symp Quant Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hori A, Toda T. 2017. Regulation of centriolar satellite integrity and its physiology. Cell Mol Life Sci 74: 213–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopes CA, Prosser SL, Romio L, Hirst RA, O’Callaghan C, et al. 2011. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci 124: 600–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, et al. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27: 163–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MF. 2016. Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev Cell 39: 424–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nigg EA, Holland AJ. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat Rev Mol Cell Biol 19: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–6 [DOI] [PubMed] [Google Scholar]
- 12.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, et al. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–207 [DOI] [PubMed] [Google Scholar]
- 13.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biology open 1: 965–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, et al. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 191: 731–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TS, Park JE, Shukla A, Choi S, Murugan RN, et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc Natl Acad Sci U S A 110: E4849–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Park JE, Kim TS, Kim JH, Kwak MJ, et al. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat Struct Mol Biol 21: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci 126: 3223–33 [DOI] [PubMed] [Google Scholar]
- 19.Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, et al. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun 5: 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Abdelaziz M, Debski J, et al. 2017. Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol 209: 863–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes CA, Jana SC, Cunha-Ferreira I, Zitouni S, Bento I, et al. 2015. PLK4 trans-Autoactivation Controls Centriole Biogenesis in Space. Dev Cell 35: 222–35 [DOI] [PubMed] [Google Scholar]
- 23.Arquint C, Gabryjonczyk AM, Imseng S, Bohm R, Sauer E, et al. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, et al. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr Biol 24: 2526–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kratz AS, Barenz F, Richter KT, Hoffmann I. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol Open 4: 370–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLamarrah TA, Buster DW, Galletta BJ, Boese CJ, Ryniawec JM, et al. 2018. An ordered pattern of Ana2 phosphorylation by Plk4 is required for centriole assembly. J Cell Biol 217: 1217–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirono M 2014. Cartwheel assembly. Philos Trans R Soc Lond B Biol Sci 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–9 [DOI] [PubMed] [Google Scholar]
- 30.van Breugel M, Wilcken R, McLaughlin SH, Rutherford TJ, Johnson CM. 2014. Structure of the SAS-6 cartwheel hub from Leishmania major. Elife 3: e01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cottee MA, Muschalik N, Johnson S, Leveson J, Raff JW, Lea SM. 2015. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. Elife 4: e07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guichard P, Hamel V, Le Guennec M, Banterle N, Iacovache I, et al. 2017. Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Nat Commun 8: 14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aydogan MG, Wainman A, Saurya S, Steinacker TL, Caballe A, et al. 2018. A homeostatic clock sets daughter centriole size in flies. J Cell Biol 217: 1233–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang WJ, Acehan D, Kao CH, Jane WN, Uryu K, Tsou MF. 2015. De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilbert M, Noga A, Frey D, Hamel V, Guichard P, et al. 2016. SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat Cell Biol 18: 393–403 [DOI] [PubMed] [Google Scholar]
- 36.Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol 23: 1360–6 [DOI] [PubMed] [Google Scholar]
- 37.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11: 825–31 [DOI] [PubMed] [Google Scholar]
- 38.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, et al. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, et al. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J 32: 1141–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJ, et al. 2013. CEP120 interacts with CPAP and positively regulates centriole elongation. J Cell Biol 202: 211–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahl KD, Sankaran DG, Bayless BA, Pinter ME, Galati DF, et al. 2015. A Short CEP135 Splice Isoform Controls Centriole Duplication. Curr Biol 25: 2591–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piperno G, Fuller MT. 1985. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol 101: 2085–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. 1990. Posttranslational glutamylation of alpha-tubulin. Science 247: 83–5 [DOI] [PubMed] [Google Scholar]
- 44.Kochanski RS, Borisy GG. 1990. Mode of centriole duplication and distribution. J Cell Biol 110: 1599–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breslow DK, Hoogendoorn S, Kopp AR, Morgens DW, Vu BK, et al. 2018. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat Genet 50: 460–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JT, Kong D, Hoerner CR, Loncarek J, Stearns T. 2017. Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, et al. 2016. Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev Cell 37: 362–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X, Ramani A, Soni K, Gottardo M, Zheng S, et al. 2016. Molecular basis for CPAP-tubulin interaction in controlling centriolar and ciliary length. Nat Commun 7: 11874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, et al. 2009. Control of centriole length by CPAP and CP110. Curr Biol 19: 1005–11 [DOI] [PubMed] [Google Scholar]
- 50.Keller LC, Geimer S, Romijn E, Yates J 3rd, Zamora I, Marshall WF 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol Biol Cell 20: 1150–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, et al. 2009. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol 185: 101–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galletta BJ, Jacobs KC, Fagerstrom CJ, Rusan NM. 2016. Asterless is required for centriole length control and sperm development. J Cell Biol 213: 435–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang CW, Hsu WB, Tsai JJ, Tang CJ, Tang TK. 2016. CEP295 interacts with microtubules and is required for centriole elongation. J Cell Sci 129: 2501–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saurya S, Roque H, Novak ZA, Wainman A, Aydogan MG, et al. 2016. Drosophila Ana1 is required for centrosome assembly and centriole elongation. J Cell Sci 129: 2514–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spektor A, Tsang WY, Khoo D, Dynlacht BD. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell 130: 678–90 [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. 2011. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 145: 914–25 [DOI] [PubMed] [Google Scholar]
- 57.Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, et al. 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 15: 187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loncarek J, Hergert P, Magidson V, Khodjakov A. 2008. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol 10: 322–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szollosi D, Calarco P, Donahue RP. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci 11: 521–41 [DOI] [PubMed] [Google Scholar]
- 60.La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol 168: 713–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, et al. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol 176: 173–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol 158: 1171–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, et al. 2015. p53 protects against genome instability following centriole duplication failure. J Cell Biol 210: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. 2007. Plk4-induced centriole biogenesis in human cells. Dev Cell 13: 190–202 [DOI] [PubMed] [Google Scholar]
- 65.Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC. 2012. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure 20: 1905–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimanovskaya E, Viscardi V, Lesigang J, Lettman MM, Qiao R, et al. 2014. Structure of the C. elegans ZYG-1 cryptic polo box suggests a conserved mechanism for centriolar docking of Plk4 kinases. Structure 22: 1090–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cunha-Ferreira I, Bento I, Pimenta-Marques A, Jana SC, Lince-Faria M, et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr Biol 23: 2245–54 [DOI] [PubMed] [Google Scholar]
- 68.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol 188: 191–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klebba JE, Buster DW, Nguyen AL, Swatkoski S, Gucek M, et al. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr Biol 23: 2255–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guderian G, Westendorf J, Uldschmid A, Nigg EA. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci 123: 2163–9 [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto S, Kitagawa D. 2018. Self-organization of Plk4 regulates symmetry breaking in centriole duplication. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montenegro Gouveia S, Zitouni S, Kong D, Duarte P, Ferreira Gomes B, et al. 2018. PLK4 is a microtubule-associated protein that self assembles promoting de novo MTOC formation. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goryachev AB, Pokhilko AV. 2008. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett 582: 1437–43 [DOI] [PubMed] [Google Scholar]
- 74.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, et al. 2009. Singularity in polarization: rewiring yeast cells to make two buds. Cell 139: 731–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goryachev AB, Leda M. 2017. Many roads to symmetry breaking: molecular mechanisms and theoretical models of yeast cell polarity. Mol Biol Cell 28: 370–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dippell RV. 1968. The development of basal bodies in paramecium. Proc Natl Acad Sci U S A 61: 461–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iftode F, Fleury-Aubusson A. 2003. Structural inheritance in Paramecium: ultrastructural evidence for basal body and associated rootlets polarity transmission through binary fission. Biol Cell 95: 39–51 [DOI] [PubMed] [Google Scholar]
- 78.O’Toole ET, Dutcher SK. 2014. Site-specific basal body duplication in Chlamydomonas. Cytoskeleton (Hoboken) 71: 108–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wloga D, Frankel J. 2012. From molecules to morphology: cellular organization of Tetrahymena thermophila. Methods Cell Biol 109: 83–140 [DOI] [PubMed] [Google Scholar]
- 80.Wong C, Stearns T. 2003. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol 5: 539–44 [DOI] [PubMed] [Google Scholar]
- 81.Kim M, O’Rourke BP, Soni RK, Jallepalli PV, Hendrickson RC, Tsou MF. 2016. Promotion and Suppression of Centriole Duplication Are Catalytically Coupled through PLK4 to Ensure Centriole Homeostasis. Cell Rep 16: 1195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsou MF, Stearns T. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442: 947–51 [DOI] [PubMed] [Google Scholar]
- 83.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell 17: 344–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. 2012. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol 22: 915–21 [DOI] [PubMed] [Google Scholar]
- 85.Lee K, Rhee K. 2012. Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle 11: 2476–85 [DOI] [PubMed] [Google Scholar]
- 86.Shukla A, Kong D, Sharma M, Magidson V, Loncarek J. 2015. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat Commun 6: 8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong D, Farmer V, Shukla A, James J, Gruskin R, et al. 2014. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J Cell Biol 206: 855–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim J, Lee K, Rhee K. 2015. PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat Commun 6: 10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang WJ, Soni RK, Uryu K, Tsou MF. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol 193: 727–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Novak ZA, Wainman A, Gartenmann L, Raff JW. 2016. Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev Cell 37: 545–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Novak ZA, Conduit PT, Wainman A, Raff JW. 2014. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr Biol 24: 1276–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Izquierdo D, Wang WJ, Uryu K, Tsou MF. 2014. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep 8: 957–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuchiya Y, Yoshiba S, Gupta A, Watanabe K, Kitagawa D. 2016. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat Commun 7: 12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu J, Lipinszki Z, Rangone H, Min M, Mykura C, et al. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat Cell Biol 18: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugioka K, Hamill DR, Lowry JB, McNeely ME, Enrick M, et al. 2017. Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lacey KR, Jackson PK, Stearns T. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci U S A 96: 2817–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283: 851–4 [DOI] [PubMed] [Google Scholar]
- 98.Matsumoto Y, Hayashi K, Nishida E. 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol 9: 429–32 [DOI] [PubMed] [Google Scholar]
- 99.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol 1: 88–93 [DOI] [PubMed] [Google Scholar]
- 100.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. 2006. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene 25: 2943–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zitouni S, Francia ME, Leal F, Montenegro Gouveia S, Nabais C, et al. 2016. CDK1 Prevents Unscheduled PLK4-STIL Complex Assembly in Centriole Biogenesis. Curr Biol 26: 1127–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spassky N, Meunier A. 2017. The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol 18: 423–36 [DOI] [PubMed] [Google Scholar]
- 103.Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ. 2013. Deuterosome-mediated centriole biogenesis. Dev Cell 27: 103–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao H, Zhu L, Zhu Y, Cao J, Li S, et al. 2013. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15: 1434–44 [DOI] [PubMed] [Google Scholar]
- 105.Al Jord A, Lemaitre AI, Delgehyr N, Faucourt M, Spassky N, Meunier A. 2014. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 516: 104–7 [DOI] [PubMed] [Google Scholar]
- 106.Zhao H, Chen Q, Huang Q, Yan X, Zhu X. 2018. Mother centrioles are dispensable for deuterosome formation and function during basal body amplification. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Al Jord A, Shihavuddin A, Servignat d’Aout R, Faucourt M, Genovesio A, et al. 2017. Calibrated mitotic oscillator drives motile ciliogenesis. Science 358: 803–06 [DOI] [PubMed] [Google Scholar]
- 108.Vladar EK, Stratton MB, Saal ML, Salazar-De Simone G, Wang X, et al. 2018. Cyclin-dependent kinase control of motile ciliogenesis. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pimenta-Marques A, Bento I, Lopes CA, Duarte P, Jana SC, Bettencourt-Dias M. 2016. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 353: aaf4866. [DOI] [PubMed] [Google Scholar]
- 110.Werner S, Pimenta-Marques A, Bettencourt-Dias M. 2017. Maintaining centrosomes and cilia. J Cell Sci 130: 3789–800 [DOI] [PubMed] [Google Scholar]
- 111.Tassin AM, Maro B, Bornens M. 1985. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 100: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Connolly JA, Kiosses BW, Kalnins VI. 1986. Centrioles are lost as embryonic myoblasts fuse into myotubes in vitro. Eur J Cell Biol 39: 341–5 [PubMed] [Google Scholar]
- 113.Conduit PT, Wainman A, Raff JW. 2015. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16: 611–24 [DOI] [PubMed] [Google Scholar]
- 114.Woodruff JB, Wueseke O, Hyman AA. 2014. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu J, Glover DM. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol 2: 120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jana SC, Marteil G, Bettencourt-Dias M. 2014. Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr Opin Cell Biol 26: 96–106 [DOI] [PubMed] [Google Scholar]
- 117.Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14: 1148–58 [DOI] [PubMed] [Google Scholar]
- 118.Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, et al. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14: 1159–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1: 965–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haren L, Stearns T, Luders J. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One 4: e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee K, Rhee K. 2011. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol 195: 1093–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, et al. 2014. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev Cell 28: 659–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Woodruff JB, Wueseke O, Viscardi V, Mahamid J, Ochoa SD, et al. 2015. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science 348: 808–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fong KW, Choi YK, Rattner JB, Qi RZ. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell 19: 115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gomez-Ferreria MA, Bashkurov M, Helbig AO, Larsen B, Pawson T, et al. 2012. Novel NEDD1 phosphorylation sites regulate gamma-tubulin binding and mitotic spindle assembly. J Cell Sci 125: 3745–51 [DOI] [PubMed] [Google Scholar]
- 127.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15: 3642–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feng Z, Caballe A, Wainman A, Johnson S, Haensele AFM, et al. 2017. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell 169: 1078–89.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, et al. 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife 3: e03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Conduit PT, Raff JW. 2015. Different Drosophila cell types exhibit differences in mitotic centrosome assembly dynamics. Curr Biol 25: R650–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laos T, Cabral G, Dammermann A. 2015. Isotropic incorporation of SPD-5 underlies centrosome assembly in C. elegans. Curr Biol 25: R648–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169: 1066–77.e10 [DOI] [PubMed] [Google Scholar]
- 133.Prosser SL, Pelletier L. 2017. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol 18: 187–201 [DOI] [PubMed] [Google Scholar]
- 134.Lambrus BG, Daggubati V, Uetake Y, Scott PM, Clutario KM, et al. 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol 214: 143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fong CS, Mazo G, Das T, Goodman J, Kim M, et al. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, et al. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol 214: 155–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bazzi H, Anderson KV. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci U S A 111: E1491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lambrus BG, Holland AJ. 2017. A New Mode of Mitotic Surveillance. Trends Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, et al. 2006. Flies without centrioles. Cell 125: 1375–86 [DOI] [PubMed] [Google Scholar]
- 140.Lattao R, Kovacs L, Glover DM. 2017. The Centrioles, Centrosomes, Basal Bodies, and Cilia of Drosophila melanogaster. Genetics 206: 33–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chan JY. 2011. A clinical overview of centrosome amplification in human cancers. International journal of biological sciences 7: 1122–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sercin O, Larsimont JC, Karambelas AE, Marthiens V, Moers V, et al. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat Cell Biol 18: 100–10 [DOI] [PubMed] [Google Scholar]
- 143.Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, et al. 2017. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev Cell 40: 313–22 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Coelho PA, Bury L, Shahbazi MN, Liakath-Ali K, Tate PH, et al. 2015. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol 5: 150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, et al. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, et al. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Silkworth WT, Nardi IK, Scholl LM, Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 4: e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Godinho SA, Picone R, Burute M, Dagher R, Su Y, et al. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510: 167–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ganier O, Schnerch D, Oertle P, Lim RY, Plodinec M, Nigg EA. 2018. Structural centrosome aberrations promote non-cell-autonomous invasiveness. EMBO J 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jayaraman D, Bae BI, Walsh CA. 2018. The Genetics of Primary Microcephaly. Annu Rev Genomics Hum Genet [DOI] [PubMed] [Google Scholar]
- 152.Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. 2014. Cortical neurogenesis in the absence of centrioles. Nat Neurosci 17: 1528–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Marjanovic M, Sanchez-Huertas C, Terre B, Gomez R, Scheel JF, et al. 2015. CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination. Nat Commun 6: 7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. 2011. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol 13: 1325–34 [DOI] [PubMed] [Google Scholar]
- 155.Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, et al. 2016. Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron 89: 83–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nachury MV, Seeley ES, Jin H. 2010. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol 26: 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reiter JF, Blacque OE, Leroux MR. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13: 608–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jensen VL, Leroux MR. 2017. Gates for soluble and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr Opin Cell Biol 47: 83–91 [DOI] [PubMed] [Google Scholar]
- 159.Garcia-Gonzalo FR, Reiter JF. 2017. Open Sesame: How Transition Fibers and the Transition Zone Control Ciliary Composition. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Benmerah A 2013. The ciliary pocket. Curr Opin Cell Biol 25: 78–84 [DOI] [PubMed] [Google Scholar]
- 161.Mitchell DR. 2017. Evolution of Cilia. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meunier A, Azimzadeh J. 2016. Multiciliated Cells in Animals. Cold Spring Harb Perspect Biol 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ishikawa T 2017. Axoneme Structure from Motile Cilia. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tucker RW, Pardee AB, Fujiwara K. 1979. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 17: 527–35 [DOI] [PubMed] [Google Scholar]
- 165.Rieder CL, Jensen CG, Jensen LC. 1979. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J Ultrastruct Res 68: 173–85 [DOI] [PubMed] [Google Scholar]
- 166.Rasi MQ, Parker JD, Feldman JL, Marshall WF, Quarmby LM. 2009. Katanin knockdown supports a role for microtubule severing in release of basal bodies before mitosis in Chlamydomonas. Mol Biol Cell 20: 379–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sorokin S 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol 15: 363–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sorokin SP. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3: 207–30 [DOI] [PubMed] [Google Scholar]
- 169.Wu CT, Chen HY, Tang TK. 2018. Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol 20: 175–85 [DOI] [PubMed] [Google Scholar]
- 170.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol 17: 228–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Knodler A, Feng S, Zhang J, Zhang X, Das A, et al. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A 107: 6346–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mukhopadhyay S, Badgandi HB, Hwang SH, Somatilaka B, Shimada IS, Pal K. 2017. Trafficking to the primary cilium membrane. Mol Biol Cell 28: 233–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pitaval A, Tseng Q, Bornens M, Thery M. 2010. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J Cell Biol 191: 303–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pitaval A, Senger F, Letort G, Gidrol X, Guyon L, et al. 2017. Microtubule stabilization drives 3D centrosome migration to initiate primary ciliogenesis. J Cell Biol 216: 3713–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Francis SS, Sfakianos J, Lo B, Mellman I. 2011. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol 193: 219–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, et al. 2010. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464: 1048–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, et al. 2010. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci 123: 1785–95 [DOI] [PubMed] [Google Scholar]
- 178.Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A 108: 2759–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. 1991. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature 349: 709–13 [DOI] [PubMed] [Google Scholar]
- 180.Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, et al. 2014. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J Cell Sci 127: 422–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Goetz SC, Liem KF Jr., Anderson KV 2012. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151: 847–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cajanek L, Nigg EA. 2014. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci U S A 111: E2841–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Watanabe T, Kakeno M, Matsui T, Sugiyama I, Arimura N, et al. 2015. TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J Cell Biol 210: 737–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Taschner M, Lorentzen E. 2016. The Intraflagellar Transport Machinery. Cold Spring Harb Perspect Biol 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, et al. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24: 2180–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nachury MV. 2018. The molecular machines that traffic signaling receptors into and out of cilia. Curr Opin Cell Biol 51: 124–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gilula NB, Satir P. 1972. The ciliary necklace. A ciliary membrane specialization. J Cell Biol 53: 494–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, et al. 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145: 513–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Williams CL, Li C, Kida K, Inglis PN, Mohan S, et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192: 1023–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reiter JF, Leroux MR. 2017. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18: 533–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Braun DA, Hildebrandt F. 2017. Ciliopathies. Cold Spring Harb Perspect Biol 9: a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ishikawa H, Marshall WF. 2017. Intraflagellar Transport and Ciliary Dynamics. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hendel NL, Thomson M, Marshall WF. 2018. Diffusion as a Ruler: Modeling Kinesin Diffusion as a Length Sensor for Intraflagellar Transport. Biophys J 114: 663–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Marshall WF, Rosenbaum JL. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol 155: 405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.He M, Subramanian R, Bangs F, Omelchenko T, Liem KF Jr., et al. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol 16: 663–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nager AR, Goldstein JS, Herranz-Perez V, Portran D, Ye F, et al. 2017. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell 168: 252–63 e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, et al. 2017. Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell 168: 264–79 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chien A, Shih SM, Bower R, Tritschler D, Porter ME, Yildiz A. 2017. Dynamics of the IFT machinery at the ciliary tip. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Parker JD, Hilton LK, Diener DR, Rasi MQ, Mahjoub MR, et al. 2010. Centrioles are freed from cilia by severing prior to mitosis. Cytoskeleton 67: 425–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Das RM, Storey KG. 2014. Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science 343: 200–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Paridaen JT, Wilsch-Brauninger M, Huttner WB. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155: 333–44 [DOI] [PubMed] [Google Scholar]
- 202.Saito M, Otsu W, Hsu KS, Chuang JZ, Yanagisawa T, et al. 2017. Tctex-1 controls ciliary resorption by regulating branched actin polymerization and endocytosis. EMBO Rep 18: 1460–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Piao T, Luo M, Wang L, Guo Y, Li D, et al. 2009. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci U S A 106: 4713–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, et al. 2015. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kim S, Lee K, Choi JH, Ringstad N, Dynlacht BD. 2015. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat Commun 6: 8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kohli P, Hohne M, Jungst C, Bertsch S, Ebert LK, et al. 2017. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep 18: 1521–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pan J, Snell WJ. 2005. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev Cell 9: 431–8 [DOI] [PubMed] [Google Scholar]
- 209.Huang K, Diener DR, Rosenbaum JL. 2009. The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J Cell Biol 186: 601–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, et al. 2011. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol 13: 402–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, et al. 2011. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol 13: 351–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Inaba H, Goto H, Kasahara K, Kumamoto K, Yonemura S, et al. 2016. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol 212: 409–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, et al. 2012. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol 197: 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, et al. 2016. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. Embo j 35: 803–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tucker RW, Scher CD, Stiles CD. 1979. Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell 18: 1065–72 [DOI] [PubMed] [Google Scholar]
- 216.Nielsen BS, Malinda RR, Schmid FM, Pedersen SF, Christensen ST, Pedersen LB. 2015. PDGFRbeta and oncogenic mutant PDGFRalpha D842V promote disassembly of primary cilia through a PLCgamma- and AURKA-dependent mechanism. J Cell Sci 128: 3543–9 [DOI] [PubMed] [Google Scholar]
- 217.Kasahara K, Aoki H, Kiyono T, Wang S, Kagiwada H, et al. 2018. EGF receptor kinase suppresses ciliogenesis through activation of USP8 deubiquitinase. Nat Commun 9: 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Spalluto C, Wilson DI, Hearn T. 2012. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur J Cell Biol 91: 675–86 [DOI] [PubMed] [Google Scholar]
- 219.Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, et al. 2012. Identification of a novel Wnt5a-CK1varepsilon-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J 31: 3104–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pan J, Wang Q, Snell WJ. 2004. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell 6: 445–51 [DOI] [PubMed] [Google Scholar]
- 221.Bradley BA, Quarmby LM. 2005. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J Cell Sci 118: 3317–26 [DOI] [PubMed] [Google Scholar]
- 222.Bangs FK, Schrode N, Hadjantonakis AK, Anderson KV. 2015. Lineage specificity of primary cilia in the mouse embryo. Nat Cell Biol 17: 113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Prosser SL, Morrison CG. 2015. Centrin2 regulates CP110 removal in primary cilium formation. J Cell Biol 208: 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. 2006. Centrosome polarization delivers secretory granules to the immunological synapse. Nature 443: 462–5 [DOI] [PubMed] [Google Scholar]
- 225.Stinchcombe JC, Randzavola LO, Angus KL, Mantell JM, Verkade P, Griffiths GM. 2015. Mother Centriole Distal Appendages Mediate Centrosome Docking at the Immunological Synapse and Reveal Mechanistic Parallels with Ciliogenesis. Curr Biol 25: 3239–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.de la Roche M, Asano Y, Griffiths GM. 2016. Origins of the cytolytic synapse. Nat Rev Immunol 16: 421–32 [DOI] [PubMed] [Google Scholar]
- 227.Finetti F, Onnis A, Baldari CT. 2015. Regulation of vesicular traffic at the T cell immune synapse: lessons from the primary cilium. Traffic 16: 241–9 [DOI] [PubMed] [Google Scholar]
- 228.Stephen LA, ElMaghloob Y, McIlwraith MJ, Yelland T, Castro Sanchez P, et al. The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK. Dev Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bangs F, Anderson KV. 2017. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nachury MV. 2014. How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci 369: 20130465–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ma M, Gallagher AR, Somlo S. 2017. Ciliary Mechanisms of Cyst Formation in Polycystic Kidney Disease. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mykytyn K, Askwith C. 2017. G-Protein-Coupled Receptor Signaling in Cilia. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pala R, Alomari N, Nauli SM. 2017. Primary Cilium-Dependent Signaling Mechanisms. Int J Mol Sci 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hilgendorf KI, Johnson CT, Jackson PK. 2016. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol 39: 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, et al. 2015. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell 35: 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, et al. 2018. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet 50: 180–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nozawa YI, Lin C, Chuang PT. 2013. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev 23: 429–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. 2013. Primary cilia are specialized calcium signalling organelles. Nature 504: 311–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, et al. 2016. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci U S A 113: 13069–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang Q, Pan J, Snell WJ. 2006. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell 125: 549–62 [DOI] [PubMed] [Google Scholar]
- 241.Klena NT, Gibbs BC, Lo CW. 2017. Cilia and Ciliopathies in Congenital Heart Disease. Cold Spring Harb Perspect Biol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bujakowska KM, Liu Q, Pierce EA. 2017. Photoreceptor Cilia and Retinal Ciliopathies. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kopinke D, Roberson EC, Reiter JF. 2017. Ciliary Hedgehog Signaling Restricts Injury-Induced Adipogenesis. Cell 170: 340–51 e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, et al. 2009. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med 15: 1055–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. 2009. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med 15: 1062–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu H, Kiseleva AA, Golemis EA. 2018. Ciliary signalling in cancer. Nat Rev Cancer [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Seeley ES, Carriere C, Goetze T, Longnecker DS, Korc M. 2009. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res 69: 422–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kim J, Dabiri S, Seeley ES. 2011. Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma. PLoS One 6: e27410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gradilone SA, Radtke BN, Bogert PS, Huang BQ, Gajdos GB, LaRusso NF. 2013. HDAC6 inhibition restores ciliary expression and decreases tumor growth. Cancer Res 73: 2259–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Menzl I, Lebeau L, Pandey R, Hassounah NB, Li FW, et al. 2014. Loss of primary cilia occurs early in breast cancer development. Cilia 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zingg D, Debbache J, Pena-Hernandez R, Antunes AT, Schaefer SM, et al. 2018. EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation. Cancer Cell 34: 69–84 e14 [DOI] [PubMed] [Google Scholar]
- 252.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, et al. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, et al. 2015. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163: 1515–26 [DOI] [PubMed] [Google Scholar]
- 254.Pusapati GV, Kong JH, Patel BB, Krishnan A, Sagner A, et al. 2018. CRISPR Screens Uncover Genes that Regulate Target Cell Sensitivity to the Morphogen Sonic Hedgehog. Dev Cell 44: 113–29 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, et al. 2017. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49: 1779–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, et al. 2017. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168: 890–903 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, et al. 2018. Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst 6: 555–68 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kim E, Dede M, Lenoir WF, Wang G, Srinivasan S, et al. 2018. Hierarchical organization of the human cell from a cancer coessentiality network. bioRxiv: 328880 [Google Scholar]
- 259.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–4 [DOI] [PubMed] [Google Scholar]
- 260.Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 30: 1520–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ishikawa H, Thompson J, Yates JR 3rd, Marshall WF 2012. Proteomic analysis of mammalian primary cilia. Curr Biol 22: 414–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Keller LC, Romijn EP, Zamora I, Yates JR 3rd, Marshall WF 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol 15: 1090–8 [DOI] [PubMed] [Google Scholar]
- 263.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH Jr., et al. 2007. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol 178: 905–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Pazour GJ, Agrin N, Leszyk J, Witman GB. 2005. Proteomic analysis of a eukaryotic cilium. J Cell Biol 170: 103–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sigg MA, Menchen T, Lee C, Johnson J, Jungnickel MK, et al. 2017. Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Dev Cell 43: 744–62 e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim DI, Roux KJ. 2016. Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26: 804–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Han S, Li J, Ting AY. 2018. Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 50: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, et al. 2015. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 163: 1484–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shi X, Garcia G 3rd, Van De Weghe JC, McGorty R, Pazour GJ, et al. 2017. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat Cell Biol 19: 1178–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Yang TT, Chong WM, Wang WJ, Mazo G, Tanos B, et al. 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun 9: 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Gambarotto D, Zwettler F, Cernohorska M, Fortun D, Borgers S, et al. 2018. Imaging beyond the super-resolution limits using ultrastructure expansion microscopy (UltraExM). bioRxiv [Google Scholar]
- 272.Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, et al. 2012. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell 151: 1029–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stepanek L, Pigino G. 2016. Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352: 721–4 [DOI] [PubMed] [Google Scholar]
- 274.Lin J, Nicastro D. 2018. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Jana SC, Mendonca S, Machado P, Werner S, Rocha J, et al. 2018. Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nat Cell Biol [DOI] [PubMed] [Google Scholar]
- 276.Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, et al. 2012. Cartwheel architecture of Trichonympha basal body. Science 337: 553. [DOI] [PubMed] [Google Scholar]