Abstract
Ostariophysans are the most diverse group of freshwater fishes and feature a pheromone-elicited fright reaction. However, the genetic basis of fright reaction is unclear. Here, we compared vomeronasal type 2 receptor-like (OlfC) genes from fishes having and lacking fright reaction, to provide insight into evolution of pheromonal olfaction in fishes. We found OlfC genes expanded remarkably in ostariophysans having fright reaction compared with fishes lacking fright reaction. Phylogenetic analysis indicates OlfC subfamily 9 expanded specifically in ostariophysans having fright reaction. Principle component and phylogenetic logistic regression analysis partitioned fishes by ecotype (having or lacking fright reaction) and identified OlfC subfamily 9 as being an important factor for fright reaction. Expression levels of expanded OlfC subfamily genes after fright reaction in zebrafish changed more than did genes that had not expanded. Furthermore, evidence of positive selection was found in the expanded OlfC proteins in ostariophysan fishes having fright reaction. These results provide new insight into the genetic basis of fright reaction in ostariophysan fish and will enable future research into the mechanism of action of OlfC proteins.
Subject terms: Evolution, Genetics
Yang et al. investigate the genetic basis of pheromone-elicited fright behavior in ostariophysan fish. They identify a subfamily of vomeronasal type 2 receptor-like (OlfC) that appears to be under strong positive selection; its transcriptional variation suggests a role in the fright behavior of ostariophysan fish.
Introduction
Ostariophysan fishes are the largest and most diverse group of primarily freshwater fishes, representing about 28% of all known fish species and 68% of the world’s freshwater fishes1. The enormous ecological and evolutionary diversity of this group as well as the restricted distribution of almost all members to freshwater habitats has made this group a focus of research in evolutionary biology2–6. Previous studies have suggested that the common ancestor of the ostariophysan fishes entered freshwater about 251 million years ago, which coincides with the global decrease in oxygen levels in marine waters caused by the large mass extinction event that occurred at the end of the Permian era4. However, fishes invading freshwater habitats are expected to have faced stronger challenges to survive compared with those remaining in seawater habitats due to the presence of a different set of predators and the greater probability of encountering predators in the smaller freshwater environments. Therefore, ostariophysan fishes must have developed a set of mechanisms to adapt to the challenging, but promising, freshwater environment.
Among the mechanisms developed, the most remarkable one is the fright reaction that is found in almost all ostariophysan fishes7–9. The fright reaction is elicited by an alarm substance, which is a pheromone that is similar or identical in all ostariophysan fishes1. When a predator damages the skin of an ostariophysan fish, even with a minor injury, an alarm substance produced by epidermal club cells is released into the surrounding water. Nearby members of the same species, or sometimes closely related species, detect this waterborne alarm substance by smell, not taste, resulting in a species-specific fright reaction, which is assumed to be a defensive behavior against predators8–10. Thus, the fright reaction has been suggested to have made a marked contribution to the biological success of the ostariophysan fishes11; however, the genetic basis underlying fright reaction in ostariophysan fishes is still unclear.
Olfaction plays a crucial role in the daily life of fishes, including kin recognition, reproduction, and aggression12. Compared to the two distinct olfactory organs (the main olfactory epithelium and the vomeronasal organ) found in mammals13, fishes only have the main olfactory organ in each nasal cavity, the olfactory rosette14. Thus, all olfaction-related receptor genes are expressed in the olfactory epithelium of the nasal cavity in fishes. The main olfactory epithelium and the vomeronasal organ in mammals employ distinct receptors and signal transduction pathways, and excite different regions of the brain to mediate olfaction13. In mammals, the main olfactory epithelium mainly detects volatile odorants while the vomeronasal organ detects pheromones15,16, although there is some functional overlap between the main olfactory epithelium and the vomeronasal organ17–21.
In mammals, vomeronasal receptors (VNRs) are specifically expressed in the vomeronasal organ and are believed to encode receptors binding pheromone, which is a secreted or excreted chemical factor triggering a social response in members of the same species22,23. The mammalian VNR family is subdivided into two evolutionarily unrelated superfamilies: the VNR family 1 (V1R) and VNR family 2 (V2R)24. It has been suggested that V1R recognize small airborne pheromones25, whereas V2R bind to water-soluble pheromones26. Recently, it has been proposed that fish V1R-like and V2R-like receptors be named ora (olfactory receptor (OR) class A-related)27 and OlfC (OR class C-related)28, respectively. As fish do not have a vomeronasal system, the corresponding VNRs are expressed in the olfactory epithelium of the nasal cavity14. Previous studies have shown that there are enormous variations in the sizes of the V1R and V2R repertoires among different species and much of this variation can be explained as adaptation by the organisms to their different environments29–31.
Since the hypothesis that fright reaction plays crucial roles in the diversification of ostariophysan fishes, is elicited by pheromones, and fish OlfC genes recognize pheromones dissolved in water, we hypothesized that OlfC receptor genes in ostariophysan fishes have important roles in the fright reaction. Notably, some ostariophysan fishes lack the fright reaction, such as cave fish (Astyanax mexicanus) and electric eel (Electrophorus electricus)32. However, ostariophysan fishes lacking the fright reaction differ markedly in their way of life compared with ostariophysan fishes with the fright reaction. For example, ostariophysan fishes without the fright reaction typically are either cave dwelling, predaceous, nocturnal, electric, armored or solitary, or occupy cryptic habitats11, which suggests that their habitats reduce the need for this defense against predation. These species, therefore, provide an opportunity to test the association between OlfC receptor genes and the fright reaction in ostariophysan fishes. To examine the contributions of OlfC genes to the fright reaction in ostariophysan fishes, we have systematically defined the OlfC gene repertoires among ostariophysan fishes, both those having and lacking a fright reaction, and compared their complexity and evolution among the different groups of fishes. Our results showed that subfamily 9 of OlfC genes expanded substantially in ostariophysan fishes having the fright reaction, and their sequences show evidence of positive selection. The levels of gene expression in this expanded subfamily are elevated after stimulation with damaged skin from zebrafish. Our findings provide insights into the genetic basis of the fright reaction in ostariophysan fishes.
Results
Expansion of the number of OlfC genes in ostariophysans
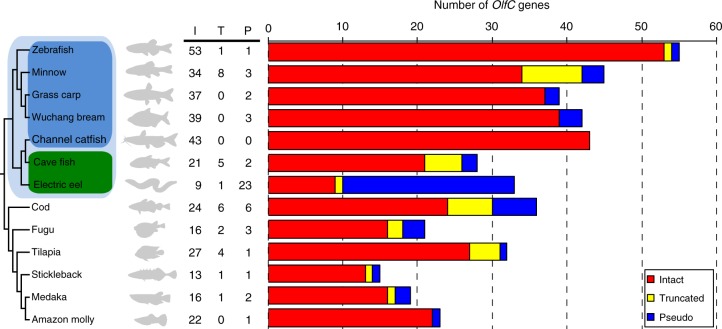
We examined the genome sequences of a total of 13 species of fish, which were divided into three groups. The first group comprised five ostariophysan fishes having the fright reaction: zebrafish, minnow, grass carp, Wuchang bream, and channel catfish. The second group comprised two ostariophysan fishes lacking the fright reaction: cave fish and electric eel. Finally, the third group was made up of six non-ostariophysan fishes: cod, fugu, tilapia, stickleback, medaka, and Amazon molly. In all, 354 OlfC genes were identified in the genomes of these 13 fish species (Fig. 1). Among the identified OlfC genes, those from the zebrafish, fugu, stickleback, and medaka are updates from previous studies33–35, while those from the minnow, grass carp, Wuchang bream, channel catfish, cave fish, electric eel, cod, tilapia, and Amazon molly are newly identified in this study. First, we compared the results from our pipeline with those from previous reports. We found that the OlfC gene counts were similar between our results and previous reports (Supplementary Table 1 and Fig. 1), suggesting that our pipeline was robust and the number of identified OlfC genes are reliable. We then classified the identified OlfC gene into three categories: intact genes (with an intact open reading frame (ORF) and complete coding region), truncated genes (with an intact ORF but partial coding region), and pseudogenes (with disrupted ORF due to nonsense or frameshift mutations in the coding region). Intact and truncated genes are possible functional genes, whereas the pseudogenes are putative nonfunctional genes. For all of the species we examined, the number of truncated genes is generally small (Fig. 1); thus, the number of intact OlfC genes is likely a good indicator of the number of functional OlfC genes in each species. The deduced protein sequences of the identified intact OlfC genes are provided in Supplementary Data 1.
Fig. 1.
Numbers of OlfC genes in the genome sequences of 13 species of fish. “I,” “T,” and “P” indicate the numbers of intact genes, truncated genes, and pseudogenes, respectively. Branch shades represent the different groups of fishes: ostariophysan fishes (light blue); ostariophysan fishes having the fright reaction (deep blue); ostariophysan fishes lacking the fright reaction (deep green). Species tree was taken from previous studies2,4,50
The total number of OlfC genes varies substantially among species (Fig. 1). Remarkably, the number of intact OlfC genes in ostariophysan fishes having the fright reaction (gene number ranges from 34 to 53) is almost twice that in ostariophysan fishes lacking the fright reaction (gene number is 9 and 21) and non-ostariophysan fishes (gene number ranges from 13 to 27). OlfC genes make up a statistically significantly higher proportion of the number of genes in the zebrafish than the cave fish genome (53/25465 = 0.21% vs. 21/23042 = 0.09%, χ2 test, P = 0.0009). To examine whether the observed increased size of the OlfC gene family in zebrafish was due to a specific increase in the size of this gene family or was part of a more general increase in sizes of gene families in zebrafish, compared with cave fish, we performed a genome-wide comparison of gene family sizes between these two species, based on the gene family annotations in Ensembl 83. We identified a total of 7360 gene families being shared between zebrafish and cave fish, with 3932 gene families having at least 2 genes in either zebrafish or cave fish retained for the subsequent analysis. We found that the ratio of the OlfC gene family repertoire size between zebrafish and cave fish of 2.52 (53/21) to be significantly higher than the mean ratio value of 1.16 for 3932 gene family sizes between the two genomes (one-sample Student’s t test, P < 2.2 × 10−16) (Supplementary Fig. 2A). When only larger gene families were used, qualitatively similar results were obtained (Supplementary Fig. 2B–D). These data suggest that the ostariophysan fishes having a fright reaction have significantly expanded the repertories of their OlfC gene family.
OlfC subfamily 9 expanded exclusively in ostariophysans
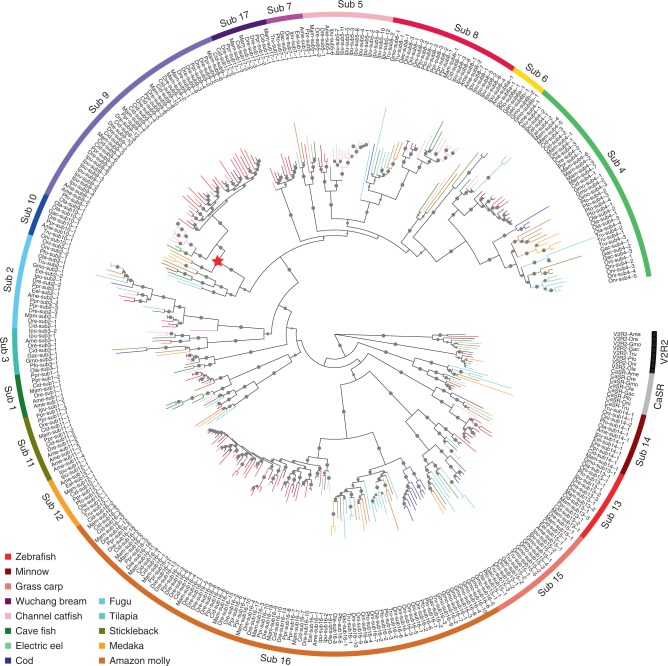
In order to examine the pattern of expansion in the OlfC gene family and to evaluate whether gain or loss of gene members in specific subfamilies might be responsible for specific functions, we constructed a maximum likelihood (ML) tree of all the aligned OlfC genes (Fig. 2 and Supplementary Fig. 3). This tree showed that there are 16 or 17 subfamilies in teleost OlfC genes, which is generally consistent with previous studies31,35,36. Most subfamilies form monophyletic groups with high bootstrap support, except subfamily 6, which is divided into two groups. The phylogenetic tree also identified many species or lineage-specific gene duplications, such as those seen in subfamilies 4, 5, 8, 9 and 16, suggesting potential functional specialization. Intriguingly, among these expanded subfamilies, only subfamily 9 was expanded exclusively within ostariophysan fishes having the fright reaction, hinting that these genes may be candidates contributing to the unique fright reaction of ostariophysan fishes (Supplementary Fig. 3, Datas 2, 3).
Fig. 2.
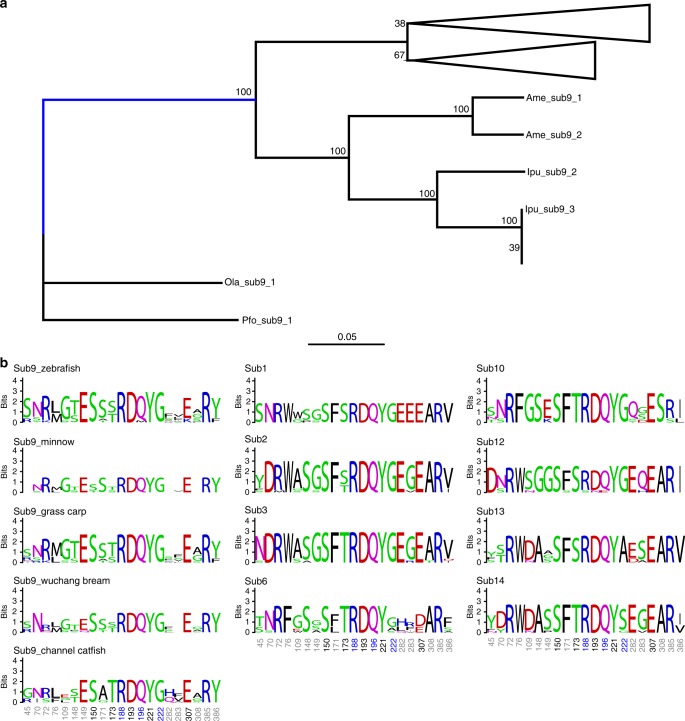
Evolutionary relationships of intact OlfC genes from 13 fish species. Tree was reconstructed using RAxML (version 8.1.17) and rooted with fish CaSR and V2R2 genes. OlfC genes from different species are indicated by different colors. The gene expansion event specific to the ostariophysan fishes having the fright reaction is represented by a red star. Dre, Danio rerio, Zebrafish; Ppr, Pimephales promelas, Minnow; Cid, Ctenopharyngodon idella, Grass carp; Mam, Megalobrama amblycephala, Wuchang bream; Ipu, Ictalurus punctatus, Channel catfish; Ame, Astyanax mexicanus, Cave fish; Eel, Electrophorus electricus, Electric eel; Gmo, Gadus morhua, Cod; Tru, Takifugu rubripes, Fugu; Oni, Oreochromis niloticus, Tilapia; Gac, Gasterosteus aculeatus, Stickleback; Ola, Oryzias latipes, Medaka; Pfo, Poecilia formosa, Amazon molly. Detailed tree with species and gene names and bootstrap values is shown in Supplementary Fig. S3
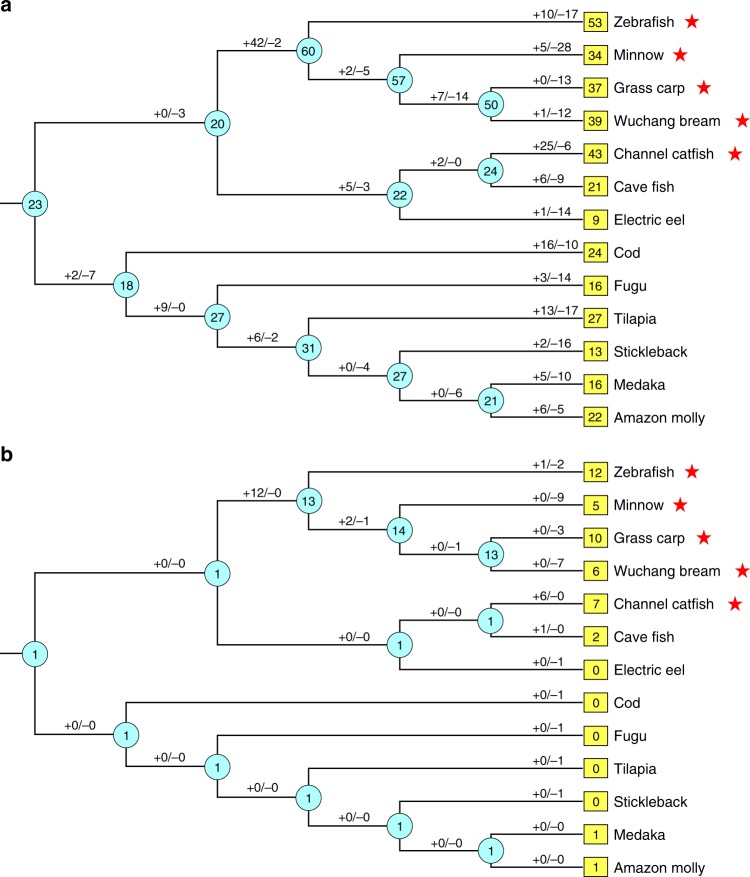
To further characterize the evolutionary dynamics of OlfC gene repertoire size among fish species, we estimated the numbers of gene birth and death events and predicted the number of ancestral gene numbers based on comparison between the gene and species trees using a reconciliation analysis37. Our analyses showed that the ancestral numbers of OlfC genes were relatively stable across the evolution of OlfC genes, implying that the difference in OlfC gene numbers among extant fishes were mostly derived from recent lineage-specific expansions and contractions (Fig. 3a). For example, among the 9 extant fish species with more than 20 OlfC genes, 7 have massive lineage- or species-specific gene gains (n > 10). Similarly, recent losses (n > 10) were observed in all 4 extant fish species with <20 OlfC genes. It should be noted that the two highest gene gains (n = 42 and n = 25) were detected in the ancestral branch leading to zebrafish, minnow, grass carp, and Wuchang bream, and the terminal branch of channel catfish, all ostariophysan fishes having the fright reaction (Fig. 3a).
Fig. 3.
Evolutionary dynamics of intact OlfC genes in fishes. Both intact OlfC genes (a) and OlfC subfamily 9 (b) are examined. The estimated numbers of OlfC gene gains and losses are shown on each branch with plus and minus signs, respectively. The estimated number of genes for the ancestral nodes is shown in the cyan circles, and the number of intact OlfC genes in extant fish species is shown in the yellow boxes. Fish species having fright reaction are marked with red stars
Considering the observation in the phylogenetic tree that the OlfC subfamily 9 expanded exclusively within ostariophysan fishes having fright reaction, we further examined gene birth and death events within OlfC subfamily 9 genes (Fig. 3b). Our results showed that OlfC subfamily 9 expanded independently in the ancestral branch for zebrafish, minnow, grass carp, and Wuchang bream, and in the terminal lineage leading to channel catfish. Considering that concerted evolution can generate a higher sequence similarity between paralogous genes than between orthologous genes38, we further test for gene conversion events among the OlfC subfamily 9 genes using Sawyer’s method, as implemented in the software GENECONV39. We indeed identified several possible events of gene conversion among OlfC subfamily 9 genes in channel catfish (Supplementary Data 4 and Table 2), suggesting that concerted evolution may result in the independent expansion of OlfC subfamily 9 genes in the ancestral branch of zebrafish, minnow, grass carp, and Wuchang bream, and in the terminal lineage leading to channel catfish. Taken together, all these analyses indicate that OlfC subfamily 9 expanded in ostariophysan fishes having the fright reaction, which supports the hypothesis that genes within OlfC subfamily 9 have an important role in the fright reaction in ostariophysan fishes.
Association of OlfC subfamily 9 with the fright reaction
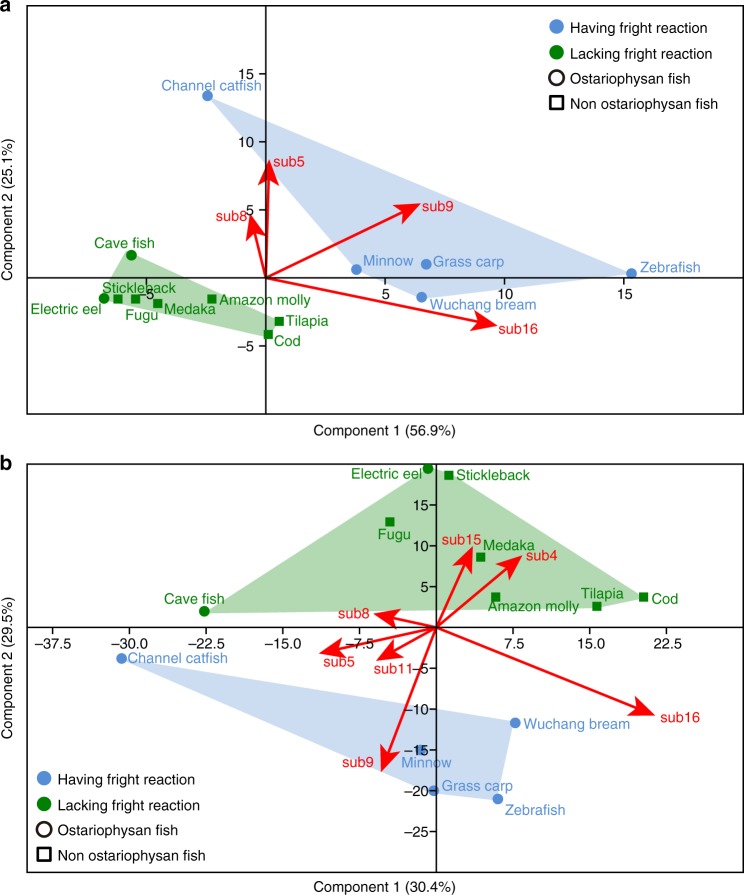
We used principal component analysis (PCA) to identify and visualize differences in the OlfC gene repertoires among ostariophysan fishes having and lacking the fright reaction and in non-ostariophysan fishes to identify OlfC subfamilies that might be these differences (Fig. 4). Our PCA results showed that fishes having the fright reaction grouped away from fishes without the fright reaction, whereas no separation was found between ostariophysan fishes and non-ostariophysan fishes (Fig. 4a and Supplementary Data 2). These results suggest that having a fright reaction or not played a role in determining the configuration of the fish OlfC gene subgenomes. The first two principal components (PCs) explained more than 82% variance of the fish OlfC gene repertoire size. An analysis of similarity (ANOSIM) showed that the fish OlfC gene repertoires varied significantly between the fishes having fright reaction and fishes lacking fright reaction (ANOSIM R = 0.73, P = 0.002), demonstrating that the repertoires of OlfC genes in each subfamily were correlated with fright reaction in ostariophysan fishes. The PCA analysis can also distinguish which OlfC gene subfamilies were driving the differences in OlfC gene repertoire among species. We found that increased size of OlfC subfamily 9 was most closely associated with fishes having fright reaction. These results are consistent both using gene count (Fig. 4a and Supplementary Data 2) and proportion (Fig. 4b and Supplementary Data 3) of the OlfC gene subfamilies (ANOSIM R = 0.73, P = 0.002, and ANOSIM R = 0.33, P = 0.02, respectively).
Fig. 4.
Scatterplots showing the results of the principal component analysis (PCA) analysis of intact OlfC genes with their respective OlfC gene subfamilies. Both OlfC gene counts (a) and proportion (b) across each subfamily are shown. First and second axes explain more than 82% and 59.9% of the variance within the data set. Blue polygons represent fishes having fright reaction and green polygons represent fishes lacking fright reaction. Red arrows represent the contribution of particular subfamilies on the positioning of each species on the plots
To further investigate whether fright reaction is associated with the number of OlfC genes while statistically controlling for phylogeny, we performed a phylogenetic logistic regression analysis, which is for binary variables40. We selected the trait “presence of a fright reaction” as the predictor variable for this analysis. According to the results from phylogenetic logistic regression analysis, having a fright reaction showed significant correlation with the number of functional OlfC genes (AIC = 8.59, P = 0.04). Taken together, all these analyses demonstrated that the presence of a fright reaction is an important factor that is associated with the number of functional OlfC genes.
Profound changes in expression of OlfC genes after fright reaction
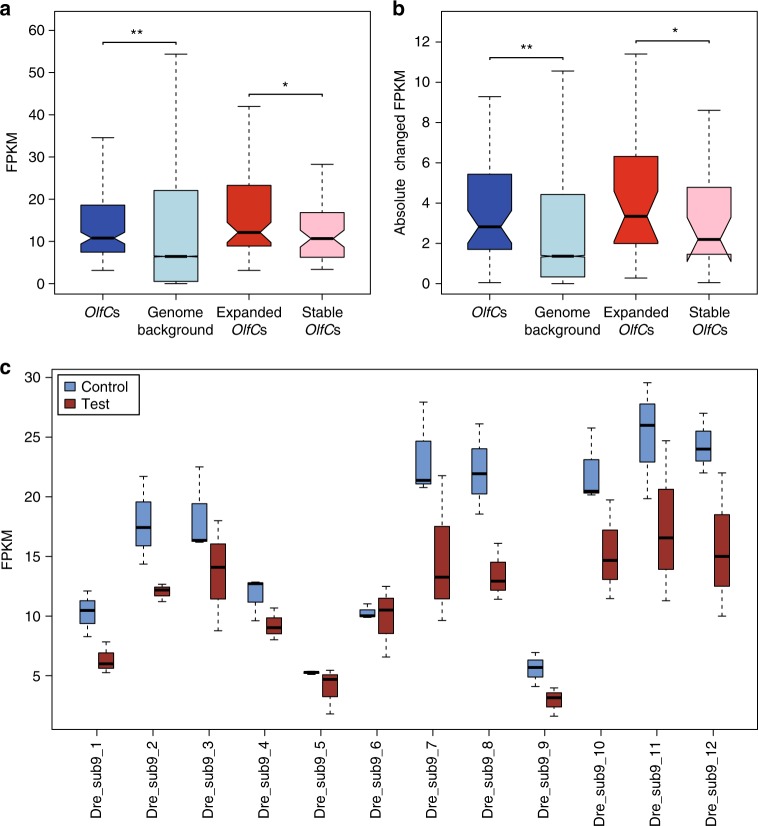
Previous studies have shown that odor stimuli in mouse could decrease the transcription levels of OR genes in activated olfactory sensory neurons41, which may represent a quick adaptation of sensory neurons to a continual stimulus42. Therefore, looking for genes whose expression decreases after fright reaction could reveal the receptors for the alarm signal. Since fishes do not have vomeronasal organs, most OlfC genes are expressed in the olfactory epithelium of the nasal cavity43. Therefore, we examined the expression patterns of OlfC genes in olfactory epithelium transcriptomes before and after fright reaction in zebrafish using RNA-sequencing (RNA-seq) with triplicates samples (Supplementary Table 3, Data 5, and Fig. 4a). The expression levels of OlfC genes were statistically significantly increased above the expression level of all the other genes across the genome in the normal control condition (Wilcoxon’s rank-sum test, P < 0.001) (Fig. 5a), confirming that the olfactory epithelium is a tissue where OlfC genes are expressed. Interestingly, our results also showed that the expression levels of the expanded OlfC genes were significantly higher than for genes in subfamilies that had not expanded (Wilcoxon’s rank-sum test, P = 0.019) (Fig. 5a). These results suggested that genes in the expanded OlfC genes might have more important functional roles than those in the subfamilies that had not expanded in detecting water-soluble pheromones in zebrafish.
Fig. 5.
Comparison of the expression patterns of OlfC genes. a Expression levels, shown as fragments per kilobase per million (FPKM), were compared between all OlfC genes and non-OlfC genes, as well as expanded OlfC genes and non-expanded OlfC genes. b Absolute change of expression levels before and after fright reaction in the zebrafish were compared between all OlfC genes and non-OlfC genes, as well as expanded OlfC genes and non-expanded OlfC genes. c Mean FPKM values of zebrafish OlfC subfamily 9 before and after fright reaction. Significant differences are represented by asterisks, based on Wilcoxon’s rank-sum tests, **P < 0.001; *P < 0.05
As the fright reaction should have substantial impact on behavioral and physiological changes in zebrafish, we hypothesized that they might be reflected by changes in gene expression. Consistent with this hypothesis, we found that the absolute difference in the expression levels of OlfC genes before and after the fright reaction in zebrafish were significantly higher than those for the other genes in the genome (Wilcoxon’s rank-sum test, P < 0.001) (Fig. 5b and Supplementary Fig. 4B). Similarly, our results also exhibited much more profound changes for expression of the expanded OlfC genes than for the non-expanded OlfC genes before and after the fright reaction (Wilcoxon’s rank-sum test, P < 0.05) (Fig. 5b). Furthermore, our results also demonstrated that the transcription levels of expanded OlfC subfamily 9 genes decreased after fright reaction (Fig. 5c), suggesting that these genes may be the receptors of the odors for fright reaction. Taken together, these analyses indicate that OlfC genes, especially the expanded OlfC genes, might play an important role in the fright reaction in zebrafish.
Expanded OlfC subfamily genes are subjected to positive selection
Previous studies have shown that positive selection serves as a major driving force for the expansion of gene families for particular functional roles44. To better understand the evolutionary dynamics of expanded OlfC subfamily 9 genes associated with flight reaction in ostariophysan fishes, we analyzed the selective pressure acting on these genes using PAML (Table 1 and Supplementary Data 6). As expected, the site model analyses showed that the selection model (M8) fitted significantly better than neutral models (i.e., M7 and M8a; Table 1, P = 1.0 × 10−10 and P = 1.7 × 10−4, respectively), indicating that functional diversification and adaptation has occurred in the expanded members of OlfC subfamily 9 genes in ostariophysan fishes having the fright reaction. To test whether the evidence for positive selection was restricted to the ancestral branch, the branch-site model was further employed. We found that the ancestral branch of OlfC subfamily 9 genes of ostariophysan fishes was significantly driven by positive selection (P = 0.02, Table 1 and Fig. 6a). Sites showing evidence of positive selection were mapped to the predicted secondary structure of zebrafish OlfC subfamily 9 protein sequences (Supplementary Fig. 5). We found that the majority of the positively selected sites were located in the N-terminal extracellular region, which is consistent with a previous study in rodents45. As the N-terminal extracellular region is thought to be the ligand-binding domain23,46,47, this result suggested that positive selection has important roles in driving changes in the binding capability of members of the expanded OlfC subfamily 9 genes in ostariophysan fishes.
Table 1.
The parameters and statistical significances of likelihood ratio tests in the OlfC subfamily 9
| Models | ln La | npb | Models compared | 2Δln Lc | d.f. | P valued |
|---|---|---|---|---|---|---|
| Site models | ||||||
| A: M1a (nearly neutral) | −36,829.61 | 82 | B vs. A | 0 | 2 | 1 |
| B: M2a (positive selection) | −36,829.61 | 84 | ||||
| C: M7 (beta) | −36,515.43 | 82 | D vs. C | 75.04 | 2 | 1.0 × 10–10 |
| D: M8 (beta and ω) | −36,477.91 | 84 | ||||
| E: M8a (beta and ωs = 1) | −36,484.98 | 83 | D vs. E | 14.14 | 1 | 1.7 × 10–4 |
| Branch-site model | ||||||
| F: Model A | −36,826.55 | 84 | ||||
| G: Null model A (ω2 = 1) | −36,829.23 | 83 | F vs. G | 5.36 | 1 | 0.02 |
aThe natural logarithm of the likelihood value
bNumber of parameters
cTwice the difference in ln L between the two models compared
dP values lower than 0.05 are shown in bold
Fig. 6.
Selective analysis of OlfC. a ML gene tree for OlfC subfamily 9 from all fishes. Branch with blue color indicates the foreground branch in branch-site model analysis. Numbers on the tree are bootstraps for each node with branches collapsed for low bootstrap support (<70). b Sequence logos for the predicted binding pocket residues in OlfC. Proximal sites predicted to be important for direct ligand “binding” are marked in black, distal binding sites responsible for binding “selectivity” are marked in gray, and the sites thought to be for structural maintenance are shown in blue. Note that the sites responsible for binding “selectivity” (marked in gray) showed much more variability in expanded OlfC subfamily 9 in ostariophysan fishes compared with other non-expanded OlfC subfamilies
Site-directed mutagenesis has found that changes in the amino acid sequence near in the N-terminal extracellular domain of OlfC genes have different functional effects on ligand binding28,48. For example, proximal sites are thought to be important for direct ligand “binding,” while distal binding sites are thought to be crucial for binding “selectivity” and structural sites were involved in structural interaction48. To test whether expansion of OlfC subfamily 9 in ostariophysan fishes with a fright reaction contributed to the functional diversification of their ligand binding, we generated sequence logos for these sequences to compare sequence conservation between OlfC subfamily 9 genes in ostariophysan fishes and the sequences from other subfamilies that did not increase subfamily size (Fig. 6b). Our results clearly show that amino acid sites essential for functions in OlfC are highly conserved, both within subfamilies that have and have not expanded in size. However, differences in the degree of sequence conservation at sites responsible for binding “selectivity” are observed. That is, OlfC subfamily 9 sequences showed much more variability in the expanded OlfC subfamily 9 in each species in ostariophysan fishes compared to other non-expanded OlfC sequences (Fig. 6b, left column vs. middle column and right column). This analysis suggested that expansion of OlfC subfamily 9 genes in ostariophysan fishes having fright reaction led to changes in ligand binding “selectivity,” which might contribute to their ability to detect various waterborne pheromones.
Discussion
Elucidating evolutionary mechanisms and selective forces shaping complex traits across the tree of life is an ultimate goal in evolutionary biology49. As the largest group of freshwater fishes, ostariophysans likely enter freshwater around 251 million years ago. This pioneering exploration was hypothesized to be driven by the global decrease in oxygen levels in seawater caused by the largest mass extinction at the end of the Permian era4. However, the first pioneers must have faced a more challenging habitat, due to the different types of predators and a smaller water area. Fortunately, most ostariophysans evolved a pheromone-mediated fright reaction to defend against predators, which is thought to contribute to their successful diversification11. Ostariophysan fishes form a monophyletic group, which included five major groups: Gonorynchiformes (milkfishes and sandfishes), Cypriniformes (minnows, loaches, and carps), Characiformes (tetras and their allies), Siluriformes (catfishes), and Gymnotiformes (electric eels)2,4,5,50,51. Some members of ostariophysan fishes do not possess a fright reaction, such as blind cave fishes, armored catfishes, and electric fishes11. Interestingly, all the ostariophysan fishes lacking the fright reaction seem to be either cave dwelling, armored, or electric, suggesting that they have evolved specific mechanisms to adapt to the freshwater environment, and do not need the fright reaction. Together, this evidence supports a hypothesis that ostariophysan fishes having a fright reaction have evolved a specific sense to detect the pheromones mediating the fright reaction11.
In accordance with above hypothesis, we found that ostariophysan fishes having a fright reaction have a larger repertoire of OlfC genes compared to ostariophysan fishes without a fright reaction and to non-ostariophysan fishes. Although it remains unclear which aspects of the pheromone detection ability the numbers of OlfC genes reflects, it is reasonable to conclude that species with higher numbers of OlfC genes have an ability to detect an expanded range of substrates. Therefore, our findings represent a perfect example of the correlation between evolutionary patterns with function for a gene family. We further found that subfamily 9 of the OlfC had specifically been expanded in ostariophysan fishes having a fright reaction. We assume that the exclusive expansion of OlfC subfamily 9 genes in ostariophysan fishes might be attributable to their fright reaction ability. Similarly, the species-rich cichlids (which are not ostariophysan fish) also expanded their OlfC gene repertoire to detect a variety of amino acids, which is thought to have contributed to their extraordinary diversification31. However, among OlfC subfamilies, subfamily 9 was not expanded in the cichlids. These observations further support the conclusion that the expansion of subfamily 9 of OlfC genes in ostariophysan fishes might have had an important role in the evolution of the fright reaction.
Although there is little direct experimental evidence addressing the function of the expanded OlfC gene family in ostariophysan fishes having fright reaction, the pattern of evolution and expression strongly indicate an important role of these OlfC genes in the fright reaction: (1) ostariophysan fishes possess a fright reaction elicited by pheromones, (2) fish OlfC genes detect pheromones dissolved in water, (3) some ostariophysan fishes lack the fright reaction, but are either cave dwelling, armored, or electric, (4) OlfC genes were specifically expanded in ostariophysan fishes having a fright reaction, but not in ostariophysan fishes without fright reaction or non-ostariophysan fishes, (5) expression levels of OlfC genes in subfamilies that have increased in number change significantly, compared to OlfC genes in non-expanded subfamilies, after the fright reaction in zebrafish, and (6) the ligand-binding region of OlfC proteins in the expanded subfamily show evidence of positive selection, especially within the ligand binding “selectivity” region that has been experimentally defined in zebrafish. Together, these lines of evidence indicate that expanded OlfC genes in ostariophysan fishes might have contributed to the fright reaction.
Previous studies have found that the evolution of chemosensory receptor genes is largely driven by genomic drift and is independent of selection30,52. In contrast, comparative genomic analyses have found that the evolution of OR repertoires have in part been shaped by natural selection, and reflects ecological adaptations53–56. Consistent with this, our results further provide new evidence that natural selection has a role in shaping the evolution of the OlfC gene repertoires. We found that the independent expansion of OlfC subfamily 9 is associated with the fright reaction in ostariophysan fishes. We speculate that, given a link between OlfC subfamily 9 and the fright reaction in ostariophysan fishes, these OlfC genes might be directly involved in the fright reaction. Further functional studies will be needed to confirm this hypothesis.
Methods
Data sources
Genome sequences from a total of 13 fish species, including 5 ostariophysan fishes having fright reaction [zebrafish (Danio rerio), minnow (Pimephales promelas), grass carp (Ctenopharyngodon idella), Wuchang bream (Megalobrama amblycephala), and channel catfish (Ictalurus punctatus)], 2 ostariophysan fishes lacking fright reaction [cave fish (Astyanax mexicanus) and electric eel (Electrophorus electricus)], and 6 non-ostariophysan fishes [cod (Gadus morhua), fugu (Takifugu rubripes), tilapia (Oreochromis niloticus), stickleback (Gasterosteus aculeatus), medaka (Oryzias latipes), and Amazon molly (Poecilia formosa)], were used in this study. Genome sequences, except minnow, grass carp, Wuchang bream, channel catfish, and electric eel, were downloaded from Ensembl version 83 (http://www.ensembl.org)57. The genome sequence of minnow was obtained from https://www.setac.org/page/fhmgenome58. The genome sequence of grass carp was obtained from http://www.ncgr.ac.cn/grasscarp/59. The genome sequence of Wuchang bream was obtained from http://gigadb.org/dataset/view/id/10030560. The genome sequence of channel catfish was obtained from http://www.ncbi.nlm.nih.gov/61. The genome sequence of electric eel was obtained from http://efishgenomics.zoology.msu.edu/62.
Gene identification
To identify OlfC genes in each of the 13 fish genomes, we followed an earlier study35 with minor modifications. First, we used previously published V2R protein sequences from vertebrates34 as queries to conduct TBLASTN63 searches against each of the 13 fish genome sequences, with an e value cutoff of 1 × 10−10. Second, redundant sequences that hit on the same genomic regions were filtered and sequences shorter than 200 nucleotides were discarded. Third, genomic sequences of homologous genes were extended in the 5′ and 3′ directions and protein-to-genomic sequence alignment with the known V2R protein sequences was conducted using GeneWise64. Finally, the protein sequences of identified putative OlfC genes were used in BLASTP against the NR database to ensure that the best hit was a V2R gene. We classified the identified OlfC genes into three categories: intact genes (I), truncated genes (T), and pseudogenes (P). Intact genes were defined as genes with an intact ORF and complete coding region. Truncated genes were defined as genes with an intact ORF but partial coding region. Pseudogenes were defined as genes with a disruptive ORF because of nonsense or frameshift mutations in the coding region.
Evolutionary analysis
A total of 354 intact OlfC genes were analyzed with CaSR and V2R2 genes used as outgroups31. All coding sequences from the OlfC, CaSR, and V2R2 genes were translated into protein sequences and subsequently aligned with the program MUSCLE65. Phylogenetic analyses were conducted using both ML and neighbor-joining (NJ) approaches. The ML tree was constructed by RAxML (version 8.1.17)66 under a JTT + G substitution model with bootstrap support values determined using 1000 replicates. The NJ tree was reconstructed using the Poisson protein distances67 and pairwise deletion of gap sites implemented in MEGA568 and was evaluated with 1000 replicates using the bootstrap method69.
To estimate gains and losses of OlfC genes across the fish phylogeny, the reconciled-tree method implemented in the program NOTUNG 2.637 was carried out by comparing the gene tree with the species tree. The gene tree topology was taken from our ML tree, while the species tree topology was taken from recent studies2,4,50. Gene gains and losses were inferred across each branch of the species tree and ancestral nodes using the incongruence between the gene and species trees and the parsimony principle.
Detection of gene conversion events
Sequence alignments were generated as described above in evolutionary analysis. The program GENECONV70 was used to identify gene conversion events, which employs permutation to detect whether gene conversion tracts are statistically significant given the distribution of mismatches in the entire sequence alignment.
PCA and analysis of similarities
PCA was conducted using the program PAST v2.17c71 on all intact OlfC genes to explore the degree of correlation between specific OlfC gene subfamilies with the fright reaction in ostariophysan fishes. The PCA algorithm used was the matrix of the OlfC gene data, which was then employed to assess patterns of variation in OlfC gene subfamily distribution between fishes having the fright reaction and fishes lacking the fright reaction. Both the number and the proportion of each OlfC subfamily were analyzed. The statistically significant difference between the above two groupings was examined using a nonparametric test for ANOSIM72 based on the Euclidean distances among all observations.
Phylogenetic logistic regression analysis
We used phylogenetic logistic regression analysis, which is for binary variables40,73, to investigate the relationship between the number of functional OlfC gene and the trait of whether having fright reaction while statistically controlling for phylogeny. We performed this analysis using R with the phylolm package (https://cran.r-project.org/web/packages/phylolm/index.html). The phylogeny of these fishes was constructed from cytb genes from these species. The trait of whether having fright reaction was coded each as 1 (having fright reaction) and 0 (lacking fright reaction).
RNA-seq data analysis
The ethics committee of the Institute of Hydrobiology, Chinese Academy of Sciences, approved all animal experiments. Two groups of adult wild-type zebrafish with the AB background were maintained in the zebrafish facility for 1 week to familiarize with the laboratory environment. One group was used as the control group, and the other group was used as a test group. In the test group, we made shallow lesions on the skin of an adult zebrafish (5 to 6 on each side) using a sharp razor, immersed the fish and washed the damaged skin with distilled water, and then dropped the water into the tank using syringe with a long tube similar to a previous study74. When the test group zebrafish showed a severe fright reaction after the alarm substances were introduced in about 10 min, they were anesthetized immediately and their olfactory mucosae dissected from them at 4 °C. Olfactory mucosae from the control group zebrafish were also collected. Olfactory mucosae were immediately frozen in liquid nitrogen. Tissues from three animals were pooled to obtain sufficient RNA for analysis. Three independent biological replicates for both the control and test groups were prepared. RNA isolation, RNA-seq library construction, and sequencing were conducted by Novogene (Beijing, China) following the approach of our previous study75. Raw reads were assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and filtered using Trim galore (version 0.3.7) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove any adaptor sequence and to trim any bases with Phred quality scores lower than 20. Only paired-end reads where both reads were longer than 50 bp after trimming were retained for the subsequent analysis. High-quality paired-end reads from each sample were separately aligned to the transcript sequences for zebrafish from Ensembl (release 83)76 using Bowtie (version 1.1.1)77, and transcript abundances were estimated using RSEM program (v1.2.20)78. Gene expression levels (fragments per kilobase per million (FPKM)) were calculated by RSEM and only genes with FPKM >1 in at least half of the samples were considered as transcriptionally active genes and used in the subsequent analysis. Raw read counts for each gene detected from RSEM were extracted and normalized to control for differences in sequencing depth using TMM method and differentially expressed genes were identified using the edgeR package79 using a minimal fold change of 2 and an adjusted P value cutoff of 0.05.
Positive selection analysis
Coding sequences of OlfC subfamily 9 that were predicted to have expanded only in ostariophysan fishes having the fright reaction were translated into their protein sequences, aligned with MUSCLE65, and then back-translated into nucleotide-coding sequences. OlfC subfamily 9 gene tree was constructed using ML methods. To detect signatures of positive selection, we employed site model and branch-site model in the codeml program (PAML 4.7 package)80 using ML gene tree as input. Specially, three pairs of paired site models were tested: M1a (nearly neutral: ω0 < 1, ω1 = 1) vs. M2a (positive selection: ω0 < 1, ω1 = 1, ω2 > 1), M7 (nearly neutral; beta distribution: 0 < ω0 < 1) vs. M8 (p positive selection; beta distribution: 0 < ω0 < 1 and ω1 > 1), and M8a (nearly neutral; beta distribution: 0 < ω0 < 1 and ω1 = 1) vs. M8 (positive selection; beta distribution: 0 < ω0 < 1 and ω1 > 1). In the branch site model (model = 2, Nsites = 2), the neutral model constrains a class of sites to have ω = 1 (fix_omega = 1, omega = 1), and the selection model allows a class of codons on the foreground branch to have ω > 1 (fix_omega = 0, omega = 1.5). Likelihood ratio tests were used to compare these nested models, where twice the log likelihood value differences with a χ2 distribution was used to identify positively selected codons. The sites under positive selection were detected by the Bayes empirical Bayes method81. Because we are focusing on the specific expanded OlfC subfamily 9, all these positive selection analyses were using OlfC subfamily 9 gene sequences after removing genes showing evidence of gene conversion. Sequence conservation was visualized using WebLogo82 for functional residues in OlfC proteins for the expanded OlfC subfamily 9 in each species from ostariophysan fishes, and for other non-expanded OlfC subfamilies in all fishes species. The protein membrane topology for OlfC subfamily 9 proteins was created using Protter83, and the locations of putative positively selected sites detected by different methods were marked by star with different colors.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary items
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31601858) and Institute of Hydrobiology, Chinese Academy of Sciences (Y55Z09 and Y85E03) to L.Y., from the Strategic Priority Research Program (XDB13020100) and the National Natural Science Foundation of China (91731301) to S.H., and from the National Natural Science Foundation of China (31702016) to Y.W. The research was supported by the Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China. We thank Prof. Wuhan Xiao to discuss with the early version of this research.
Author contributions
S.H. and T.J.N. lead the project. L.Y. conceived and designed the project. L.Y. performed all computational analyses. L.Y., H.J., Y.W., Y.L., J.C., N.S., W.L. and C.W. performed the experiments. L.Y. analyzed the data, and wrote and revised the paper.
Data availability
The sequencing data have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. SRP154651).
Code availability
The custom scripts have been deposited in Github (https://github.com/yangliandong/OlfC_evolution).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thomas J. Near, Email: thomas.near@yale.edu
Shunping He, Email: clad@ihb.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0479-2.
References
- 1.Nelson JS. Fishes of the World. 4th edn. New York: Wiley; 2006. [Google Scholar]
- 2.Chen WJ, Lavoue S, Mayden RL. Evolutionary origin and early biogeography of otophysan fishes (Ostariophysi: Teleostei) Int. J. Org. Evol. 2013;67:2218–2239. doi: 10.1111/evo.12104. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh K, Miya M, Inoue JG, Ishiguro NB, Nishida M. Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. J. Mol. Evol. 2003;56:464–472. doi: 10.1007/s00239-002-2417-y. [DOI] [PubMed] [Google Scholar]
- 4.Nakatani M, Miya M, Mabuchi K, Saitoh K, Nishida M. Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol. Biol. 2011;11:177. doi: 10.1186/1471-2148-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink SV, Fink WL. Interrelationships of the Ostariophysan Fishes (Teleostei) Zool. J. Linn. Soc. 1981;72:297–353. doi: 10.1111/j.1096-3642.1981.tb01575.x. [DOI] [Google Scholar]
- 6.Briggs JC. The biogeography of otophysan fishes (Ostariophysi: Otophysi): a new appraisal. J. Biogeogr. 2005;32:287–294. doi: 10.1111/j.1365-2699.2004.01170.x. [DOI] [Google Scholar]
- 7.Smith RJF. The evolution of chemical alarm signals in fishes. Chem. Signals Vertebr. 1986;4:99–115. [Google Scholar]
- 8.Frisch KV. Zur Psychologie des Fisch-Schwarmes. Naturwissenschaften. 1938;26:601–606. doi: 10.1007/BF01590598. [DOI] [Google Scholar]
- 9.Frisch KV. Die Bedeutung des Geruchsinnes im Leben der Fische. Naturwissenschaften. 1941;29:321–333. doi: 10.1007/BF01481736. [DOI] [Google Scholar]
- 10.Smith RJF. Alarm signals in fishes. Rev. Fish Biol. Fish. 1992;2:33–63. doi: 10.1007/BF00042916. [DOI] [Google Scholar]
- 11.Pfeiffer W. The distribution of fright reaction and alarm substance cells in fishes. Copeia. 1977;4:653–665. doi: 10.2307/1443164. [DOI] [Google Scholar]
- 12.Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res. Brain Res. Rev. 2001;36:46–59. doi: 10.1016/S0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- 13.Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat. Rev. Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- 14.Grus WE, Zhang J. Origin and evolution of the vertebrate vomeronasal system viewed through system-specific genes. BioEssays. 2006;28:709–718. doi: 10.1002/bies.20432. [DOI] [PubMed] [Google Scholar]
- 15.Buck LB. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/S0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- 16.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 17.Baxi KN, Dorries KM, Eisthen HL. Is the vomeronasal system really specialized for detecting pheromones? Trends Neurosci. 2006;29:1–7. doi: 10.1016/j.tins.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Yoon HY, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123:669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 19.Sam M, et al. Neuropharmacology—odorants may arouse instinctive behaviours. Nature. 2001;412:142–142. doi: 10.1038/35084137. [DOI] [PubMed] [Google Scholar]
- 20.Boehm U, Zou ZH, Buck LB. Feedback loops link odor and pheromone signaling with reproduction. Cell. 2005;123:683–695. doi: 10.1016/j.cell.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Mandiyan VS, Coats JK, Shah NM. Deficits in sexual and aggressive behaviors in Cnga2 mutant mice. Nat. Neurosci. 2005;8:1660–1662. doi: 10.1038/nn1589. [DOI] [PubMed] [Google Scholar]
- 22.Stowers L, Kuo TH. Mammalian pheromones: emerging properties and mechanisms of detection. Curr. Opin. Neurobiol. 2015;34:103–109. doi: 10.1016/j.conb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/S0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- 24.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 25.Boschat C, et al. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- 26.Leinders-Zufall T, et al. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- 27.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: candidate chemosensory receptors for amino acids. BMC Genom. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi P, Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 31.Nikaido M, et al. Lineage-specific expansion of vomeronasal type 2 receptor-like (OlfC) genes in cichlids may contribute to diversification of amino acid detection systems. Genome Biol. Evol. 2013;5:711–722. doi: 10.1093/gbe/evt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer W. Alarm substances. Experientia. 1963;19:113–123. doi: 10.1007/BF02171582. [DOI] [PubMed] [Google Scholar]
- 33.Hashiguchi Y, Furuta Y, Nishida M. Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes. PLoS ONE. 2008;3:e4083. doi: 10.1371/journal.pone.0004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong D, Jin K, Wu X, Zhong Y. CRDB: database of chemosensory receptor gene families in vertebrate. PLoS ONE. 2012;7:e31540. doi: 10.1371/journal.pone.0031540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashiguchi Y, Nishida M. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol. Biol. 2006;6:76. doi: 10.1186/1471-2148-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnstone KA, et al. Genomic organization and evolution of the vomeronasal type 2 receptor-like (OlfC) gene clusters in Atlantic salmon, Salmo salar. Mol. Biol. Evol. 2009;26:1117–1125. doi: 10.1093/molbev/msp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 2000;7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- 38.Liao D. Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 40.Ives AR, Garland T. Phylogenetic logistic regression for binary dependent variables. Syst. Biol. 2010;59:9–26. doi: 10.1093/sysbio/syp074. [DOI] [PubMed] [Google Scholar]
- 41.von der Weid B, et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci. 2015;18:1455–1463. doi: 10.1038/nn.4100. [DOI] [PubMed] [Google Scholar]
- 42.Nishizumi H, Sakano H. Decoding and deorphanizing an olfactory map. Nat. Neurosci. 2015;18:1432–1433. doi: 10.1038/nn.4121. [DOI] [PubMed] [Google Scholar]
- 43.Saraiva LR, et al. Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Sci. Rep. 2015;5:11487. doi: 10.1038/srep11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu DD, Wang GD, Irwin DM, Zhang YP. A profound role for the expansion of trypsin-like serine protease family in the evolution of hematophagy in mosquito. Mol. Biol. Evol. 2009;26:2333–2341. doi: 10.1093/molbev/msp139. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/S0092-8674(00)80536-X. [DOI] [PubMed] [Google Scholar]
- 47.Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/S0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 48.Luu P, Acher F, Bertrand HO, Fan JH, Ngai J. Molecular determinants of ligand selectivity in a vertebrate odorant receptor. J. Neurosci. 2004;24:10128–10137. doi: 10.1523/JNEUROSCI.3117-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storz JF, et al. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3:e45. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Near TJ, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl Acad. Sci. USA. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarty P, et al. Phylogenomic systematics of ostariophysan fishes: ultraconserved elements support the surprising non-monophyly of Characiformes. Syst. Biol. 2017;66:881–895. doi: 10.1093/sysbio/syx038. [DOI] [PubMed] [Google Scholar]
- 52.Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc. Natl Acad. Sci. USA. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayden S, et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayden S, et al. A cluster of olfactory receptor genes linked to frugivory in bats. Mol. Biol. Evol. 2014;31:917–927. doi: 10.1093/molbev/msu043. [DOI] [PubMed] [Google Scholar]
- 55.Khan I, et al. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol. Biol. Evol. 2015;32:2832–2843. doi: 10.1093/molbev/msv155. [DOI] [PubMed] [Google Scholar]
- 56.Vandewege MW, et al. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol. Evol. 2016;8:470–480. doi: 10.1093/gbe/evw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burns FR, et al. Sequencing and de novo draft assemblies of a fathead minnow (Pimephales promelas) reference genome. Environ. Toxicol. Chem. 2016;35:212–217. doi: 10.1002/etc.3186. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015;47:625–631. doi: 10.1038/ng.3280. [DOI] [PubMed] [Google Scholar]
- 60.Liu H, et al. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. GigaScience. 2017;6:1–13. doi: 10.1093/gigascience/gix039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat. Commun. 2016;7:11757. doi: 10.1038/ncomms11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gallant JR, et al. Nonhuman genetics. Genomic basis for the convergent evolution of electric organs. Science. 2014;344:1522–1525. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 68.Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Felsenstein J. Confidence-limits on phylogenies—an approach using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 70.Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 71.Hammer O, Harper DAT, Ryan PD. PAST. Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:1–9. [Google Scholar]
- 72.Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 73.Pollux BJ, Meredith RW, Springer MS, Garland T, Reznick DN. The evolution of the placenta drives a shift in sexual selection in livebearing fish. Nature. 2014;513:233–236. doi: 10.1038/nature13451. [DOI] [PubMed] [Google Scholar]
- 74.Mathuru AS, et al. Chondroitin fragments are odorants that trigger fear behavior in fish. Curr. Biol. 2012;22:538–544. doi: 10.1016/j.cub.2012.01.061. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, Yang L, Wu B, Song Z, He S. Transcriptome analysis of the plateau fish (Triplophysa dalaica): implications for adaptation to hypoxia in fishes. Gene. 2015;565:211–220. doi: 10.1016/j.gene.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 76.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 81.Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 82.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary items
Data Availability Statement
The sequencing data have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession no. SRP154651).
The custom scripts have been deposited in Github (https://github.com/yangliandong/OlfC_evolution).