Abstract
Honey bee larval food jelly is a secretion of the hypopharyngeal and mandibular glands of young worker bees that take care of the growing brood in the hive. Food jelly is fed to all larvae (workers, drones and queens) and as royal jelly to the queen bee for her entire life. Up to 18% of the food jelly account for proteins the majority of which belongs to the major royal jelly protein (MRJP) family. These proteins are produced in the hypopharyngeal glands at a pH value of 7.0. Before being fed to the larvae, they are mixed with the fatty acids secreted by the mandibular glands of the worker bees resulting at a pH of 4.0 in the food jelly. Thus, MRJPs are exposed to a broad pH range from their site of synthesis to the actual secreted larval food. We therefore determined the pH-dependent stability of MRJP1, MRJP2 and MRJP3 purified from royal jelly using differential scanning fluorimetry. All MRJPs were much more stable at acidic pH values compared to neutral ones with all proteins showing highest stability at pH 4.0 or 4.5, the native pH of royal jelly.
Subject terms: Protein folding, Proteins, Entomology
Introduction
Honey bees (Apis sp.) feed their growing larvae with a special food jelly, a secretion produced by the hypopharyngeal and mandibular glands of nurse worker bees that take care of the brood in the hive1,2. The food jelly provides all nutrients that are necessary to develop into an adult and is particularly rich in proteins (11–18% w/w)3–5. Up to 90% of the total proteins belong to the major royal jelly protein (MRJP) family6 which comprises ten different proteins (MRJP1-10)7,8. However, only MRJP1-3 and 5 are secreted in larger amounts into the food jelly6. As a peculiarity, MRJP1 can occur as a monomer (monoMRJP1) or as an oligomer in complex with apisimin (oligoMRJP1/apisimin)9–12. The functions of MRJPs reach from providing essential nutrients6 over having antibacterial effects13,14 and binding RNA15 to increasing the viscosity of the food jelly by fibril formation of oligoMRJP1/apisimin at acidic pH 4.016,17. MRJPs share a high amino acid sequence similarity, and thus pH might also play an important role for the other MRJPs as they do indeed encounter a large pH range in the course of their lifetime: The proteins are synthesized in the hypopharyngeal glands of nursing bees18,19 as secretory proteins and are thus directly translated into the endoplasmic reticulum of the secretory cells at a pH value of around 7.020. Next, the proteins are stored in secretory vesicles at a pH of 5.5 to 5.120–22. After being secreted from the hypopharyngeal glands, the proteins are exposed to the acidic mandibular gland secretions (pH 3.9 ± 0.1) composed of fatty acids resulting in a final pH of 4.0 in food jelly21. Yet MRJPs are by no means limited to the food jelly and can be found in manifold organs and body fluids of honey bees; including the venom23,24 (pH 4.5 to 5.225,26), the brain27,28 and the hemolymph29,30 (pH 6.831). If MRJPs have specific biotic functions they need to be stable within the wide pH range from 4.0 to 7.0 at temperatures normally ranging from 20 °C (bee cluster in winter at outside temperatures of down to −10 °C) to 35 °C (bee cluster during brood rearing)32. In addition, extreme temperatures reach from 6 °C (bee in the periphery of the winter cluster)33 to 46 °C (bees with activated flight muscles attacking a wasp)34. These extreme temperatures might be irrelevant for MRJPs in the food jelly, as the brood is raised at constant temperatures of 34 °C32, but might become important for MRJPs being present in the bee venom, the brain and the hemolymph. In addition, most proteins do have their stability optimum at a pH of 7.0 or 7.5 and only very few proteins are stable at a pH below 4.535. However, exactly at these acidic pH values MRJPs need to be stable as they reach their highest incidence in royal jelly at pH 4.0.
The stability of proteins can be assessed by experimentally denaturing their native structure36. Transitions between the native and the disordered state are typically induced by denaturing agents, e.g. guanidinium chloride or heat. We determined the melting temperature (Tm), where the fraction of folded and unfolded protein is equal36,37, of purified oligoMRJP1/apisimin, monoMRJP1, MRJP2 and MRJP3 in terms of their pH-dependent stability using differential scanning fluorimetry (ThermoFluor)37–39. All tested MRJPs exhibited within their natural pH range (4.0 to 7.0) Tm values (≥43 °C) above the maximum temperature of 35 °C normally occurring in the hive. In addition, all tested MRJPs were much more stable at pH 4.0, the native pH of the food jelly, compared to pH 7.0 (ΔTm = 14.3–18.6 °C) ensuring that MRJPs in the food jelly are not denatured by heat even at elevated ambient temperatures.
Results
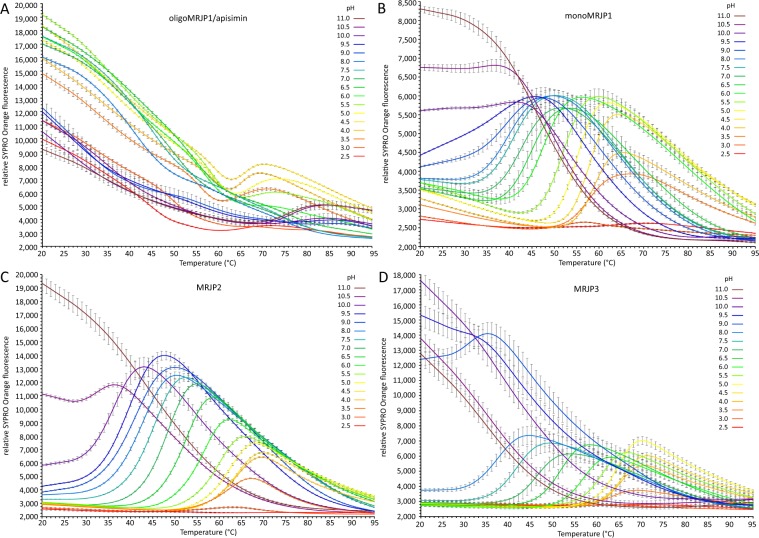
pH dependent stability of oligoMRJP1/apisimin, monoMRJP1, MRJP2 and the MRJP3 isoforms was determined covering a pH range from 2.5 to 11. At the beginning of the experiment, at low temperature, a low fluorescence is expected as an indicator of a well folded protein. Within the physiological pH range for MRJPs (pH 4.0–7.0), monoMRJP1, MRJP2 and the MRJP3 isoforms possess at 20 °C a rather low fluorescence around 3,000–3,500 relative fluorescence units (RFU) (Fig. 1B–D). Contrary to that, the complex of oligoMRJP1/apisimin exhibited at the same pH values already at 20 °C a fluorescence between 15,000–20,000 RFU (Fig. 1A) indicating exposed hydrophobic regions. For pH 4.0 to 6.0, this fluorescence declines continuously until approximately 60 °C, followed by a further fluorescence increase peaking between 67 to 74 °C. For pH 6.5 and 7.0, the decline in fluorescence was constant. In addition, MRJP2 has a higher fluorescence intensity increase than monoMRJP1, and MRJP3 within the physiological pH range (Δfluorescence: monoMRJP1, ~3,000; MRJP2, ~10,000; MRJP3, ~4,000) implying a tighter packed hydrophobic core for MRJP2.
Figure 1.
Thermal unfolding curves determined with differential scanning fluorimetry. Measurements were performed at 2 µM protein concentration and 1:1000 SYPRO Orange in four replicates. Curves are exemplarily only shown for one RJ. Measurements from pH 2.5 to 8.0 were performed in 50 mM Na2HPO4/citric acid and measurements from pH 9.0 to 11.0 in 50 mM Na2CO3/NaHCO3. (A) oligoMRJP1/apisimin. (B) monoMRJP1. (C) MRJP2. (D) MRJP3 isoforms.
At pH 2.5 and 3.0, the curves recorded for monoMRJP1, MRJP2 and MRJP3 did either not show any increase in fluorescence at all or just a very slight one of approximately 500 RFU (Fig. 1B–D). Either the proteins do not unfold at these acidic pH values or the increased solvent polarity at decreased pH values leads to lower fluorescence intensity or shifting of the maximum fluorescence emission to other wavelengths as shown for other fluorescent dyes than SYPRO Orange40. Again, the complex of oligoMRJP1/apisimin exhibited also at these pH values an increased starting fluorescence (~10,000–12,000 RFU) which decreased at elevated temperatures (Fig. 1A).
At pH values above 9.0, starting fluorescence gradually increased with higher pH also for monoMRJP1, MRJP2 and MRJP3 indicating exposed hydrophobic residues and protein unfolding even at lower temperatures (monoMRJP1 pH ≥10.0, MRJP2 pH ≥10.5, MRJP3 ≥9.0).
Due to the increased starting fluorescence for oligoMRJP1/apisimin, transition midpoints (Tm) were only determined for monoMRJP1, MRJP2 and MRJP3. All proteins were much more stable at pH 4.0 than at pH 7.0 (ΔTm: 14.3–18.6 °C) (Table 1; Fig. S2) (Kruskal-Wallis ANOVA (n = 360; H = 351; P < 0.001), monoMRJP1: P = 0.016; MRJP2: P = 0.004; MRJP3: P < 0.001) and all MRJPs showed maximal thermal stability around the native pH of the food jelly (4.0–4.5). Therefore, monoMRJP1, MRJP2 and MRJP3 belong to the ~5% of proteins that have their pH-optimum of stability at pH 4.5 or less35.
Table 1.
Transition midpoints (Tm) of the differential scanning fluorimetry unfolding curves.
| pH | monoMRJP1 | MRJP2 | MRJP3 |
|---|---|---|---|
| 3.5 | 58.6 ± 0.7 °Ca,b | 61.3 ± 1.3 °Ca,b | 62.4 ± 0.4 °Ca,b |
| 4.0 | 59.3 ± 0.5 °Ca | 63.2 ± 1.3 °Ca | 64.2 ± 0.3 °Ca |
| 4.5 | 58.0 ± 0.6 °Ca,b | 63.6 ± 1.3 °Ca | 64.8 ± 0.4 °Ca |
| 5.0 | 55.4 ± 0.6 °Ca–c | 62.3 ± 1.3 °Ca | 63.5 ± 0.3 °Ca |
| 5.5 | 52.2 ± 0.9 °Ca–c | 59.7 ± 1.4 °Ca–c | 60.1 ± 0.2 °Ca,b |
| 6.0 | 48.5 ± 1.3 °Ca–c | 56.4 ± 1.3 °Ca–d | 56.1 ± 0.1 °Ca–c |
| 6.5 | 45.7 ± 1.4 °Ca–c | 52.8 ± 1.4 °Ca–d | 50.8 ± 0.2 °Cb–d |
| 7.0 | 43.3 ± 2.4 °Cb,c | 48.9 ± 1.5 °Cb–d | 45.6 ± 0.3 °Cc,d |
| 7.5 | 40.9 ± 2.1 °Cc | 46.0 ± 1.6 °Cc,d | 41.5 ± 0.4 °Cc,d |
| 8.0 | 39.3 ± 1.7 °Cc | 43.3 ± 1.9 °Cd | 38.1 ± 1.3 °Cd |
| 9.0 | 36.1 ± 1.3 °C | 42.8 ± 1.7 °C | largely unfolded |
| 9.5 | largely unfolded | 40.8 ± 1.8 °C | unfolded |
| 10.0 | unfolded | 37.0 ± 1.4 °C | unfolded |
| 10.5 | unfolded | largely unfolded | unfolded |
| 11.0 | unfolded | unfolded | unfolded |
Values are means ± standard deviations (3 biological replicates (proteins purified from three different royal jellies), 4 technical replicates per protein). Statistics were performed only from pH 3.5 to 8.0 where values for all three proteins were present (Kruskal-Wallis ANOVA, n = 360; H = 351; p < 0.001). a–dTms in the same row with different superscripts are significantly different (P < 0.05). The highest Tm for each protein is highlighted in bold.
Discussion
Transition midpoints could be determined for all proteins except for the complex of oligoMRJP1 and apisimin, which showed elevated starting fluorescence already at low temperatures indicating exposed hydrophobic regions. As monoMRJP1 does not show any peculiarity regarding hydrophobicity, the high hydrophobicity of oligoMRJP1/apisimin might be attributed to several reasons: (1) Either to the bound apisimin itself, which consists of ~40% hydrophobic amino acids, or (2) to hydrophobic residues of MRJP1 which are exposed only after apisimin binding. Hydrogen/deuterium exchange experiments indeed revealed that the N-terminal part of MRJP1 within oligoMRJP1/apisimin is highly disordered11 but a comparison to the structure of monoMRJP1 is missing. (3) Only very recently it has been revealed that the complex of oligoMRJP1 and apisimin binds in addition eight molecules of 24-methylenecholesterol41 which explains most likely the high hydrophobicity. However, also a combination of the reasons mentioned might be possible. Still, the melting curves of oligoMRJP1/apisimin have marked transitions points at pH values between 4.0 and 6.0 (Tm ~ 66–70 °C) which might be caused by the unfolding of the structured part of the complex. OligoMRJP1/apisimin (pH 4.0–6.0; Tm ~ 66–70 °C) appears to have an increased stability compared to monoMRJP1 (pH 4.0–6.0; Tm = 59.3–48.5 °C), irrespective of the fact that some hydrophobic residues are already exposed at low temperatures. At neutral pH, fluorescence of oligoMRJP1/apisimin constantly decreased and no evaluation of the curves was possible in this study. However, it was shown that incubation of oligoMRJP1/apisimin in phosphate buffered saline (pH 7.5) at 56 °C for 30 min did not lead to a dissociation of the complex42 and that the heat treatment did not impair proliferation activity of the complex on human lymphoid cells43. Circular dichroism spectroscopy in 2 mM HEPES, pH 7.0 showed even a slight structural gain from 20 °C to 95 °C44. This suggests that oligoMRJP1/apisimin is still folded at elevated temperatures at near neutral pH and illustrates that differential scanning fluorimetry is not an appropriate method to analyze the stability of the complex of oligoMRJP1 and apisimin. In addition, no strong difference was observed between fluorescence curves recorded above and below pH 5.0 although oligoMRJP1/apisimin starts to assemble into fibrillary structures below pH 5.016,17.
MonoMRJP1, MRJP2 and the MRJP3 isoforms were all much more stable at pH 4.0 than at neutral pH (ΔTm: 14.3–18.6 °C). At elevated alkaline pH (≥9.0), hydrophobic residues were already exposed at 20 °C, residues which were buried at lower pH values. Indeed MRJPs have been shown to be more stable against limited proteolysis at acidic rather than alkaline conditions45. The different MRJPs showed at pH 4.0 a remarkable stability with melting temperatures ranging between 59.3 to 64.2 °C. This is in accord with accelerated protein degradation in fresh RJ above 65 °C46 as the proteins unfold at these temperatures.
Minimum solubility often coincides with the isoelectric point (pI) of proteins47 where the electrostatic forces are at minimum and the proteins might precipitate out of solution. MRJPs exhibit theoretical pIs between 5.0 and 6.7 (monoMRJP1 and oligoMRJP1/apisimin–5.0, MRJP2–6.7, MRJP3–6.5) and should thus show lowest solubility at a pH of 5.0 (monoMRJP1 and oligoMRJP1/apisimin) and ~6.5 (MRJP2 and MRJP3). In the worker bee, all MRJPs reach their highest concentration in the hypopharyngeal gland secretion, before being mixed with the mandibular gland secretion to produce the final food jelly. Interestingly, the hypopharyngeal gland secretion has a pH of 5.1 ± 0.121, which is exactly at the pI of monoMRJP1 and oligoMRJP1/apisimin. Thus, the increased protein stability at the acidic pH values is essential to ensure that MRJPs, and especially oligoMRJP1/apisimin, do not precipitate in the hypopharyngeal gland secretion as this would lead to blocking of the glands and prevent further food jelly secretion.
Materials and Methods
Royal jelly samples and protein purification
Three fresh royal jelly (RJ) samples (Apis mellifera), used for protein purification, were acquired in 2015 from three different beekeepers in Romania (RJ1 - Cluj-Napoca, Cluj County – July/RJ2 - Bratca, Bihor County – August/RJ3 - Cluj-Napoca, Cluj County - June). All RJ samples were stored at −20 °C. OligoMRJP1/apisimin, monoMRJP1 and MRJP2 were purified via cation exchange chromatography using SP Sepharose Fast Flow (GE Healthcare, Little Chalfont, UK) according to previous studies45,48. MRJP3 and 5 that co-eluted from the SP Sepharose column with 50 mM Na2HPO4/NaH2PO4, 200 mM NaCl, pH 7.0 were rebuffered into 50 mM Na2HPO4/NaH2PO4, 2 M (NH4)2SO4, pH 7.0 using PD-10 columns (GE Healthcare). The rebuffered sample (~2 ml) with an absorption at 280 nm of maximum 4.5, as measured with a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), was loaded onto a 1 ml Butyl-Sepharose 4 Fast Flow (GE Healthcare) column. The column was washed with 10 ml 50 mM Na2HPO4/NaH2PO4, 2 M (NH4)2SO4, pH 7.0 and elution of MRJP3 was achieved with 10 ml 50 mM Na2HPO4/NaH2PO4, 0.75 M (NH4)2SO4, pH 7.0. Fractions of 1 ml were collected and protein purity was verified with sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE)49 and native PAGE at pH 7.050. Pure protein fractions were combined (Fig. S1).
Due to a repetitive region with length polymorphisms51, MRJP3 occurs in different isoforms showing as multiple bands on SDS PA gels between 60 and 70 kDa15,45 (Fig. S1, lane 6). Buffer exchange for further experiments was performed with PD-10 desalting columns. Protein concentrations were determined via UV spectroscopy. Molar extinction coefficients were calculated with ProtParam52 as 56,185 M−1 cm−1 for oligoMRJP1/apisimin and monoMRJP1, 51,590 M−1 cm−1 for MRJP2 and 47,580 M−1 cm−1 for MRJP3.
Thermal unfolding using ThermoFluor
To monitor the pH dependency of MRJP unfolding, ThermoFluor experiments37–39 were conducted. This method is based on monitoring the fluorescence of specific dyes, e.g. Sypro Orange, which is quenched in aqueous solutions but is highly fluorescent in presence of hydrophobic sites of unfolded proteins37,39. Thus, upon proceeding protein unfolding, fluorescence intensity increases and can be plotted as function of temperature. Experiments were performed in Hard-Shell 96-well microplates sealed with Microseal ‘B’ seals in a CFF Connect Real-Time System (all Bio-Rad, Hercules, CA, USA). Measurements were performed (3 biological replicates (proteins purified from three different royal jellies), 4 technical replicates per protein) in a total volume of 20 µl with 2 µM oligoMRJP1/apisimin, monoMRJP1, MRJP2 and MRJP3 and SYPRO Orange (Sigma-Aldrich, St. Louis, MO, USA) at a dilution of 1:1000 in 50 mM Na2HPO4/citric acid pH 2.5–8.0 or 50 mM Na2CO3/NaHCO3 pH 9.0–11.0. Samples were heated from 20 °C to 95 °C at 1 °C per min and fluorescence intensity of SYPRO Orange was measured every degree using the FRET channel of the Real-Time System (excitation: 450–490 nm, detection: 560–580 nm). To determine the melting temperature (Tm - temperature at which the concentrations of folded and unfolded protein are equal), fluorescence intensities (FI) were plotted as a function of temperature (T) and fitted with Origin 5.0 (Microcal Software Inc., Northampton, MA, USA) according to the Boltzmann equation.
Supplementary information
Acknowledgements
This project was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft - DFG, Grant MO 373/32-1 to RFAM). AB was funded during manuscript writing by the institutional strategy ‘The Synergetic University’ of the Technische Universität Dresden financed by the Excellence Initiative of the German federal and state governments. We are very grateful to Robin F.A. Moritz for providing infrastructural support and we thank Josephine Buchholz for help with lab work. We acknowledge support by the Open Access Publication Funds of the SLUB/TU Dresden.
Author Contributions
Conceptualization, A.B. Methodology, A.B. Software, C.M. and A.B. Formal Analysis, C.M. and A.B. Investigation, C.M. Writing-Original Draft Preparation, C.M. and A.B. Writing-Review & Editing, C.M. and A.B. Supervision, A.B. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45460-0.
References
- 1.Callow RK, Johnston NC, Simpson J. 10-Hydroxy-Δ2-decenoic acid in the honeybee (Apis mellifera) Experientia. 1959;15:421–422. doi: 10.1007/BF02157689. [DOI] [PubMed] [Google Scholar]
- 2.Schiemenz P. Über das Herkommen des Futtersaftes und die Speicheldrüsen der Biene nebst einem Anhange über das Riechorgan. Z. Wiss. Zool. 1883;38:71–135. [Google Scholar]
- 3.von Planta A. Über den Futtersaft der Bienen. Z. Physiol. Chem. 1888;12:327–354. doi: 10.1515/bchm1.1888.12.4.327. [DOI] [Google Scholar]
- 4.von Planta A. Über den Futtersaft der Bienen. II. Z. Physiol. Chem. 1889;13:552–561. doi: 10.1515/bchm1.1889.13.6.552. [DOI] [Google Scholar]
- 5.Wang Y, et al. Comparison of the nutrient composition of royal jelly and worker jelly of honey bees (Apis mellifera) Apidologie. 2016;47:48–56. doi: 10.1007/s13592-015-0374-x. [DOI] [Google Scholar]
- 6.Schmitzová J, et al. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell. Mol. Life Sci. 1998;54:1020–1030. doi: 10.1007/s000180050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drapeau MD, Albert Š, Kucharski R, Prusko C, Maleszka R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helbing S, Lattorff HMG, Moritz RFA, Buttstedt A. Comparative analyses of the major royal jelly protein gene cluster in three Apis species with long amplicon sequencing. DNA Res. 2017;24:279–287. doi: 10.1093/dnares/dsw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bíliková K, et al. Apisimin, a new serine-valine-rich peptide from honeybee (Apis mellifera L.) royal jelly: purification and molecular characterization. FEBS Lett. 2002;528:125–129. doi: 10.1016/S0014-5793(02)03272-6. [DOI] [PubMed] [Google Scholar]
- 10.Kamakura M, Fukuda T, Fukushima M, Yonekura M. Storage-dependent degradation of 57-kDa protein in royal jelly: a possible marker for freshness. Biosci. Biotech. Biochem. 2001;65:277–284. doi: 10.1271/bbb.65.277. [DOI] [PubMed] [Google Scholar]
- 11.Mandacaru SC, et al. Characterizing the structure and oligomerization of major royal jelly protein 1 (MRJP1) by mass spectrometry and complementary biophysical tools. Biochemistry. 2017;56:1645–1655. doi: 10.1021/acs.biochem.7b00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Šimúth J. Some properties of the main protein of honeybee (Apis mellifera) royal jelly. Apidologie. 2001;32:69–80. doi: 10.1051/apido:2001112. [DOI] [Google Scholar]
- 13.Bíliková K, et al. Towards functional proteomics of minority component of honeybee royal jelly: the effect of post-translational modifications on the antimicrobial activity of apalbumin 2. Proteomics. 2009;9:2131–2138. doi: 10.1002/pmic.200800705. [DOI] [PubMed] [Google Scholar]
- 14.Vezeteu TV, Bobiş O, Moritz RFA, Buttstedt A. Food to some, poison to others - honeybee royal jelly and its growth inhibiting effect on European Foulbrood bacteria. Microbiology Open. 2017;6:e00397. doi: 10.1002/mbo3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maori E, et al. A Secreted RNA Binding Protein Forms RNA-Stabilizing Granules in the Honeybee Royal Jelly. Mol. Cell. 2019;74:598–608. doi: 10.1016/j.molcel.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buttstedt A, et al. How honeybees defy gravity with royal jelly to raise queens. Curr. Biol. 2018;28:1095–1100. doi: 10.1016/j.cub.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Kurth T., Kretschmar S., Buttstedt A. Royal jelly in focus. Insectes Sociaux. 2018;66(1):81–89. doi: 10.1007/s00040-018-0662-3. [DOI] [Google Scholar]
- 18.Hanes J, Šimúth J. Identification and partial characterization of the major royal jelly protein of the honey bee (Apis mellifera L.) J. Apicult. Res. 1992;31:22–26. doi: 10.1080/00218839.1992.11101256. [DOI] [Google Scholar]
- 19.Patel NG, Haydak MH, Gochnauer TA. Electrophoretic components of the proteins in honeybee larval food. Nature. 1960;186:633–634. doi: 10.1038/186633a0. [DOI] [PubMed] [Google Scholar]
- 20.Demaurex N. pH homeostasis of cellular organelles. News Physiol. Sci. 2002;17:1–5. doi: 10.1152/physiologyonline.2002.17.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann I. Untersuchungen über die Herkunft von Komponenten des Königinnenfuttersaftes der Honigbienen. Z. Bienenforsch. 1960;5:101–111. [Google Scholar]
- 22.Knecht D, Kaatz HH. Patterns of larval food production by hypopharyngeal glands in adult worker honey bees. Apidologie. 1990;21:457–468. doi: 10.1051/apido:19900507. [DOI] [Google Scholar]
- 23.Matysiak J, Hajduk J, Mayer F, Hebeler R, Kokot ZJ. Hyphenated LC-MALDI-ToF/ToF and LC-ESI-QToF approach in proteomic characterization of honeybee venom. J. Pharmaceut. Biomed. 2016;121:69–76. doi: 10.1016/j.jpba.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Peiren N, et al. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Acta. 2005;1752:1–5. doi: 10.1016/j.bbapap.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Fischer FG, Neumann WP. Das Gift der Honigbiene. Trennung und chemische Charakterisierung der beiden Hauptfraktionen. Biochem. Z. 1953;324:447–464. [PubMed] [Google Scholar]
- 26.Lensky Y, Schindler H. Motility and reversible inactivation of honeybee spermatozoa in vivo and in vitro. Ann. Abeille. 1967;10:5–16. doi: 10.1051/apido:19670101. [DOI] [Google Scholar]
- 27.Garcia L, et al. Proteomic analysis of honey bee brain upon ontogenetic and behavioral development. J. Proteome Res. 2009;8:1464–1473. doi: 10.1021/pr800823r. [DOI] [PubMed] [Google Scholar]
- 28.Kucharski R, Maleszka R, Hayward DC, Ball EE. A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain. Naturwissenschaften. 1998;85:343–346. doi: 10.1007/s001140050512. [DOI] [PubMed] [Google Scholar]
- 29.Ararso Z, et al. Proteome comparisons between hemolymph of two honeybee strains (Apis mellifera ligustica) reveal divergent molecular basis in driving hemolymph function and high royal jelly secretion. J. Proteome Res. 2018;17:402–419. doi: 10.1021/acs.jproteome.7b00621. [DOI] [PubMed] [Google Scholar]
- 30.Randolt K, et al. Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee worker larvae and adults. Arch. Insect Biochem. 2008;69:155–167. doi: 10.1002/arch.20269. [DOI] [PubMed] [Google Scholar]
- 31.Bishop GH. Body fluid of the honey bee larva. I. Osmotic pressure, specific gravity, pH, O2 capacity, CO2 capacity, and buffer value, and their changes with larval activity and metamorphosis. J. Biol. Chem. 1923;58:543–565. [Google Scholar]
- 32.Gates BN. The temperature of the bee colony. Bull. US Depart. Agric. 1914;96:1–29. [Google Scholar]
- 33.Stabentheiner A, Pressl H, Papst T, Hrassnigg N, Crailsheim K. Endothermic heat production in honeybee winter clusters. J Exp. Biol. 2003;206:353–358. doi: 10.1242/jeb.00082. [DOI] [PubMed] [Google Scholar]
- 34.Stabentheiner A, Kovac H, Schmaranzer S. Thermal behavior of honeybees during aggressive interactions. Ethology. 2007;113:995–1006. doi: 10.1111/j.1439-0310.2007.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talley K, Alexov E. On the pH-optimum of activity and stability of proteins. Proteins. 2010;78:2699–2706. doi: 10.1002/prot.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Privalov PL. Stability of proteins. Small globular proteins. Adv. Protein Chem. 1979;33:167–241. doi: 10.1016/S0065-3233(08)60460-X. [DOI] [PubMed] [Google Scholar]
- 37.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 38.Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic Stability of Carbonic Anhydrase: Measurements of Binding Affinity and Stoichiometry Using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 39.Pantoliano MW, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 40.Baruah M, et al. Solvent and pH dependent fluorescent properties of a dimethylaminostyryl borondipyrromethene dye in solution. J. Phys. Chem. A. 2006;110:5998–6009. doi: 10.1021/jp054878u. [DOI] [PubMed] [Google Scholar]
- 41.Tian W, et al. Architecture of the native majr royal jelly protein 1 oligomer. Nature Communications. 2018;9:3373. doi: 10.1038/s41467-018-05619-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura S, et al. Molecular characteristics and physiological functions of major royal jelly protein 1 oligomer. Proteomics. 2009;9:5534–5543. doi: 10.1002/pmic.200900541. [DOI] [PubMed] [Google Scholar]
- 43.Moriyama T, Ito A, Omote S, Miura Y, Tsumoto H. Heat resistant characteristics of major royal jelly protein 1 (MRJP1) oligomer. PLoS One. 2015;10:e0119169. doi: 10.1371/journal.pone.0119169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz GCN, et al. Calcium effect and pH-dependence on self-association and structural stability of the Apis mellifera major royal jelly protein 1. Apidologie. 2011;42:252–269. doi: 10.1007/s13592-011-0025-9. [DOI] [Google Scholar]
- 45.Mureşan CI, Schierhorn A, Buttstedt A. The fate of major royal jelly proteins during proteolytic digestion in the human gastrointestinal tract. J. Agr. Food Chem. 2018;66:4164–4170. doi: 10.1021/acs.jafc.8b00961. [DOI] [PubMed] [Google Scholar]
- 46.Lazarevska S, Makreski P. Insights into the infrared and Raman spectra of fresh and lyophilized royal jelly and protein degradation IR spectroscopy study during heating. Maced. J. Chem. Chem. En. 2015;34:87–93. [Google Scholar]
- 47.Svedberg T, et al. General Discussion. T. Faraday Soc. 1930;26:737–740. doi: 10.1039/TF9302600737. [DOI] [Google Scholar]
- 48.Buttstedt A, Ihling CH, Pietzsch M, Moritz RFA. Royalactin is not a royal making of a queen. Nature. 2016;473:478–483. doi: 10.1038/nature19349. [DOI] [PubMed] [Google Scholar]
- 49.Laemmli UK. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 50.Gallagher SR. One-Dimensional Electrophoresis Using Nondenaturing Conditions. Curr. Protoc. Mol. Biol. 1999;47:10.2B.1–10.2B.11. doi: 10.1002/0471142727.mb1002bs47. [DOI] [PubMed] [Google Scholar]
- 51.Albert Š, Klaudiny J, Šimuth J. Molecular characterization of MRJP3, highly polymorphic protein of honeybee (Apis mellifera) royal jelly. Insect Biochem. Mol. Biol. 1999;29:427–434. doi: 10.1016/S0965-1748(99)00019-3. [DOI] [PubMed] [Google Scholar]
- 52.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.