Abstract
The late-infantile Batten disease or late-infantile neuronal ceroid lipofuscinosis (LINCL) is an autosomal recessive lysosomal storage disorder caused by mutations in the Cln2 gene leading to deficiency of lysosomal enzyme tripeptidyl peptidase 1 (TPP1). At present, available options for this fatal disorder are enzyme replacement therapy and gene therapy, which are extensively invasive and expensive. Our study demonstrates that 3-hydroxy-(2,2)-dimethyl butyrate (HDMB), a brain endogenous molecule, is capable of stimulating TPP1 expression and activity in mouse primary astrocytes and a neuronal cell line. HDMB activated peroxisome proliferator-activated receptor-α (PPARα), which, by forming heterodimer with Retinoid × receptor-α (RXRα), transcriptionally upregulated the Cln2 gene. Moreover, by using primary astrocytes from wild type, PPARα−/− and PPARβ−/− mice, we demonstrated that HDMB specifically required PPARα for inducing TPP1 expression. Finally, oral administration of HDMB to Cln2 heterozygous (Cln2+/−) mice led to a marked upregulation of TPP1 expression in the motor cortex and striatum in a PPARα-dependent fashion. Our study suggests that HDMB, a brain endogenous ligand of PPARα, might have therapeutic importance for LINCL treatment.
Keywords: 3-Hydroxy-(2,2)-dimethyl butyrate; TPP1; LINCL; PPARα
Introduction
The Neuronal ceroid lipofuscinosis (NCL) comprises a group of clinically related hereditary lysosomal storage diseases that primarily occurs during childhood. They exhibit severe neurodegenerative conditions and collectively represent the most predominant neurodegenerative disorders in children with age of onset ranging from birth through early adulthood (1,2). These fatal disorders are clinically characterized by progressive cognitive decline, mental deterioration, impairment of motor function, retinopathy leading to loss of vision and seizures (1,3–5). In spite of having diverse biochemical etiology, a spectrum of symptoms and disease severity, all forms of NCLs share common histopathological features marked by excessive accumulation of autofluorescent ceroid lipopigments in neurons and other cells (1,5). Classically NCLs were grouped based on age of onset, type of storage material; however recently, NCLs have been classified into 14 categories (CLN1–14) based on genetic origin (6,7).
The classic late-infantile NCL (LINCL), formerly known as Janský-Bielschowsky disease (OMIM #204500), is caused by mutations in the CLN2 gene resulting in deficiency in the activity of the lysosomal enzyme tripeptidyl peptidase 1 (TPP1) (EC 3.4.14.9) (8). The Cln2 gene (Ceroid lipofuscinosis, neuronal, 2) (MIM #607998) is a 6.65 kb gene comprised of 13 exons and 12 introns mapped to chromosome 11p15.5. (9,10) and encodes a 563-amino acid containing TPP1 preproenzyme which following removal of signal peptide, glycosylation and cleavage yields the 367 residue long 46 kD mature active TPP1 enzyme (11,12). The protein TPP1 progressively eliminates tripeptides from the N-terminus of small polypeptides, one of which is the subunit c of mitochondrial ATP synthase (SCMAS), the major constituent of the storage granules in LINCL (13,14). Classical LINCL manifests at 2 to 4 years of age followed by the disease course involving seizures, ataxia, myoclonus, developmental delay, speech impairment, progressive neuronal death leading to deterioration of locomotor functions and vision making the patients reach a vegetative state and ultimately death by 8 to 12 years of age (12,15–17). At the ultrastructural level, curvilinear autofluorescent ceroid-lipofuscin granules accumulate in the lysosomes of neurons causing extensive neuronal death by apoptosis (12,15,18).
Gene therapy, enzyme-replacement therapy and stem cell transplantation have been widely explored in animal models as well as human patients for treatment of LINCL. Recently, enzyme replacement therapy using intraventricular infusion of cerliponase alfa (recombinant human TPP1) in children with CLN2 disease has demonstrated reduced impairment of locomotor and language functions; however the mode of treatment was fairly invasive and was associated with a battery of adverse side effects (19). Therefore, at present, there is a requirement for novel, less invasive treatment options for LINCL. Studies have suggested that residual TPP1 activity is retained in LINCL patients, suggesting that a limited number of functional copies of the Cln2 gene are present in these patients (3,20,21). Therefore, drug-mediated therapeutic strategies targeted towards augmenting the expression and residual activity of TPP1 could be of potential benefit for LINCL treatment.
Our lab has demonstrated that FDA-approved fibrate drugs Gemfibrozil and Fenofibrate upregulate TPP1 in brain cells via activation of peroxisome proliferator-activated receptor α (PPARα) (22). Interestingly, our recent study identified a novel endogenous ligand of PPARα, 3-hydroxy-(2,2)-dimethyl butyrate (HDMB), from mouse hippocampus and described a role for HDMB in inducing synaptic functions (23). Given the previous evidence that HDMB activates PPARα, we examined whether HDMB could stimulate TPP1 in mouse brain cells and in a CLN2 mouse model. Here, we demonstrate that HDMB upregulated TPP1 expression in MN9D neuronal cells as well as mouse primary astrocytes via PPARα. Furthermore, oral administration of HDMB enhanced TPP1 levels in the motor cortex and striatum of Cln2+/−, but not Cln2+/− PPARα−/−, mice. Finally, we delineate the underlying mechanism that HDMB stimulates PPARα to form a transcriptional complex on the Cln2 gene promoter to upregulate the expression of Cln2. Therefore, upregulation of TPP1 by a brain endogenous molecule, HDMB, might have therapeutic implications for LINCL.
Materials and methods:
Reagents
DMEM 1x, DMEM F-12 50/50 1x, Hanks’ balanced salt solution (HBSS 1x), and 0.05% trypsin were obtained from Mediatech (Washington, D. C.). Fetal bovine serum (FBS) was purchased from Atlas Biologicals (Fort Collins, CO). Neurobasal media and B27 supplement were purchased from Invitrogen (San Diego, CA). Antibiotic-antimycotic and other cell culture reagents were obtained from Sigma. 3-hydroxy-(2,2)-dimethyl butyrate (HDMB) was obtained from Santacruz Biotechnology Inc. For details on antibodies, please see Table 1.
Table 1: Antibodies, sources, applications, and dilutions used in this paper.
| Target | Antibody (Clone) | Epitope/Immunogen | App lication/D ilution | Source; Catalog |
|---|---|---|---|---|
| β-Actin | Mouse monoclonal (AC-15) | a.a. 1–15 of Xenopus laevis β-actin | WB - 1:5000 | Abcam; ab6276 |
| TPP1 | Rabbit monoclonal EPR16537 | aa 350–500 Recombinant fragment within Human TPP1. | WB - 1:5000 IF - 1:1000 | Abcam; ab195234 |
| GFAP | Rabbit polyclonal | Synth peptide to cow GFAP | IHC - 1:2000 | Dako; z0334 |
| TH | Rabbit polyclonal | SDS denatured from rat TH | IF - 1:1500 | PelFreez Biologicals; P40101–150 |
| PPARα | Mouse monoclonal (3B6) | Recombinant full-length protein | ChIP - 1:200 | Abcam; ab2779 |
| PPARβ | Mouse monoclonal (F-10) × TransCruz antibody | Amino acids 2–75 of PPARβ of mouse origin | ChIP - 1:50 | Santa cruz Biotechnology Inc, sc- 74517 |
| PPARγ | Rabbit monoclonal (81B8) | Synthetic peptide corresponding to residues surrounding His494 of human PPARγ | ChIP - 1:100 | Cell signaling Technology, 2443S |
| PGC1 α | Mouse monoclonal (4C1.3) | Recombinant protein consisting of amino acids 1–120 of mouse PGC-1α | ChIP - 1:1000 | EMD Millipore, ST1202 |
| RXR α | Rabbit monoclonal (D6H10) | Synthetic peptide corresponding to residues near the amino terminus of human RXRα protein | ChIP - 1:100 | Cell Signaling Technology Inc., 3085 |
TPP1, tripeptidyl peptidase I; TH, tyrosine hydroxylase; GFAP, glial fibrillary acidic protein; PPAR, peroxisome proliferator-activated receptor; RXR, retinoid × receptor; PGC1α, PPARγ coactivator 1 alpha; WB, Western blot; IF, immunofluorescence; ChIP, chromatin immunoprecipitation
Animals
Animal maintaining and experiments were in accordance with National Institute of Health guidelines and were approved by the Institutional Animal Care and Use committee (IACUC) of the Rush University of Medical Center, Chicago, IL. Animals exhibiting mild seizures and tremors were fed and watered through animal feeding needles. However, if any mouse came to the moribund stage, it was decapitated after anesthesia with ketamine/xylazine injectables. Conditions for moribund were as follows: Central nervous system disturbance (Head tilt, Seizures, Tremors, Circling, Spasticity, and Paresis); Inability to remain upright; Evidence of muscle atrophy; Chronic diarrhea or constipation; Rough coat and distended abdomen; Spreading area of alopecia caused by disease; Coughing, rales, wheezing and nasal discharge; Distinct jaundice and/or paleness (anemia); Markedly discolored urine, polyuria or anuria; Frank bleeding from any orifice; Persistent self-induced trauma.
Cln2(+/−) animals were kindly provided by Dr. Peter Lobel (Center for Advanced Biotechnology and Medicine, Robert Wood Johnson Medical School, Piscataway, New Jersey, USA). These animals were inbred and subsequent generations were screened by RT-PCR to further obtain Cln2+/+ and Cln2+/− strains.
Treatment of Cln2+/− mice with HDMB
Age- and sex-matched Cln2+/+ mice from the same background were used as wild type (WT) controls and Cln2+/− animals were used in different treatment groups. HDMB (5 mg/kg body wt/day) was dissolved in 0.1% methyl cellulose (MeC). Cln2+/− mice were either not fed (control group), gavaged with 0.1% MeC (vehicle-treated group) or gavaged with HDMB (HDMB-treated group) for 2 weeks for biochemical studies.
Cells
MN9D neuronal cell line was obtained from Dr. A. Heller (University of Chicago, Chicago, IL, USA) and maintained in the lab. Cells were grown in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum and allowed to differentiate in neurobasal media containing 2% B27, glutamine and 1% antibiotic–antimycotic solution (Sigma). For experiments, prior to HDMB treatment, cells were incubated in neurobasal media containing B27 minus antioxidants.
Isolation of Mouse Primary Astroglia
Mouse primary astroglia were isolated from mixed glial cultures as previously described (24,25) and following the procedure of Giulian and Baker (26). Mixed glial cultures isolated from pups were maintained in Dulbecco’s modified Eagle’s medium/F-12–50/50. On day 9, the cultures are subjected to shaking at 240 rpm for 2 h at 37 °C on a rotary shaker for the elimination of microglia. On day 11, the shaking was performed for 24 h for the elimination of oligodendroglia. This ensures the complete elimination of all non-astroglial cells from the culture. The attached cells were astroglia. For experiments, cells were seeded onto new plates and grown for 1 to 2 days before treatment.
Semi-quantitative Reverse Transcriptase-coupled PCR (RT-PCR)
Total RNA was isolated from mouse primary astrocytes and MN9D neuronal cells using the Qiagen RNA-Easy kit (Valencia, CA) following the manufacturer’s protocol. Semi-quantitative reverse-transcriptase PCR was performed according to standard procedure (27,28). Isolated RNA was converted to cDNA using oligo(dT)12–18 as primer and MMLV RT (Moloney murine leukemia virus reverse transcriptase) (Invitrogen) in a 20-μl reaction mixture. The cDNA was appropriately amplified by PCR reactions using Promega Master Mix (Madison, WI) and the following primers (Invitrogen) for murine genes: mouse Cln1, sense 5′-ACACAGAGGACCGCCTGGGG-3′ and antisense 5′-TCATGCACGGCCCACACAGC-3′ mouse Cln2, sense 5′-CACCATCCAGTTACTTCAATGC-3′ and antisense 5′-CTGACCCTCCACTTCTTCATTC-3′ mouse Cln3, sense 5′-TGCTGCCCTGCCATCGAGTG-3′ and antisense 5′-GGCAGCGCTCAGCATCACCA-3′ and mouse GAPDH, sense 5′-GCACAGTCAAGGCCGAGAAT-3′ and antisense 5′-GCCTTCTCCATGGTGGTGAA-3′. Amplified PCR products were electrophoresed on 2% agarose (Invitrogen) gels and detected by ethidium bromide (Invitrogen). The expression of Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control.
Quantitative Real-Time PCR
As described before (22), quantitative real-time PCR was performed using the ABI-Prism7700 sequence detection system (Applied Biosystems, Foster City, CA) using SYBR green. The mRNA expression of the target gene was normalized to the expression of GAPDH mRNA. The fold change in expression of the treated with respect to the untreated control was calculated and represented.
Immunostaining of Cells
Immunocytochemistry was performed as described before (27,28). Briefly, MN9D cells or primary astrocytes were seeded onto square cover glass in 6-well plates and cultured to 70–80% confluency. Following treatment, cells were fixed using chilled methanol (Fisher) overnight, followed by two brief wash with filtered PBS. Cells were blocked using 2% BSA (Fisher) in PBSTT (PBS containing Tween 20 (Sigma) and Triton X-100 (Sigma)) for 30 min and further incubated with primary antibody (in PBS) at room temperature under gentle shaking conditions for 2 h [primary antibodies: TPP1 (1:200; Santa Cruz Biotechnology); GFAP (1:100; Santa Cruz Biotechnology)]. Next, cells were washed with filtered PBS (15min washes, 4times) and incubated with Cy2- or Cy5-labeled secondary antibodies (1:200; Jackson ImmunoResearch) for 1 h under gentle shaking conditions. Following wash in PBS (15min washes, 4times), cells were further incubated with 4′,6-diamidino-2-phenylindole (DAPI, 1:10,000; Sigma) for 4 to 5 min. The cover glass was mounted on glass slides, ran through ethanol and xylene (Fisher) gradient and visualized under Olympus BX41 fluorescence microscope.
Immunostaining of Tissue Sections
It was performed as described previously (22,29). After 14 days of HDMB treatment, mice were perfused using PBS. Hemibrains were incubated in 4% paraformaldehyde followed by 30% sucrose and embedded. Free floating sections were made from different brain regions. For immunohistochemistry, following blocking in 2% BSA, sections were incubated with TPP1 (1:200) and NeuN (1:100) primary antibodies. The sections were mounted, ran through ethanol and xylene gradient and observed under Olympus BX41 fluorescence microscope.
Immunoblotting
Western blotting was conducted as described in earlier studies (22,30,31) with minor modifications. Briefly, cells were scraped in PBS and lysed in RIPA buffer. Proteins were electrophoresed on 12% SDS/PAGE gels and transferred onto nitrocellulose membrane (Bio-Rad) using the Thermo-Pierce Fast Semi-Dry Blotter. Next, the membrane was blocked for 1 h using Li-Cor Odyssey blocking solution (Li-COR, Lincoln, NE) and incubated overnight with primary antibody at 4 °C under shaking conditions [primary antibodies: TPP1 (1:250, Abcam) and β-actin (1:800; Abcam)] (Table 1). The following day, membranes were washed in PBST four times (15min each) and incubated in secondary antibodies against the primary antibody hosts (all 1:10,000; Li-COR) for 1 h at room temperature. Next, the membrane is washed for 1 hour with PBST and visualized under the Odyssey Infrared Imaging System (Li-COR, Lincoln, NE).
TPP1 Activity Assay
It was performed as described by Vines and Warburton (32) with modifications (22). Briefly, cells were homogenized and 20 μg protein was mixed with substrate (40 μl) in individual wells of a polystyrene 96-well plate (Nalge Nunc International). The substrate solution consisted of 250 μmol/liter Ala-Ala-Phe 7-amido-4-methylcoumarin (catalog no. A3401; Sigma; diluted freshly from a 25 mmol/liter stock dissolved in dimethyl sulfoxide and stored at −20 °C) in substrate dilution buffer (0.15 mol/liter NaCl, 1 g/liter Triton X-100, 0.1 mol/liter sodium acetate, adjusted to pH 4.0). Following addition, the plate was briefly centrifuged to remove bubbles and placed in 37°C. Before taking the reading, the plate was mixed for 10 sec. The plate was read in Victor X2 microplate reader (Perkin Elmer) from the bottom using 360/20 nm excitation and 460/25 nm emission filters. Optimum substrate and protein concentrations were determined using different concentrations prior to the assay.
Chromatin Immunoprecipitation (ChIP) Assay
It was performed using the method described by Nelson et al. (33), with modifications (22). Briefly, MN9D cells were stimulated by 10 μM and 20 μM HDMB for 2 h followed by fixing with formaldehyde (1.42% final volume) and quenching with 125 mM glycine. Next, the cells were pelleted and lysed in IP buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, Nonidet P-40 (0.5% v/v), Triton X-100 (1.0% v/v). For 500 ml, add 4.383 g of NaCl, 25 ml of 100 mM EDTA (pH 8.0), 25 ml of 1 M Tris-HCl (pH 7.5), 25 ml of 10% (v/v) Nonidet P-40, and 50 ml of 10% (v/v) Triton X-100 containing the following inhibitors: 10 μg/ml leupeptin, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 30 mM p-nitrophenyl phosphate, 10 mM NaF, 0.1 mM Na3VO4, 0.1 mM Na2MoO4, and 10 mM β-glycerophosphate) (22). Following a wash with 1.0 ml IP buffer, the pellet was resuspended in 1 ml IP buffer (containing all inhibitors), and sonicated. The sheared chromatin was divided into two fractions (one for using as Input). The remaining fraction was incubated with 5 μg of primary antibodies (anti-PPARα, anti-PPARβ, anti-PPARγ, and anti-PGC1α) and normal IgG overnight under rotation at 4 °C followed by incubation with protein G-agarose (Santa Cruz Biotechnology) for 2 h at 4 °C under rotating conditions. Next, beads were washed five times with cold IP buffer, and 100 μl of 10% Chelex (10 g/100 ml H2O) was added directly to the beads and vortexed. Following 10 min boiling, the Chelex/protein G bead suspension was allowed to cool to room temperature. Proteinase K (100 μg/ml) was added, and the beads were incubated for 30 min at 55 °C while shaking, followed by another round of boiling for 10 min. Next, the suspension was centrifuged, and the supernatant was collected. The Chelex/protein G beads fraction was vortexed with another 100 μl of water, centrifuged again, and the first and the second supernatants were combined. The eluate was used directly as a template for PCR. The following primers were used for PCR reactions to amplify fragments flanking the RXR-binding element on the mouse Cln2 promoter: Set1, sense 5′-CAG CTG CCA TGT CCC CCA GC-3′ and antisense 5′-TGC GCA GCT CTG TGT CAT CCG-3′; Set2, sense 5′-GCT CCC TCT CCT CAG CTG CCA-3′ and antisense 5′-CAT CCG GAG GCT CCA GGC CA-3′(34). The PCR reaction was standardized by using varying cycle numbers and different amounts of templates so that the results were in the linear range of PCR (34,35).
Densitometric Analysis
Immunoblots blots were analyzed using ImageJ (National Institutes of Health, Bethesda) and bands were normalized with their respective β-actin loading controls. Data represents the average fold change with respect to untreated control for three independent experiments.
Statistics
Statistical analyses were performed with Student’s t test (for two-group comparisons) and one-way ANOVA, followed by Tukey’s multiple-comparison tests, as appropriate (for multiple groups comparison), using Prism 7 (GraphPad Software). Data represented as mean±SD or mean±SEM as stated in figure legends. A level of p<0.05 was considered statistically significant.
Results
HDMB upregulates TPP1 expression in MN9D neuronal cells
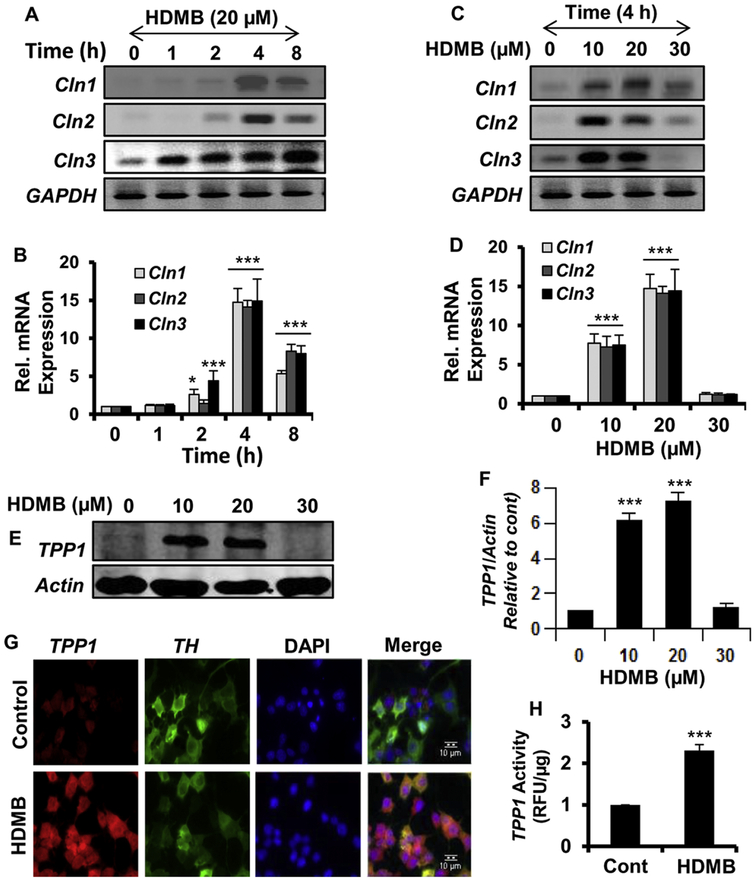
We first examined whether HDMB could upregulate TPP1 levels in MN9D cells, a mouse neuronal cell line. In this regard, we analyzed the mRNA expression of different Cln genes, Cln1, Cln2 and Cln3, which, when mutated, cause infantile, late infantile and juvenile forms of NCL respectively. Semi-quantitative RT-PCR data showed that HDMB, at 20 μM dose, remarkably upregulated the expression of Cln1, Cln2 and Cln3 mRNA in a time-dependent manner (fig 1A). This was further confirmed by a similar pattern of increase in the expression of these genes in real-time PCR (fig 1B). Since HDMB showed the highest effect at 4h time point, we conducted a dose-dependent study with 4h treatment of different doses (10, 20 and 30 μM) of HDMB in MN9D cells. Interestingly, semi-quantitative and real-time PCR data showed that HDMB was able to increase Cln1, Cln2 and Cln3 mRNA expression at 10 and 20 μM concentrations; however the effect decreased at 30 μM HDMB (fig 1C, D). Next, we checked the protein expression of TPP1, the lysosomal enzyme encoded by the Cln2 gene. Parallel to our previous results, HDMB was able to upregulate the protein levels of TPP1 in a dose-dependent manner in MN9D cells (fig 1E, F). Similarly, immunocytochemistry revealed a marked increase in TPP1 expression in TH-positive MN9D cells following 20 μM HDMB treatment (fig 1G). Finally, we evaluated whether HDMB-mediated upregulation of TPP1 mRNA and protein also results in enhanced functionality of this protease. We performed an enzymatic assay for TPP1 in MN9D cells and observed a profound increase in the activity of TPP1 with 20 μM HDMB treatment (fig 1H). Taken together, these results indicate that HDMB augments the expression and activity of TPP1 in MN9D neuronal cells.
Figure 1. HDMB upregulates TPP1 expression in MN9D neuronal cells.
(A-B) For time-point analysis, MN9D cells were treated with 20 μM HDMB in neurobasal media without B27 for 1, 2, 4, and 8 h followed by monitoring the mRNA expression of Cln1, Cln2, and Cln3 by semi-quantitative RT-PCR (A) and real-time qPCR (B). (C-D) For dose-dependent study, MN9D cells were treated with different concentrations of HDMB (10, 20, 30uM) for 4 h under the same culture conditions followed by monitoring the mRNA expression of Cln1, Cln2 and Cln3 by semi-quantitative RT-PCR (C) and real-time qPCR (D). (E-F) MN9D cells were treated with different concentrations of HDMB for 18h under the same culture conditions followed by immunoblot analysis for TPP1 (E) and densitometric analysis of TPP1 expression relative to β-actin (F). (G) MN9D cells were treated with 20 μM HDMB under similar culture conditions and were double-labeled for TPP1 (red) and TH (green); Scale bar10μm. (H) MN9D cells were treated with 20μM HDMB under the same culture conditions for 18 h followed by TPP1 activity assay using cell extract containing 20 μg of total protein. All results are represented as mean ± SD of at least three independent experiments. *p < 0.05 & ***p < 0.001 vs control.
HDMB enhances TPP1 expression in mouse primary astrocytes
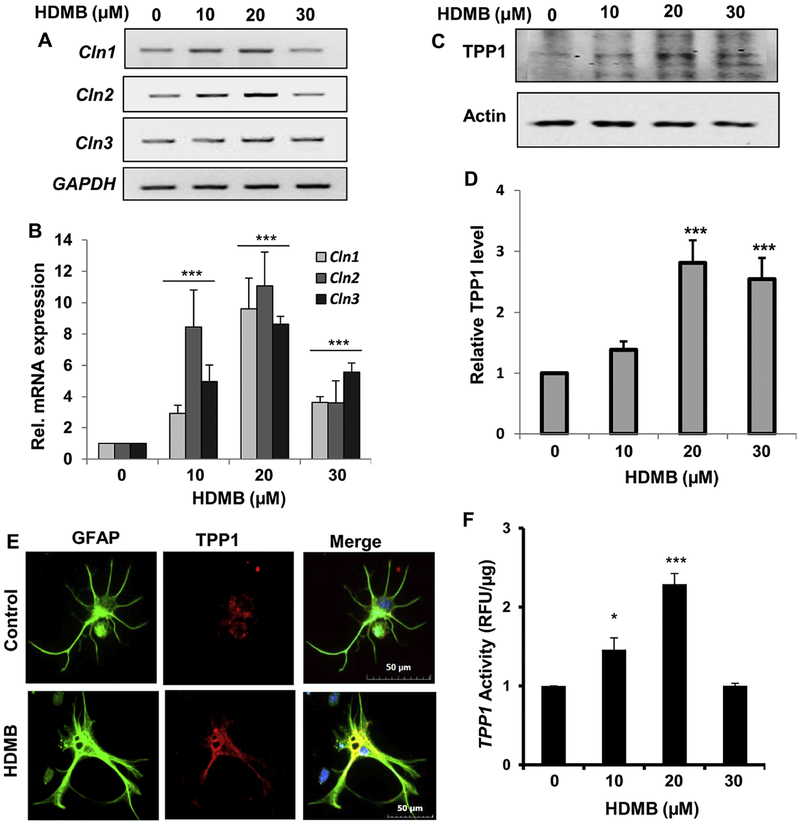
Next, we checked the effect of HDMB on TPP1 expression in wild type (WT) mouse primary astrocytes. Similar to our findings in MN9D neuronal cells, we observed that in primary astrocytes, HDMB distinctly increased the mRNA expression of Cln1, Cln2 and Cln3 in a dose-dependent fashion as demonstrated by semi-quantitative RT-PCR and real-time PCR (fig 2A, B). We further analyzed the expression of TPP1 with different doses of HDMB treatment and observed that HDMB was able to significantly upregulate TPP1 protein levels with 20 μM showing the highest increase (fig 2C, D). This was further validated by immunocytochemistry which revealed marked upregulation with 20 μM HDMB treatment (fig 2E). Next, we analyzed the activity of the TPP1 enzyme following HDMB treatment. Consistent with our earlier observation in MN9D cells, data showed that 20 μM HDMB was able to significantly augment the activity of TPP1 in primary astrocytes. Collectively, these data suggest that HDMB increases TPP1 levels and activity in mouse primary astrocytes.
Figure 2. HDMB enhances TPP1 expression in mouse primary astrocytes.
(A-B) Wild Type (WT) mouse primary astrocytes were treated with different concentrations of HDMB in serum-free DMEM/F12 50/50 media for 4 h followed by monitoring the mRNA expression of Cln1, Cln2 and Cln3 by semi-quantitative RT-PCR (A) and real-time qPCR (B). (C-D) WT Mouse primary astrocytes were treated with different concentrations of HDMB for 18 h under the same culture conditions followed by immunoblot analysis for TPP1 (C) and densitometric analysis of TPP1 expression relative to β-actin (D). (E) Mouse primary astrocytes were treated with 20 μM HDMB for 24 h under similar culture conditions and were double-labeled for TPP1 (red) and GFAP (green). DAPI was used for staining nuclei. Scale bar = 10 μm. (F) Mouse primary astrocytes were treated with different concentrations of HDMB (10, 20, 30 μM) in serum-free DMEM/F12 50/50 media for 18 h followed by the TPP1 activity assay using cell extract containing 20 μg of total protein. All results represent mean ± SD of at least three independent experiments. *p < 0.05 & ***p < 0.001 vs control.
HDMB requires PPARα for increasing TPP1 expression
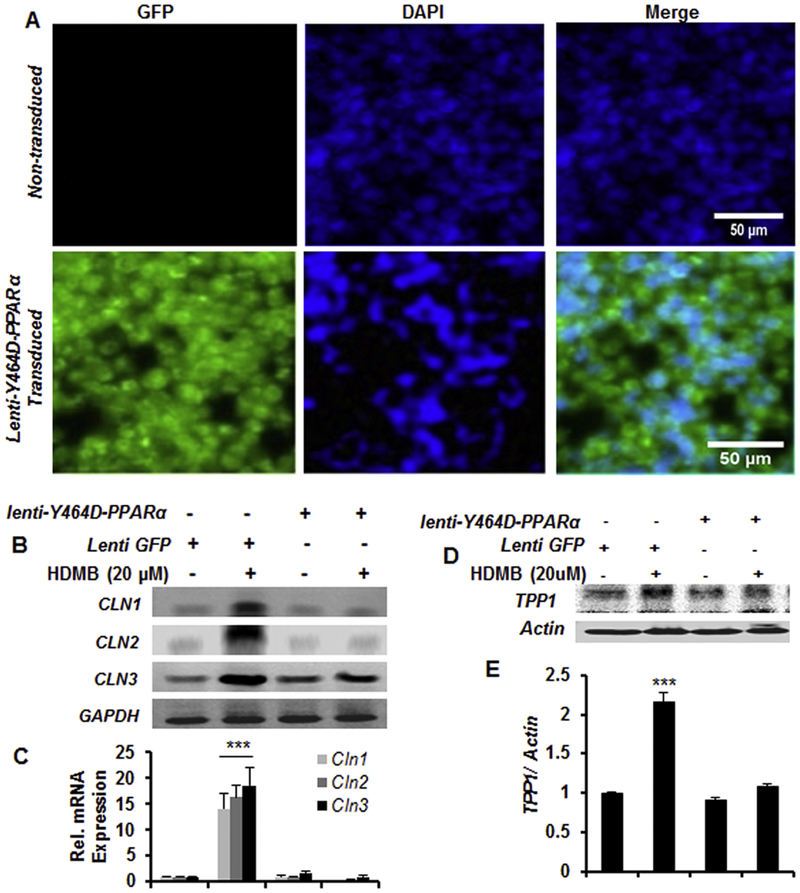
After establishing the role of HDMB in upregulation of TPP1, we intended to delineate the upstream mechanisms. In a previous study, we had established that PPARα agonist gemfibrozil increases TPP1 expression in brain cells via forming heterodimer with retinoid × receptor-α (22). Indeed, PPARs are known to form transcriptional complex with RXR and regulate the expression of target genes (36). Based on the previous evidence that HDMB is an endogenous ligand of PPARα (23) and PPARα agonist enhances TPP1 expression (22), we speculated that HDMB might activate PPARα to upregulate TPP1. To explore this, we transduced MN9D neuronal cells with lentivirus containing a dominant-negative construct of PPARα, Y464D (fig 3A), or with lentivirus containing GFP which served as a control. The Tyrosine 464 residue is a key site for binding of HDMB to PPARα ligand-binding domain and the dominant-negative construct Y464D inhibits the binding (23). Following transduction, we analyzed the expression of different Cln genes by semi-quantitative and real-time PCR. As expected, HDMB significantly upregulated the mRNA expression of Cln1, Cln2 and Cln3 in the Lenti-GFP-transduced cells; however, the effect of HDMB was abrogated in the Lenti-Y464D-PPARα-transduced MN9D cells (fig 3B, C), indicating that HDMB is not able to increase the expression of these genes in the absence of PPARα. Immunoblot analysis also revealed similar pattern of results where HDMB-mediated upregulation of TPP1 expression was drastically abolished in the presence of Lenti-Y464D-PPARα construct (fig 3D, E). Together, these results demonstrate that HDMB requires PPARα for increasing TPP1 expression in MN9D neuronal cells.
Figure 3. HDMB requires PPARα for increasing TPP1 expression.
(A-E) MN9D cells were transduced with Lenti-Y464D-PPARα construct or Lenti-GFP for 48h. Figure 3A shows the transduction efficiency in MN9D cells compared to non-transduced cells; DAPI was used to stain the nuclei (A). Transduced cells were treated with 20μM HDMB for 4h followed by monitoring the mRNA expression of Cln1, Cln2, and Cln3 by semi-quantitative RT-PCR (B) and real-time qPCR (C). Lenti-Y464D-PPARα or Lenti-GFP transduced cells were treated with 20μM of HDMB for 18 h followed by immunoblot for TPP1 (D) and densitometric analysis of TPP1 expression relative to β-actin (E). All results are represented as mean ± SD of at least three independent experiments. ***p < 0.001 vs control.
Upregulation of TPP1 by HDMB is dependent on PPARα
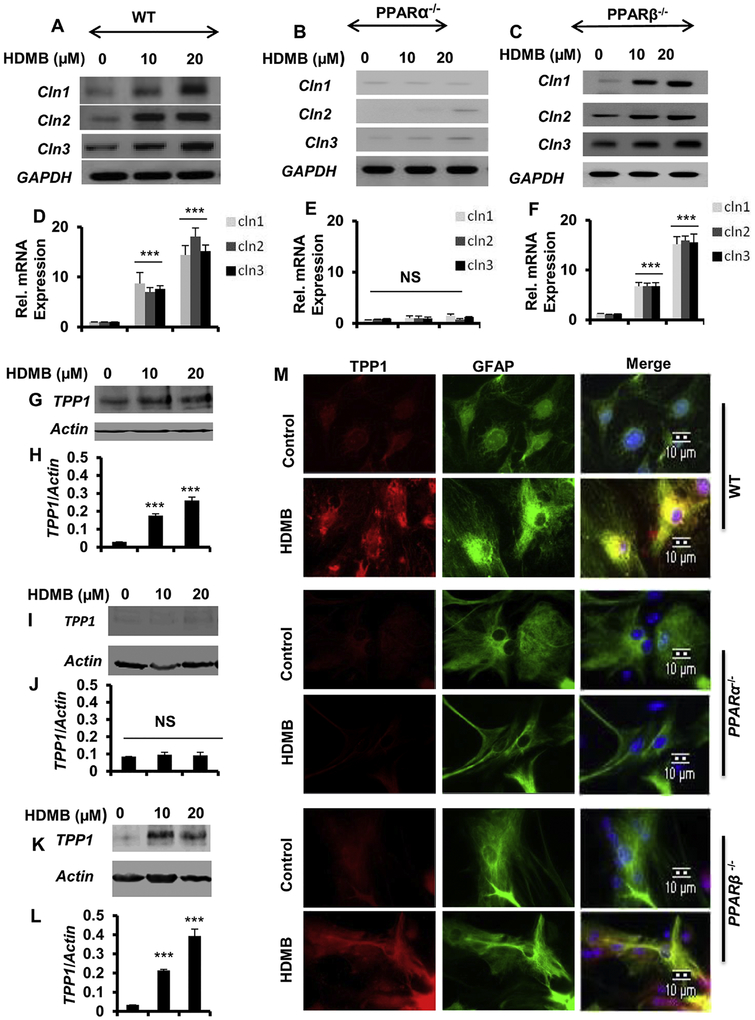
To further confirm the involvement of PPARα, we analyzed the effect of HDMB on TPP1 expression in primary astrocytes isolated from wild type (WT), PPARα−/− and PPARβ−/− mice. We first checked the mRNA expression of different Cln genes following HDMB treatment. Interestingly, HDMB upregulated Cln1, Cln2 and Cln3 gene expressions in astrocytes isolated from WT (fig 4A, D) and PPARβ−/− (fig 4C, F), but not PPARα−/−, mice (fig B, E), indicating that HDMB specifically requires PPARα for the upregulation of these Cln genes. Next, we checked the protein levels of TPP1 in WT, PPARα−/− and PPARβ−/− astrocytes by immunoblotting and observed that HDMB increases TPP1 levels in WT (fig 4G, H) and PPARβ−/− (fig 4K, L), but not in PPARα−/−, astrocytes (fig 4I, J), confirming that HDMB upregulates TPP1 via PPARα. Immunofluorescence study also demonstrated that HDMB markedly enhanced TPP1 expression in WT and PPARβ−/− astrocytes; however, no effect of HDMB was observed in PPARα−/− astrocytes (fig 4M). Collectively, these results establish that HDMB requires PPARα to upregulate TPP1.
Figure 4. Upregulation of TPP1 by HDMB is dependent on PPARα.
(A-F) Mouse primary astrocytes isolated from Wild Type (WT), PPARα−/− and PPARβ−/− mice were treated with different concentrations of HDMB in serum-free DMEM/F-12, 50/50 media for 4 h followed by monitoring the mRNA expression of Cln1, Cln2 and Cln3 genes by semi-quantitative RT-PCR (A, B, C) and real-time qPCR (D, E, F). (G-L) WT, PPARα−/− and PPARβ−/− mouse primary astrocytes were treated with different concentrations of HDMB under serum free conditions for 18 h followed by immunoblot analysis of TPP1 and densitometric analysis of TPP1 expression relative to β-actin (G,H for WT; I,J for PPARα−/− and K,L for PPARβ−/−). (M) Mouse primary astrocytes isolated from WT, PPARα−/− and PPARβ−/− mice were treated with 20 μM HDMB in serum-free DMEM/F-12, 50/50 media for 18 h and double-labeled for TPP1 (red) and GFAP (green). DAPI was used for staining nuclei; Scale bar=10 μm. (M). All results are represented as mean ± SD of at least three independent experiments. ***p < 0.001 vs control
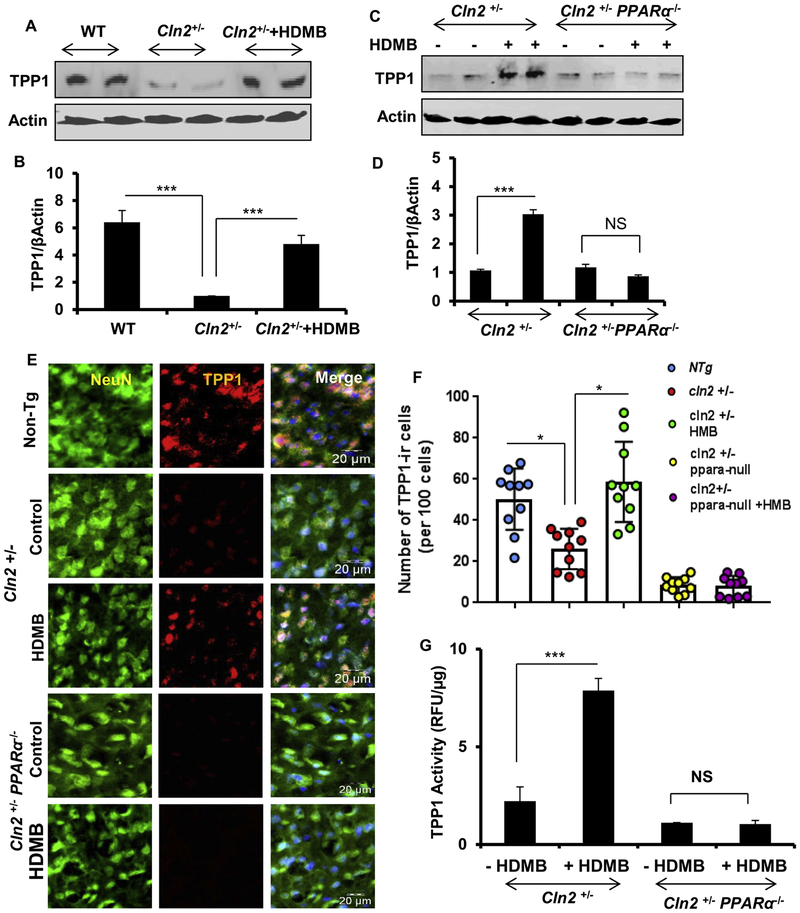
Administration of HDMB increases TPP1 levels in vivo in the brain of Cln2 +/− mice
After demonstrating the effect of HDMB in vitro, we next explored whether HDMB could augment TPP1 expression in the brain of transgenic mice heterozygous for Cln2 (Cln2 +/− mice). The Cln2+/− mice retain one intact copy of the Cln2 gene which makes it possible to test for potential therapeutic agents for their ability to enhance TPP1 levels in this model. Cln2+/− mice (8-weeks old) were treated with HDMB (5 mg/kg body weight/day) or vehicle (0.1% methylcellulose) for 14 days via oral gavage. First, we analyzed the mRNA expression of Cln1, Cln2 and Cln3 genes in the cortex and striatum of these mice. Semi-quantitative RT-PCR and real-time PCR data showed that HDMB treatment distinctly increased the mRNA expression of Cln1, Cln2 and Cln3 in both motor cortex (fig 5A, B) and striatum (fig 5C, D) whereas vehicle treated group did not exhibit any such effect. Next, we checked the protein levels of TPP1 and observed a remarkable increase with HDMB treatment compared to the untreated control and the vehicle-treated group both in motor cortex (fig 5E, F) and striatum (fig 5G, H). These results were further confirmed by double-labeling of TPP1 and NeuN in free-floating sections that showed distinct increase in TPP1 expression in the motor cortex (fig 5I) and striatum (fig 5J) of HDMB-treated mice. Subsequently, we analyzed the TPP1 activity in the motor cortex of these mice. HDMB-treated mice exhibited significantly higher TPP1 activity relative to the untreated control and vehicle-treated mice (fig 5K) indicating that oral administration of HDMB enhances the functional activity of TPP1 in Cln2+/− mice. Therefore, our results demonstrate that oral administration of HDMB augments both level and function of TPP1 in the brain of Cln2 +/− mice.
Figure 5. Administration of HDMB increases TPP1 levels in vivo in the brain of Cln2 +/− mice.
(A-D) 8-weeks old Cln2+/− mice (n = 5 in each group) were treated with 5 mg/kg body weight/day HDMB (dissolved in 0.1% methylcellulose) or vehicle (0.1% methylcellulose) via oral gavage. After 14 days of treatment, untreated, vehicle-treated and HDMB-treated mice were sacrificed and the motor cortex and striatum were dissected. The mRNA expression of Cln1, Cln2 and Cln3 were monitored by semi-quantitative RT-PCR and real-time qPCR in cortex (A, B) and striatum (C, D). (E-H) Immunoblot and densitometric analysis of TPP1 expression in the motor cortex (E, F) and striatum (G, H). (I-J) Double labeling of TPP1 and NeuN in Cortical (I) and Striatal (J) free floating sections; Data represent analysis of three cortical and striatal sections from each of four different mice per group. Scale bar=100 μm. (K) TPP1 was assayed from mouse cortical homogenate as described under Methods section and presented as relative to control. Control TPP1 activity represents 0.9+0.2 nmol/mg protein. Results are mean + SEM of five mice per group. *p < 0.05,**p < 0.01 & ***p < 0.001 vs untreated control
HDMB-mediated upregulation of TPP1 is abrogated in the absence of PPARα
Next, we explored whether HDMB-mediated upregulation of TPP1 is dependent on PPARα. In this regard, we generated a mouse model which was heterozygous for Cln2 and null for PPARα (Cln2+/−PPARα−/−) by crossing Cln2−/− mice with PPARα−/− mice. We used two months old Cln2+/− PPARα−/− mice (n=5/group) for HDMB treatment. As expected, Cln2+/− mice presented significantly less amount of TPP1 in the motor cortex compared to the WT mice; however, oral administration of HDMB significantly upregulated TPP1 levels in the motor cortex of Cln2+/− mice (fig 6A, B). Interestingly, when we compared Cln2+/− and Cln2+/−PPARα−/− mice, we observed that unlike Cln2+/− mice, HDMB was unable to upregulate TPP1 in the motor cortex of Cln2+/−PPARα−/− mice (fig 6C, D), indicating that HDMB does not exhibit any effect in the absence of PPARα. In parallel to our immunoblot results, double-labeling of TPP1 and NeuN in the motor cortex revealed that Cln2+/− mice have distinctly lower number of TPP1 positive cells compared to the WT control. Importantly, oral administration of HDMB remarkably increased the TPP1 levels in Cln2+/−, but not in Cln2+/−PPARα−/−, mice (fig 6E, F), further confirming that the effect of HDMB is abolished in the absence of PPARα. Accordingly, HDMB treatment also increased the activity of TPP1 in the motor cortex of Cln2+/−, but not in Cln2+/−PPARα−/−, mice (fig 6G). Taken together, these results establish that HDMB upregulates TPP1 in the brain of Cln2+/− mice via PPARα. We did not notice any side effect (e.g. hair loss, weight loss, diarrhea, untoward infection, etc.) in any of the mice during HDMB treatment.
Figure 6. HDMB-mediated upregulation of TPP1 is abrogated in the absence of PPARα.
(A-G) 8-weeks old Cln2+/− and Cln2+/−PPARα−/− mice (n = 5 in each group) were treated with 5 mg/kg body weight/day HDMB (dissolved in 0.1% methylcellulose) or vehicle (0.1% methylcellulose) for 14 days via oral gavage. Untreated WT mice were used as a control for basal endogenous levels of TPP1. Immunoblot analysis for TPP1 expression in the motor cortex of WT and Cln2+/− untreated control and Cln2+/− HDMB treated mice (B) and densitometric analysis (relative to β-actin) (C). Immunoblot analysis of TPP1 expression in untreated and HDMB treated Cln2+/− and Cln2+/−PPARα−/− mice (D) and densitometric analysis (E). Double-labeling of motor cortex from untreated and HDMB treated Cln2+/− and Cln2+/−PPARα−/− mice with TPP1 (red) and NeuN (green). DAPI was used for staining the nuclei. Scale bar = 20μm (F). Quantification of number of TPP1-immunoreactive cells per 100 cells in the motor cortex (G). Data represents analysis of three cortical sections from each of five different mice per group. TPP1 activity assay from cortex of untreated and treated Cln2+/− and Cln2+/−PPARα−/− mice (H). Results are mean ± SEM of five mice per group. *p < 0.05 & ***p < 0.001.
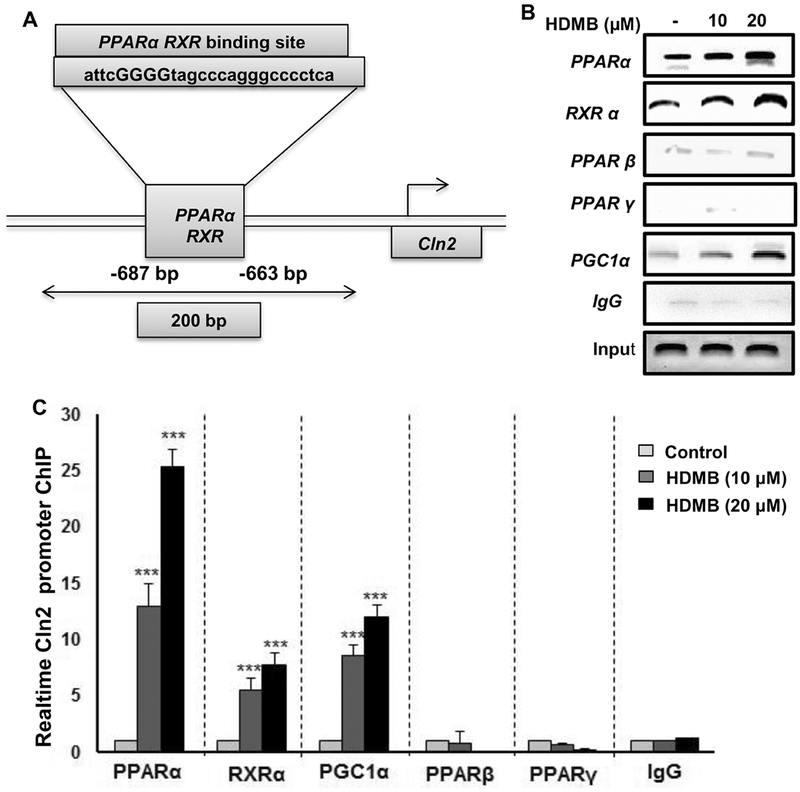
HDMB treatment stimulates the recruitment of PPARα, but neither PPARβ nor PPARγ, to the Cln2 gene promoter
Finally, we intended to characterize the mechanism how exactly activation of PPARα by HDMB regulates Cln2 expression. Previously, we reported the presence of a RXR-binding site on the Cln2 gene promoter (depicted in fig 7A) and established that the PPARα-RXRα heterodimer transcriptionally upregulates the expression of Cln2 (22). Hence, we checked whether HDMB treatment recruits PPARα to the Cln2 promoter by performing chromatin immunoprecipitation in MN9D neuronal cells. Following 2h of treatment with HDMB, the sheered chromatin was immunoprecipitated with antibodies against PPARα, RXRα, PPARβ, PPARγ, and PGC1α. Semi-quantitatve PCR data showed that HDMB selectively recruited PPARα, RXRα and PGC1α, but neither PPARβ nor PPARγ, to the Cln2 promoter (fig 7B). Real-time PCR data showed a similar pattern of increase in PPARα, RXRα and PGC1α (fig 7C), further confirming that HDMB promotes the formation of a transcriptional complex containing PPARα, RXRα and PGC1α on the Cln2 promoter which positively regulates the expression of the Cln2 gene. These data directly demonstrate that HDMB-mediated PPARα activation transcriptionally upregulates TPP1.
Figure 7. HDMB stimulates the recruitment of PPARα to the Cln2 gene promoter.
MN9D neuronal cells were treated with 10 and 20 μM HDMB under serum free conditions for 2h following which the nuclear extract was subjected to chromatin immunoprecipitation. Recruitment of PPARα, RXRα, PPARβ, PPARγ, and PGC1α on the RXR-binding site of Cln2 promoter (A) was monitored by semi-quantitative PCR (B) and real-time qPCR (C). IgG was used as a control. All results are represented as mean ± SD of at least three independent experiments. ***p < 0.001 vs control.
Discussion:
Current therapeutic approaches for the treatment of NCLs are focused on delivery of functional copy of the gene, stem cells or recombinant enzyme to the brain which, although have promising outcomes, are extensively invasive and expensive. Hence, there is a necessity for a novel, minimally or non-invasive and economic treatment strategies for these fatal childhood disorders. The classic late-infantile form of NCL (LINCL) is characterized by mutations in the Cln2 gene, which encodes a lysosomal enzyme TPP1. Several reports have indicated that residual activity of TPP1 is retained in LINCL (3,20,21) prompting a new field of research directed towards enhancing the function of residual TPP1 under disease scenario. In this study, we demonstrate that HDMB, an endogenous ligand of PPARα, can upregulate TPP1 expression and activity in mouse neuronal cell line and primary brain cells. We also established that the effect of HDMB was specifically mediated by activation of the nuclear receptor PPARα. Importantly, administration of HDMB to the heterozygous Cln2+/− mice markedly enhanced TPP1 levels in motor cortex and striatum in a PPARα-dependent fashion. Finally, we delineate that recruitment of PPARα and formation of a transcriptional complex on the Cln2 gene promoter is the underlying mechanism of HDMB-mediated TPP1 upregulation. Therefore, administration of HDMB might be beneficial for the treatment of LINCL patients.
PPARs are ligand inducible transcription factors that act as lipid sensors. One of the isoforms, PPARα, is primarily involved in energy homeostasis although it has been reported to modulate several other physiological processes (36,37). Our lab has established that PPARα is involved in memory improvement via targeting the CREB signaling pathway (38). We have also demonstrated that acivation of PPARα induces lysosomal biogenesis in brain cells through transcriptional upregulation of the master regulator TFEB (39). Importantly, activation of PPARα by fibrate drugs was observed to stimulate TPP1 expression via forming heterodimer with RXRα (22). Owing to the immense implications of this hormone receptor in physiological and pathological processes, endogenous ligands of PPARα could be highly beneficial as therapeutic agents. In this study, we demonstrate that HDMB induces TPP1 expression in brain cells as well as in vivo in the brain of a disease model for LINCL (Cln2+/−) via PPARα.
In a recent study by our group, HDMB was detected as an endogenous molecule that binds to the ligand-binding domain of PPARα in the mouse hippocampus and was reported to stimulate synaptic functions (23). The Tyr464 (Y464) was identified as an active site residue in the ligand-binding domain of PPARα which was important for binding of HDMB (23). In this study, as expected, when a dominant-negative construct of PPARα (Y464D) was used, the effect of HDMB on TPP1 upregulation was abolished. This confirmed that HDMB specifically requires the binding to the PPARα ligand-binding domain for inducing TPP1 expression.
While examining the mechanisms underlying HDMB-mediated upregulation of TPP1, we observed that HDMB transcriptionally enhances Cln2 expression via PPARα. PPARs contain four functional domains including a ligand-binding domain and a highly conserved DNA-binding domain (36,37). Under unstimulated conditions, PPARs are sequestered in the cytosol by binding to corepressor molecules. Activation of the receptor upon ligand-binding leads to dissociation from the corepressor complex, binding of coactivators and formation of heterodimer with RXRs. The PPAR-RXR transcriptional complex binds to the PPRE (peroxisome-proliferator response elements) sequence on the promoter of target gene to regulate gene expression (36,37). Promoter analysis of mouse Cln2 gene revealed no PPRE site; however, RXR-binding site was observed in the promoter region upstream of the transcription start site (−687 to −663 bp) (22). We have previously demonstrated that PPARα agonist gemfibrozil leads to recruitment of PPARα-RXRα complex to the RXR-binding site on the Cln2 gene promoter (22). In this study, we observed that HDMB treatment leads to the recruitment of PPARα, RXRα and PGC1α to the RXR-binding site on Cln2 promoter to transcriptionally upregulate Cln2 gene expression.
Our cell culture studies revealed that along with Cln2, HDMB also enhanced the expression of lysosomal genes Cln1 and Cln3 in the MN9D neuronal cell line and in mouse primary astrocytes. The gene Cln1, located on chromosome 1p34.2, encodes the enzyme PPT1 (palmitoyl protein thioesterase 1) which eliminates palmitate groups from its substrate proteins. Mutations in Cln1 lead to PPT1 deficiency resulting in classic infantile NCL or Haltia-Santavuori disease (1,5,40). The Cln3 gene is mapped to 16p11.2 and encodes a membrane protein Cln3, which when mutated causes juvenile NCL or Batten-Spielmeyer-Sjögren disease (1,5,40). We have observed that HDMB increases the mRNA expression of both Cln1 and Cln3 in a dose-dependent manner. These results suggest that HDMB might have therapeutic implications for Infantile NCL (INCL) and Juvenile NCL (JNCL) as well.
Several mutations have been associated with the Cln2 gene that results in loss or deficiency of TPP1 activity. The three most common mutations observed in most LINCL cases are an intron G to C transversion in the 3’ splice junction of intron 5 (3556G-C); an exon 6 C to T transversion that prematurely stops translation (3670C-T; Arg 208Stop); and a missense mutation G to C in exon 10 (5271G-C; Gln422His) (12,13). Many of the missense mutations observed in TPP1 are associated with abnormal folding of the mutant protein resulting in extended half-life of pro-TPP1 or inefficient trafficking to the lysosomes resulting in lower activity of the enzyme (21). However, several TPP1 variants have been observed to response to permissive temperature and chemical chaperone treatment indicating that folding enhancement could be an effective strategy to rescue the function of some variant of TPP1 (21).
Mounting evidence indicate that residual TPP1 activity is retained in LINCL patients and can be targeted for therapeutic strategies (3,20,21). Indeed, low levels of TPP1 activity was observed in the lymphocytes from LINCL patients by a capillary electrophoresis technique (20). Similar to this, residual TPP1 activity was also reported in biological samples from LINCL patients as well as different animal model using an enzyme-based assay (14). An Arg447His missense mutation associated with late age of onset and clinically protracted course of the disease suggests that restoration of low levels of normal TPP1 activity might be adequate and result in a much milder form of the disease (3,12). An interesting study on CLN2 mutant mice expressing variant amounts of residual TPP1 activity found that 3% of regular TPP1 activity is sufficient to delay the onset and double the lifespan whereas 6% normal TPP1 activity results in attenuation of the disease and remarkably improved lifespan of the mice indicating 6% functional TPP1 activity. Therefore, therapies targeted at achieving as low as 6% of TPP1 functionality in the CNS might be beneficial for LINCL patients (41).
The Cln2 targeted mutant mice (Cln2−/−) successfully recapitulates many of the LINCL characteristics including tremor, ataxia, neuronal pathology marked by lysosomal storage material, axonal degeneration and reduced lifespan (42). In a previous study, we demonstrated that Gemfibrozil, a FDA approved PPARα agonist, increased the lifespan of Cln2−/− model of LINCL (43). Gemfibrozil-treated mice had reduced burden of storage granules, enhanced expression of anti-inflammatory and anti-apoptotic factors and lowered neuronal apoptosis compared to vehicle treated Cln2−/− mice (43). In this study, we used Cln2 heterozygous mice (Cln2+/−) which have one normal copy of the Cln2 gene providing opportunity to test drug-mediated regulation of TPP1 in this model. Marked increase in TPP1 in the motor cortex of Cln2+/−, but not Cln2+/−PPARα−/−, mice by HDMB suggests that HDMB requires PPARα to upregulate the expression of TPP1 in Cln2+/− mice.
Gene therapy, enzyme replacement therapy and stem cell transplantation have shown promising results for the treatment of LINCL (15,16,44–50). These therapies are targeted at delivery of active TPP1 in adequate amount to counter the cell loss observed in LINCL (12). In vivo gene therapy involves delivery of a vector containing the human Cln2 cDNA to the CNS and retinal cells in LINCL patients. Although it has demonstrated efficacy in animal models and human patients (15,46–49), several factors makes it challenging to evolve as a widely applicable therapy. In order to succeed, firstly, substantial amount of the active TPP1 protein has to be produced to reach the therapeutic levels. Secondly, since LINCL is a genetic disorder, the active TPP1 protein has to be produced for long duration and will need subsequent delivery of the vector. Thirdly, the gene delivery has to target an extensive brain region so that the protein can spread through the affected cells (12,46). In enzyme augmentation therapy, recombinant TPP1 protein is directly infused into the brain of LINCL patients. However, purification of recombination enzyme and the need for continuous administration of the protein pose several difficulties (12). A recent study showed that enzyme augmentation therapy via intraventricular infusion of cerliponase alfa (recombinant human pro-TPP1) reduces the impairment of locomotor and language functions in children affected with CLN2 disease (19). The study involved infusion of the recombinant protein every 2 weeks for 240 weeks in children of 3 to 16 years age. Although this is the first FDA-approved therapy for LINCL, the repeated administration of intraventricular device is fairly invasive and expensive. Moreover, the treatment led to adverse side effects including development of infections from the device (19). Therefore, a drug-mediated therapeutic strategy using HDMB could be advantageous than prospective treatments for LINCL due to several factors. HDMB is an endogenous molecule and therefore will be well-tolerated by the body and have minimal toxicities. Finally, HDMB can be delivered orally which is the least painful route for treatment and is suitable for children.
In summary, this study demonstrates that HDMB, an endogenous ligand of PPARα, can upregulate TPP1. We delineate that HDMB activates the nuclear receptor PPARα to enhance TPP1 expression. Furthermore, we showed that HDMB recruits the PPARα-RXRα heterodimer complex to the Cln2 gene promoter to transcriptionally regulate the expression. Moreover, in Cln2+/− mice, oral administration of HDMB leads to enhanced TPP1 expression in the motor cortex and striatum. Therefore, HDMB may have therapeutic potential for LINCL.
Highlights.
3-hydroxy-(2,2)-dimethyl butyrate (HDMB), a brain endogenous molecule, stimulates Cln1, Cln2 and Cln3 in brain cells.
HDMB stimulates Cln1, Cln2 and Cln3 in astrocytes via PPARα.
Oral administration of HDMB increases TPP1 in the brain of Cln2+/− mice via PPARα.
HDMB increases the transcription of Cln2 via PPARα:RXRα:PGC1α.
ACKNOWLEDGEMENTS
This study was supported by a grant from National Institutes of Health (AG050431) and a merit award from Veteran Affairs (1I01BX003033).
Abbreviations:
- LINCL
Late-infantile neuronal ceroid lipofuscinosis
- Cln2
Ceroid lipofuscinosis, neuronal 2
- TPP1
Tripeptidyl peptidase 1
- HDMB
3-Hydroxy-(2,2)-dimethyl butyrate
- PPARα
Peroxisome proliferator-activated receptor α
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nita DA, Mole SE, and Minassian BA (2016) Neuronal ceroid lipofuscinoses. Epileptic Disord 18, 73–88 [DOI] [PubMed] [Google Scholar]
- 2.Schulz A, Kohlschutter A, Mink J, Simonati A, and Williams R (2013) NCL diseases - clinical perspectives. Biochim Biophys Acta 1832, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sleat DE, Gin RM, Sohar I, Wisniewski K, Sklower-Brooks S, Pullarkat RK, Palmer DN, Lerner TJ, Boustany RM, Uldall P, Siakotos AN, Donnelly RJ, and Lobel P (1999) Mutational analysis of the defective protease in classic late-infantile neuronal ceroid lipofuscinosis, a neurodegenerative lysosomal storage disorder. Am J Hum Genet 64, 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hachiya Y, Hayashi M, Kumada S, Uchiyama A, Tsuchiya K, and Kurata K (2006) Mechanisms of neurodegeneration in neuronal ceroid-lipofuscinoses. Acta Neuropathol 111, 168–177 [DOI] [PubMed] [Google Scholar]
- 5.Warrier V, Vieira M, and Mole SE (2013) Genetic basis and phenotypic correlations of the neuronal ceroid lipofusinoses. Biochim Biophys Acta 1832, 1827–1830 [DOI] [PubMed] [Google Scholar]
- 6.Williams RE, and Mole SE (2012) New nomenclature and classification scheme for the neuronal ceroid lipofuscinoses. Neurology 79, 183–191 [DOI] [PubMed] [Google Scholar]
- 7.Mole SE, and Cotman SL (2015) Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim Biophys Acta 1852, 2237–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sleat DE, Donnelly RJ, Lackland H, Liu CG, Sohar I, Pullarkat RK, and Lobel P (1997) Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science 277, 1802–1805 [DOI] [PubMed] [Google Scholar]
- 9.Liu CG, Sleat DE, Donnelly RJ, and Lobel P (1998) Structural organization and sequence of CLN2, the defective gene in classical late infantile neuronal ceroid lipofuscinosis. Genomics 50, 206–212 [DOI] [PubMed] [Google Scholar]
- 10.Haines JL, Boustany RM, Alroy J, Auger KJ, Shook KS, Terwedow H, and Lerner TJ (1998) Chromosomal localization of two genes underlying late-infantile neuronal ceroid lipofuscinosis. Neurogenetics 1, 217–222 [DOI] [PubMed] [Google Scholar]
- 11.Golabek AA, Kida E, Walus M, Wujek P, Mehta P, and Wisniewski KE (2003) Biosynthesis, glycosylation, and enzymatic processing in vivo of human tripeptidyl-peptidase I. J Biol Chem 278, 7135–7145 [DOI] [PubMed] [Google Scholar]
- 12.Sondhi D, Hackett NR, Apblett RL, Kaminsky SM, Pergolizzi RG, and Crystal RG (2001) Feasibility of gene therapy for late neuronal ceroid lipofuscinosis. Arch Neurol 58, 1793–1798 [DOI] [PubMed] [Google Scholar]
- 13.Crystal RG, Sondhi D, Hackett NR, Kaminsky SM, Worgall S, Stieg P, Souweidane M, Hosain S, Heier L, Ballon D, Dinner M, Wisniewski K, Kaplitt M, Greenwald BM, Howell JD, Strybing K, Dyke J, and Voss H (2004) Clinical protocol. Administration of a replication-deficient adeno-associated virus gene transfer vector expressing the human CLN2 cDNA to the brain of children with late infantile neuronal ceroid lipofuscinosis. Hum Gene Ther 15, 1131–1154 [DOI] [PubMed] [Google Scholar]
- 14.Sohar I, Sleat DE, Jadot M, and Lobel P (1999) Biochemical characterization of a lysosomal protease deficient in classical late infantile neuronal ceroid lipofuscinosis (LINCL) and development of an enzyme-based assay for diagnosis and exclusion of LINCL in human specimens and animal models. J Neurochem 73, 700–711 [DOI] [PubMed] [Google Scholar]
- 15.Worgall S, Sondhi D, Hackett NR, Kosofsky B, Kekatpure MV, Neyzi N, Dyke JP, Ballon D, Heier L, Greenwald BM, Christos P, Mazumdar M, Souweidane MM, Kaplitt MG, and Crystal RG (2008) Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum Gene Ther 19, 463–474 [DOI] [PubMed] [Google Scholar]
- 16.Xu S, Wang L, El-Banna M, Sohar I, Sleat DE, and Lobel P (2011) Large-volume intrathecal enzyme delivery increases survival of a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol Ther 19, 1842–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haltia M (2003) The neuronal ceroid-lipofuscinoses. J Neuropathol Exp Neurol 62, 1–13 [DOI] [PubMed] [Google Scholar]
- 18.Lane SC, Jolly RD, Schmechel DE, Alroy J, and Boustany RM (1996) Apoptosis as the mechanism of neurodegeneration in Batten’s disease. J Neurochem 67, 677–683 [DOI] [PubMed] [Google Scholar]
- 19.Schulz A, Ajayi T, Specchio N, de Los Reyes E, Gissen P, Ballon D, Dyke JP, Cahan H, Slasor P, Jacoby D, Kohlschutter A, and Group CLNS (2018) Study of Intraventricular Cerliponase Alfa for CLN2 Disease. N Engl J Med 378, 1898–1907 [DOI] [PubMed] [Google Scholar]
- 20.Viglio S, Marchi E, Wisniewski K, Casado B, Cetta G, and Iadarola P (2001) Diagnosis of late-infantile neuronal ceroid lipofuscinosis: a new sensitive method to assay lysosomal pepstatin-insensitive proteinase activity in human and animal specimens by capillary electrophoresis. Electrophoresis 22, 2343–2350 [DOI] [PubMed] [Google Scholar]
- 21.Walus M, Kida E, and Golabek AA (2010) Functional consequences and rescue potential of pathogenic missense mutations in tripeptidyl peptidase I. Hum Mutat 31, 710–721 [DOI] [PubMed] [Google Scholar]
- 22.Ghosh A, Corbett GT, Gonzalez FJ, and Pahan K (2012) Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor alpha: implications for late infantile Batten disease therapy. J Biol Chem 287, 38922–38935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy A, Kundu M, Jana M, Mishra RK, Yung Y, Luan CH, Gonzalez FJ, and Pahan K (2016) Identification and characterization of PPARalpha ligands in the hippocampus. Nat Chem Biol 12, 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahmachari S, and Pahan K (2007) Sodium benzoate, a food additive and a metabolite of cinnamon, modifies T cells at multiple steps and inhibits adoptive transfer of experimental allergic encephalomyelitis. J Immunol 179, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha RN, and Pahan K (2007) Differential regulation of Mn-superoxide dismutase in neurons and astroglia by HIV-1 gp120: Implications for HIV-associated dementia. Free Radic Biol Med 42, 1866–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giulian D, and Baker TJ (1986) Characterization of ameboid microglia isolated from developing mammalian brain. J Neurosci 6, 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khasnavis S, Jana A, Roy A, Mazumder M, Bhushan B, Wood T, Ghosh S, Watson R, and Pahan K Suppression of nuclear factor-kappaB activation and inflammation in microglia by physically modified saline. J Biol Chem 287, 29529–29542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khasnavis S, and Pahan K Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates neuroprotective Parkinson disease protein DJ-1 in astrocytes and neurons. J Neuroimmune Pharmacol 7, 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta S, Jana M, Zhou Y, Fung YK, Ghosh S, and Pahan K (2004) Antineuroinflammatory effect of NF-kappaB essential modifier-binding domain peptides in the adoptive transfer model of experimental allergic encephalomyelitis. J Immunol 173, 1344–1354 [DOI] [PubMed] [Google Scholar]
- 30.Corbett GT, Roy A, and Pahan K Gemfibrozil, a lipid-lowering drug, upregulates IL-1 receptor antagonist in mouse cortical neurons: implications for neuronal self-defense. J Immunol 189, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha RN, Liu X, and Pahan K (2006) Up-regulation of BDNF in astrocytes by TNF-alpha: a case for the neuroprotective role of cytokine. J Neuroimmune Pharmacol 1, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vines DJ, and Warburton MJ (1999) Classical late infantile neuronal ceroid lipofuscinosis fibroblasts are deficient in lysosomal tripeptidyl peptidase I. FEBS Lett 443, 131–135 [DOI] [PubMed] [Google Scholar]
- 33.Nelson JD, Denisenko O, and Bomsztyk K (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 34.Ghosh A, Corbett GT, Gonzalez FJ, and Pahan K Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor alpha: implications for late infantile Batten disease therapy. J Biol Chem 287, 38922–38935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh A, Rangasamy SB, Modi KK, and Pahan K Gemfibrozil, Food and Drug Administration-approved lipid-lowering drug, increases longevity in mouse model of Late Infantile Neuronal Ceroid Lipofuscinosis. J Neurochem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heneka MT, and Landreth GE (2007) PPARs in the brain. Biochim Biophys Acta 1771, 1031–1045 [DOI] [PubMed] [Google Scholar]
- 37.Heneka MT, Reyes-Irisarri E, Hull M, and Kummer MP (2011) Impact and Therapeutic Potential of PPARs in Alzheimer’s Disease. Curr Neuropharmacol 9, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Jana M, Corbett GT, Ramaswamy S, Kordower JH, Gonzalez FJ, and Pahan K (2013) Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep 4, 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh A, Jana M, Modi K, Gonzalez FJ, Sims KB, Berry-Kravis E, and Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem 290, 10309–10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haltia M (2006) The neuronal ceroid-lipofuscinoses: from past to present. Biochim Biophys Acta 1762, 850–856 [DOI] [PubMed] [Google Scholar]
- 41.Sleat DE, El-Banna M, Sohar I, Kim KH, Dobrenis K, Walkley SU, and Lobel P (2008) Residual levels of tripeptidyl-peptidase I activity dramatically ameliorate disease in late-infantile neuronal ceroid lipofuscinosis. Mol Genet Metab 94, 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sleat DE, Wiseman JA, El-Banna M, Kim KH, Mao Q, Price S, Macauley SL, Sidman RL, Shen MM, Zhao Q, Passini MA, Davidson BL, Stewart GR, and Lobel P (2004) A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J Neurosci 24, 9117–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh A, Rangasamy SB, Modi KK, and Pahan K (2017) Gemfibrozil, food and drug administration-approved lipid-lowering drug, increases longevity in mouse model of late infantile neuronal ceroid lipofuscinosis. J Neurochem 141, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang M, Cooper JD, Sleat DE, Cheng SH, Dodge JC, Passini MA, Lobel P, and Davidson BL (2008) Intraventricular enzyme replacement improves disease phenotypes in a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol Ther 16, 649–656 [DOI] [PubMed] [Google Scholar]
- 45.Vuillemenot BR, Katz ML, Coates JR, Kennedy D, Tiger P, Kanazono S, Lobel P, Sohar I, Xu S, Cahayag R, Keve S, Koren E, Bunting S, Tsuruda LS, and O’Neill CA (2011) Intrathecal tripeptidyl-peptidase 1 reduces lysosomal storage in a canine model of late infantile neuronal ceroid lipofuscinosis. Mol Genet Metab 104, 325–337 [DOI] [PubMed] [Google Scholar]
- 46.Sondhi D, Hackett NR, Peterson DA, Stratton J, Baad M, Travis KM, Wilson JM, and Crystal RG (2007) Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh.10 rhesus macaque-derived adeno-associated virus vector. Mol Ther 15, 481–491 [DOI] [PubMed] [Google Scholar]
- 47.Sondhi D, Peterson DA, Edelstein AM, del Fierro K, Hackett NR, and Crystal RG (2008) Survival advantage of neonatal CNS gene transfer for late infantile neuronal ceroid lipofuscinosis. Exp Neurol 213, 18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabrera-Salazar MA, Roskelley EM, Bu J, Hodges BL, Yew N, Dodge JC, Shihabuddin LS, Sohar I, Sleat DE, Scheule RK, Davidson BL, Cheng SH, Lobel P, and Passini MA (2007) Timing of therapeutic intervention determines functional and survival outcomes in a mouse model of late infantile batten disease. Mol Ther 15, 1782–1788 [DOI] [PubMed] [Google Scholar]
- 49.Hackett NR, Redmond DE, Sondhi D, Giannaris EL, Vassallo E, Stratton J, Qiu J, Kaminsky SM, Lesser ML, Fisch GS, Rouselle SD, and Crystal RG (2005) Safety of direct administration of AAV2(CU) hCLN2, a candidate treatment for the central nervous system manifestations of late infantile neuronal ceroid lipofuscinosis, to the brain of rats and nonhuman primates. Hum Gene Ther 16, 1484–1503 [DOI] [PubMed] [Google Scholar]
- 50.Selden NR, Al-Uzri A, Huhn SL, Koch TK, Sikora DM, Nguyen-Driver MD, Guillaume DJ, Koh JL, Gultekin SH, Anderson JC, Vogel H, Sutcliffe TL, Jacobs Y, and Steiner RD (2013) Central nervous system stem cell transplantation for children with neuronal ceroid lipofuscinosis. J Neurosurg Pediatr 11, 643–652 [DOI] [PubMed] [Google Scholar]