Abstract
Succession of gut microbial community structure for newborns is highly influenced by early life factors. Many preterm infants cared for in the NICU are exposed to parent-infant separation, stress and pain from medical care procedures. The purpose of the study was to investigate the impact of early life stress on the trajectory of gut microbial structure. Stool samples from very preterm infants were collected weekly for 6 weeks. NICU stress exposure data was collected daily for 6 weeks. V4 region of the 16S rRNA gene was amplified by PCR and sequenced. Zero-inflated Beta regression model with random effects was used to assess the impact of stress on gut microbiome trajectories. Week of sampling was significant for: Escherichia, Staphylococcus, Enterococcus, Bifidobacterium, Proteus, Streptococcus, Clostridium_butyricum and Clostridium_perfringens. Antibiotic usage was significant for Proteus, Citrobacter and Clostridium_perfringens. Gender was significant for Proteus. Stress exposure occurring 1 and 2 weeks prior to sampling had a significant effect on Proteus and Veillonella. NICU stress exposure had a significant effect on Proteus and Veillonella. There was an overall dominance of Gammaproteobacteria. Findings suggest early life NICU stress may significantly influence the developing gut microbiome, with important NICU practice and future microbiome research considerations.
Keywords: preterm infants, NICU, stress, chronic stress, early life stress, gut microbiome
1. Introduction
Gastrointestinal and neurodevelopmental systems are highly vulnerable in preterm infants. Some of the most devastating complications experienced by preterm infants include diseases that originate in these systems (i.e. necrotizing enterocolitis and intraventricular hemorrhage) (Bolisetty et al., 2014; Christian et al., 2015; Fitzgibbons et al., 2009; Kidokoro et al., 2014; Stoll, Hansen, Bell, & et al., 2015). How stress alters the gastrointestinal system remains largely unknown. Interestingly, a powerful bidirectional system of communication exists between the gut and brain, but we know little about this in preterm infants.
An important factor influencing health is the commensal gut microbiota that coexist in a mutualistic relationship with the host (Sampson & Mazmanian, 2015). Of particular interest is the general enterotype of commensal and potentially virulent microbial species and their relative proportions, that may play a role in general health as well as mental health (La Rosa et al., 2014; Liu, 2017; Sharon et al., 2014). Preliminary relationships are emerging that link the composition of the gut microbiome to increased risk for anxiety and depression (Kaplan, Rucklidge, Romijn, & McLeod, 2015). For former preterm infants, anxiety and depression are some of the internalizing behaviors they are at-risk for in adulthood (Mathewson et al., 2017; Pyhälä et al., 2017).
After birth, newborns are colonized with trillions of bacteria, the specific composition of bacteria contributing to the programming of gut function and development, epithelial barrier function, immune system and overall gut homeostasis (Hooper, 2004). For healthy term infants, the major first source of colonizing bacteria is of maternal origin (Dominguez-Bello et al., 2010; Dominguez-Bello et al., 2016; Mueller, Bakacs, Combellick, Grigoryan, & Dominguez-Bello, 2015). Previous research has shown a developmental sequence of phyla blooms occurs after birth, ultimately leading to one’s microbial signature throughout adulthood (Lan, Kriete, & Rosen, 2013). NICU infants are often deprived of the “normal colonization” process, resulting in potential delays in the normal taxa succession (Bentley et al., 2016; Lax & Gilbert, 2015; Raveh-Sadka et al., 2016; Shin, Whon, & Bae, 2015). This delay can lead to a state of dysbiosis with short and long-term pathological implications.
Clinicians and researchers have been hopeful that this newer area of science will lead to discoveries about the underlying pathology of preterm infant illness and disease. However, studies investigating the developing microbiome of preterm infants have revealed conflicting and sometimes counterintuitive results. For the first 3 months of life, Brown et al. (2018) examined the gut microbiome of 35 preterm infants and found high variability within infants. In subsequent sampling, some infants had microbial community abundance that was unique from earlier samples as those from other infants and no species were strongly associated with necrotizing enterocolitis (NEC) (Brown et al., 2018). Dobbler et al. (2017) identified low microbial diversity and chaotic succession for infants who developed NEC. The Norwegian Microbiota Study reported duration of NICU care to be a significant mediator of diversity, but not breastfeeding or history of antibiotic use (Dahl et al., 2018). After adjusting for antibiotic and breastmilk exposure and delivery mode, Chernikova et al. (2018) found differences in diversity based upon birth gestational age, with extremely preterm infants having less diversity than moderate-late preterm infants (Chernikova et al., 2018). Questions remain if this association is because of inherent gut maturation, also leading to immune effects, or influences from the NICU environment.
Early life stress, an environmental exposure, has the potential to leave lasting effects. While some stressors during the lifespan may promote growth and adaptation, other stressors may become biologically embedded, to the extent that they alter future health trajectories. For many hospitalized preterm infants, early life exposure to cumulative and intense stressors may be perceived by the infant as potentially toxic. Toxic stress is defined as strong, frequent or prolonged activation of the body’s stress response systems, without the buffering protection of parental support, which has the potential to alter health trajectories. Of particular importance to the developing preterm infants is brain development and future regulation of stress (Jensen, Dickie, Schwartz, & et al., 2015; Shonkoff et al., 2012; Westfall & Nemeroff, 2015). Toxic stress has been investigated in a number of populations, however little is known about how toxic stress effects the developing preterm infants (Barker, 1998; Gunnar, Wewerka, Frenn, Long, & Griggs, 2009). A number of studies now suggest that stress can affect the gut microbiome and lead to dysbiosis, as well as interrupt activation of the innate immune system in response to stress (Dantzer, Cohen, Russo, & Dinan, 2018; Karl et al., 2018).
Despite the patient-focused manner caregivers seek to provide care, the NICU is largely a medically focused environment driven by protocols and caregiver schedules. The environment places preterm infants under a great deal of stress as infants are exposed to prolonged separation from parents, chronic and extreme stressors, and painful life-saving medical procedures (D’Agata, Young, Cong, Grasso, & McGrath, 2016). Furthermore, preterm infants are unable to articulate to caregivers their burden of stress. The cumulative and multifaceted experiences of high-level stress, coupled with the preverbal stage of development and inability to self-advocate, potentially leads to toxic stress exposure. The frequent or prolonged activation of the body’s stress response systems, in the absence of buffering protection of a supportive adult relationship, leads to alterations in the child’s biologic functioning (Gunnar et al., 2009; Shonkoff et al., 2012). Neurodevelopmental health trajectories are of utmost concern for infants as this period of brain development has been identified for infants and young children as a functionally sensitive period in which experience and environmental stimuli exert great influence (Sale, Berardi, & Maffei, 2014).
An alternative, yet highly relevant, factor of influence is the commensal gut microbiota that coexists in a mutualistic relationship with its host (Sampson & Mazmanian, 2015). The bidirectional axis between the highly complex systems of gut-brain has been suggested to significantly influence health (Clarke et al., 2014; De Palma, Collins, Bercik, & Verdu, 2014; M. W. Groer et al., 2014; Lu et al., 2015; Stilling et al., 2015). Animal studies have demonstrated the gut microbiome exerts an influence on systems outside of the gastrointestinal tract, including brain circuits that control stress response, anxiety-like behavior and cognitive function (Arboleya et al., 2016; Clarke et al., 2013; Gilbert et al., 2016; Heijtz et al., 2011; Neufeld, Kang, Bienenstock, & Foster, 2011; Sudo et al., 2004).
Compelling evidence has demonstrated that intense experiences may become biologically embedded, potentially affecting outcomes. Care provided in the NICU, however well intended, is an intense early life experience for many infants who require medical care after birth. Currently, a gap in the literature exists about the relationship between early life stress from routine NICU care and longitudinal microbial communities for very low birth weight (VLBW) infants. We hypothesized that the microbial community of the preterm infant gut would be altered by the NICU stress experience. To understand how early life NICU stress exposure may contribute to preterm infant gut microbiome development, we conducted a retrospective, longitudinal data analysis of stool samples sequenced for microbial abundance and diversity and NICU stress exposure during the first 6 weeks of life.
2. Methods
2.1. Participants
This study was conducted using secondary data. VLBW infant participants were part of a larger longitudinal study of long-term growth, health and neurodevelopmental effects (Groer, Ashmeade, Duffy, Morse, & Zaritt, 2016; Groer, Ashmeade, Louis-Jacques, Beckstead, & Ji, 2016; Ho et al., 2018). Infants were admitted to the Neonatal Intensive Care Unit at Tampa General Hospital in Tampa, Florida between 2011 and 2015. Exclusion criteria for entry into the parent study included birth weight greater than 1500 grams, major congenital anomalies, moribund infants and infants of HIV-infected mothers. The study was approved by the University of South Florida Institutional Review Board. Infants were enrolled after informed consent was obtained from their parent or guardian. A total of 82 VLBW infants were eligible to participate in this study.
2.2. Stress Data
Stress data were collected using the Neonatal Infant Stressor Scale (NISS), a ranked quantitative instrument used to collect information concerning daily stress experiences of infants during care in the NICU (D’Agata et al., 2017; Newnham, Inder, & Milgrom, 2009). This instrument measures interventions performed, not infant response. Common NICU interventions are ranked in categories on a 4-point scale as a little stressful, moderately stressful, very stressful and extremely stressful. Each procedure attempt is counted as one stress event. Examples of NICU care procedures and their corresponding scoring include: intubation attempt is extremely stressful and weighted with a score of 5; suctioning attempt is very stressful, score of 4; diaper change is moderately stressful, score of 3; and aspiration of nasogastric tube is a little stressful, score of 2.
For preterm infants who receive NICU care, care is both individualized and standardized. Individualized interventions are directed at the specific illness experienced by the infant, while standardized interventions are interventions performed more routinely, by virtue of being hospitalized. Examples of standardized interventions may include daily weight, diaper changes and position changes. For many preterm infants, typical care also includes some type of respiratory support and gavage feedings. Many of these interventions are performed repeatedly each day and at regular intervals. Higher intensity interventions, such as intubation, chest tube insertion, blood draws and suctioning, occur with greater variability, thus tend to be more individualized.
To develop a core score of standardized interventions, those interventions typically performed routinely on VLBW infants, we analyzed approximately 850 days of previously collected NISS data from NICU infants in another state (D’Agata et al., 2017). From 850 days of NISS data, we extrapolated and averaged the a little stressful and moderately stressful category data (D’Agata et al., 2017). This provided a core score of standardized and lower intensity stress interventions to then add to the higher intensity interventions collected from each infant’s electronic medical record. From the electronic medical record, we collected the actual attempted and completed higher intensity stress events, very stressful and extremely stressful categories. These actual daily events were then added to the core score measure to arrive at a daily stress score. The core score of 109 reflected an average of 40 daily stress events.
To validate the core score, we randomly selected 15 participants from the current parent study and collected the same a little stressful and moderately stressful data over 42 days (approximately 630 days of data) to compare findings. Despite the comparison of data for these two cohorts being 2 different states in different regions of the United States, the same core score was identified.
Stress data were collected daily from NICU admission through 6 weeks of care or discharge, whichever came first. The total daily stress scores were totaled by week for weekly stress scores. The weekly stress scores were used as an independent variable in the analyses.
2.3. Microbiome Data
Stool samples were collected for 6 consecutive weeks from the diapers of VLBW infants while they were admitted to the NICU. The purpose of sample collection in the parent study was to study the succession of the microbiome, and the relationships of microbiome to early and later health outcomes. Weekly stool samples were collected from these infants and preserved at −80⁰ C for microbiome analysis. Samples available for this study primarily included weeks 3–6 as inadequate sample volumes were available for weeks of life 1 and 2 after other stool assays were done in the parent study (Groer, Ashmeade, Louis-Jacques, et al., 2016).
The Mo Bio Power Fecal DNA extraction kit (Qiagen) was used to extract total DNA from 100–250 mg of thawed stool samples. The V4 region of the 16S rRNA gene was amplified by polymerase chain reaction and isolated, then the amplicon library was generated following a standardized protocol using Illumina Miseq (Illumina, San Diego, CA, USA) at Argonne National Laboratory (Chicago, IL). Two hundred base pairs DNA sequence reads were demultiplexed, merged, and imported into QIIME™ for identification of bacterial operational taxonomic units (OTUs). Miseq data were reported to the genus level, with some OTUs reported at species level. Species was used to differentiate genus with same naming convention.
2.4. Statistical Analysis
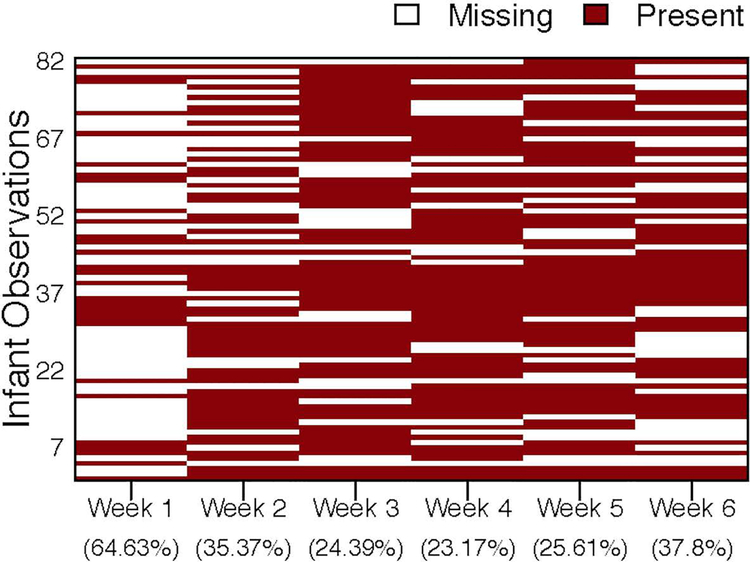
We report samples on a weekly basis. Stool sample collection was attempted at least once per week while infants were hospitalized in the NICU however, some samples were collected inadvertently several times in one week and no sample collected in the subsequent week. Figure 1 shows the missing data pattern of the microbiome compositional data by week. Due to the large amount of missing data in the first two weeks of enrollment, we focused our study of the longitudinal bacterial relative abundances at the genus level from week 3 to week 6, which had the maximum number of infants (116 samples with 29 infants) with complete data. We then considered the subset of infants which had only one missing value between week 3 and week 6 exclusively. The Little’s MCAR test was applied to the subset of infants and showed that the microbiome compositional data at each week were missing completely at random (MCAR), where the probability of missingness does not depend on the values of data (Little, 1988). Under this assumption and because genera abundances are fairly stable across time, cubic spline interpolation approach (Steinway, Biggs, Loughran, Papin, & Albert, 2015) was applied to infants with only one missing value between week 3 to week 6 exclusively. After the imputation, we had a total of 188 samples with 47 infants.
Figure 1.
Plot of missing data pattern and missing data percentage for microbiome data by week, where red color indicates the data are present and white color indicates the data are missing.
Exploratory data analysis was conducted to describe characteristics of each baseline variable (antibiotic, infant gender, birth weight, and gestational age), and the time-dependent variable (cumulative weekly stress) during each week, using summary statistics for the 47 infants.
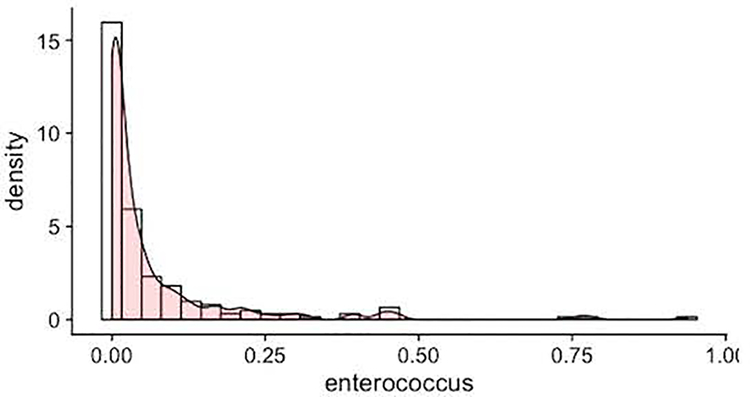
After removing the low abudant genus, 21 bacterial genera remained (Kostic et al., 2015). Since the relative abundances of each bacterial genera were bounded in [0, 1), highly skewed and zero inflated (Figure 2), the zero-inflated Beta regression model with random effects (ZIBR) was used to assess the impact of stress on the trajectories of the gut microbiome compositional data, adjusting for the baseline covariates effects (Chen & Li, 2016). ZIBR model is a mixture of the Binary model and Beta models. The Binary part models the presence of the bacterial genera and the Beta part models the non-zero relative abundance of the bacterial genera. A subject-specific random effect is specified in each part of the model to capture the dependence of the longitudinal abundance measures over time. For both parts of the model, we considered the time effect (week3 as baseline, week4, week5, week6), antibiotic usage, infant gender, birth weight, gestational age, stress 2 weeks prior to sample collection (stresst-2), and stress one week prior to sample collection (stresst-1). Stresst-2 and stresst-1 represent 7-day total NISS scores from the respective week. For example, to study the relative abundance at week 3 (t=3), we used 7-day total NISS scores at week 1 (stress1) and 7-day total NISS score at week 2 (stress2) as covariates. All the continuous variables were standardized.
Figure 2.
Example of a genus from the gut microbiome data. Bars represent the histogram and the curve represents the kernel density estimate of Enterococcus.
We applied the ZIBR to each taxon individually and used the Benjamini-Hochberg approach (Benjamini & Hochberg, 1995) to control the false discovery rate (FDR=5%). P-values are computed for each part (Binary and Beta) of the ZIBR model. To be more specific, p-values from the Binary part measure the strength of association between the presence of genera and covariates, while p-values from the Beta part measure the strength of association between the non-zero relative abundance of genera and covariates (Chen & Li, 2016). In addition, we calculate the overall p-values from ZIBR model, which measure the strength of overall association between genera and covariates by integrating the covariate effects from both Binary and Beta parts. All the analyses were run in SAS version 9.4 and R version 3.4.1 using R package ZIBR.
3. Results
A total of 82 infants were born between 24–37 weeks postmenstrual age and enrolled in the parent study. After adjusting the sample to only those infants with at most one missing sample within weeks 3–6, the sample was reduced to 47 infants. For the 47 infants included in these analyses, there was complete data for all the covariates. Mean birth weight was 1075 grams (range 953–1200), infants were 55% female, mean gestational age at birth was 28 weeks and the majority of infants (72%) were delivered by cesarean section. See Table 1 for additional sample characteristics. The count and percentage of microbiome samples for each week was: week1= 31(37.80), week2= 53(64.63), week3= 69(84.15), week4= 66(80.49), week5= 63(76.83) and week6= 55(67.07).
Table 1.
Descriptive statistics of study sample, n = 47. Shown as percentage or median (inter-quartile range).
| Delivery Method | Vaginal (0) | 13 | 27.66 |
| C-section (1) | 34 | 72.34 | |
| Antibiotic | No (0) | 24 | 51.06 |
| Yes (1) | 23 | 48.94 | |
| Baby Gender | Male (0) | 21 | 44.68 |
| Female (1) | 26 | 55.32 | |
| Birth Weight (g) | 1075.00 | (952.50, 1200.00) | |
| Gestational Age (day) | 27.60 | (26.50, 29.00) | |
| Cumulative Weekly Stress Score | Week 1 | 937.00 | (875.00, 1018.00) |
| Week 2 | 943.00 | (847.00, 1026.00) | |
| Week 3 | 915.00 | (827.00, 977.00) | |
| Week 4 | 883.00 | (805.00, 975.00) | |
| Week 5 | 824.00 | (778.00, 923.00) | |
| Week 6 | 808.00 | (763.00, 878.00) |
We report in Table 2 the estimates from the Binary (Panel A) and Beta (Part B) parts of the ZIBR model, respectively. A positive estimate in Panel A, with significant p-value, indicates that the gene is more likely to be present in the stool sample with larger value of the covariate. Similarly, a positive estimate in Panel B, with significant p-value, indicates that the gene is more likely to have larger non-zero relative abundance with larger value of the covariate.
Table 2.
Panel A. Estimates from the Binary part of ZIBR model for 11 bacterial genera.
| Species | Week4 | Week5 | Week6 | Antibiotic | Baby Gender |
Birth Weight |
Gestational Age |
Stresst-2 | Stresst-1 |
|---|---|---|---|---|---|---|---|---|---|
| Klebsiella | 0.359 | 0.025 | 0.818 | −1.659 | −3.808 | 4.073 | 1.174 | 0.012 | −0.485 |
| Escherichia | 2.609 | 3.486 | 4.802 | −1.941 | 0.259 | 1.129 | −2.368 | −0.110 | −0.783 |
| Staphylococcus | −0.975 | −1.822 | −1.591 | 0.501 | −0.176 | 0.455 | −0.042 | 0.151 | −0.111 |
| Enterococcus | 1.849 | 2.594 | 3.899 | −2.285 | −2.937 | −1.154 | 0.941 | −1.122 | −0.304 |
| Bifidobacterium | 1.135 | 2.492 | 3.052 | −3.428 | 1.210 | 3.063 | −2.644 | 0.178 | −0.782 |
| Proteus | 5.092 | 3.480 | 5.089 | 2.217 | −5.328 | −4.582 | 2.959 | −1.954 | −1.841 |
| Citrobacter | 0.368 | 1.549 | 1.077 | −1.446 | −4.263 | 0.619 | 2.869 | −0.228 | 0.539 |
| Streptococcus | 1.287 | 2.105 | 2.541 | 0.176 | −1.383 | 0.598 | −0.339 | −0.176 | 0.119 |
| Clostridium. butyricum |
1.126 | 1.534 | 1.855 | −0.075 | −1.749 | −0.007 | 1.015 | −0.185 | 0.434 |
| Clostridium. perfringens |
1.514 | 1.858 | 2.847 | −2.633 | −0.850 | 0.842 | 0.585 | 0.471 | 0.169 |
| Veillonella | −0.159 | −0.033 | 1.826 | −4.205 | −5.926 | −0.528 | 0.364 | −0.646 | −2.223 |
Table 2.
Panel B. Estimates from the Beta part of ZIBR model for 11 bacterial genera.
| Species | Week4 | Week5 | Week6 | Antibiotic | Baby Gender |
Birth Weight |
Gestational Age |
Stresst-2 | Stresst-1 |
|---|---|---|---|---|---|---|---|---|---|
| Klebsiella | 0.438 | −0.160 | −0.408 | 0.159 | 0.504 | −0.080 | −0.102 | 0.000 | −0.018 |
| Escherichia | 0.145 | 0.218 | −0.156 | 0.229 | −0.738 | −0.381 | −1.089 | −0.267 | −0.020 |
| Staphylococcus | −0.290 | −0.218 | −0.357 | 0.061 | −0.102 | −0.001 | −0.005 | −0.037 | 0.092 |
| Enterococcus | 0.163 | 0.531 | 0.341 | −0.425 | 0.144 | 0.141 | −0.205 | −0.053 | −0.165 |
| Bifidobacterium | 0.286 | 0.299 | 0.422 | −0.218 | 0.228 | −0.162 | 0.050 | 0.292 | 0.140 |
| Proteus | 1.749 | 0.210 | 0.642 | −3.057 | −0.586 | −0.086 | −0.906 | −0.487 | 1.380 |
| Citrobacter | −0.565 | −0.477 | −0.388 | 1.285 | 0.324 | −0.123 | 0.184 | 0.066 | −0.075 |
| Streptococcus | 0.304 | 0.365 | 0.032 | −0.090 | 0.490 | 0.438 | −0.221 | 0.051 | 0.069 |
| Clostridium. butyricum |
−0.006 | 0.008 | −0.211 | −0.051 | −0.224 | −0.098 | −0.020 | 0.184 | 0.212 |
| Clostridium. perfringens |
0.060 | −0.089 | −1.409 | 1.847 | 1.333 | −0.525 | 0.232 | −0.286 | 0.940 |
| Veillonella | −0.328 | −0.527 | −0.505 | −0.448 | −0.243 | 0.210 | 0.045 | 0.086 | 0.047 |
All of the following p-values are FDR-adjusted p-values. In Table 3, we report the overall p-values from ZIBR model. Table 3 shows that stress 2 weeks prior (stresst-2) had a significant effect only on Proteus (overall p-value = 0.015). In addition, we observed that stress 1 week prior (stresst-1) had a significant effect on two genera, including Proteus (overall p-value < 0.001) and Veillonella (overall p-value = 0.033). Larger stress scores were associated with smaller probabilities of Proteus and Veillonella being present. However, when Proteus and Veillonella were present, larger stress scores were associated with larger values of relative abundances for both genera.
Table 3.
Overall p-values from ZIBR model for 11 bacterial genera.
| Species | Week4 | Week5 | Week6 | Antibiotic | Baby Gender | Birth Weight | Gestational Age | Stresst-2 | Stresst-1 |
|---|---|---|---|---|---|---|---|---|---|
| Klebsiella | 0.582 | 0.888 | 0.273 | 0.534 | 0.247 | 0.037* | 1.000 | 1.000 | 0.970 |
| Escherichia | 0.424 | 0.060 | 0.004** | 0.691 | 0.575 | 0.287 | 0.090 | 0.693 | 0.928 |
| Staphylococcus | 0.483 | 0.018* | 0.017* | 0.691 | 0.934 | 0.461 | 1.000 | 0.990 | 0.970 |
| Enterococcus | 0.424 | 0.010* | 0.003** | 0.099 | 0.575 | 0.237 | 0.280 | 0.349 | 0.896 |
| Bifidobacterium | 0.672 | 0.059 | 0.004** | 0.136 | 0.563 | 0.237 | 0.255 | 0.693 | 0.872 |
| Proteus | 0.000*** | 0.010* | 0.003** | 0.001** | 0.034* | 0.048* | 0.104 | 0.015* | 0.000*** |
| Citrobacter | 0.868 | 0.236 | 0.350 | 0.022* | 0.331 | 0.533 | 0.700 | 0.990 | 0.928 |
| Streptococcus | 0.483 | 0.044* | 0.006** | 0.982 | 0.331 | 0.287 | 0.894 | 0.990 | 0.970 |
| Clostridium. butyricum |
0.612 | 0.169 | 0.042* | 0.982 | 0.221 | 0.863 | 0.255 | 0.867 | 0.872 |
| Clostridium. perfringens |
0.483 | 0.169 | 0.003** | 0.005** | 0.113 | 0.100 | 0.290 | 0.532 | 0.052 |
| Veillonella | 0.893 | 0.499 | 0.059 | 0.099 | 0.221 | 0.287 | 0.767 | 0.773 | 0.033* |
p-value < 0.05;
p-value < 0.01;
p-value < 0.001
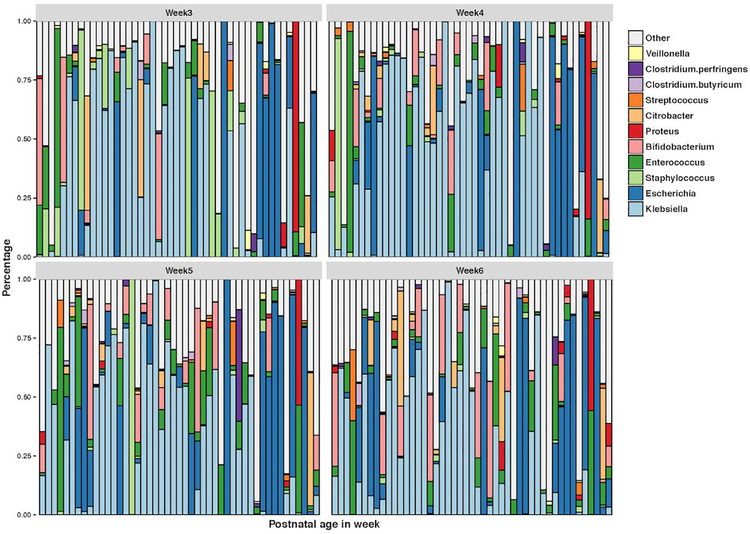
Figure 3 shows the relative abundance of the significant genera by week. Table 3 shows the overall bacteria genera p-value. The model identified 8 genera for the week effect: Escherichia (week6: overall p-value = 0.004); Staphylococcus (week5: overall p-value = 0.018; week6: overall p-value = 0.017); Enterococcus (week5: overall p-value = 0.010; week6: overall p-value = 0.003); Bifidobacterium (week6: overall p-value = 0.004); Proteus (week4: overall p-value < 0.001; week5: overall p-value = 0.010; week6: overall p-value = 0.003); Streptococcus (week5: overall p-value = 0.044; week6: overall p-value = 0.006); Clostridium.butyricum (week6: overall p-value=0.042); Clostridium.perfringens (week6: overall p-value = 0.003).
Figure 3.
Relative abundance of significant genera by week of life.
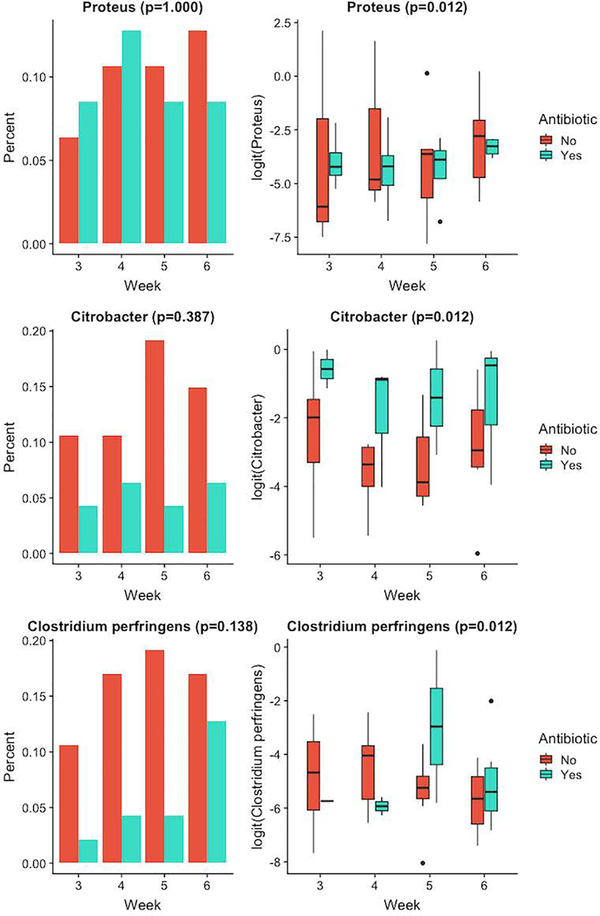
Figure 4 shows the relative abundances of the three genera with significant overall antibiotic effect, by week and antibiotic usage. Antibiotic did not lead to different probabilities of observing Proteus (p-value = 1.000), Citrobacter (p-value = 0.387) or Clostridium.perfringens (p-value = 0.138) but led to different values of non-zero relative abundances: Proteus (p-value = 0.012), Citrobacter (p-value = 0.012) and Clostridium.perfringens (p-value = 0.012), adjusting for the other covariates.
Figure 4.
Relative abundance of genera with significant effect for antibiotic (0= no antibiotic usage, 1= yes antibiotic usage). Left panel shows the percentage of samples in the two antibiotic groups where the genus was present. Right panel shows the logit-transformed non-zero relative abundance in the two groups.
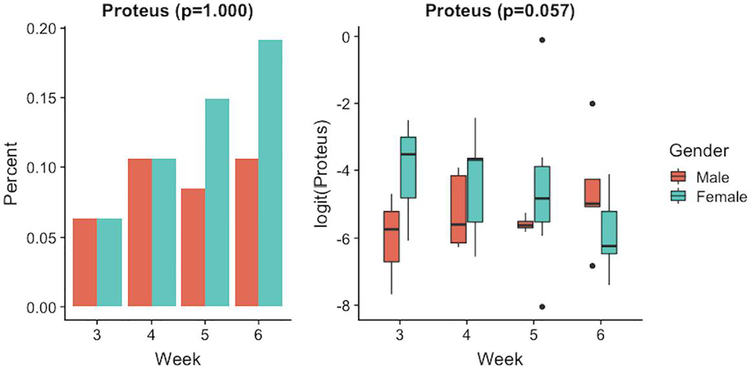
Proteus was the only genus that had a significant overall gender effect (overall p-value = 0.034), see Figure 5. However, if we consider each part of the model separately, gender did not lead to different probabilities of observing Proteus (p-value = 1.000) or different values of relative abundances for Proteus (p-value = 0.057) given Proteus was present.
Figure 5.
Relative abundance of genus with significant effect for gender (0= male and 1= female). Left panel shows the percentage of samples in the two gender groups where the genus was present. Right panel shows the logit-transformed non-zero relative abundance in the two groups.
Two genera were significantly affected by the birth weight, including Klebsiella and Proteus (overall p-value = 0.037; overall p-value = 0.048). Gestational age was not a significant factor for all the genera considered in this model.
4. Discussion
Stress exposure has been reported to alter biological mechanisms in preterm infants who require neonatal intensive care. Understanding the role NICU stress exposure plays in the developing gut microbiome however requires further investigation. By examining the microbial abundance and diversity of preterm infant stool during the delivery of intensive care, this study sought to explore if higher intensities of stress exposure alter the gut microbiome composition during the first 6 weeks of life. Several differences in bacterial genera during this time period. We found an overall dominance from the Gammaproteobacteria taxon. In particular, sampling weeks 4, 5, 6, antibiotic usage, gender, birth weight and stress exposure during weeks 1 and 2 prior to sampling had significant effects on the dominance of Proteus. Other significant effects of Gammaproteobacteria included birth weight on Klebsiella, week6 of sampling on Escherichia, antibiotic usage on Citrobacter (see Table 3).
In this study, we were particularly interested in understanding what role stress exposure from NICU care may play in the developing gut microbiome. Using the NISS, daily stress exposure was summed into weekly totals. Our findings indicate that stress exposure during 1 and/or 2 weeks prior to sampling had a significant effect on both presence and relative abundance of Proteus and Veillonella, with Clostridium_perfringens approaching significance. Furthermore, the estimates indicate that if these genera are present, infants with higher stress exposure tend to have higher relative abundances of Proteus and Veillonella. In the animal model, stress has been linked to bowel disease (Chow, Tang, & Mazmanian, 2011; Medel‐Matus, Shin, Dorfman, Sankar, & Mazarati, 2018; Reber et al., 2016). Additionally, the presence of Proteobacteria pathobionts species have been shown to profoundly influence vulnerability to gut pathology (Chow et al., 2011; Langgartner et al., 2017). In humans, connections are emerging between the microbiota and brain health (Jiang et al., 2018; Pulikkan et al., 2018; Shen et al., 2018; Winter, Hart Robert, Charlesworth Richard, & Sharpley Christopher, 2018). As research continues to elucidate the microbiota-brain-gut axis (Foster, Rinaman, & Cryan, 2017), it will be important to understand how early life environmental stress exposure and dysbiosis effect preterm infant short and long-term neurodevelopment (Groer et al., 2014).
Gammaproteobacteria are Gram-negative rod-shaped facultative anaerobes. There are many potentially virulent organisms in this family (e.g. Escherichia, Klebsiella, Serratia, Enterobacter and Proteus) and many of these are related to common NICU morbidities (Mithal, Palac, Yogev, Ernst, & Mestan, 2017; Patel et al., 2016). Veillonella are Gram-negative commensal obligate anaerobes; however, Veillonella has been implicated in intestinal pathology (Bajer et al., 2017). Preterm infant guts are believed to follow a nonrandom process of convergence to Clostridia by initial dominance of Bacilli followed by a surge of Gammaproteobacteria blooms, with a gradual increase in Clostridia. Zwittink et al (2017) found GA and associated intensity of NICU care (respiratory support and antibiotic treatment) to be associated with Bifidobacterium abundance whereas, extremely preterm infants demonstrated lower abundance of Bifidobacterium and higher abundance of facultative anaerobes (Zwittink et al., 2017). In addition to the present study’s aforementioned dominance of Gammaproteobacteria, our other dominant taxa included Bacilli (Staphylococcus, Streptococcus, Enterococcus) and Clostridia (Veillonella, Clostridium) genera.
A time series abundance study of a single preterm infant’s gut colonization revealed week 3 of life to represent dynamic changes in microbial community structure. Change were marked by dominance of the facultative anaerobe E-coli early in week 3, then transitioning to obligate anaerobes (Streptococcus, Clostridium_butyricum and Veillonella) by the end of the same week (Brown et al., 2013). Researchers suggest these changes may reflect fermentation based metabolism, as there were no clinical indicators to account for the observed changes (Brown et al., 2013). In our study, week5 had a significant effect on Staphylococcus, Enterococcus and Streptococcus. Additionally, week6 had a significant effect on Escherichia, Staphylococcus, Enterococcus, Bifidobacterium, Streptococcus, Clostridium_butyricum, Clostridium_perfringens. Findings suggest that some of our time effect significance may be due to expected microbial succession.
Our only binary covariates included antibiotics and gender. Infants with antibiotic use had significantly less Proteus and significantly more Citrobacter and Clostridium_perfringens when these genera were present, see Figure 4. Additionally, female infants had significantly less Proteus microbes when Proteus was present, see Figure 5.
In our study, intertwined effects were noted in which some covariates had an effect on particular OTUs, which may be important to preterm infant health and gut composition. Proteus and Veillonella were significantly affected by stress exposure (Clostridium_perfringens approached significance) while, Proteus and Clostridium_perfringens were significantly affected by antibiotic usage (Veillonella approached significance). When considering opportunities for future causal research, these findings beg the question, did these preterm infants experience more stress because they were sick and required antibiotics, thus creating an environment for potentially virulent bacteria or did antibiotic usage cause an increased risk through overabundance which then is additive with gender, GA and stress? Investigating the functional significance of the dysbiosis will be important for clinical practice. Of note, effects of stress experienced by preterm infants in the NICU have not been well studied. Continued interrogation of what role early life NICU stress, as well as nurturing experiences, play in preterm infant outcomes and how to best measure them are critically important. Other considerations for future research include sample size, stress exposure and missing data suggestions. First, given our small sample size, it will be important to replicate this study in a larger sample size with comparable NICU stress exposure data. Second, investigating if particular NICU stressors or interventions have more of an effect on the presence and abundance of certain OTUs will be important for both research and clinical practice. Third, understanding if there may be a metabolic interaction between Proteus and Veillonella will be important when considering bacterial risk. Finally, future studies are needed that develop new statistical methods to handle missing compositional data when the missing completely at random assumption is violated.
In summary, the process of infant gut colonization is dynamic and influenced by many environmental factors. For vulnerable preterm infants who experience high NICU stress exposure, our data suggests these infants also experience increased microbial abundances of Proteus and Veillonella. As a species of Gammaproteobacteria, dominance of Proteus may pose an increased risk for immunocompromised preterm infants. Antibiotic usage, gender, birth weight and week of life were also significant for multiple genera. The factors found to be significant in this study touch almost all NICU infants. Thus, continued investigations of how stress and other environmental factors potentially pose risks to the developing gut microbiome represent exciting opportunities for future research.
Acknowledgements:
The authors wish to thank the parents and infants who participated in this study. Thank you to Tracy Burke and Dionna Stibila for their data collection support and tireless work on behalf of NICU infants and their families.
Funding: This work was supported by the National Institutes of Health [R01NR015446]; Little Giraffe Foundation, Chicago, IL.
Contributor Information
Amy L. D’Agata, College of Nursing, University of South Florida.
Jing Wu, Computer Science and Statistics, University of Rhode Island.
Manushi K. V. Welandawe, Computer Science and Statistics, University of Rhode Island.
Samia V. O. Dutra, College of Nursing, University of South Florida.
Bradley Kane, College of Nursing, University of South Florida.
Maureen W. Groer, College of Nursing, University of South Florida.
References
- Arboleya S, Sánchez B, Solís G, Fernández N, Suárez M, Hernández-Barranco AM, … Gueimonde M (2016). Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: A functional inference study. International Journal of Molecular Sciences, 17(5), 649. doi: 10.3390/ijms17050649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, … Drastich P (2017). Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World Journal of Gastroenterology, 23(25), 4548–4558. doi: 10.3748/wjg.v23.i25.4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP (1998). In utero programming of chronic disease. Clinical Science, 95(2), 115–128. doi: 10.1042/cs0950115 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Bentley JP, Simpson JM, Bowen JR, Morris JM, Roberts CL, & Nassar N (2016). Gestational age, mode of birth and breastmilk feeding all influence acute early childhood gastroenteritis: a record-linkage cohort study. BMC Pediatrics, 16(1), 1–10. doi: 10.1186/s12887-016-0591-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolisetty S, Dhawan A, Abdel-Latif M, Bajuk B, Stack J, & Lui K (2014). Intraventricular hemorrhage and neurodevelopmental outcomes in extreme preterm infants. Pediatrics, 133(1), 55–62. doi: 10.1542/peds.2013-0372 [DOI] [PubMed] [Google Scholar]
- Brown CT, Sharon I, Thomas BC, Castelle CJ, Morowitz MJ, & Banfield JF (2013). Genome resolved analysis of a premature infant gut microbial community reveals a Varibaculum cambriense genome and a shift towards fermentation-based metabolism during the third week of life. Microbiome, 1, 30–30. doi: 10.1186/2049-2618-1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Xiong W, Olm MR, Thomas BC, Baker R, Firek B, … Banfield JF (2018). Hospitalized premature infants are colonized by related bacterial strains with distinct proteomic profiles. mBio, 9(2), e00441–00418. doi: 10.1128/mBio.00441-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EZ, & Li H (2016). A two-part mixed-effects model for analyzing longitudinal microbiome compositional data. Bioinformatics, 32(17), 2611–2617. doi: 10.1093/bioinformatics/btw308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova DA, Madan JC, Housman ML, Zain-ul-abideen M, Lundgren SN, Morrison HG, … Hoen AG (2018). The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatric Research. doi: 10.1038/s41390-018-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Tang H, & Mazmanian SK (2011). Pathobionts of the gastrointestinal microbiota and inflammatory disease. Current Opinion in Immunology, 23(4), 473–480. doi: 10.1016/j.coi.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian EA, Jin DL, Attenello F, Wen T, Cen S, Mack WJ, … McComb JG (2015). Trends in hospitalization of preterm infants with intraventricular hemorrhage and hydrocephalus in the United States, 2000–2010. Journal of Neurosurgery: Pediatrics, 17(3), 260–269. doi: 10.3171/2015.7.PEDS15140 [DOI] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, … Cryan JF (2013). The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Molecular Psychiatry, 18(6), 666–673. doi: 10.1038/mp.2012.77 [DOI] [PubMed] [Google Scholar]
- Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, & Dinan TG (2014). Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol, 28(8), 1221–1238. doi: 10.1210/me.2014-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agata AL, Walsh S, Vittner D, Cong X, McGrath JM, & Young EE (2017). FKBP5 genotype and early life stress exposure predict neurobehavioral outcomes for preterm infants. Developmental Psychobiology, 59(3), 410–418. doi: 10.1002/dev.21507 [DOI] [PubMed] [Google Scholar]
- D’Agata AL, Young EE, Cong X, Grasso DJ, & McGrath JM (2016). Infant medical trauma in the neonatal intensive care unit (IMTN): A proposed concept for science and practice. Adv Neonatal Care. doi: 10.1097/anc.0000000000000309 [DOI] [PubMed] [Google Scholar]
- Dahl C, Stigum H, Valeur J, Iszatt N, Lenters V, Peddada S, … Eggesbø M (2018). Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. International Journal of Epidemiology, dyy064–dyy064. doi: 10.1093/ije/dyy064 [DOI] [PubMed] [Google Scholar]
- Dantzer R, Cohen S, Russo SJ, & Dinan TG (2018). Resilience and immunity. Brain Behav Immun. doi: 10.1016/j.bbi.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G, Collins SM, Bercik P, & Verdu EF (2014). The microbiota-gut-brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both? J Physiol, 592(14), 2989–2997. doi: 10.1113/jphysiol.2014.273995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, & Knight R (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America, 107(26), 11971–11975. doi: 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, … Clemente JC (2016). Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med, 22(3), 250–253. doi:10.1038/nm.4039http://www.nature.com/nm/journal/v22/n3/abs/nm.4039.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbons SC, Ching Y, Yu D, Carpenter J, Kenny M, Weldon C, … Jaksic T (2009). Mortality of necrotizing enterocolitis expressed by birth weight categories. Journal of Pediatric Surgery, 44(6), 1072–1076. doi: 10.1016/j.jpedsurg.2009.02.013 [DOI] [PubMed] [Google Scholar]
- Foster JA, Rinaman L, & Cryan JF (2017). Stress & the gut-brain axis: Regulation by the microbiome. Neurobiology of Stress, 7, 124–136. doi: 10.1016/j.ynstr.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, … Knight R (2016). Microbiome-wide association studies link dynamic microbial consortia to disease. Nature, 535(7610), 94–103. doi: 10.1038/nature18850 [DOI] [PubMed] [Google Scholar]
- Groer M, Ashmeade T, Duffy A, Morse S, & Zaritt J (2016). Changes in the Immune Components of Preterm Human Milk and Associations With Maternal and Infant Characteristics. J Obstet Gynecol Neonatal Nurs, 45(5), 639–648. doi: 10.1016/j.jogn.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Groer M, Ashmeade T, Louis-Jacques A, Beckstead J, & Ji M (2016). Relationships of Feeding and Mother’s Own Milk with Fecal Calprotectin Levels in Preterm Infants. Breastfeed Med, 11, 207–212. doi: 10.1089/bfm.2015.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, & Gilbert JA (2014). Development of the preterm infant gut microbiome: a research priority. Microbiome, 2, 38–38. doi: 10.1186/2049-2618-2-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnar MR, Wewerka S, Frenn K, Long JD, & Griggs C (2009). Developmental changes in hypothalamus–pituitary–adrenal activity over the transition to adolescence: Normative changes and associations with puberty. Development and psychopathology, 21(1), 69–85. doi: 10.1017/S0954579409000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, … Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America, 108(7), 3047–3052. doi: 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TTB, Groer MW, Kane B, Yee AL, Torres BA, Gilbert JA, & Maheshwari A (2018). Dichotomous development of the gut microbiome in preterm infants. Microbiome, 6, 157. doi: 10.1186/s40168-018-0547-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV (2004). Bacterial contributions to mammalian gut development. Trends in Microbiology, 12(3), 129–134. doi: 10.1016/j.tim.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Jensen SG, Dickie EW, Schwartz DH, & et al. (2015). EFfect of early adversity and childhood internalizing symptoms on brain structure in young men. JAMA Pediatrics, 169(10), 938–946. doi: 10.1001/jamapediatrics.2015.1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. y., Zhou Y. y., Zhou G. l., Li Y. c., Yuan J, Li X. h., & Ruan B (2018). Gut microbiota profiles in treatment-naïve children with attention deficit hyperactivity disorder. Behavioural Brain Research, 347, 408–413. doi: 10.1016/j.bbr.2018.03.036 [DOI] [PubMed] [Google Scholar]
- Kaplan BJ, Rucklidge JJ, Romijn A, & McLeod K (2015). The emerging field of nutritional mental health: Inflammation, the microbiome, oxidative stress, and mitochondrial function. Clinical Psychological Science, 3(6), 964–980. doi: 10.1177/2167702614555413 [DOI] [Google Scholar]
- Karl JP, Hatch AM, Arcidiacono SM, Pearce SC, Pantoja-Feliciano IG, Doherty LA, & Soares JW (2018). Effects of Psychological, Environmental and Physical Stressors on the Gut Microbiota. Front Microbiol, 9, 2013. doi: 10.3389/fmicb.2018.02013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro H, Anderson PJ, Doyle LW, Woodward LJ, Neil JJ, & Inder TE (2014). Brain injury and altered brain growth in preterm infants: predictors and prognosis. Pediatrics, 134(2), e444–453. doi: 10.1542/peds.2013-2336 [DOI] [PubMed] [Google Scholar]
- Kostic Aleksandar D., Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, … Xavier Ramnik J. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell host & microbe, 17(2), 260–273. doi: 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, … Tarr PI (2014). Patterned progression of bacterial populations in the premature infant gut. Proceedings of the National Academy of Sciences of the United States of America, 111(34), 12522–12527. doi: 10.1073/pnas.1409497111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kriete A, & Rosen GL (2013). Selecting age-related functional characteristics in the human gut microbiome. Microbiome, 1, 2–2. doi: 10.1186/2049-2618-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langgartner D, Peterlik D, Foertsch S, Füchsl AM, Brokmann P, Flor PJ, … Reber SO (2017). Individual differences in stress vulnerability: The role of gut pathobionts in stress-induced colitis. Brain Behav Immun, 64, 23–32. doi: 10.1016/j.bbi.2016.12.019 [DOI] [PubMed] [Google Scholar]
- Lax S, & Gilbert JA (2015). Hospital-associated microbiota and implications for nosocomial infections. Trends in molecular medicine, 21(7), 427–432. doi: 10.1016/j.molmed.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Little RJA (1988). A Test of Missing Completely at Random for Multivariate Data with Missing Values. Journal of the American Statistical Association, 83(404), 1198–1202. doi: 10.1080/01621459.1988.10478722 [DOI] [Google Scholar]
- Liu RT (2017). The microbiome as a novel paradigm in studying stress and mental health. American Psychologist, 72(7), 655–667. doi: 10.1037/amp0000058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Yu Y, Guo Y, Wang Y, Chang EB, & Claud EC (2015). Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model. PLoS ONE, 10(4), e0124504. doi: 10.1371/journal.pone.0124504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathewson KJ, Chow CHT, Dobson KG, Pope EI, Schmidt LA, & Van Lieshout RJ (2017). Mental health of extremely low birth weight survivors: A systematic review and meta-analysis. Psychological Bulletin, 143(4), 347–383. doi: 10.1037/bul000009110.1037/bul0000091.supp (Supplemental) [DOI] [PubMed] [Google Scholar]
- Medel‐Matus JS, Shin D, Dorfman E, Sankar R, & Mazarati A (2018). Facilitation of kindling epileptogenesis by chronic stress may be mediated by intestinal microbiome. Epilepsia Open, 3(2), 290–294. doi: 10.1002/epi4.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithal LB, Palac HL, Yogev R, Ernst LM, & Mestan KK (2017). Cord blood acute phase reactants predict early onset neonatal sepsis in preterm infants. PLoS ONE, 12(1), e0168677. doi: 10.1371/journal.pone.0168677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NT, Bakacs E, Combellick J, Grigoryan Z, & Dominguez-Bello MG (2015). The infant microbiome development: mom matters. Trends in molecular medicine, 21(2), 109–117. doi: 10.1016/j.molmed.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, & Foster JA (2011). Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterology & Motility, 23(3), 255–e119. doi: 10.1111/j.1365-2982.2010.01620.x [DOI] [PubMed] [Google Scholar]
- Newnham CA, Inder TE, & Milgrom J (2009). Measuring preterm cumulative stressors within the NICU: The neonatal infant stressor scale. Early Human Development, 85(9), 549–555. doi: 10.1016/j.earlhumdev.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Patel AL, Mutlu EA, Sun Y, Koenig L, Green S, Jakubowicz A, … Keshavarzian A (2016). Longitudinal survey of microbiota in hospitalized preterm very low birth weight infants. Journal of pediatric gastroenterology and nutrition, 62(2), 292–303. doi: 10.1097/MPG.0000000000000913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan J, Maji A, Dhakan DB, Saxena R, Mohan B, Anto MM, … Sharma VK (2018). Gut microbial dysbiosis in indian children with autism spectrum disorders. Microbial Ecology. doi: 10.1007/s00248-018-1176-2 [DOI] [PubMed] [Google Scholar]
- Pyhälä R, Wolford E, Kautiainen H, Andersson S, Bartmann P, Baumann N, … Räikkönen K (2017). Self-reported mental health problems among adults born preterm: A meta-analysis. Pediatrics, 139(4). [DOI] [PubMed] [Google Scholar]
- Raveh-Sadka T, Firek B, Sharon I, Baker R, Brown CT, Thomas BC, … Banfield JF (2016). Evidence for persistent and shared bacterial strains against a background of largely unique gut colonization in hospitalized premature infants. Isme j. doi: 10.1038/ismej.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SO, Siebler PH, Donner NC, Morton JT, Smith DG, Kopelman JM, … Lowry CA (2016). Immunization with a heat-killed preparation of the environmental bacterium <em>Mycobacterium vaccae</em> promotes stress resilience in mice. Proceedings of the National Academy of Sciences, 113(22), E3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale A, Berardi N, & Maffei L (2014). Environment and brain plasticity: Towards an endogenous pharmacotherapy. Physiological Reviews, 94(1), 189–234. doi: 10.1152/physrev.00036.2012 [DOI] [PubMed] [Google Scholar]
- Sampson TR, & Mazmanian SK (2015). Control of brain development, function, and behavior by the microbiome. Cell host & microbe, 17(5), 565–576. doi: 10.1016/j.chom.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, & Mazmanian SK (2014). Specialized metabolites from the microbiome in health and disease. Cell metabolism, 20(5), 719–730. doi: 10.1016/j.cmet.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J, … Liang Y (2018). Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: A cross-sectional study. Schizophrenia Research. doi: 10.1016/j.schres.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Shin N-R, Whon TW, & Bae J-W (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in Biotechnology, 33(9), 496–503. doi: 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Shonkoff JP, Garner AS, Siegel BS, Dobbins MI, Earls MF, Garner AS, … Wood DL (2012). The lifelong effects of early childhood adversity and toxic stress. Pediatrics, 129(1), e232–e246. doi: 10.1542/peds.2011-2663 [DOI] [PubMed] [Google Scholar]
- Steinway SN, Biggs MB, Loughran TP Jr., Papin JA, & Albert R (2015). Inference of network dynamics and metabolic interactions in the gut microbiome. PLOS Computational Biology, 11(6), e1004338. doi: 10.1371/journal.pcbi.1004338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, … Cryan JF (2015). Microbes & neurodevelopment--Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun, 50, 209–220. doi: 10.1016/j.bbi.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Stoll BJ, Hansen NI, Bell EF, & et al. (2015). Trends in care practices, morbidity, and mortality of extremely preterm neonates, 1993–2012. JAMA, 314(10), 1039–1051. doi: 10.1001/jama.2015.10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, … Koga Y (2004). Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol, 558(Pt 1), 263–275. doi: 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall NC, & Nemeroff CB (2015). The preeminence of early life trauma as a risk factor for worsened long-term health outcomes in women. Current Psychiatry Reports, 17(11), 1–18. doi: 10.1007/s11920-015-0625-6 [DOI] [PubMed] [Google Scholar]
- Winter G, Hart Robert A, Charlesworth Richard PG, & Sharpley Christopher F (2018). Gut microbiome and depression: what we know and what we need to know Reviews in the Neurosciences (Vol. 0). [DOI] [PubMed] [Google Scholar]
- Zwittink RD, van Zoeren-Grobben D, Martin R, van Lingen RA, Groot Jebbink LJ, Boeren S, … Knol J (2017). Metaproteomics reveals functional differences in intestinal microbiota development of preterm infants. Molecular & Cellular Proteomics, 16(9), 1610–1620. doi: 10.1074/mcp.RA117.000102 [DOI] [PMC free article] [PubMed] [Google Scholar]