Abstract
In the plant response to pathogen infection, many genes’ expression is temporally induced, while few spatially induced expression genes have been reported. Here, we show that GhBOP1 can autonomously expand expression from restrained tissue when Gossypium hirsutum plants are attacked by Verticillium dahliae, which is considered to be spatially induced expression. Loss- and gain-of-function analyses show that GhBOP1 is a positive regulator in the modulation of plant resistance to V. dahliae. Yeast two-hybrid assays, luciferase complementation imaging and GUS reporting show that GhBOP1 interaction with GhTGA3 promotes its activation activity, regulating the expression of down-stream defence-related genes. Moreover, the induced spatial expression of GhBOP1 is accompanied by GhBP1 repression. Both antagonistically regulate the lignin biosynthesis, conferring cotton plants enhanced resistance to V. dahliae. Taken together, these results demonstrate that GhBOP1 is an economic positive regulator participating in plant defence through both the GhBOP1-GhTGA3 module and lignin accumulation.
Subject terms: Plant stress responses, Plant molecular biology
Zhennan Zhang, Peng Wang, Xiaoli Luo et al. show that BOP1 from cotton plants is able to induce gene expression in tissues affected by the fungal pathogen Verticillium dahlia, resulting in increased disease resistance. They propose a model involving TGA3 and lignin accumulation.
Introduction
Plants stand and face various biotic and abiotic stresses; therefore, they have evolved a wide spectrum of mechanisms to constantly defend themselves against these stresses, especially pathogen infestation. The mechanisms in response to pathogen attack are divided into two groups, pre-existing and induced. The pre-existing mechanisms mainly involve physical and chemical barriers for protecting plants from pathogen infestation at the first line, including the plant cuticle, cell wall, and antimicrobial compounds1. Salicylic acid (SA) crosstalking with auxin, ethylene, and jasmonates (JA) is crucial for the inducible response of plants to pathogens infection including systemic acquired resistance and induced systemic resistance2–5. In disease resistance induced mechanism, NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) is regarded as a major knotting component to regulate plant defence, required for SA perception and other hormones6,7. NPR1 is a BTB-ankyrin protein including two conserved motifs: a BTB/POZ (for Broad Complex, Tramtrack, and Bric-a-brac/POX virus and Zinc finger) domain at the N-terminus and four ankyrin motifs near the C-terminus. These inducible responses of plant to pathogens attack involve in expression change of lots of genes in temporal mode. However, gene expression change in spatial mode remains to be less known.
BTB-ankyrin proteins are a small family in plants. In Arabidopsis, there are six BTB-ankyrin proteins8. NPR1, NPR3, and NPR4 through SA perception have a role in defence7,9,10, and NPR2 has been described to have a secondary role in SA perception11. BLADE-ON-PETIOLE1 (BOP1) and BOP2 are the other two BTB-ankyrin proteins, the expression of which is generally restrained in lateral-organ boundaries (LOBs). BOP1/2 is described to have an important role in the development of the leaf and inflorescence architecture12,13. A meta-analysis of sequencing data showed that BTB-ankyrin proteins originated prior to the emergence of land plants14. All land plants sequenced genomes, including primitive mosses, encode homologs of both NPRs and BOP1/2, indicating that the ancestral BTB-ankyrin proteins may have held functions in both defence and development15. The initial characterization of the bop1bop2 mutant showed no change in resistance to pathogens16. However, a recent report has indicated that BOP1/2 participates in plant defence through the JA signalling pathway17. Thus, the function of BOP1/2 in resistance to pathogens needs to be further evaluated.
BOP1/2 and NPR1 share homologous functional domains, BTB/POZ and ankyrin repeats, that potentially support a similar mode of action. Thus, the NPR1 signalling mechanism potentially serves as a paradigm for BOPs. BTB-ankyrin proteins including NPR1 and BOP1 can interact with defence-related TGA bZIP transcription factors to exert functions in disease resistance16,17. TGA bZIP proteins are a distinct subclade in the bZIP superfamily18. In Arabidopsis, these defence-related TGAs contain three clades: Class I comprises TGA1 and TGA4; Class II comprises TGA2, TGA5, and TGA6; and Class III comprises TGA3 and TGA719. NPR1 interaction with TGAs participates in plant defence, which should represent a similar interaction mode for the function of BOPs in pathogen resistance. Weak physical interactions have been detected between BOPs and most TGAs in a yeast heterologous system16,20,21. A recent report has shown that the change of BOP interacts with TGAs in yeast indirectly caused by co-expression of npr1-114,17. Subsequently, the methyl jasmonate-induced resistance is abolished in bop1bop2 mutants and enhanced in plants overexpressing BOP1 or BOP2, possibly through the ability of NPR1 to disrupt BOP interactions with TGAs14,17. Thereby, the BOP-TGA interaction is considered to have confirmed roles in defence and development.
Verticillium wilt is a highly destructive vascular disease caused by Verticillium sp, a soil-borne fungus, which infects a wide range of plants22,23. Mounting evidence has confirmed that verticillium wilt resistance is directly associated with lignin accumulation in plants. In cotton, the series of lignin synthesis enzymes are upregulated when the plants are infected by V. dahliae, resulting in lignin accumulation22,24–27. For example, transcript levels of lignin metabolism-related genes are increased in cotton plants after inoculation with V. dahliae24. GbERF1-like regulates lignin metabolism-related gene expression for lignin accumulation, increasing the resistance to V. dahliae infection26. In Arabidopsis, plants inoculated with V. longisporum promote novel vascular formation and lignin synthesis28. The Ve-mediated resistance response of tomato to V. dahliae also involves lignin and PAL gene expression29. These reports document that lignin plays important roles in plant defence. Interestingly, it has been reported that BOPs participate in lignin synthesis by regulating lignin metabolism-related gene expression to show phenotypes in bops mutants and overexpression plants13. However, it remains unclear whether BOPs participate in disease resistance through lignin synthesis.
BOPs function in lignin synthesis in association with some gene regulation, including BP1 in Arabidopsis13,16. When BP1 expression is repressed in stem, BOP1/2 exhibits ectopic expression outside of LOBs, suggesting that BOPs and BP1 have antagonistic roles in regulating lignin biosynthesis13,30. Seven lignin biosynthesis genes with upregulated expression in bp1 mutants (PAL1, C4H1, 4CL1, C3H1, CCoMT1, CAD5, and PRXR9) can be restored to near wild-type levels by bop1bop2 mutation; similar to this result, in BOP overexpression plants, five of the seven genes are dramatically upregulated, which is consistent with the promotion of lignin biosynthesis13,30. Therefore, BOPs and BP1 together regulate lignin synthesis, potentially participating in plant disease resistance.
In the present study, we found that cotton BOP1 expression was upregulated and autonomously expanded outside of LOBs coupled with BP1 repression during V. dahliae infestation. The results of genetic and biochemistry experiments showed that GhBOP1 was an economical regulator participating in plant defence against V. dahliae, resulting from the interaction with GhTGA3 and lignin accumulation. Taken together, these data shed light on the molecular mechanisms of BOPs function in plant defence, including BOP1-TGA3, similar to the NPR1-TGA module and antagonistic roles of BOP1 and BP1 in lignin synthesis.
Results
GhBOP1 autonomously expanding expression infected by fungus
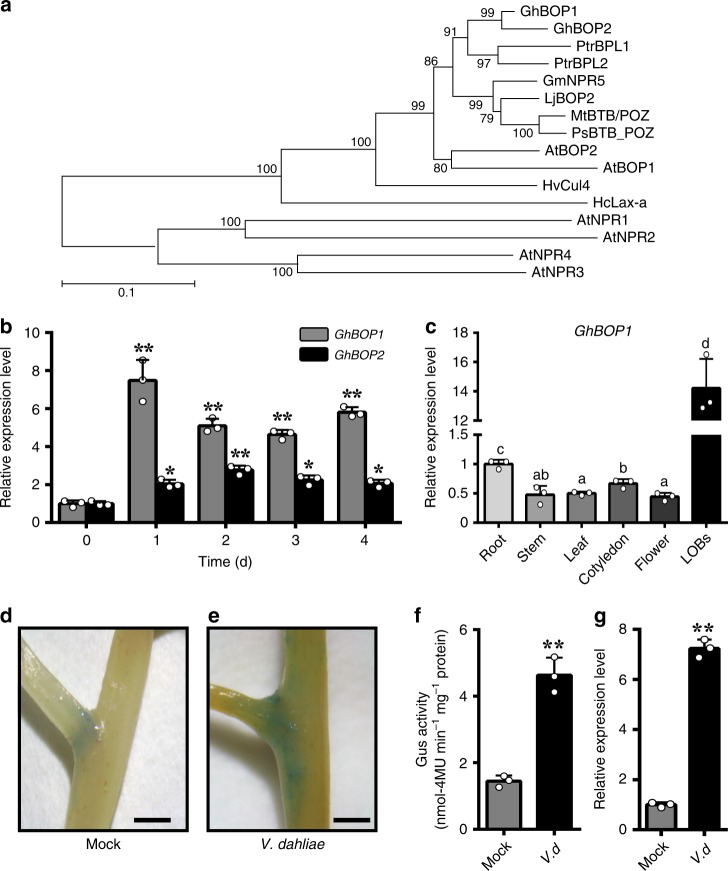
In cotton RNA-sequencing of the plant response to V. dahliae infection, the NPR1-like (BOPs) expression level was significantly induced31, and BOPs possessed the same domains and a similar structure as NPR1 (Supplementary Fig. 1a), which led us to dissect the function of BOPs in cotton plant response to pathogen infection. In G. hirsutum, there are two homologues of BOP proteins, GhBOP1 and GhBOP2. GhBOP1 has two copies, Gh_A09G1115 and Gh_D09G1120 located in At and Dt subgenomes, respectively, sharing 99.6% and 99.4% similarities in coding sequences and amino acid sequences, respectively (Supplementary Figs. 2 and 3). Thus, Gh_A09G1115 were chosen for researching characterization of GhBOP1. GhBOP1 and GhBOP2 (Gh_A01G1644) are highly conserved with other plant BOPs (Supplementary Fig. 1b). GhBOP1/2 proteins have a strong relationship with two BOPs in Populus Trichocarpa belonging to BTB-ankyrin protein family, as well as NPRs (Fig. 1a). GhBOP1 expression in roots was significantly induced after V. dahliae inoculation, while the GhBOP2 expression level was moderately increased (Fig. 1b). The results of the virus-induced gene silencing (VIGS) analysis showed that GhBOP1-silenced plants had an increased sensitivity to V. dahliae infection, while GhBOP2-silenced plants were similar to the control with respect to disease symptoms (Supplementary Fig. 4). Thereby, GhBOP1 was considered to potentially participate in plant defence against V. dahliae.
Fig. 1.
Analysis of GhBOP1 specific expression in the lateral-organ boundaries and autonomously expanding expression in response to V. dahliae infection. a Phylogenetic tree of BOP proteins from G. hirsutum and other species. The complete amino acid sequences of BOPs were aligned using ClustalX and assessed with MEGA 5.0 using the neighbour-joining method with 1000 bootstrap replicates. The numbers next to each node represent confidence percentages. Branch lengths are proportional to the amount of inferred evolutionary change. b Expression patterns of GhBOP1 and GhBOP2 induced by V. dahliae. Total RNA was extracted from roots at 0, 1, 2, 3, and 4 day post-inoculation. GhUB7 served as an internal control. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences compared to respective 0 d, as determined by Student’s t-test (*P < 0.05, **P < 0.01). c Transcript levels of GhBOP1 in different tissues of cotton. Total RNA was extracted from root, stem, leaf, cotyledon, flower, and LOBs. GhUB7 served as an internal control. Error bars represent the SD of three biological replicates. The different letters indicate statistically different means at P < 0.05 (one-way ANOVA with a Duncan post-hoc test). d, e, f GUS staining of the LOBs of GhBOP1pro:GUS transgenic plants at 0 day (d) and 3 day (e) after V. dahliae inoculation, and GUS activity analysis (f), respectively. The scale bars indicate 1 mm. Error bars represent the SD (n = 12) of three biological replicates. Double asterisks indicate statistically significant differences, as determined by the Student’s t-test (**P < 0.01). g GhBOP1 expression analysis of the LOBs of seedlings at 3 day after V. dahliae inoculation. Error bars represent the SD of three biological replicates. Statistical analysis was performed using the Student’s t-test (**P < 0.01)
In Arabidopsis, previous reports have shown that BOP gene expression is restrained in LOBs14,16,20,32–34. Using qPCR analysis, GhBOP1 was dominantly expressed at the LOBs compared with other tissues (Fig. 1c). Analysis of a GhBOP1pro:GUS reporter gene showed that the GUS staining spots were mainly focused at the boundary between the stem and brunch, suggesting that GhBOP1 expression was restrained in LOBs (Fig. 1d).
Transgenic plants carrying the GhBOP1pro:GUS reporter gene were also analysed in response to fungal inoculation. The GUS staining colour clearly deepened at the boundary between the stem and branch of plants inoculated with V. dahliae compared with the mock control, which was consolidated by the GUS quantitative analysis (Fig. 1e and f). More interestingly, GUS staining spots expanded beyond the LOBs by V. dahliae induction (Fig. 1e and f). Compatible with the GUS staining analysis, the GhBOP1 expression level in the zones around the boundary was 7.5-fold higher than in the mock control (Fig. 1g). Additionally, the GUS staining color in root after inoculated with V. dahliae was darker than that of the mock treatment (Supplementary Fig. 5), indicating the GhBOP1 expression level in roots was induced by the fungi. These data suggested that the expression of the GhBOP1 gene was spatially induced by V. dahliae.
GhBOP1 positively regulates plant defence against V. dahliae
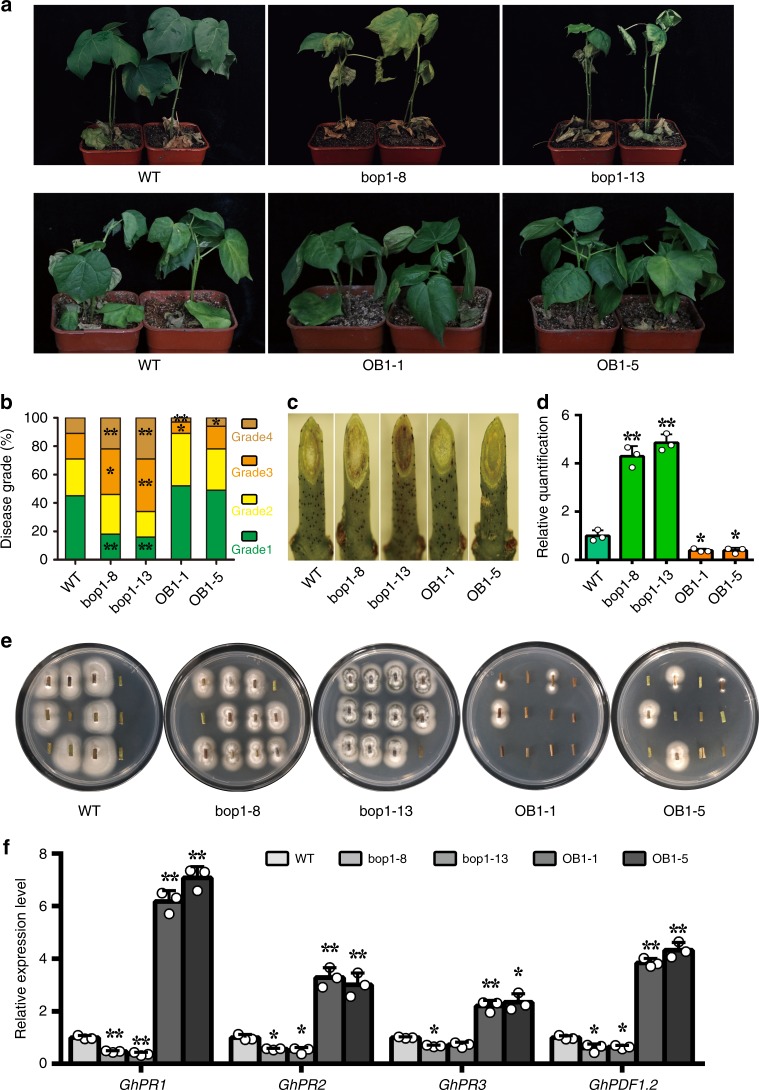
To evaluate the function of GhBOP1 in plant response to pathogen, GhBOP1 RNAi and gain-of-function plants were generalized using A. tumefaciens-mediated transformation methods. Two out of 24 independent GhBOP1 RNAi transformants, bop1-8 and bop1-13, showed lower GhBOP1 transcriptional levels, (Supplementary Fig. 6a). The bop1-8 and bop1-13 contained one T-DNA copy in the genome by southern blot identification (Supplementary Fig. 6b). These adult RNAi plants were slightly slender in architecture compared with the wild-type (WT) plants (Supplementary Fig. 6c). The knockdown seedlings were more sensitive to V. dahliae infection, with more severe defoliation and yellowing symptoms compared with the WT plants (Fig. 2a). The RNAi plants displayed a larger ratio of disease grade 3 and 4 than the WT plants (Fig. 2b). The brown spots in the vascular tissue of the RNAi stem slope sections were more intense compared to the WT (Fig. 2c). Additionally, the GhBOP1-knockdown plants had a greater fungal biomass than in the WT (Fig. 2d). The fungal recovery assay showed that the stem sections from RNAi plants inoculated with V. dahliae had more fungal growth than the inoculated WT (Fig. 2e). These data indicated that GhBOP1 positively regulated plant resistance to V. dahliae.
Fig. 2.
GhBOP1 positively regulates plant defence against V. dahlia. a The disease symptoms of WT, GhBOP1-RNAi, and -OE plants inoculated with V. dahliae. Images were obtained at 18 day after pathogen inoculation. b Disease grade analysis of the infected plants at 18 day after pathogen inoculation. The asterisks indicate statistically significant differences compared to corresponding disease grade of WT, as determined by the Student’s t-test (*P < 0.05, **P < 0.01). c The oblique sections of stems revealed the disease symptoms in the vascular tissue of WT, GhBOP1-RNAi, and -OE plants. d Relative quantification of the fungal biomass in infected stems. qPCR analysis was conducted to compare the DNA contents between the ITS gene (measure of the fungal biomass) of V. dahliae and the UB-7 gene of cotton (for equilibration) at 18 day post-inoculation. Error bars represent the SD of the three biological replicates. The asterisks indicate statistically significant differences, as determined by the Student’s t-test (*P < 0.05, **P < 0.01). e Fungal recovery assay. The stem segments of inoculated plants were placed on PDA medium, and photographs were obtained at 4 day after culture. f Relative expression analysis of four resistance-related genes in WT and transgenic plants after V. dahliae inoculation. GhUB7 served as an internal control. Error bars represent the SD of three biological replicates. The asterisks indicate statistically significant differences compared to corresponding gene expression level in WT, as determined by the Student’s t-test (*P < 0.05, **P < 0.01)
To further investigate GhBOP1 function, two out of 26 independent GhBOP1 overexpression transformants, OB1-1 and OB1-5, with higher GhBOP1 transcriptional levels and one T-DNA copy insertion in the genome were selected (Supplementary Fig. 6a and b). The GhBOP1-overexpressing adult plants showed a slightly shorter plant height than the WT (Supplementary Fig. 6c). The GhBOP1 gain-of-function plants inoculated with V. dahliae exhibited an obvious resistance phenotype with less severe defoliation and yellowing symptoms compared with WT (Fig. 2a). Compared to WT, OB1-1 and OB1-5 showed lower proportion of disease grade 3 and 4, less brown colour intensity in the vascular tissue, less extent of fungal recovery and lower fungal biomass (Fig. 2a–e). Collectively, these results show that GhBOP1 is a positive regulator in the plant response to V. dahliae infestation.
We then analysed the expression levels of genes associated with the SA- or JA-mediated defence pathways in transgenic roots at 3 day after V. dahliae inoculation. As shown in Fig. 2f, the expression levels of the SA-mediated genes GhPR1 and GhPR2 were dramatically decreased in GhBOP1-RNAi plants compared with WT. Interestingly, the expression levels of the JA-mediated genes GhPR3 and GhPDF1.2 were also extremely reduced in knockdown plants. However, in overexpression plants, the expression levels of SA- and JA-mediated genes were clearly increased. These results indicate that GhBOP1 possibly participates in both SA- and JA-mediated defence pathways in cotton.
Interaction of GhBOP1 with GhTGA3
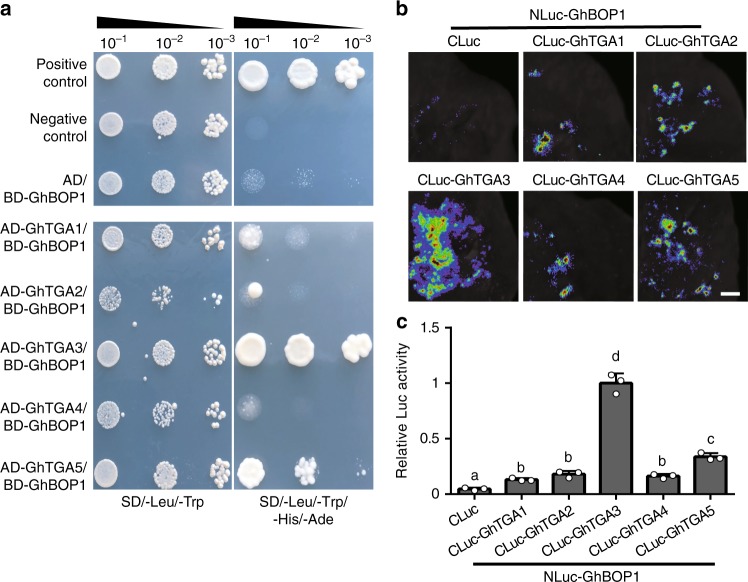
Given that BOP1 protein is similar to NPR1 in domain and structure, we considered the GhBOP1 defence function to be directly related its defence interaction partners in the TGA family, including TGA1 and TGA4 (subgroups I), TGA2 and TGA5 (II), and TGA3 (III). Therefore, the yeast two-hybrid was employed to examine the GhBOP1 interaction with GhTGA1, GhTGA2, GhTGA3, GhTGA4, or GhTGA5. As shown in Supplementary Fig. 7, self-activation of GhTGAs was not observed. The yeast cells co-transfected with vectors harbouring GhBOP1-BD/AD and the negative control (Lam) did not grow on medium containing SD/–Ade/–His/–Leu/–Trp, while the yeast cells co-transfected with the positive control (p53) exhibited normal growth. The yeast cells with GhBOP1-BD/GhTGA3-AD grew normally on medium containing SD/–Ade/–His/–Leu/–Trp, whereas the diluted 10−3 yeast cells with GhBOP1-BD/GhTGA1-AD, GhBOP1-BD/GhTGA2-AD, GhBOP1-BD/GhTGA4-AD, or GhBOP1-BD/GhTGA5-AD hardly grew (Fig. 3a).
Fig. 3.
Interaction of GhBOP1 with GhTGA3. a Yeast two-hybrid assay to detect the interaction between GhBOP1 and GhTGA1/2/3/4/5. The yeast strains containing two correct plasmids were grown on SD/–Leu/–Trp plates and SD/–Leu/–Trp/–His/–Ade plates for 4 day. b Luciferase complementation imaging analysis of the interaction of GhBOP1 with GhTGA1/2/3/4/5. Agrobacterium strains containing the indicated vector were co-expressed in N. benthamiana. The luminescent imagines were taken at 36 h after infiltration. The scale bars indicate 2 mm. c Relative Luc activities in tobacco leaves measured with luminescence intensity by IndiGo software. In present NLuc-GhBOP1, the Luc activity of the CLuc-GhTGA3 in tobacco leaves was set to 1. Error bars represent the SD (n = 12) of three biological replicates. The different letters indicate statistically different means at P < 0.05 (one-way ANOVA with a Duncan post-hoc test)
To consolidate the interaction of GhBOP1 with GhTGA3, a luciferase (Luc) complementation imaging assay was performed. As expected, the tobacco leaves co-injected with of NLuc-GhBOP1/CLuc-GhTGA3 exhibited strong fluorescence intensity (Fig. 3b). However, co-injection of NLuc-GhBOP1/CLuc-GhTGA1, NLuc-GhBOP1/CLuc-GhTGA4, NLuc-GhBOP1/CLuc-GhTGA2, or NLuc-GhBOP1/CLuc-GhTGA5 exhibited poor fluorescence, indicating that GhBOP1 might not interact with them or had a weak interaction. The same results were obtained for the quantity of fluorescence intensity of luciferase (Luc) complementation imagines calculated by pixel, as shown in Fig. 3c. These results show that GhBOP1 can strongly interact with GhTGA3 in cells.
Dependence of GhBOP1 cellular distribution on GhTGA3
Given the interactions between GhBOP1-GhTGA3 and considering the elements of the NPR1-TGAs mechanism as a framework in Arabidopsis35,36, we sought evidence in support of GhBOP1-GhTGA3 in participating in plant defence. We first examined the subcellular localization of GhBOP1 and GhTGA3 in Nicotiana benthamiana epidemic cells. As shown in Fig. 4a, green fluorescence was retained in the nuclei of cells transiently expressing GhTGA3-GFP fusion protein, consistent with the subcellular localization of TGAs in Arabidopsis, while the GhBOP1-GFP fusion protein was found in both the cytosol and the nucleus, which is linked to Arabidopsis BOP1/2 localization16.
Fig. 4.
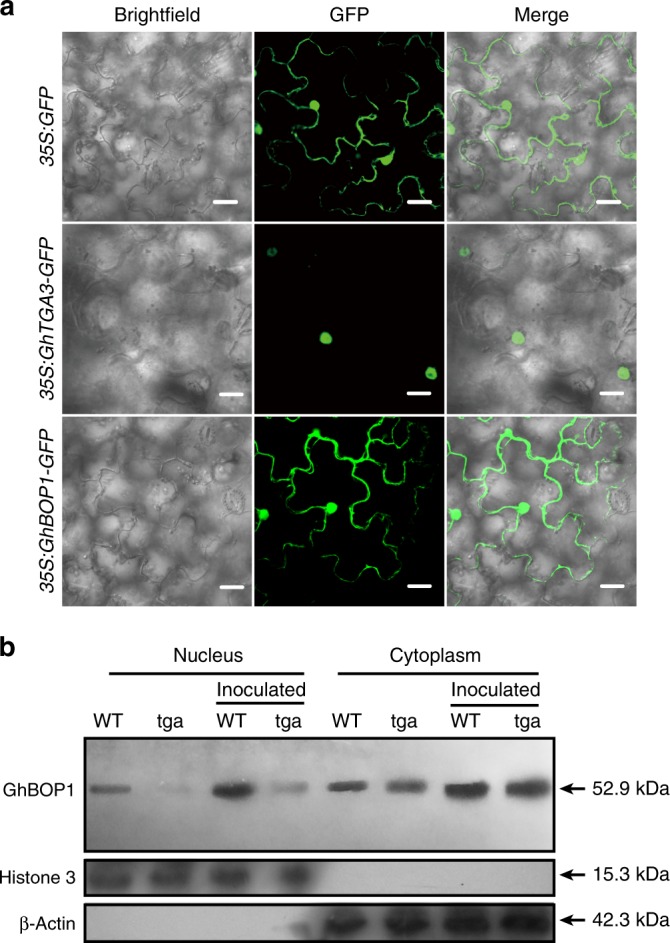
GhBOP1 cellular distribution possibly dependent on GhTGA3. a Subcellular localization of GhGBOP1 and GhTGA3 in N. benthamiana. The scale bars indicate 20 μm. b The distribution of GhBOP1 between the nucleus and cytoplasm depended on the presence of GhTGA3. The nuclear and cytoplasm proteins were extracted from the roots of WT or GhTGA3-silenced plants after pathogen infection or mock treatment. Histone 3, the nuclear protein marker. β-Actin, the cytoplasm protein marker
To elucidate whether GhBOP1 localization was affected by GhTGA3 interactions, immune blot analysis was employed to assess the levels of the two proteins in the nucleus and/or cytoplasm. As shown in Fig. 4b, GhBOP1 proteins were present in both the nucleus and cytoplasm of cotton root cells, consistent with the subcellular localization of the GhBOP1-GFP fusion. However, in GhTGA3-silenced plants, the amount of GhBOP1 proteins in the nucleus was remarkably lower than in WT plants, whereas the protein content in the cytoplasm was comparable. To validate the dependence of the GhBOP1 nuclear distribution on GhTGA3, we also included a parallel experiment in which the plants were infected by the pathogen. After challenging with V. dahliae, the GhBOP1 protein contents in both the nucleus and cytoplasm of root cells increased compared with the unchallenged plants due to pathogen induction of GhBOP1 expression. In GhTAG3-silenced plants, the abundance of GhBOP1 proteins in the nucleus was much lower than in the control. These results indicate that the distribution or accumulation of GhBOP1 proteins in the nucleus is dependent on the presence of GhTGA3 proteins.
GhBOP1 promotes GhTGA3 activation activity
To check the DNA-binding activity of TGA3 and whether this binding could be enhanced by GhBOP1, electrophoretic mobility shift assay (EMSA) experiments were conducted with the recombinant protein TGA3 from Escherichia coli and the synthetic probe with the two core sequences TGACG (hereafter called the TGACG element)37. The EMSA experimental results showed that the retarded band of GhTGA3 protein binding the labelled probe was present, and the signal was gradually decreased by the addition of increasing amounts of unlabelled probe (Fig. 5a), demonstrating that GhTGA3 could bind specifically to the TGACG element in vitro.
Fig. 5.
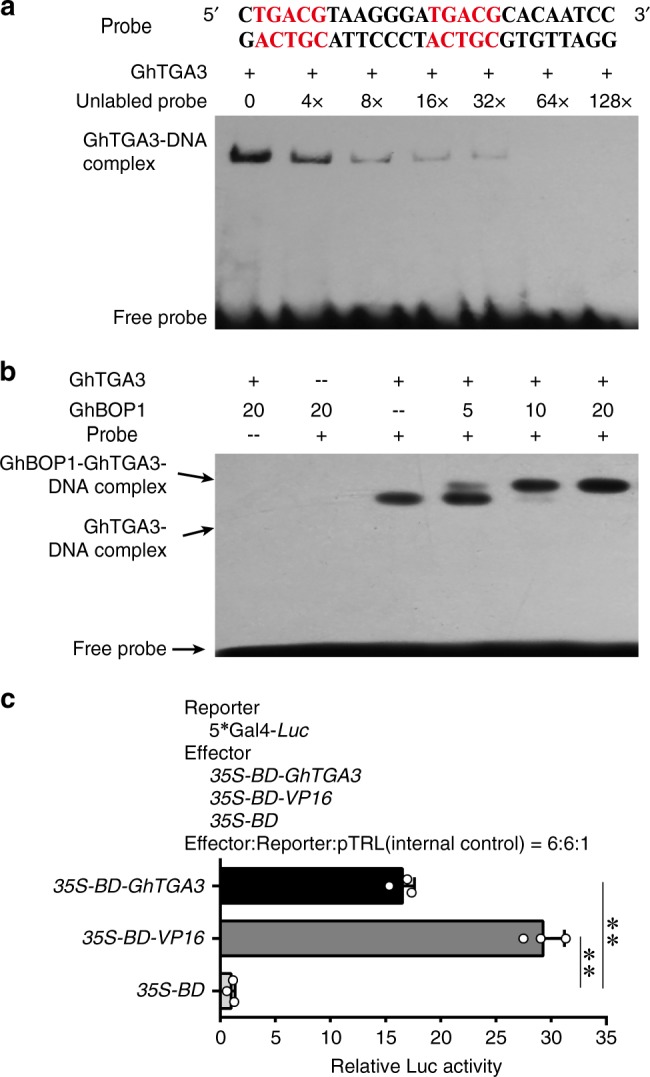
GhTGA3 activation activity and GhBOP1 promotion potential. a EMSA analysis of the specific binding of GhTGA3 to the probe containing the TGACG motif. GhBOP1 proteins were incubated with the biotin-labelled DNA probe in the reaction mixture for 30 min. The indicated amounts of unlabelled probe were used in the competition assay. The TGACG motif sequences are highlighted in red. b EMSA analysis of the efficiency of GhBOP1 in the binding activity of GhTGA3 to the TGACG motif. GhTGA3 proteins were incubated with the biotin-labelled probe, which contain two TGACG motifs in the present of GhBOP1. c Dual-luciferase reporter assay of the activation of GhTGA3 in Arabidopsis protoplasts. The effector vectors pRT-BD and pRT-BD-VP16 were used as negative and positive controls, respectively. Error bars represent the SD of three biological replicates. Each sample consisted of three technical repeats. Double asterisks indicate statistically significant differences, as determined by the Student’s t-test (**P < 0.01)
To assess whether the interaction of GhBOP1 with GhTGA3 may have an effect on its binding activity, the binding analysis of GhTGA3 to the TGACG element was performed in the presence of recombinant GhBOP1 from E. coli. As shown in Fig. 5b, GhBOP1 alone did not bind to the probe, while an up-shifted band appeared when GhBOP1 was present in the reaction. Only the up-shifted band could be viewed when GhBOP1 was added up to 20 pM. These results further confirm that GhBOP1 can interact with GhTGA3, potentially enhancing transcription factor TGACG-binding activity in vitro.
To examine the transcription factor activity of GhTGA3, a dual-luciferase reporter assay system in Arabidopsis protoplasts was performed38,39. As shown in Fig. 5c, the relative value of GhTGA3 activating reporter expression was significantly higher than the control, demonstrating that GhTGA3 possesses high activation ability in vivo.
GhBOP1 promotes GhTGA3 activation activity for GhPR1
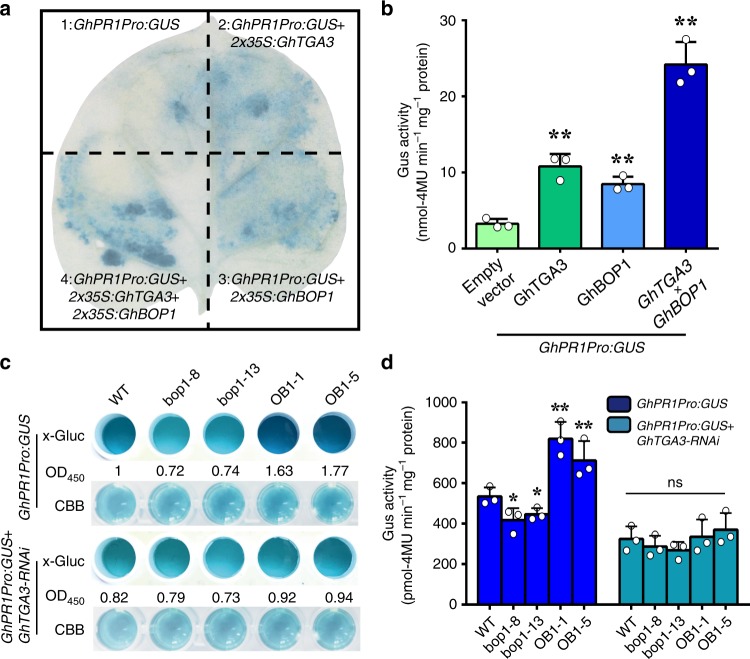
It is known that PR1 expression can be regulated by TGAs through LS5 and LS7 (containing the TGACG element) of the promoter40. In cotton, Gh_D09G0971 was identified as GhPR1 based on the CottonGen database (https://www.cottongen.org/), the promoter of GhPR1 (Gh_D09G0971), which contains a typical TGACG element at 846 bp upstream of the start codon according to the Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Supplementary Table 1). In the loss- and gain-of-function of GhBOP1 plants, GhPR1 transcriptional levels showed striking changes (Fig. 2f). Thus, we chose this gene as a target to assess the roles of GhTGA3 and GhBOP1 interactions using a transient expression assay in vivo. A promoter sequence of GhPR1 replaced the CaMV35S promoter of vector pBIN121 to drive the GUS reporter gene. The Agrobacterium cells harbouring the indicated plasmids (Fig. 6a) were simultaneously injected into tobacco leaves. Two days later, the expression of GhBOP1 and GhTGA3 was confirmed by RT-PCR (Supplementary Fig. 8), and GUS expression was examined. As shown in Fig. 6a and b, the GhPR1 promoter itself drove GUS expression weakly, and the GUS staining intensity was ~3-fold higher when co-transformed with 35 S:GhTGA3. The staining intensity increased when 35 S:GhBOP1 was co-transformed with GhPR1pro:GUS, likely caused by the presence of tobacco TGA3 homolog(s), which might act in concert with GhBOP1 and stimulate GhPR1 promoter activity. When cells containing 35 S:GhTGA3, 35 S:GhBOP1, and GhPR1pro:GUS were simultaneously injected, the GUS staining intensity was much higher and reached an ~2-fold higher level than that in the cells co-transformed with GhPR1pro:GUS and 35 S:GhTGA3. These results demonstrate that GhTGA3 can activate GhPR1 promoter activity, and GhBOP1 can in turn enhance this activity of GhTGA3 in vivo. To evaluate that the expression of GhPR1 might rely on the functions of GhBOP1 and GhTGA3, GUS expression driven by the GhPR1 promoter was tested in cotyledons of GhBOP1-RNAi and -overexpressing plants through transient expression. The protein in cotyledons agro-infiltrated with GhPR1pro:GUS was isolated, and GUS enzyme activity was monitored using two substrates, x-Gluc and 4-MUG. As shown in Fig. 6c, the blue colour in the bop1-8 and bop1-13 reaction solutions with x-Gluc was lighter than in WT, while it was deeper in OB1-1 and OB1-5. When the GhTGA3-RNAi vector was co-injected in cotyledons with the GhPR1pro:GUS vector, the GUS enzyme activities of GhBOP1-overexpressing plants and WT significantly decreased compared with the cotyledon injected exclusively with GhPR1pro:GUS vector, while the enzymatic activity of GhBOP1-RNAi plants was comparable to those injected only with GhPR1pro:GUS vector (Fig. 6c). The relative blue intensity calculated by the absorbance at OD450 confirmed that GhBOP1 promoted the expression of downstream genes regulated by GhTGA3 (Fig. 6c). Consistent with these data, the results of the 4-MUG analyses also confirmed that GhBOP1 could enhance GhTGA3 transcriptional activity (Fig. 6d). These data support that both GhBOP1 and GhTGA3 function in the transcriptional regulation of downstream genes, including GhPR1, in cotton cells, implying that the transcriptional activation activity of GhTGA3 is coupled to the function of GhBOP1.
Fig. 6.
GhBOP1 enhances GhTGA3 activating activity in plants. a GUS staining analysis of GhBOP1 enhances GhTGA3 transcriptional activation to the GhPR1 promoter in tobacco leaf at 48 h after Agrobacterium infiltration with the indicated vectors. b Quantitative analysis of GUS activities in a. Error bars represent the SD (n = 18) of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (**P < 0.01). c GUS staining analysis of GhBOP1 enhancing GhTGA3 transcriptional activation to the GhPR1 promoter in WT and transgenic cotton cotyledon at 48 h after Agrobacterium infiltration with GhPR1pro:GUS or GhPR1pro:GUS and GhTGA3-RNAi, respectively. CBB (Coomassie brilliant blue) was used to normalize the protein extracted from tobacco leaves. The values indicate the relative blue densities of corresponding wells tested by OD450, the blue density of WT was normalized as 1. d GUS activity analysis in c. Error bars represent the SD (n = 18) of three biological replicates. Asterisks indicate statistically significant differences compared to WT, as determined by Student’s t-test (*P < 0.05, **P < 0.01)
To further evaluate whether the positive regulation by GhBOP1 involving GhTGA3 in plant defence, the GhTGA3-silenced plants were produced by the VIGS method and challenged by V. dahliae. As shown in Supplementary Fig. 9, the GhTGA3-silenced plants exhibited increased sensitivity to the pathogens compared with the WT plants, showing serious disease symptom that included a higher disease rate, more fungal recovery potential and a larger fungal biomass.
GhBOP1 is a positive regulator of lignin synthesis
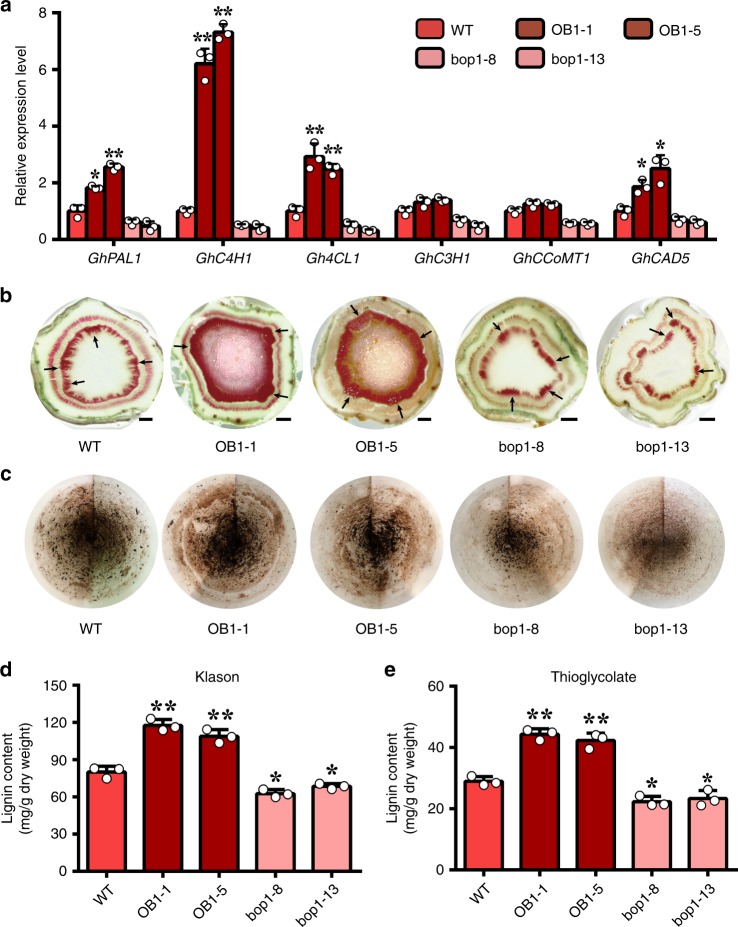
In Arabidopsis, BOP1/2 promotes the expression of lignin biosynthetic genes, while, BP1 represses the expression of these lignin genes41. Recently, mounting evidence has shown that lignin accumulation confers an increased cotton plant resistance to V. dahliae22,24–26,42. Thus, it will be interesting to confirm biochemically and genetically whether GhBOP1 promotes the expression of lignin biosynthetic genes to participate in cotton plant resistance. As shown in Fig. 7a, the lignin biosynthetic genes, PAL1, C4H1, 4CL1, C3H1, CCoMT1, and CAD5, showed significantly upregulation expression in overexpression plants compared with WT stems. In contrast, in RNAi plants, all these lignin biosynthesis genes showed downregulated expression, similar to previously reported results for plants inoculated with V. dahliae22,24,25,27,42.
Fig. 7.
GhBOP1 positively regulates lignin deposition. a The expression levels of lignin synthesis-related genes in WT and transgenic cotton. Total RNA was extracted from the root of WT and transgenic cotton seedlings. Error bars represent the SD of three biological replicates. The asterisks indicate statistically significant differences compared to corresponding gene expression levels of WT plants, as performed by the Student’s t-test (*P < 0.05, **P < 0.01). b Phloroglucinol-HCl staining of stem sections at the same position in WT and transgenic cottons. The black arrows indicate vascular bunldes in stem across sections. Scale bars = 0.2 mm. c Acid-insoluble lignin residues remained on quantitative filter paper after the Klason extraction. d, e The lignin content of WT and transgenic cotton was measured by the Klason method (d) and thioglycolate (e) analysis, respectively. Error bars represent the SD (n = 18) of three biological replicates. Asterisks indicate statistically significant differences in comparison to WT, as determined by the Student’s t-test (*P < 0.05, **P < 0.01)
To further investigate the function of GhBOP1 in lignin accumulation, we stained cotton stems with phloroglucinol-HCl. Compared with the wild-type stems, the GhBOP1-overexpressing stems displayed more intense red staining (darker red) and a larger staining area, suggesting the presence of a relatively higher amount of lignin. In contrast, the RNAi plant stem sections showed the faintest red staining (pink) and a smaller staining area (Fig. 7b). To analyse the content of lignin in plant stems, we employed both the Klason and thioglycolate methods to measure the lignin content. As shown in Fig. 7c, we clearly observed different amounts of insoluble lignin (Klason lignin) from WT and transgenic stems. The mean dry weight of Klason lignin was 80.1 mg/g for wild type, 117.6 and 108.8 mg/g for OB1-1 and OB1-5, and 62.7 and 68.6 mg/g for bop1-8 and bop1-13, respectively (Fig. 7d). The results of lignin contents from the thioglycolate method were similar to results of Klason lignin (Fig. 7e).
Additionally, we tested the lignin accumulation of GhBP1-silenced plants, which provided similar results to GhBOP1-overexpressing plants with the two methods, as shown in Supplementary Fig. 10. These results document that GhBOP1 is a positive regulator in lignin deposition and that its accumulation may be involved in the negative regulation of GhBP1, consistent with the Arabidopsis lignin deposition13. Thus, the expanded GhBOP1 expression out of LOBs confers enhanced plant resistance to V. dahliae, partially due to the promotion of both lignin accumulation as well as GhTGA3 transcriptional activity.
Expanding GhBOP1 expression out of LOB by GhBP1 repression
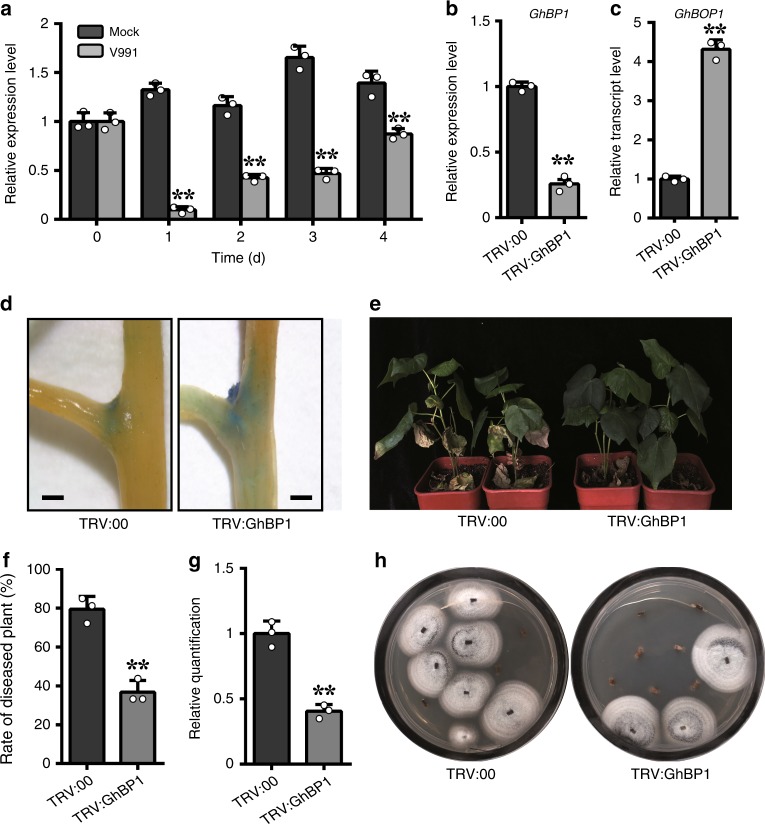
The BOP-induced spatial expression during fungal infestation was similar to the phenotype observed in Arabidopsis bp1 mutants, reminiscent of the modulation of BP1 repression13. Thus, we tested the expression levels of the cotton BP1 gene in plants inoculated with V. dahliae. The GhBP1 expression levels were significantly decreased post-inoculation compared with the control (Fig. 8a). These results indicated that GhBP1 expression was extremely repressed during fungus infection, which possibly resulted in ectopic GhBOP1 expression.
Fig. 8.
GhBP1 repression of the expanded GhBOP1 expression beyond the lateral-organ boundary. a GhBP1 expression patterns in cotton roots at 0, 1, 2, 3, and 4 day post-inoculated with V. dahliae. GhUB7 served as an internal control. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences compared to corresponding expression level of mock treated plants, as performed by the Student’s t-test (*P < 0.05, **P < 0.01). b GhBP1 relative expression levels in roots of TRV:00 and TRV:GhBP1 plants. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences, as determined by the Student’s t-test (**P < 0.01). c GhBOP1 relative expression levels in roots of TRV:00 and TRV:GhBP1 plants. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences, as determined by the Student’s t-test (**P < 0.01). d GUS staining analysis of the GhBOP1pro:GUS transgenic stems with TRV:00 or TRV:GhBP1 treatment. e The disease symptoms of TRV:00 and TRV: GhBP1 plants inoculated with the V. dahliae. The photographs were taken at 18 day after V. dahliae inoculation. f The diseased rate of TRV:00 and TRV:GhBP1 cotton plants at 18 day after inoculated with the V. dahliae. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences compared to TRV:00, as determined by Student’s t-test (**P < 0.01). g The relative quantification of the fungal biomass in the infected stems. Error bars represent the SD of three biological replicates. Asterisks indicate statistically significant differences compared to TRV:00, as determined by Student’s t-test (**P < 0.01). h Fungal recovery assay of TRV:00 and TRV:GhBP1 plants at 18 day after V. dahliae inoculation. The stem segments at the same position of the plants were placed on PDA medium, and taken photographs at 4 day after culture
To further characterize the function of GhBP1 in the expanded expression of GhBOP1, the expression profiles in GhBP1-silenced plants were tested. The GhBP1 expression level was significantly decreased in the GhBP1-silenced plants and showed markedly higher GhBOP1 expression levels in stems, reaching 4.3-fold higher levels than in the control plants (Fig. 8b and c). When GhBP1 was silenced in GhBOP1pro:GUS plants, the GUS staining area almost expanded to the stem and petiole around the LOBs, while GUS staining in the control was retained in the LOBs (Fig. 8d). Thereby, the results of the GUS analysis showed that GhBP1 knockdown could trigger ectopic expression of GhBOP1, similar to the results obtained for V. dahliae infection.
To assess the relationship of the GhBOP1 defence function and the expanding expression by GhBP1 repression, the GhBP1-silenced plants were challenged with V. dahliae. These gene-knockdown plants showed higher resistance with a lower rate of diseased plants, fungal biomass and frequency of fungal recovery compared with the control, as shown in Fig. 8e–h, in line with the disease symptoms of the GhBOP1-overexpressing plants. Taken together, those results show that GhBP1 and GhBOP1 antagonistically regulate plant defence, possibly due to the expanded GhBOP1 expression beyond the LOBs, which was strictly restricted by GhBP1.
Discussion
BOP1 and BOP2 redundantly regulate the plant architecture, including development of the leaf, internode, inflorescence, nodule identity, and abscission zone13,14,16,20,21,43–49. Notably, BOP1 and BOP2 proteins are extremely similar to NPR1 in domains and structure, a major component in the plant defence pathway, especially in the SA signalling pathway50. Thus, it is worthy to investigate whether BOPs participate in plant resistance against the pathogens. In the present study, we found that cotton BOP1 could autonomously expand its expression out of LOBs when the plants were infected by V. dahliae, which may be regarded as spatially induced expression. The spatially induced expression of GhBOP1 could confer economic increases plant resistance to fungi by promoting TGA3 transcriptional activity and lignin accumulation.
BOP1 expression in cotton is specific to LOBs, mainly focusing on the abscission zone between branches and petioles attached to the stem, potentially playing important roles in plant development. When the plants were challenged with V. dahlia, the GhBOP1 gene showed autonomously expanded expression out of LOBs, increasing plant resistance to this fungus. Thereby, GhBOP1 in normal plants may regulate development; while the plants are infected by the pathogens, GhBOP1 shows spatially induced expression to confer increases in plant defence. Compatible with this result, GhBOP1-overexpressing plants also showed greater resistance to V. dahliae. In Arabidopsis, ectopic expression of BOP1/2 in the stem or pedicle from LOBs functions in the plant architecture14. These data indicate that the spatially induced expression of BOPs beyond the LOBs can confer some novel functions including plant defence functions, which is considered an economic model in which only one protein plays roles that are transmit from development to defence according to environmental stresses.
In the present study, we found that GhBOP1 could interact with GhTGA3 and then promote transcriptional levels of downstream disease resistance genes carrying a TGACG element in their promoters. GhTGA3 affected the distribution of GhBOP1 in the cytoplasm and nucleus and GhBOP1 could promote GhTGA3 transcriptional activity, together regulating plant resistance to V. dahliae. Additionally, SA defence-related genes showed upregulated and downregulated expression in GhBOP1 overexpression and RNAi plants, respectively, indicating that the function of GhBOP1-GhTGA3 in plant defence involved the SA signalling pathway. In Arabidopsis, researches have indicated that GhTGAs interacting with NPR1 participate in plant defence mainly through the SA signalling pathway35,36. Thereby, GhBOP1-GhTGA3 could have same model of action in cotton plant defence as Arabidopsis NPR1-TGAs.
In GhBOP1-overexpressing plants, the expression level of lignin synthesis-related genes was upregulated in the stem. The amounts of lignin deposited in the vasculature of GhBOP1-overexpressing and GhBP1-silenced stems. In the plants treated with V. dahliae, the expression patterns of GhBP1 and GhBOP1 showed antagonistic trends, in agreement with the results in Arabidopsis. Khan et al.13 showed that the expression levels of lignin synthesis-relative genes in bp1 mutant and BOP1/2-overexpressing plants were higher than in WT plants, resulting in greater lignin accumulation in the vascular tissue of the stem. These results suggest that plant BP1 and BOPs can antagonistically regulate lignin synthesis, and BP1 is possibly an upstream regulator of BOPs, which has been proposed in some reports13,14. It is regrettable that we did not observe the direct interaction of GhBOP1 with GhBP1. Cotton lignin content is positively related to plant resistance to V. dahliae, which has been confirmed in many recent studies22,24–29. Thereby, GhBOP1 positively regulates cotton plant defence, partial by the promotion of lignin synthesis.
GhBOP1 mainly participates in cotton plant defence against V. dahliae infection through induced expression, VIGS analysis, confirmation of gain- and loss-of-function plants. While GhBOP2 expression mildly increased in plants infected by V. dahliae, and GhBOP2-silenced plants showed similar susceptibility to the control. In Arabidopsis, single bop1 and bop2 mutants showed the same disease resistance as the wild-type, but bop1bop2 mutant showed significant susceptibility to Pseudomonas syringe (Canet et al., 2012). Therefore, GhBOP1 and GhBOP2 do not show obvious functional redundant in cotton plant disease resistance, which is distinguished from Arabidopsis.
Disease resistance function of NPR1 in Arabidopsis is documented by different npr1 mutants including npr1-1 and npr1-336,51–53. The NPR1 in SA perception enters the nucleus and promotes TGAs transcriptional activity, resulting in defence in the attack sites and system acquired resistance in other organs uninfected by pathogens6. At the same time, NPR1 has been reported to be involved in the resistance-inducing ability of JA2. This JA-inducing resistance is considered to come from a cytosolic function of NPR1, likely crucial in cross-talk between SA and JA signalling36,51–54. While Canet et al.17 reported that NPR1 disrupted interaction of BOPs and TGAs, likely attenuating JA-inducing resistance. GhBOP1, as a homolog of NPR1, takes part in cotton plant resistance against V. dahliae possibly associated in SA and JA signalling pathways. Addtionally, in overexpression and RNAi plants, JA-dependent genes, GhPR3 and GhPDF1.2, showed different expression from the wild-type, indicating that the role of GhBOP1 in disease resistance is associated with JA signalling pathway.
Distribution of NPR1 between cytoplasm and nucleus is crucial to confer plant disease resistance, since NPR1 in SA perception becomes an active monomer, which enters into the nucleus and interacts with TGA transcriptional factor. It has been proposed that BOPs locate in both cytoplasm and nucleus in multiple species like NPR1 distribution in the cell16,21,55. In this study, GhBOP1 was also distributed in cytoplasm and nucleus in plants and V. dahliae-inoculated plants. And GhBOP1 distribution or accumulation in nucleus depended on the presence of GhTGA3. Additionally, since GhBOP1 seems to lack a canonical nuclear localization sequence unlike NPR1 protein and is larger protein in size than the protein for passive diffusion of proteins through nuclear pores56, we speculate that there is a potential mechanism for GhBOP1 to shuttle between cytoplasm and nucleus. Collectively, GhBOP1in disease resistance is a similar manner to that of NPR1, and in response to pathogen infection, may accumulate in the nucleus and/or become activated so that they may complex with TGA3 to promote the transcription of defence-related genes.
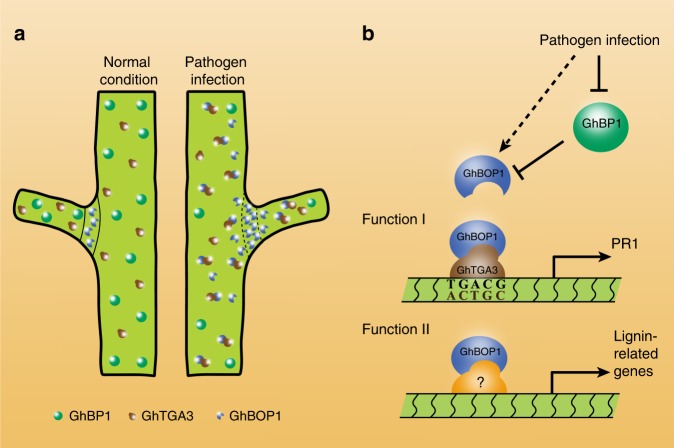
In plants, BOP expression is limited to the LOB, regulating developmental process14. Here we found that GhBOP1 expression could be spatially induced out of LOBs by V. dahliae infection, resulting in increased plant defence. GhBOP1 interaction with GhTGA3 promoted its activation activity, which increased the transcriptional level of downstream resistant-related genes. Additionally, the expanded GhBOP1 expression out of LOBs could upregulate the expression of lignin biosynthesis genes coupled with GhBP1 repression during V. dahliae infection, promoting lignin accumulation to confer increasing plant resistance to this fungus. Figure 9a diagrammatically shows that GhBOP1 expression was normally retained in LOBs, functioning in plant development, while GhBOP1 autonomously expanded out of LOBs when plants encountered pathogen attack. Overall, GhBOP1 is an economically positive regulator that participates in plant defence possibly due to the GhBOP-GhTGA3 complex, similar to the NPR1-TGA module in Arabidopsis, and lignin accumulation by co-regulation with GhBP1.
Fig. 9.
GhBOP1 autonomously expanding expression beyond LOBS increases cotton plant defence against V. dahliae. a Schematic diagram showing GhBOP1 can expand expression out of LOBs in cotton stem after pathogen infection. The area of solid or point lines surrounding represent LOBs. The green, brown, and blue spheres represent GhBP1, GhTGA3, and GhBOP1, respectively. b Working model of GhBOP1 in plant defence process. GhBP1 expression is repressed under the fungus infection, resulting in GhBOP1 ectopic expression. GhBOP1 acts as a positive regulator participating in plant defence through two functions. Function I, GhBOP1 interacts GhTGA3 and promotes GhTGA3 activation activity for GhPR1 promoter. Function II, GhBOP1 interacts with an unknown protein, which could be binding the cis-element of lignin synthesis-related genes, enhancing lignin accumulation
Materials and methods
Plant materials, growth conditions, and transformation
The seeds of G. hirsutum cv. CCR35 cotton plants were used in this article and reaped by self-pollination. The cotton plants were grown in soil (roseite: organic matter soil = 1:3) or Hoagland nutrient solution under an illumination incubator with an 8-h/16-h dark/light photoperiod at 25 °C and a relative humidity of 60%.
The tobacco (N. benthamiana) was cultivated in soil in the illumination incubator with an 8-h/16-h dark/light photoperiod at 25 °C and a relative humidity of 60%.
The CDS sequence of GhBOP1 was cloned into the pBIN438 vector to generate GhBOP1 overexpression vector, named pBIN438-GhBOP1. The specific sequence of GhBOP1 was inserted into RNAi vector pHANNIBAL to generate the GhBOP1-RNAi vector. The recombinant vectors were used to transform into G. hirsutum cv. CCR35 by A.tumefaciens (LBA4404)-mediated transformation according to previous methods57, respectively. The primers used for transformation were listed in Table 1.
Table 1.
Primers used in this study
| Gene name | Purpose | Forward primer | Reverse primer |
|---|---|---|---|
| GhBOP1 | pHANNIBAL-1; RNAi | CGGGAATTCCAGAAACAAACCACAGCACTCC | CGGGGTACCTAGGGTTGTCTTTCACGAGTAAA |
| GhBOP1 | pHANNIBAL-2; RNAi | CGGTCTAGACAGAAACAAACCACAGCACTCC | CGCGGATCCTAGGGTTGTCTTTCACGAGTAAA |
| GhBOP1 | pBin438; OE | CGCGGATCCATGAGTAGCTTTGAGGAGTCCTTAAG | ACGCGTCGACCTAGAAATCATGAGAGTGATGGTACATT |
| ProGhBOP1 | pBIN121 | CCCAAGCTTTAGGCTTCACTTTGGCTTCGAT | CGCGGATCCTTTAGGGTTGTTTTTCACGAGAAAAAG |
| GhBOP1 | pGBKT7 | CGGGAATTCATGAGTAGCTTTGAGGAGTCCTTAAG | CGCGGATCCCTAGAAATCATGAGAGTGATGGTACATT |
| GhTGA1 | pGADT7 | CGGGAATTCATGGCGACCCAACACTTGG | CGCGGATCCCTAAGGCAAGGAAGGTTCAGGA |
| GhTGA2 | pGADT7 | CGGGAATTCATGGGCAGTAGAACGTTGAAAAAT | CGCGGATCCTCACTCGCGAGGTCTAGCCAG |
| GhTGA3 | pGADT7 | CGGGAATTCATGACAATATACGAGCAACTAAACC | CGCGGATCCTCAGAGTGAACTGAGAGCTCGG |
| GhTGA4 | pGADT7 | CGGGAATTCATGGCTAATGGGCTCATACTCCT | CGCGGATCCTTATGCAGGTTCACGAGCACAAG |
| GhTGA5 | pGADT7 | CGGGAATTCATGCCGGGTTTTGACTCACA | CGCGGATCCTCACTCTCGGGGTCGGGC |
| GhBOP1 | pCambia-NLuc | CGCGGATCCATGAGTAGCTTTGAGGAGTCCTTAAG | ACGCGTCGACCTAGAAATCATGAGAGTGATGGTACATT |
| GhTGA1 | pCambia-CLuc | CGGGGTACCATGGCGACCCAACACTTGG | ACGCGTCGACCTAAGGCAAGGAAGGTTCAGGA |
| GhTGA2 | pCambia-CLuc | CGGGGTACCATGGGCAGTAGAACGTTGAAAAAT | ACGCGTCGACTCACTCGCGAGGTCTAGCCAG |
| GhTGA3 | pCambia-CLuc | CGGGGTACCATGACAATATACGAGCAACTAAACC | ACGCGTCGACTCAGAGTGAACTGAGAGCTCGG |
| GhTGA4 | pCambia-CLuc | CGGGGTACCATGGCTAATGGGCTCATACTCCT | ACGCGTCGACTTATGCAGGTTCACGAGCACAAG |
| GhTGA5 | pCambia-CLuc | CGGGGTACCATGCCGGGTTTTGACTCACA | ACGCGTCGACTCACTCTCGGGGTCGGGC |
| GhBOP1 | pPZP111; gene-GFP | CGCGGATCCATGAGTAGCTTTGAGGAGTCCTTAAG | GTGGCCGCGGTAGAAATCATGAGAGTGATGGTACATT |
| GhTGA3 | pPZP111; gene-GFP | CGCGGATCCATGACAATATACGAGCAACTAAACC | GTGGCCGCGGCAGAGTGAACTGAGAGCTCGG |
| GhBOP1 | pMAL-p2X; MBP-GhBOP1 | CGCGGATCCATGAGTAGCTTTGAGGAGTCCTTAAG | CCCAAGCTTCTAGAAATCATGAGAGTGATGGTACATT |
| GhTGA3 | pGEX6P-1; GST-GhTGA3 | CGCGGATCCATGACAATATACGAGCAACTAAACC | CGCGAATTCTCAGAGTGAACTGAGAGCTCGG |
| ProGhPR1 | pBIN121 | CCCAAGCTTTCACATAGATTTGGTGGGTAGGG | CGCGGATCCTTTGAAGTTTGATTCTATAAGAATATTGC |
| GhTGA3 | pHANNIBAL-1; RNAi | CGGGAATTCCTGAGCGTTTCTTCCATTGGAT | CGGGGTACCGCTCGGAGACGGTTGAAGTAA |
| GhTGA3 | pHANNIBAL-2; RNAi | CGGTCTAGACTGAGCGTTTCTTCCATTGGAT | CGCGGATCCGCTCGGAGACGGTTGAAGTAA |
| GhBOP1 | pTL156; VIGS | CGGGGTACCCAGAAACAAACCACAGCACTCC | CGCGGATCCTAGGGTTGTCTTTCACGAGTAAA |
| GhBOP2 | pTL156; VIGS | CGGGGTACCCTAAAGTTTTCTCTTTTCTCTGTCTATC | CGCGGATCCGGTTGTTTTCCCAGAGGAATAC |
| GhBP1 | pTL156; VIGS | CGGGGTACCCGGGAAGAACTAACGAGGCC | CGCGGATCCGTTGATTCAGCCAATGCCACC |
| GhTGA3 | pTL156; VIGS | CGGGGTACCCTGAGCGTTTCTTCCATTGGAT | CGCGGATCCGCTCGGAGACGGTTGAAGTAA |
| NtHistone 3 | RT-PCR | TCCTGGGCAATTTCACGAACAAGC | TGCCCGTAAATCTACTGGAGGCAA |
| GhUB-7 | qPCR | GAAGGCATTCCACCTGACCAAC | CTTGACCTTCTTCTTCTTGTGCTTG |
| GhBOP1 | qPCR | AACCCACCAACTTCAACTGCGA | TCACCCTCCATTCTCGAACCCA |
| GhBOP2 | qPCR | AGAGCTTGGTGCAGCTGATGTT | TGGTGGTCTAAAAGCACCGCAA |
| ITS | qPCR | AAAGTTTTAATGGTTCGCTAAGA | CTTGGTCATTTAGAGGAAGTAA |
| GhPAL1 | qPCR | TCCAGGACAAATTGAGGCAGCG | CCAAGCCACTGTGGAGAAGTCC |
| GhC4H1 | qPCR | CCGAACCCGACACCCATAAGC | GCAGGGATGTCATACCCACCAAG |
| Gh4CL1 | qPCR | GTGTCTTGCCTTTATTCCACATTTAC | TTCTTAGCCAACAACACCACCAAC |
| GhC3H1 | qPCR | ACTATTATTGGACTTCTTTGGGACAT | ATCAGTTTCAGACATCACCCTTTC |
| GhCCoMT1 | qPCR | CACCTGGGTCACAATCCCTTAC | CCAACTGTGGCACGGCAAT |
| GhCAD5 | qPCR | CCGACCATGCGACTCAGACAAT | CTTGGGTGGGTTTTCCGTCAGT |
| GhPR1 | qPCR | GCTCTTGTAGGTGCTCTTGTTCTTCCCT | CTGGTTGTGAACCCTTAGATAATCTTGTGG |
| GhPR2 | qPCR | CAATCTCCCTTGCTCGTGAATCTCTACC | CGTTATCAACAGTGGACTGGGCGG |
| GhPR3 | qPCR | TTAACGGCCTCCTCGAAGCTGCTATTT | CGCAACATAAACAGTGAAACATCATTGGAA |
| GhPDF1.2 | qPCR | CAAGTGGGACATGGTCAGGGGTT | CACTTGTGTGCTGGGAAGACATAGTTGC |
| GhBP1 | qPCR | AGCGGCTCCAACGTGAGTAAT | GCTTCCAAGAGGTTAGAGTACTGAGG |
Solid lines under sequences indicate restriction enzyme cutting sites
Phylogenetic analysis
The neighbour-joining method in MEGA programme version 5.0 was used to generate the phylogenetic tree of the BOPs58. Sequence alignment was performed using DNAMAN 7.0 software for BOP homologue analysis.
RNA and RT/qRT-PCR reaction
Total RNA of cotton or tobacco plants was extracted using the Plant Total RNA Extraction Kit (Sangon Biotech, Shanghai, China). For first-strand cDNA synthesis, 1 µg of total RNA was carried out the reverse-transcription reaction using TransScript First-Strand cDNA Synthesis kit (TransGen). All cDNA samples were diluted by 20-fold and stored at −70 °C until further analysis. RT/qRT-PCR was employed to monitor related genes expression level in plant tissues and treatment samples. For qRT-PCR analysis, the SYBR Green Real-Time PCR Master Mix (TsingKe, Beijing, China) was used on the CFX96 TouchTM Real-Time PCR detection systems (Bio-Rad, Foster City, United States). The Ntactin and GhUb-7 gene were used as internal control for normalization, respectively. All reactions were performed in three biological repeat with three technical repeats. The primers used are shown in Table 1.
DNA extraction and Southern blotting
The cotton genomic DNA was extracted from leaves by Plant Genome Extraction Kit (TianGen, Beijing, China) according to the instruction manual. To determine the T-DNA inserted copy numbers of GhBOP1 transgenic plants, a southern blot analysis was performed according to a previously described method59. Briefly, a total of 20 μg genomic DNA was digested by Hind III for 24 h, then separated by electrophoresis on a 0.8% agarose gel. After that, transfer the separated DNA to the nylon membrane (Hybond-N+; Amersham, Buckinghamshire, UK). The amplicon of NPTII fragment was used as a probe, which was labeled with α-[32P] through a random primer labeling kit (Promega, Madison, WI, United States).
Pathogen infection and disease assay
V. dahliae strain V991, a highly aggressive defoliating isolate, was stored in 20% glycerol at −80 °C. For activation, 10 µL conidia in glycerol suspension were cultured on potato dextrose agar plates for 3 day at 25 °C in the dark, and then the activity of the fungus was transferred into Czapek’s medium for culturing under 200 rpm and 25 °C. Three days later, we collected the fungal conidia and adjusted the concentration to 105 conidia/mL with sterile distilled water containing 5% sucrose. The roots of 3-week-old plants were washed with tap-water, and then the clean roots were soaked in the V. dahliae conidia buffer for 1 min24. The treated plants were planted in soil and incubated in a chamber.
To investigate plant defence in response to V. dahliae infection, the disease grade was surveyed according to a previous report60. The fungal recovery assay and fungal biomass quantification were employed at 18 day post-inoculation according to a previously described method61,62.
Histochemical assay of GUS activity
The histochemical assay of GUS activity was performed according to a previously described method63. In brief, detached fresh tissues were immediately incubated in 95% acetone for 3 h at 4 °C. The incubated tissues were washed two times with 100 mM sodium phosphate buffer (pH 7.0) and then transferred into staining solution [50 mM PBS, 2 mM K4Fe(CN)6, 2 mM K3Fe(CN) 6, 10 mM EDTA, 0.1% Triton-100 and 1 mg/mL 5-bromo-4-chloro-3-indolylglucuronide] for 18–36 h at 37 °C. Next, the stained tissues were washed with buffer containing a gradually increasing content of ethanol from 35 to 95% for 10 min, and photographed with a stereomicroscope (DM2500; Leica, Germany). For quantitative analysis of GUS activity, proteins were extracted in lysis buffer (100 mM PBS, 10 mM EDTA, 0.1% Triton-100, 0.1% SDS and 10 mM β-mercaptoethanol). The 2 mM 4-MUG was added to the reaction buffer to start the GUS reaction, and finally, 200 mM Na2CO3 was added to stop the reaction at 0, 10, 20, 30, and 40 min, respectively. The fluorescence of 4-MU was measured at an excitation wavelength of 365 nm and emission wavelength of 455 nm using the Infinite 200 PRO multimode reader (Tecan, Switzerland).
VIGS mediated by Agrobacterium
Cotton VIGS was conducted according to previously described methods64. In brief, the low similarity 5’UTR sequences of GhBOP1 and GhBOP2 were used as target to silence GhBOP1 and GhBOP2 by VIGS (Supplementary Fig. 11). The specific fragments of the GhBOP1, GhBOP2, GhTGA3, and GhBP1 genes were amplified by PCR and cloned into the tobacco rattle virus (TRV) vector pTRV2, named pTRV2-GhBOP1, pTRV2-GhBOP2, pTRV2-GhTGA3, and pTRV2-GhBP1, respectively. Primers used in VIGS were listed in Table 1. The pTRV2-gene and assistant vector pTRV1 were transformed into the A. tumefaciens GV3101 strains by electroporation, respectively. Agrobacteria containing different vectors were cultured in LB medium at 28 °C for 2 d, collected and resuspended in MMA solution. The suspension concentration was adjusted to an OD600 value of 1.2. For VIGS infiltration, each Agrobacterium containing the pTRV2-gene was equally mixed with the assistant vector pTRV1 strain, respectively. Then, the mixed cultures were incubated at 25 °C for ~3 h in the dark and then injected into cotyledons of 6-day-old cotton seedlings through a 1-mL needleless syringe.
Yeast two-hybrid assay
For the directed yeast two-hybrid assays, the Matchmaker Gold System was employed according to the manufacturer’s instructions (Clontech, Mountain View, CA). The coding region of GhBOP1 was cloned into the BD vector pGBKT7, named BD-GhBOP1. The coding regions of the GhTGA1/2/3/4/5 genes were cloned into the AD vector pGADT7 to produce AD-GhTGA1/2/3/4/5, respectively. The recombinant plasmid BD-GhBOP1 and each AD-gene construct were co-transformed into the yeast strain AH109.
Transient expression in N. benthamiana
To activate A. tumefaciens, each strain was grown in LB medium at 28 °C overnight. The activated strains were then cultured in 20 mL LB liquid medium with 50 µg/mL kanamycin and 40 µg/mL rifampicin at 200 rpm and 28 °C. The cultures were centrifuged, and then resuspended in MMA buffer (10 mM MgCl2, 10 mM MES and 200 μM acetosyringone), and finally adjusted the OD600 value to 1.2. Subsequently, the resuspended cultures were incubated at room temperature for 3 h. For the co-expression analysis, two or three strains carrying appropriate reconstructs were equally mixed and infiltrated into N. benthamiana young leaves using a 1-mL needleless syringe. Leaf samples were collected at 36 h after agro-infiltration to analyse the GUS activity and gene expression. Agro-infiltration was carried out at least three times for the same experiment.
Immunoblot analysis
The roots of wild-type and GhTGA3-silenced cotton plants challenged with V. dahliae or mock were ground into fine powders in liquid nitrogen. The proteins of nucleus and cytoplasm were extracted using Plant Nuclear and Cytoplasmic Protein Extraction Kit (Bestbio, Shanghai, China). Both nuclear and cytoplasmic proteins were quantified by Bradford assay, and 40 µg of the proteins was subjected to SDS-PAGE. The immunoblot experiment was performed using the antibodies (1:2000 dilution) raised against GhBOP1, β-Actin (1:5000 dilution; EarthOx, San Francisco, CA) and Histone 3 (1:5000 dilution; EarthOx) as the primary antibodies, and the horseradish peroxidase conjugated goat anti-rabbit/mouse IgG (1:3000 dilution; Sungene Biotechnology, Tian Jin, China) as the secondary antibody. The uncropped blots were presented in Supplementary Fig. 12.
Electrophoretic mobility shift assay
The coding region of GhBOP1 was inserted into the pMAL-p2X vector to generate a fusion protein: maltose-binding protein (MBP)-GhBOP1. The coding region of GhTGA3 was cloned into the pGEX6P-1 vector to produce a fusion protein: glutathione S-transferase (GST)-GhTGA3. The recombinant proteins were expressed in E. coli strain BL21 and purified using corresponding affinity columns. For the EMSA assay, the biotin-labelled probes and a Pierce LightShift Chemiluminescent EMSA kit (Thermo, Rockford, IL) were used. The binding reaction was performed in a 20 μL reaction mixture at room temperature for 30 min and then separated on a native 6% polyacrylamide gel in 0.5 × Tris-borate/EDTA buffer. The labelled probes were detected based on the manufacturer’s instructions. The uncropped blots were presented in Supplementary Fig. 12.
Dual-luciferase reporter assay
The coding region of GhTGA3 was cloned into the expression vector pRT-BD to construct the BD-GhTGA3 effecter plasmid. The 5 × Gal4-Luc and Renilla Luc gene were used as the reporter and internal control, respectively. The dual-luciferase reporter assay was conducted according to the method described by Ohta et al.38. The samples of co-transformed Arabidopsis protoplasts were cultured in the dark for 12 h at 25 °C, as described by He et al.39. The Luc assay was carried out using the Promega dual-luciferase reporter assay system and measured using a GloMax 20/20 luminometer (Bio-Rad).
Firefly luciferase complementation imaging assay
The luciferase complementation imaging assay is usually employed to validate protein interactions. In this study, the luciferase complementation imaging assay was conducted as described previously23. In brief, the coding sequences of GhBOP1 cloned into the pCAMBIA-NLuc vector to generate NLuc-GhBOP1 and the coding sequences of GhTAG1/2/3/4/5 were inserted into the pCAMBIA-CLuc vector to produce GhTAGs-CLuc, individually. Agrobacterium carrying the constructs was co-infiltrated into the leaves of N. benthamiana. Two days later, the treated leaves were detached and infiltrated with 1 mM luciferin (Promega) and placed in the dark for 10 min. Next, we started to capture the Luc signal and measured the relative Luc activity using a low-light cooled charge-coupled device camera (Night owl LB985, Berthold Technologies, Germany).
Histochemical staining and total lignin content analysis
Histochemical staining was used to visualize lignin deposition using Wiesner reagent according to Xu et al.24. Cotton stems of transgenic plants and WT were crosscut into sections by hand. These stem sections were dipped in phloroglucinol solution (2% in 95% ethanol) for 10 min and then transferred into 18% HCl for 20 min and photographed using a stereomicroscope (DM2500; Leica, Germany). The total lignin content of the stem was determined using the Klason and thioglycolate methods as described previously65. The same experiment was conducted with three biological replicates.
Statistical analysis
Statistical analyses were performed by one-way ANOVA with Duncan’s post-hoc test or two-way ANOVA with student’s t-test and P < 0.05 was considered as significant.
Accession numbers
Arabidopsis genes sequence invoved in this work can be found in the TAIR (The Arabidopsis Information Resource, http://www.arabidopsis.org/), accession numbers as following: AtBOP1 (AT3G57130), AtBOP2 (AT2G41370), AtNPR1 (AT1G64280), AtNPR2 (AT4G26120), AtNPR3 (AT5G45110), AtNPR4 (AT4G19660).
Cotton genes sequnce informations obtained from the CottonGen database (https://www.cottongen.org/), accession numbers as following: GhBOP1 (Gh_A09G1115), GhBOP2 (Gh_A01G1644), GhTGA1 (Gh_D11G2225), GhTGA2 (Gh_A11G2749), GhTGA3 (Gh_D02G0824), GhTGA4 (Gh_D10G0995), GhTGA5 (Gh_A10G1678), GhBP1 (Gh_A05G1857), GhPR1 (Gh_D09G0971).
The other species genes sequence were download from NCBI database (https://www.ncbi.nlm.nih.gov/), accession numbers as following: PtrBPL2 (XP_002308905.1), PtrBPL1 (XP_002323261), NtBOP4 (AFK30390.1), LjBOP2 (AEM62768.1), GmNPR5 (XP_003521136.1), PsBTB/POZ (AET34790.1), MtBTB/POZ (AET34786.1), HvCul4 (BAJ91942.1).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (31771848 and 31471544), the State Key Laboratory of Cotton Biology Open Fund (CB2017B04).
Author contributions
J.W. and G.X. conceived the study and designed the experiments. Z.Z., P.W., X.L., C.Y., Y.T., and Z.W. performed the experiments. G.H., X.G., and J.W. analyzed the data. Z.Z. and J.W. wrote the manuscript. All of the authors read and approved the final manuscript.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information. Source data underlying the graphs presented in the main figures is available in Supplementary Data 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhennan Zhang, Peng Wang, Xiaoli Luo
Contributor Information
Guixian Xia, Email: xiagx@im.ac.cn.
Jiahe Wu, Email: wujiahe@im.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0468-5.
References
- 1.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 2.Pieterse CM, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katagiri F, Thilmony R, He S. The Arabidopsis thaliana-pseudomonas syringae interaction. Arab. Book. 2002;1:e0039. doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez MA, Bannenberg G, Castresana C. Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 2008;11:420–427. doi: 10.1016/j.pbi.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 6.Dong X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Ding Y, et al. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 2018;173:1454–1467. doi: 10.1016/j.cell.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006;48:647–656. doi: 10.1111/j.1365-313X.2006.02903.x. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Holub EB, Alonso JM, Ecker JR, Fobert PR. An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 2005;41:304–318. doi: 10.1111/j.1365-313X.2004.02296.x. [DOI] [PubMed] [Google Scholar]
- 10.Fu ZQ, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canet JV, Dobon A, Roig A, Tornero P. Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 2010;33:1911–1922. doi: 10.1111/j.1365-3040.2010.02194.x. [DOI] [PubMed] [Google Scholar]
- 12.Ha CM, et al. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- 13.Khan M, et al. Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 2012;158:946–960. doi: 10.1104/pp.111.188573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan M, Xu H, Hepworth SR. BLADE-ON-PETIOLE genes: setting boundaries in development and defense. Plant Sci. 2014;215-216:157–171. doi: 10.1016/j.plantsci.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Lewis LA, McCourt RM. Green algae and the origin of land plants. Am. J. Bot. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- 16.Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canet JV, Dobón A, Fajmonová J, Tornero P. The BLADE-ON-PETIOLE genes of Arabidopsis are essential for resistance induced by methyl jasmonate. BMC Plant Biol. 2012;12:199. doi: 10.1186/1471-2229-12-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakoby M, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 19.Gatz C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. plant-microbe Interact. 2013;26:151–159. doi: 10.1094/MPMI-04-12-0078-IA. [DOI] [PubMed] [Google Scholar]
- 20.Xu M, et al. Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 2010;63:974–989. doi: 10.1111/j.1365-313X.2010.04299.x. [DOI] [PubMed] [Google Scholar]
- 21.Wu XM, et al. The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 2012;159:835–850. doi: 10.1104/pp.112.193482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, et al. Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 2014;65:6679–6692. doi: 10.1093/jxb/eru393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang CL, et al. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant. 2015;8:399–411. doi: 10.1016/j.molp.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 24.Xu L, et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011;62:5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, et al. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim Biophys. Sin. (Shanghai) 2012;44:555–564. doi: 10.1093/abbs/gms035. [DOI] [PubMed] [Google Scholar]
- 26.Gao W, et al. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell Proteom. 2013;12:3690–3703. doi: 10.1074/mcp.M113.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, et al. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016;91:305–318. doi: 10.1007/s11103-016-0467-6. [DOI] [PubMed] [Google Scholar]
- 28.Reusche M, et al. Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell. 2012;24:3823–3837. doi: 10.1105/tpc.112.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gayoso C, Pomar F, Novo Uzal E, Merino F, de Ilárduya ÓM. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010;10:232–250. doi: 10.1186/1471-2229-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mele G, Ori N, Sato Y, Hake S. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 2003;17:2088–2093. doi: 10.1101/gad.1120003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 2013;14:637. doi: 10.1186/1471-2164-14-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1361–1370. doi: 10.1093/pcp/pch201. [DOI] [PubMed] [Google Scholar]
- 33.Norberg M, Holmlund M, Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005;132:2203–2213. doi: 10.1242/dev.01815. [DOI] [PubMed] [Google Scholar]
- 34.McKim SM, et al. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development. 2008;135:1537–1546. doi: 10.1242/dev.012807. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Tessaro MJ, Lassner M, Li X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell. 2003;15:2647–2653. doi: 10.1105/tpc.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spoel SH. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam E, Benfey PN, Gilmartin PM, Fang RX, Chua NH. Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl Acad. Sci. USA. 1989;86:7890–7894. doi: 10.1073/pnas.86.20.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/tpc.13.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He P, Shan L, Sheen J. The use of protoplasts to study innate immune responses. Methods Mol. Biol. 2007;354:1–9. doi: 10.1385/1-59259-966-4:1. [DOI] [PubMed] [Google Scholar]
- 40.Lebel E, et al. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith HM, Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu W, et al. GhUMC1, a blue copper-binding protein, regulates lignin synthesis and cotton immune response. Biochem. Biophys. Res. Commun. 2018;504:75–81. doi: 10.1016/j.bbrc.2018.08.128. [DOI] [PubMed] [Google Scholar]
- 43.Ding L, et al. HANABA TARANU (HAN) Bridges Meristem and Organ Primordia Boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during Flower Development in Arabidopsis. PLoS Genet. 2015;11:e1005479. doi: 10.1371/journal.pgen.1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavakol E, et al. The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 2015;168:164–174. doi: 10.1104/pp.114.252882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couzigou JM, et al. The legume NOOT-BOP-COCH-LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol. 2016;209:228–240. doi: 10.1111/nph.13634. [DOI] [PubMed] [Google Scholar]
- 46.Jost M, et al. A homolog of Blade-On-Petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 2016;171:1113–1127. doi: 10.1104/pp.16.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izhaki A, et al. The tomato BLADE ON PETIOLE and TERMINATING FLOWER regulate leaf axil patterning along the proximal-distal axes. Front. Plant Sci. 2018;9:1126. doi: 10.3389/fpls.2018.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magne K, et al. MtNODULE ROOT1 and MtNODULE ROOT2 are essential for indeterminate nodule identity. Plant Physiol. 2018;178:295–316. doi: 10.1104/pp.18.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magne K, et al. Lotus japonicus NOOT-BOP-COCH-LIKE1 is essential for nodule, nectary, leaf and flower development. Plant J. 2018;94:880–894. doi: 10.1111/tpj.13905. [DOI] [PubMed] [Google Scholar]
- 50.Withers J, Dong X. Posttranslational modifications of NPR1: a single protein playing multiple roles in plant immunity and physiology. PLoS Pathog. 2016;12:e1005707. doi: 10.1371/journal.ppat.1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glazebrook J, et al. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313X.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- 52.Leon-Reyes A, et al. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009;149:1797–1809. doi: 10.1104/pp.108.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramírez V, et al. OCP3 is an important modulator of NPR1-mediated jasmonic acid-dependent induced defenses in Arabidopsis. BMC Plant Biol. 2010;10:199. doi: 10.1186/1471-2229-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansson A, Staal J, Dixelius C. Early responses in the Arabidopsis-Verticillium longisporum pathosystem are dependent on NDR1, JA- and ET-associated signals via cytosolic NPR1 and RFO1. Mol. plant-microbe Interact. 2006;19:958–969. doi: 10.1094/MPMI-19-0958. [DOI] [PubMed] [Google Scholar]
- 55.Jun JH, Ha CM, Fletcher JC. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell. 2010;22:62–76. doi: 10.1105/tpc.109.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haasen D, Kohler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313X.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- 57.Jin S, et al. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 2006;50:519–524. doi: 10.1007/s10535-006-0082-5. [DOI] [Google Scholar]
- 58.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Luo X, Zhang X, Shi Y, Tian Y. Development of insect-resistant transgenic cotton with chimeric TVip3A* accumulating in chloroplasts. Transgenic Res. 2011;20:963–973. doi: 10.1007/s11248-011-9483-0. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, et al. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 2017;215:368–381. doi: 10.1111/nph.14537. [DOI] [PubMed] [Google Scholar]
- 61.Ellendorff U, Fradin EF, de Jonge R, Thomma BP. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, et al. Functional characterization of a novel jasmonate ZIM-domain interactor (NINJA) from upland cotton (Gossypium hirsutum) Plant Physiol. Biochem. 2017;112:152–160. doi: 10.1016/j.plaphy.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Deng F, et al. GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 2012;158:890–904. doi: 10.1104/pp.111.186742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pang J, et al. Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One. 2013;8:e73211. doi: 10.1371/journal.pone.0073211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han LB, et al. The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell. 2013;25:4421–4438. doi: 10.1105/tpc.113.116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information. Source data underlying the graphs presented in the main figures is available in Supplementary Data 1.