Abstract
Hearing loss is often due to the absence or the degeneration of hair cells in the cochlea. Understanding the mechanisms regulating the generation of hair cells may therefore lead to better treatments for hearing disorders. To elucidate the transcriptional control mechanisms specifying the progenitor cells (i.e. prosensory cells) that generate the hair cells and support cells critical for hearing function, we compared chromatin accessibility using ATAC-seq in sorted prosensory cells (Sox2-EGFP+) and surrounding cells (Sox2-EGFP−) from E12, E14.5 and E16 cochlear ducts. In Sox2-EGFP+, we find greater accessibility in and near genes restricted in expression to the prosensory region of the cochlear duct including Sox2, Isl1, Eya1 and Pou4f3. Furthermore, we find significant enrichment for the consensus binding sites of Sox2, Six1 and Gata3—transcription factors required for prosensory development—in the open chromatin regions. Over 2,200 regions displayed differential accessibility with developmental time in Sox2-EGFP+ cells, with most changes in the E12-14.5 window. Open chromatin regions detected in Sox2-EGFP+ cells map to over 48,000 orthologous regions in the human genome that include regions in genes linked to deafness. Our results reveal a dynamic landscape of open chromatin in prosensory cells with potential implications for cochlear development and disease.
Subject terms: Developmental neurogenesis, Cochlea
Introduction
Hearing is mediated by a specialized sensory epithelium, the organ of Corti, within the cochlea of the inner ear. In the organ of Corti, rows of hair cells interdigitated with support cells sense sound1. Hair cell loss or dysfunction underlies most sensorineural hearing loss. Hearing loss is broadly affected by genetics and perhaps also by epigenetics. Genetic factors contribute to 68% of hearing loss at birth—affecting 1–2 newborns per 1000—as well as to progressive hearing loss in 2.7 children per 1000 by adolescence2. In addition to congenital hearing loss, significant genetic contributors have been identified for all major acquired forms of hearing loss including presbycusis3–8, noise-induced hearing loss9–18 and ototoxic medication-induced hearing loss19–21. Hearing loss-causing variants in 153 genes have been reported including variants that map to introns and other noncoding regions of the genome22. Detailed characterization of the regulatory genome in the inner ear will likely lead to a better understanding of how genetic differences influence the etiology of congenital and acquired hearing loss.
In other tissues and cell types, concerted efforts have characterized a wide range of gene regulatory elements by eQTL analysis23 and by genome-wide sequencing of open chromatin, histone-mark enrichment, transcription factor-binding and other epigenomic features24. Tissue- and cell-specific ‘maps’ of regulatory elements are now crucial resources for systems studies of gene regulation and gene networks. Such studies promise greater understanding of the function of genetic variants in noncoding regions and the identification of enhancers suitable for the experimental and therapeutic goals of targeted transgenesis.
Despite the vast potential utility of mapping the regulatory genome of inner ear cell types, little progress has been made in this regard. Elements having enhancer activity in otic tissues have been identified based on evolutionary conservation25 and on histone marks associated with gene-activation in other tissues26. Recent studies detected enhancers using ATAC-seq and MethylC-seq in whole mouse cochlea27,28. Mapping of open chromatin in specific cell types of the cochlea using ATAC-seq can further elucidate the regulatory genome and related gene networks of cochlear development.
Hair and support cells differentiate from a region of the floor of the cochlear duct called the prosensory domain. The cells of the prosensory domain exit the mitotic cycle between E12 and E16 in an apical to base sweep29,30. They begin their differentiation as hair cells or support cells within a few days first at the base and lastly at the apex29–33.
The regulation of specification of prosensory cells involves Notch-, Bmp- and Fgf-signaling34–45 as well as the transcription factors Gata3, Sox2 and Six146–53. Differentiation of support cells requires Fgf-signaling and Notch-signaling54,55; differentiation of hair cells requires Wnt-signaling, Atoh1 and Pou4f356–58. To better understand the interactions among these signaling molecules and transcriptional regulation of cochlear development, we used ATAC-seq to map the open chromatin regions in cells of the prosensory domain (Sox2-EGFP+) as well as the surrounding nonsensory cells (Sox2-EGFP−) isolated from the embryonic mouse cochlear duct.
This approach detected over 65,000 open chromatin regions in Sox2-EGFP+ cells from embryonic cochlear duct, including many regions mapping to known and putative gene regulatory regions. Motif enrichment analysis of these open chromatin regions identifies classes of transcription factors not previously implicated in inner ear development and provides strong evidence for a prosensory-specific landscape of open chromatin. Differential accessibility analysis across three developmental stages elucidated epigenetic dynamics in prosensory cell development. These datasets not only provide insights into mechanisms of prosensory development in the cochlea, but also represent a significant resource for the field of epigenomic research in the inner ear and future mechanistic studies of cis-regulation of deafness genes.
Results
Sox2-EGFP expression in prosensory cells of the embryonic cochlea
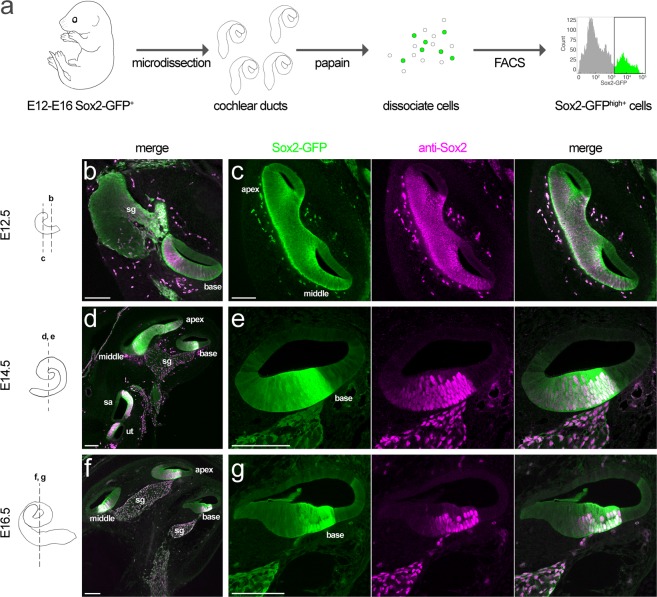
To selectively characterize the accessible chromatin in prosensory cells in the developing cochlea, we carried out ATAC-seq on FACS isolated EGFP+ cells from Sox2-EGFP mice, a strain having EGFP knocked-into the Sox2 coding region59. To determine whether EGFP specifically marks cochlear prosensory cells in heterozygous Sox2-EGFP mice, Sox2-EGFP expression in prosensory cells was compared to Sox2 immunofluorescence at several developmental stages along the cochlea spiral in vibratome sections of E12-16 temporal bones. Similar to endogenous Sox2 expression, the highest level of Sox2-EGFP immunofluorescence is evident in the prosensory cells of the cochlear duct as well as the glia of the spiral ganglion in the E12, E14 and E16 cochleae (Fig. 1). In the following study, cochlear ducts were carefully dissected to remove developing spiral ganglion neurons and associated glia.
Figure 1.
Sox2-EGFP expression in prosensory cells of the embryonic cochlea. (a) Shows the tissue isolation and FACS pipeline used to generate the cells. (b–f) Show Sox2-EGFP expression (green) in vibratome sections of cochlea at the indicated stages of embryonic development. Sox2 immunofluorescence (magenta) demonstrates both the prosensory cells in the cochlear duct and the glia of the spiral ganglion. Note that Sox2-EGFP expression corresponds to Sox2 immunofluorescence and that Sox2-EGFP developmental dynamics mirror those of endogenous Sox2 expression. For example, the Sox2+/Sox2-EGFP+ field of cells in the floor of the cochlear duct narrows between E12.5-16.5—first at the base of the duct, then apically (compare b,d and f) and first at the lateral side of the duct, then medially (compare c, e and g). The localization of Sox2-EGFP expression shown here is representative of that in at least three temporal bones. Scale bars = 100 μm. sg, spiral ganglion; sa, saccule; ut, utricle.
Developmental dynamics in Sox2-EGFP expression mirror those of endogenous Sox2 expression; the cells expressing the highest level of EGFP (Sox2-EGFPhigh+) correspond to the Sox2+ immunolabeled cells. For example, the anti-Sox2+/Sox2-EGFPhigh+ domain initially occupies the entire floor of the cochlear duct (Fig. 1b,c) and becomes progressively restricted between E12.5-16.5, first at the base of the duct, then apically (compare Fig. 1b,d,f). Furthermore, the anti-Sox2+/Sox2-EGFPhigh+ field of cells in the floor of the cochlear duct narrows first at the lateral side of the duct, then medially (compare Fig. 1c,e,g). Laterally, p75/Ngfr immunofluorescence in Claudius cells flanks anti-Sox2+/Sox2-EGFPhigh+ expression at E16 (Supplementary Fig. 1). Similar to the embryonic cochlea, Sox2-EGFPhigh+ corresponds with endogenous Sox2 in E12-16 utricle, saccule and cristae (Supplementary Figs 2 and 3).
Although low and intermediate levels of Sox2-EGFP were observable in some nonsensory cells of the cochlear duct, these findings support a strategy to isolate Sox2-EGFPhigh+ prosensory cells from dissociated cochlear duct cells by gating for cells having the highest Sox2-EGFP signal in FACS (Fig. 1a). FACS demonstrated that Sox2-EGFPhigh+ cells represent ~70% of the E12 cochlear duct, ~50% of the E14.5 cochlear duct and ~25% of the E16 cochlear duct (Supplementary Fig. 4), which reflects the size of the Sox2-EGFPhigh+ regions observed in midmodiolar sections (Fig. 1).
Open chromatin regions in Sox2-EGFPhigh+ cells of the embryonic cochlear duct map to genes and gene-regulatory regions including known otic enhancers
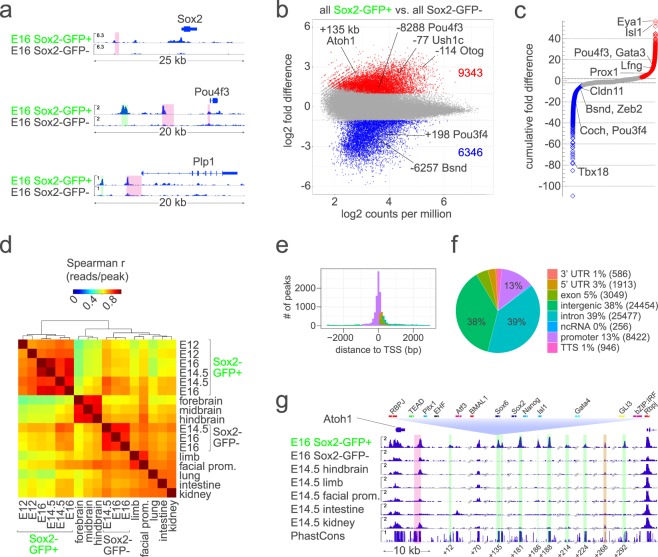
We carried out ATAC-seq on FACS purified Sox2-EGFPhigh+ and Sox2-EGFP− cells, we sequenced and mapped the reads, and then identified peaks with MACS260. We found >65,000 replicated peaks in the E12-16 Sox2-EGFPhigh+ cochlear duct cells (e.g. Fig. 2a, Supplementary Data 1). To determine whether ATAC-seq detects known otic enhancers, we curated a comprehensive list of otic enhancers and promoters (i.e. driving gene expression in vivo) from published literature on all species and the VISTA Enhancer database61, then compared the enhancers and promoters to ATAC-seq peaks detected in Sox2-EGFPhigh+ and Sox2-EGFP− cells (Supplementary Data 2). ATAC-seq peaks mapped to 41 of the 56 enhancers and promoters that were previously reported to have otic activity, including those of Sox2, Pou4f3, Plp1 and Atoh1 (Fig. 2a,g and Supplementary Data 2).
Figure 2.
ATAC-seq detects gene regulatory features that include known otic enhancers in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. (a) Shows ATAC-seq signal in E16 Sox2-EGFPhigh+ cochlear cells relative to that in E16 Sox2-EGFP− cochlear cells collected in parallel at the Sox2, Pou4f3, and Plp1 loci. Highlighted in pink are known enhancer regions. Highlighted in green are novel open chromatin regions detected only in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. (b) Shows fold difference relative to the normalized read counts for ATAC-seq peaks determined to be significantly increased (red), significantly decreased (blue) or unchanged (grey) in Sox2-EGFPhigh+ cells from E12-16 cochlear duct versus Sox2-EGFP− cells from E14.5-16 cochlear duct. (c) Shows the cumulative fold differences (i.e. the sum of the fold differences of all peaks nearest to each gene) in accessibility of each gene in Sox2-EGFPhigh+ cells from E12-16 cochlear duct versus Sox2-EGFP− cells from E14.5-16 cochlear duct. In the highlighted genes, accessibility corresponds to known differential patterns of gene expression. (d) shows a clustered Spearman correlation matrix of the reads in peaks from ATAC-seq of FACS-sorted cochlear cells and of various E14.5 tissues. Note that reads in cochlear ATAC-seq samples are highly correlated. (e) Shows the frequency of ATAC-seq peaks relative to the distance to transcription start sites. (f) Shows the percentages of all replicated peaks detected in Sox2-EGFPhigh+ cells of the E12-16 cochlear duct mapping to the indicated genomic features. (g) Shows ATAC-seq signal at the Atoh1 locus in E16 Sox2-EGFPhigh+ cochlear cells aligned to that in E16 Sox2-EGFP− cochlear cells collected in parallel; several other E14.5 tissues, and PhastCons 30-way vertebrate conservation. As an example of transcription factor binding motifs in peaks, we show those predicted in the +135 kb peak. Highlighted in pink is the known 3′ enhancer. Highlighted in green are 7 open chromatin regions specific to Sox2-EGFPhigh+ cells of the embryonic cochlear duct downstream of Atoh1. Highlighted in gold is a region that increased in accessibility in E14.5 vs. E12 Sox2-EGFPhigh+ cells of the embryonic cochlear duct. Transcription factor families having similar bindings motifs are color-coded. Highest scoring members of each family are indicated.
To better resolve quantitative differences in accessibility at the peaks found in Sox2-EGFPhigh+ and Sox2-EGFP− cells, we carried out a differential accessibility analysis using edgeR62 that calculates significance and fold-differences based on normalized reads per peak in replicate samples. We detected 9,343 peaks that show significantly greater accessibility in Sox2-EGFPhigh+ cells than in surrounding Sox2-EGFP− cells and 6346 peaks showing significantly less accessibility in Sox2-EGFPhigh+ cells than in Sox2-EGFP− cells (Fig. 2b, Supplementary Data 3). Several genes known to be differentially expressed in prosensory and nonsensory cells such as Isl163 and Bsnd64, respectively, showed corresponding differences in accessibility (Fig. 2c). This can be seen in Fig. 2c as the cumulative fold difference in accessibility of all peaks nearest to each gene. We further confirmed the efficacy and specificity of ATAC-seq by comparing the Sox2-EGFPhigh+ peaks with previously reported unmethylated regions and low methylated regions of the E16.5 cochlear sensory epithelium65 (Supplementary Fig. 5). We find that most ATAC-Seq peaks in cochear duct cells correspond to unmethylated and low methylated regions.
To further test for the specificity of the peaks in the Sox2-EGFPhigh+ cells, we carried out a correlation analysis of reads present per ATAC peak in E12-16 ATAC-seq samples of Sox2-EGFPhigh+ cochlear duct cells to reads present per ATAC peak in ATAC-seq samples from other tissues (Fig. 2d). We combined peaks from several studies called by MACS2 (>90,000) and examined the correlation of reads in peaks in every sample. We found high correlation among cochlear Sox2-EGFPhigh+ samples, highlighting the unique epigenomic landscape of prosensory cells. Similar to accessible chromatin in other tissues66,67, the Sox2-EGFPhigh+ ATAC-seq peaks increased in frequency near transcription start sites (Fig. 2e) and primarily mapped to introns and potential cis-regulatory intragenic regions (Fig. 2f and Supplementary Fig. 6). Furthermore, replicated peaks detected in E12-16 Sox2-EGFPhigh+ cochlear duct cells showed over 30-fold enrichment for CpG islands, 5′ untranslated regions and promoters (13,225, 13,069 and 14,026 peaks, respectively; Supplementary Fig. 6). Of the >65,000 peaks called in the Sox2-EGFPhigh+ cells, over 29,000 peaks were detected only in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. These peaks were not present in E14.5-16 Sox2-EGFP− cochlear duct cells, or in the other embryonic tissues we analyzed, and include regions near known regulators of inner ear development. As an example of this Fig. 2g depicts the Atoh1 locus and these open chromatin regions are highlighted in green (coordinates in Supplementary Data 1). We did not successfully prepare ATAC-seq libraries from E12 Sox2-GFP− cells, which are a small percentage of the cochlear duct (Supplementary Fig. 4), so we did not include this stage of Sox2-EGFP− cells in our analysis.
Differential enrichment of transcription factor-binding motifs in Sox2-EGFPhigh+ cells of the embryonic cochlear duct
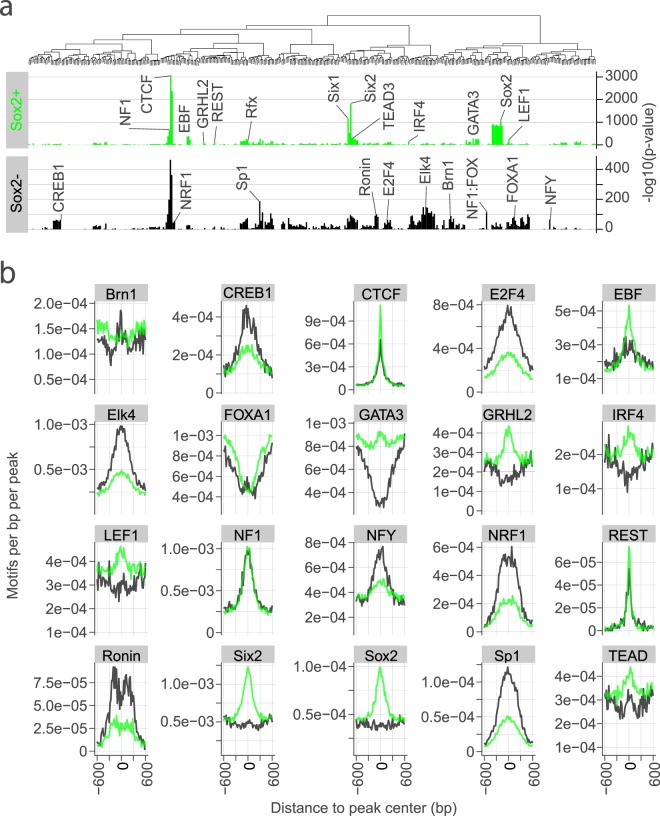
To better understand the regulatory circuits controlling cochlear development in the prosensory region, we carried out an analysis of transcription factor binding motifs in the Sox2-EGFPhigh+ peaks. Motif enrichment in replicated ATAC-seq peaks detected in Sox2-EGFPhigh+ cells of the E12-16 cochlea duct and in Sox2-EGFP− cells of the E14.5-16 cochlea duct were compared using the p-value of enrichment for all motifs in the HOMER library (Fig. 3a) and the frequency of motifs (Fig. 3b). To group similar motifs in Fig. 3a, motifs were clustered based on the correlation of motif matrices (Supplementary Fig. 7). Scales vary in Fig. 3b with differences in motif abundance.
Figure 3.
Motif enrichment in open chromatin regions in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. (a) Plots qualitative differences in motif enrichment in Sox2-EGFPhigh+ cells from E12-16 cochlear duct and Sox2-EGFP− cells from E14.5-16 cochlear duct as −log10 of the p-value in replicated peaks. Motifs are clustered based on similarity. The most highly enriched members of transcription factor families/clusters are selectively labeled based on differential enrichment in the samples. Differences in enrichment might be indicative of context-specific transcription factor activity. Scales in (a) vary due to differences in peak number and coverage. (b) Plots quantitative differences in enrichment of representative motifs as the frequency of motifs relative to peak center. Note that high enrichment (a) often corresponds to greater central localization in peaks (b).
Open chromatin regions in Sox2-EGFPhigh+ cochlear duct cells showed greater enrichment for motifs corresponding to factors in the Six, Sox, Gata, Ebf and Tead families as well as motifs for Grhl2, Lef1, Irf4 and Rest (Fig. 3, Supplementary Data 4). Motif enrichment analysis in the ‘prosensory-specific’ subset (i.e., the 29,000 peaks detected only in Sox2-EGFPhigh+ cells of the embryonic cochlear duct) was similar to that in peaks of Sox2-EGFPhigh+ cells (Fig. 4a,b) with some differences. For example, ‘prosensory-specific’ ATAC regions showed 2–3-fold enrichment for Six2 and Sox2 motifs and enrichment also for the Zeb1 motif (Fig. 4b). Relative to that in Sox2-EGFPhigh+ cells, open chromatin regions in Sox2-EGFP− cochlear duct cells showed greater enrichment for motifs corresponding to the factors in the Forkhead, E2f, Ets, Sp/Klf, bZip and Pou families, as well as Nfy, Nrf1 and Ronin. Ctcf, Rfx and Nf1 were similarly enriched for in Sox2-EGFPhigh+ and Sox2-EGFP− cells.
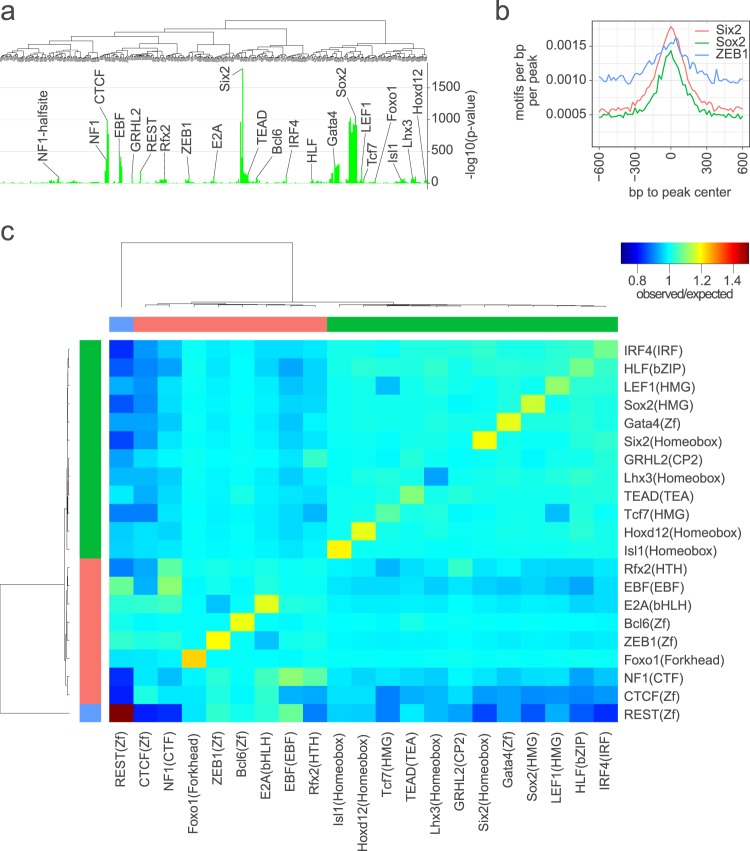
Figure 4.
Motif enrichment and co-occurrence in prosensory-specific open chromatin regions of the embryonic cochlear duct. (a) Shows the motif enrichment as -log10 of the p-value in the subset of replicated peaks detected only in Sox2-EGFPhigh+ cells from E12-16 cochlear duct (i.e., not detected in Sox2-EGFP- cells or other ENCODE datasets examined). (b) Plots quantitative differences in enrichment of representative motifs as the frequency of motifs relative to peak center. (c) Plots clustered matrix of the ratio of the observed frequency of co-occurrence of each combination of enriched motifs in the prosensory-specific peaks relative to the expected frequency of co-occurrence.
To look for evidence of combinatorial transcription factor activity in prosensory open chromatin, we analyzed motif co-occurrence in the ‘prosensory-specific’ subset (i.e., the 29,000 peaks detected only in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. Examining the ratio of observed co-occurrence in the prosensory-specific peaks, relative to baseline co-occurrence in the genome, highlights that some combinations of motifs occur in prosensory open chromatin regions at frequencies greater than random chance (Fig. 4c and Supplementary Data 7). Motif co-occurrence analysis showed that many motifs occur multiple times in the same open chromatin regions (see diagonal in Fig. 4c).
Chromatin dynamics in Sox2-EGFPhigh+ cells of the embryonic cochlear duct
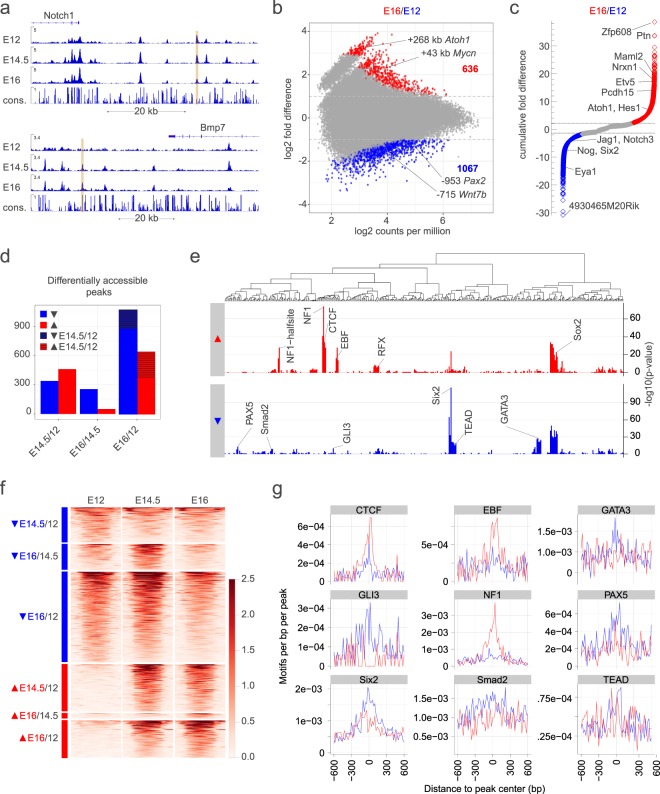
To determine whether any of the peaks present in the Sox2-EGFPhigh+ cells were differentially accessible at different ages of cochlear development, we compared the ATAC-seq peaks from the three different ages using the differential accessibility analysis described above. We detected over 2,200 peaks in Sox2-EGFPhigh+ cells that change between E12 and E16. Many of the regions that show age-dependent differential accessibility are near regulatory genes, deafness genes and components of pathways of known significance to cochlear and prosensory development (Fig. 5a). For example, one peak +268 kb from the Atoh1 TSS increased significantly in accessibility in E14.5 when compared with E12 Sox2-EGFPhigh+ cells (yellow in Fig. 2g). The +268 kb differentially accessible region downstream of Atoh1 is ~300 bp and contains putative binding sites for members of the Ets, MADS, Zf and Homeobox families as well as 5 consensus binding motifs for the bHLH transcription factor family (not shown).
Figure 5.
Developmentally-dynamic open chromatin in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. (a) Shows examples of developmentally dynamic ATAC-seq peaks (gold) in Sox2-EGFPhigh+ cells of the E12-16 cochlear duct at the indicated loci. (b) Shows the fold difference relative to the normalized read counts for ATAC-seq peaks determined to be significantly increased (red), significantly decreased (blue) or unchanged (grey) in Sox2-EGFPhigh+ cells from E16 cochlear duct versus those from E12 cochlear duct. (c) Shows the cumulative fold differences (i.e. the sum of the fold differences of all peaks nearest to each gene) in accessibility of each gene in Sox2-EGFPhigh+ cells from E16 cochlear duct versus those from E12 cochlear duct. (d) Shows the numbers of differentially accessible peaks detected in Sox2-EGFPhigh+ cells at the indicated stages. Note that most significant changes were detected in the E12-14.5 window. (e) Shows differential motif enrichment in peaks increasing in accessibility in E12-16 Sox2-EGFPhigh+ cells from the cochlear duct relative to those decreasing in accessibility. (f) Plots normalized reads in the differentially accessible regions. (g) Plots the frequency of differentially enriched motifs relative to peak center.
Changes in cumulative accessibility in Sox2-EGFPhigh+ cells showed gene set enrichment for known processes in prosensory development. For example, at E14.5 versus E12, ‘positive regulation of synapse structure or activity’ was the most enriched process (p = 3.4e-4; Supplementary Fig. 8). The genes having the highest differential accessibility and therefore greatest contribution to the enrichment score for ‘positive regulation of synapse structure or activity’ in E14.5 versus E12 Sox2-EGFPhigh+ cells (i.e. the leading edge genes) included Nrxn1, Adgrl2, Flrt2, Wnt7a, Bhlhb9 and Nlgn1. At E12 versus E14.5, ‘cochlear development’ was the most enriched process (p = 4.4e-4; Supplementary Fig. 8) and the leading edge genes included Pax2, Fgfr3, Gli2 and Wnt5a.
Overall, accessibility in most differentially accessible peaks in Sox2-EGFPhigh+ cells diminished with developmental time (Fig. 5b,d,f; raw data in Supplementary Data 5). There was a greater difference in the number of differentially accessible peaks between E14.5 and E12 than between E14.5 and E16 in Sox2-EGFPhigh+ cells. Developmental changes in the cumulative accessibility of several genes including Atoh1 and Hes1 corresponded to known dynamics in gene expression68–71 (Fig. 5c).
Motif enrichment analysis of peak subsets that increased in accessibility across development showed significant enrichment for motifs of the Six, Rfx, Ctcf and Sox transcription factor families, as well as specific enrichment for some motifs not corresponding to any known regulators of cochlear development: Ebf and Nf1 (Fig. 5e,g, raw data in Supplementary Data 6). Motif enrichment analysis of peak subsets that decreased in accessibility across development also showed significant enrichment for motifs of the Six and Sox families as well as specific enrichment for Tead, Gata, Smad, Gli and Pax transcription factor families (Fig. 5e,g).
Open chromatin regions in Sox2-EGFPhigh+ cells of the embryonic cochlear duct map to SNPs in human deafness genes
To determine whether open chromatin regions detected in Sox2-EGFPhigh+ cells of the embryonic cochlear duct in mouse have potential significance in the regulation of human deafness genes, mouse open chromatin regions were first mapped to the human genome (Hg19) using UCSC liftOver72. Variants in deafness-associated genes have recently been curated in the Deafness Variation Database (DVD)22. Over 48,000 of 65,129 ATAC-seq peaks detected in E12-16 Sox2-EGFPhigh+ cochlear duct cells mapped to regions of >70% sequence similarity in the human genome (Supplementary Data 8). Over 20,000 SNPs in the DVD overlap the open chromatin regions detected in mouse (Fig. 6a). Most SNPs in the DVD found to overlap with mouse open chromatin regions are intronic and of unknown significance to the pathogenesis of deafness (Fig. 6a). Some SNPs in human deafness genes coincide with transcription factor binding motifs in mouse open chromatin regions. For example, Fig. 6b shows two SNPs in a SIX motif overlapping an open chromatin region detected in Sox2-EGFPhigh+ cells of the embryonic mouse cochlear duct of high conservation within the first intron of MYO7A.
Figure 6.
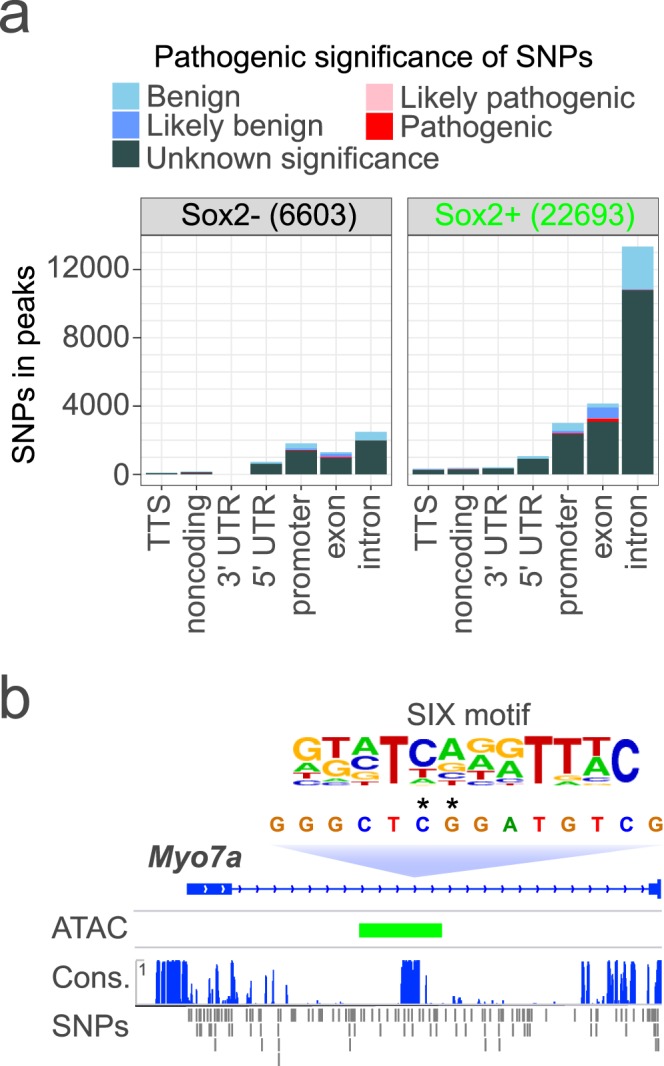
Human deafness gene SNPs in orthologous regions to ATAC-seq peaks detected in Sox2-EGFPhigh+ cells of the embryonic cochlear duct. Numbers of SNPs in the Deafness Variation Database (DVD) overlapping regions orthologous to ATAC-seq peaks in the indicated genomic features are shown in (a). Colors indicate pathogenicity as classified in the DVD. In (b), asterisks indicate two SNPs of unknown significance in MYO7A intron 1 that potentially affect binding at a SIX motif in a region of high evolutionary conservation (Cons.) that is orthologous to an ATAC-seq peak (ATAC) detected in Myo7a in embryonic mouse Sox2-EGFPhigh+ cochlear duct cells.
Discussion
Our analysis of open chromatin in prosensory cells (Sox2-EGFPhigh+) adds considerable insight into gene regulation during early sensory development. During the development of the cochlea, hair cells are derived from Sox2+ progenitors in the prosensory domain of the floor of the cochlear duct39,47,49,55,73. Open chromatin regions detected in prosensory cells map to both experimentally validated and putative gene-activating regions (Fig. 2, Supplementary Fig. 6, Supplementary Data 2). Furthermore, over 29,000 open chromatin regions were found only in Sox2-EGFPhigh+ cochlear duct cells (Fig. 2g, Supplementary Data 1), suggesting a unique signature of open chromatin in these cells. By extension, we speculate that a unique epigenetic mechanism contributes to the specification of the prosensory lineage. Motif enrichment in prosensory open chromatin provides confirmatory evidence for the roles of Sox2, Six1 and Gata346–48 in prosensory development (Fig. 3) and also implicates several transcription factor families in cochlear development that have not previously been reported to have a role in cochlear development, such as Tead, Ebf, Nfi, Klf/Sp and Ets families of transcription factors (Fig. 3). These findings imply that the regulatory factors identified may influence chromatin structure and cochlear cell reprogramming through pioneer activity74. In that regard, our analysis provides evidence for centering and increased density of transcription factor binding motifs in open chromatin peaks—two phenomena associated with pioneering activity75—of the transcription factor binding motifs enriched in prosensory open chromatin (Figs 3 and 4).
A recent report showed that Ctcf is required for cochlear development but not for prosensory gene expression76. Some previous evidence supports the possibility that Tead, Ebf, Nfi, Klf/Sp and Ets transcription factors could also influence cochlear development. For example, inhibition of the Yap–Tead interaction reduces supporting cell proliferation in the utricle77 and yap1a inhibition reduces neuromast formation in the zebrafish lateral line78, consistent with a possible role for the Hippo/Yap/Tead pathway in cochlear development. Previous findings in our lab showed that the inhibition of Fgf-signaling in cochlear explant cultures abolishes sensory specification as well as the expression of the downstream targets Etv4 and Etv534, suggesting a role for the Ets family in sensory specification in the cochlea. Mutations in Klf/Sp, Ebf and Nfi family members have been associated with neurodevelopmental defects79–88. Furthermore, defects observed in Nfix mutants include hearing loss84,86. Our findings show enrichment of the Ebf1-4 motif in the open chromatin of Sox2-EGFPhigh+ cells (Fig. 3) and in regions that displayed an increase in accessibility in the Sox2-EGFPhigh+ cells between E12 and E16 (Fig. 4). Detailed transcriptomic analysis of isolated cochlear prosensory cells will be needed to further deduce the specific transcription factors of importance in prosensory development as each enriched motif corresponds to transcription factor families comprising multiple members.
Our findings provide insight into the epigenetic regulation of the prosensory cell during development. For example, accessibility in over 2,200 regions changed during development, largely between E12 and E14.5 and most dynamic sites diminished in accessibility. Single cell ATAC-seq has recently been developed89 and has the potential to better resolve the complexity of the developing prosensory population and the extent to which the early steps of hair cell and support cell specification relate to epigenetic dynamics in cochlear prosensory cells.
Findings in the present study add to knowledge of the epigenetic mechanisms of Atoh1 regulation. Atoh1 is not only necessary for hair cell formation56 but also sufficient to induce hair cell differentiation in a limited population of cochlear cells90. Epigenetic regulation of Atoh1 expression therefore has implications for hair cell regeneration strategies. Previous studies demonstrated a critical 3′ enhancer and showed association of this region as well as the promoter and an exonic region with activation-associated histone marks26,91. We find that these regions are accessible in Sox2-EGFPhigh+ cells of E12 cochlear duct, suggesting that the onset of Atoh1 expression at E14.556 is perhaps primed but unrelated to coincident changes in the accessibility of these regions. Rather, a + 268 kb distal region containing 5 bHLH binding motifs increased significantly in accessibility in E14.5 vs. E12 Sox2-EGFPhigh+ cells (Fig. 2g).
We were surprised to find greater cumulative accessibility in otic fibroblast marker genes such as Coch, Tbx18 and Pou3f4 than in roof epithelial marker genes such as Cldn11 and Bsnd (Fig. 2c) in Sox2-EGFP− vs. Sox2-EGFPhigh+. Likewise, high expression of otic fibroblast markers was found in Lfng-GFP−, Pou4f3-GFP− and Math1-GFP− cells isolated in RNA-seq studies92–94. This could imply that all these GFP− samples are enriched with otic fibroblasts. Otic fibroblasts and melanocytes are closely associated with the roof of the cochlear duct at prosensory stages95,96 and we therefore speculate that the enrichment for otic fibroblasts—despite microdissection away of most surrounding mesenchyme in these samples—could be related to the relative efficiency of dissociating mesenchymal cells versus epithelial cells. An alternative explanation for the otic fibroblast gene expression and accessibility evident in these samples is that fibroblasts or mesenchymally-derived epithelial cells are perhaps in the cochlear duct. Lineage tracing using the Wnt1-CreER demonstrated neural crest contributions to the cochlear duct epithelium in one study97, but not in a more recent study using a different reporter98. Regardless, it is possible that the FACS sorted cells contain otic fibroblasts even though the majority of these cells were removed by microdissection.
Most sensorineural hearing loss relates to the loss or dysfunction of hair cells and most emerging therapies for hearing loss therefore seek to protect or replace hair cells99. Functionally significant regeneration of cochlear hair cells is not robust in mammalian models of hearing loss. The strategies showing greatest promise for the regeneration of hair cells have altered the activity of Sox2, Atoh1 and other transcription factors to achieve the conversion of limited populations of cochlear cells to hair cells35,90,100–107 and also the differentiation vestibular-like hair cells from embryonic stem cells108,109. Findings from our study implicate new transcription factor classes in prosensory development that merit consideration as possible regulators of support cell and hair cell formation.
In summary, ATAC-seq analysis of Sox2-EGFPhigh+ cells of the embryonic cochlear duct implicates novel classes of transcription factors in inner ear development and provides strong evidence for a prosensory-specific landscape of open chromatin as well as epigenetic dynamics during prosensory cell development. These datasets (in Supplementary Data 1, 3, 5 and 8 and summarized in Supplementary Fig. 9) not only provide insights into the gene regulatory networks guiding prosensory development in the cochlea but also represent a significant resource for the mechanistic study of the cis-regulation of deafness genes and genes of interest for regeneration studies in the inner ear.
Materials and Methods
Mice
Sox2-EGFP knockin mice59 from (Jackson Stock: 017592) were bred to generate E12-16 litters. Mice were housed in the University of Washington Department of Comparative Medicine. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Washington and performed in accordance with NIH guidelines. Sox2-EGFP+ embryos were identified by epifluorescence. Stages were verified by Theiler’s criteria.
Vibratome sections of Sox2-EGFP+ embryonic temporal bone
E12-16 Sox2-EGFP+ heterozygous embryos were decapitated in Hank’s buffered salt solution (Cat. No. 14025-92; Thermo Fisher Scientific; Waltham, MA) and fixed overnight at 4 °C in 4% paraformaldehyde (Cat. No. 15710; Electron Microscopy Sciences; Hatfield, PA) in phosphate-buffered saline (PBS; Cat. No. BP399; Thermo Fisher Scientific). After 3 × 30 minute washes in PBS, temporal bones were isolated by microdissection, embedded in in 4% agarose and sectioned at 100 μm with a vibratome.
Sox2 and EGFP immunolabeling
Sections were immunolabeled essentially as described by Hartman et al.110. Sections were permeabilized (2% Triton-X-100/PBS) for 30 minutes, then blocked (10% donkey serum/0.5% Triton-X-100/PBS) for 2–4 hours. Sections were incubated overnight in 1:200 goat anti-Sox2 (Y-17; Santa Cruz) and 1:250 chick anti-GFP (ab13970; Abcam) diluted in block. After 3 × 1 hour washes in 0.5% Triton-X-100/PBS, sections were incubated overnight in block, then overnight in 1:400 Alexa488-conjugated donkey anti-goat and 1:400 Alexa647-conjugated donkey anti-chick diluted in block. After 3 × 1 hour washes in 0.5% Triton-X-100/PBS, sections were cleared by 3 × 10 minute incubations in 60% glycerol/PBS, then imaged.
Confocal microscopy
Z-stacks of Sox2 and EGFP immunofluorescence were collected on a LSM880 with Airyscan using 10X/0.45 (1.1 μm steps) and 20X/0.8 M27 (0.3 μm steps) Plan-Apochromat objectives (Ziess, Jena, Germany). Zeiss Zen Black v14.0.0.0 was used to acquire micrographs and to perform Airyscan processing, maximum intensity projections (i.e. of 2–5 optical sections) and linear adjustments to black and white points.
Isolation of Sox2-EGFPhigh+ cochlear duct cells
Cochlear ducts were dissected away from mesenchyme after collagenase digestion as described previously34. Care was taken to dissect away all cochlear nerve tissue to remove the potentially confounding population of Sox2-EGFP+ glial cells. Cochlear duct cells were then dissociated using papain at 37 °C and sorted to isolate live Sox2-EGFPhigh+ cells using a BD Aria III (85 μm nozzle, flow rate 1–3) as shown in Fig. 1a. All dissection, sorting, spins and washes were performed in ice-cold media. For comparative ATAC-seq analysis, Sox2-EGFP− cells were isolated in parallel in some cases. Gating was centered on the highest and lowest peaks in the histogram and EGFPlow cells were excluded.
ATAC-seq
Sox2-EGFPhigh+ cells were isolated from 2 litters per embryonic stage (i.e. 1 litter per biological replicate of 6–12 pooled cochlear ducts at E12, E14.5 and E16). ATAC-seq sample preparation, transposition and library amplification were performed as described by Buenrostro and colleagues111. Briefly, adapters for library amplification and sequencing were inserted into open chromatin via transposase in vitro and libraries were prepared using the Illumina Nextera kit (Cat. No. FC-121-1030). The Genomics & Bioinformatics Shared Resource at the Fred Hutchinson Cancer Research Center (Illumina Hi-seq 2500) and Seattle Genomics (Illumina Next Seq. 500) performed deep sequencing. Adapters were trimmed from reads and low-quality sequences (Phred <33) were removed using Trim Galore!112,113. Reads were aligned to mm9 using Bowtie2114 (option:–very-sensitive). Duplicate reads were marked using Picard ‘MarkDuplicates’ (http://broadinstitute.github.io/picard/). Duplicate and mitochondrial reads were removed using SAMtools115. Bigwigs were generated using a custom script (https://rpubs.com/achitsaz/98857) and visualized in IGV116. Supplementary Table 1 describes samples and coverage. Percentages of reads in peaks were found using ‘plotEnrichment’ in deepTools2117. ATAC-seq data has been deposited in NCBI GEO database under the accession code GSE131775.
Peak calling and filtering
Peaks were called using MACS260 (options:–nomodel–shift -100–extsize 200). Irreproducible peaks were filtered using BEDOPS118 (command: ‘bedops -e 1 replicate1.bed replicate2.bed’). Overlapping and nearby ATAC-seq peaks were merged using BEDtools119 (command: bedtools merge -i replicatedpeaks.bed -d 100). Peaks in regions previously found by ENCODE to produce artefactual signal120 (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/) were removed using BEDOPS command: -n 1. Only reproducible peaks (i.e. peaks replicated in at least one other dataset of the same sample type) were used for genomic annotation, motif enrichment and other analysis. To find prosensory-specific peaks, BEDOPS118 (command: -n 1) was used to isolate peaks of Sox2-EGFPhigh+ cochlear duct cells not overlapping with any ATAC-seq peaks called as described above in Sox2-EGFP− cochlear duct cells as well as in the following ENCODE ATAC-seq datasets: G1E mouse embryonic stem cells (ENCSR280ZDP), E14.5 C57Bl/6 mouse facial prominence (ENCSR876SYO), forebrain (ENCSR810HQR), midbrain (ENCSR384JBF), hindbrain (ENCSR798FDL), neural tube (ENCSR700QBR), limb (ENCSR460BUL), kidney (ENCSR758IRM), intestine (ENCSR150EOO), liver (ENCSR032HKE), lung (ENCSR335VJW), stomach (ENCSR618HDK) and heart (ENCSR068YGC). ENCODE datasets were aligned and peaks were called using the pipeline described above.
Annotation and genomic feature enrichment analysis
Annotation of ATAC-seq peaks and genomic annotation enrichment analysis were performed using HOMER121, which assigns peaks to the nearest TSS then annotates peaks based on UCSC refGene annotations122 for mm9.
Curation of previously reported otic enhancers and promoters
To determine whether ATAC-seq detects known otic enhancers, we mapped ATAC-seq peaks to known otic enhancers. As no comprehensive list for otic enhancers and promoters driving gene-activation in vivo existed, a list had to be generated through a comprehensive review. To generate the list, we considered our knowledge, entries in the VISTA Enhancer Browser61 listing ‘ear’ expression patterns as well as all articles published up to March 1, 2018 found using the Pubmed search: (“ear”[MeSH Terms]) AND regulatory sequences, nucleic acid[MeSH Terms]. This research yielded 56 otic enhancers and promoters described in sufficient detail to map to mm9 and determine overlap with ATAC-seq data. Regions identified in other systems (e.g. human cell lines, chick and zebrafish) were mapped to orthologous regions in mm9 using BLAST and liftOver. The list is available in Supplementary Data 2.
Differential accessibility analysis in ATAC-seq peaks
Differential accessibility across ATAC-seq sample groups was determined as detailed in the edgeR62 users guide. To define the regions for differential accessibility analysis, coordinates of all overlapping and nearby (<100 bp) intervals of replicated ATAC-seq peaks detected in all compared groups was determined using ‘merge’ in BEDtools119 (option: -d 100). A matrix of counts per million for all samples was then generated using the ‘featureCounts’ in the Rsubread package (options: isPairedEnd = TRUE, maxFragLength = 2000). The counts matrix was filtered to select rows having at least 1 count per million in n − 1 samples to minimize the influence of variability at the threshold for sensitivity on the analysis. Cumulative fold differences in accessibility at each gene were calculated as the sum of the fold differences of all peaks nearest to each gene.
Correlation analysis of ATAC-seq reads in peaks
Using deepTools2117, matrix of read counts in each sample was first computed using ‘multiBamSummary‘. A Spearman correlation matrix of reads in peaks was then generated ‘plotCorrelation’. A distance matrix for the correlations was generated using ‘dist’ (https://www.rdocumentation.org/packages/stats/versions/3.5.2; mode = Euclidean). Clustering of the correlation matrix was performed in RStudio using ‘hclust’ in the fastCluster package123 (mode = Ward.D2). A heatmap of the correlation matrix was generated using ‘heatmap.2’ (https://www.rdocumentation.org/packages/gplots).
Intersect analysis of ATAC-seq and Methyl-seq data
The ‘venn’ command in Intervene124 was used to calculate the intersect sizes for ATAC-seq peak and Methyl-seq regions and plot the venn diagrams.
Motif-enrichment analysis
Motif-enrichment analysis in ATAC-seq peaks was performed using ‘findmotifsgenome.pl’ in HOMER command (options: -size given -mask) and the HOMER library of consensus binding motifs determined in vivo by ChIP-seq. Frequencies of motifs per base pair per peak (Fig. 3b) were calculated using ‘annotatepeaks.pl’ in HOMER.
Correlation analysis of motif matrices
To group similar motifs in Fig. 3a, ‘comparemotifs.pl’ in HOMER was used to generate a correlation matrix of the motif matrices in the HOMER motif library. Clustering of the correlation matrix and plotting was performed as described above.
Co-occurrence of motifs in ATAC peaks
The co-occurrence of significantly enriched motifs (Fig. 4c and Data 7) was calculated using ‘annotatepeaks.pl’ in HOMER. Clustering of the co-occurrence ratio matrix and plotting was performed as described above.
Gene set enrichment analysis
Gene set enrichment analysis125 for GO Biological Processes (http://www.go2msig.org/cgi-bin/prebuilt.cgi?taxi=10090) was performed on the cumulative fold differences in accessibility using the fgsea package126 (options: minSize = 25, maxSize = 500, nperm = 10000). Enriched gene sets were reduced to independent gene sets using ‘collapsePathways’ in fgsea.
Mapping mouse ATAC-seq peaks to the human genome
The UCSC liftOver127 command line tool was used to convert mm9 coordinates of ATAC-seq peaks to orthologous regions in the human genome Hg19 (option: -minMatch = 0.7). The human SNPs in the Deafness Variation Database v8.1 (http://deafnessvariationdatabase.org/public/) overlapping the conserved ATAC-seq regions were identified using ‘findOverlaps’ in the GRanges128 package. Transcription factor binding motifs in the conserved regions were visualized in IGV using the file available at http://homer.ucsd.edu/homer/index.html.
Plotting and other statistical analysis
Cumulative accessibility was plotted in Prism v5.0 f. For Fig. 4f, a counts matrix was generated using ‘computeMatrix’, then plotted using ‘plotHeatmap’ in deepTools2117. Venn diagrams were plotted using Intervene124. Other plots were generated in RStudio v1.1.456. Scatterplots, bar plots and line plots were generated using the ggplot2 package129. Correlation heatmaps were generated using the gplots package (https://www.rdocumentation.org/packages/gplots). Figure compositions were performed in Adobe Illustrator and Photoshop CC 2017 (San Jose, CA). To test for differences in cell counts across development, one-way ANOVA and Tukey’s HSD multiple comparisons tests were performed in R.
Supplementary information
Acknowledgements
This work was supported by 5R21DC014768 (OBM and TAR), 5F32DC016480 and 5T32DC005361 (BAW), T32GM007270 and T32HD007183 (LSV). We thank Isabel Lee for assistance in immunofluorescence, Heather Zebroski for help with animal husbandry and Stefanie Wohl and Donna Prunkard for help with FACS.
Author Contributions
O.B.M., T.R., M.W. and B.W. and conceived of and designed the study. B.W. and L.S.V. prepared ATAC-seq samples. B.W., A.C. and L.S.V. performed bioinformatics analysis and data visualization. B.W. and O.B.M. interpreted the data and wrote the manuscript. All authors reviewed and commented on the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45515-2.
References
- 1.Hudspeth AJ. Integrating the active process of hair cells with cochlear function. Nature Reviews Neuroscience. 2014;15:600. doi: 10.1038/nrn3786. [DOI] [PubMed] [Google Scholar]
- 2.Morton CC, Nance WE. Newborn hearing screening–a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 3.Van Laer L, et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum Mol Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- 4.Oonk AM, et al. Progressive hereditary hearing impairment caused by a MYO6 mutation resembles presbyacusis. Hear Res. 2013;299:88–98. doi: 10.1016/j.heares.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Manche SK, Jangala M, Putta P, Koralla RM, Akka J. Association of oxidative stress gene polymorphisms with presbycusis. Gene. 2016;593:277–283. doi: 10.1016/j.gene.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 6.Newman DL, et al. GRM7 variants associated with age-related hearing loss based on auditory perception. Hear Res. 2012;294:125–132. doi: 10.1016/j.heares.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falah M, et al. Association of genetic variations in the mitochondrial DNA control region with presbycusis. Clinical interventions in aging. 2017;12:459–465. doi: 10.2147/cia.S123278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutai H, Watabe T, Kosaki K, Ogawa K, Matsunaga T. Mitochondrial mutations in maternally inherited hearing loss. BMC medical genetics. 2017;18:32. doi: 10.1186/s12881-017-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Laer L, et al. The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Human mutation. 2006;27:786–795. doi: 10.1002/humu.20360. [DOI] [PubMed] [Google Scholar]
- 10.Konings A, et al. Variations in HSP70 genes associated with noise-induced hearing loss in two independent populations. Eur J Hum Genet. 2009;17:329–335. doi: 10.1038/ejhg.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konings A, et al. Candidate gene association study for noise-induced hearing loss in two independent noise-exposed populations. Annals of human genetics. 2009;73:215–224. doi: 10.1111/j.1469-1809.2008.00499.x. [DOI] [PubMed] [Google Scholar]
- 12.Li X, et al. PON2 and ATP2B2 gene polymorphisms with noise-induced hearing loss. Journal of thoracic disease. 2016;8:430–438. doi: 10.21037/jtd.2016.02.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Association between genetic variations in GRHL2 and noise-induced hearing loss in Chinese high intensity noise exposed workers: a case-control analysis. Industrial health. 2013;51:612–621. doi: 10.2486/indhealth.2012-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen H, et al. A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One. 2014;9:e89662. doi: 10.1371/journal.pone.0089662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grondin Y, et al. Genetic Polymorphisms Associated with Hearing Threshold Shift in Subjects during First Encounter with Occupational Impulse Noise. PLoS One. 2015;10:e0130827. doi: 10.1371/journal.pone.0130827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: a case- control study. Environmental health: a global access science source. 2015;14:77. doi: 10.1186/s12940-015-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Xiangrong, Yang Qiuyue, Jiao Jie, He Lihua, Yu Shanfa, Wang Jingjing, Gu Guizhen, Chen Guoshun, Zhou Wenhui, Wu Hui, Li Yanhong, Zhang Huanling. Genetic Variation in POU4F3 and GRHL2 Associated with Noise-Induced Hearing Loss in Chinese Population: A Case-Control Study. International Journal of Environmental Research and Public Health. 2016;13(6):561. doi: 10.3390/ijerph13060561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong S, et al. Association of polymorphisms in heat shock protein 70 genes with the susceptibility to noise-induced hearing loss: A meta-analysis. PLoS One. 2017;12:e0188195. doi: 10.1371/journal.pone.0188195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedemann L, et al. Megalin genetic polymorphisms and individual sensitivity to the ototoxic effect of cisplatin. The pharmacogenomics journal. 2008;8:23–28. doi: 10.1038/sj.tpj.6500455. [DOI] [PubMed] [Google Scholar]
- 20.Langer T, Am Zehnhoff-Dinnesen A, Radtke S, Meitert J, Zolk O. Understanding platinum-induced ototoxicity. Trends in pharmacological sciences. 2013;34:458–469. doi: 10.1016/j.tips.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman E, Lahav A. Ototoxicity in preterm infants: effects of genetics, aminoglycosides, and loud environmental noise. Journal of perinatology: official journal of the California Perinatal Association. 2013;33:3–8. doi: 10.1038/jp.2012.105. [DOI] [PubMed] [Google Scholar]
- 22.Azaiez H, et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. The American Journal of Human Genetics. 2018;103:484–497. doi: 10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science348, 648 (2015). [DOI] [PMC free article] [PubMed]
- 24.Yue F, et al. A Comparative Encyclopedia of DNA Elements in the Mouse Genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Developmental cell. 2003;4:509–519. doi: 10.1016/S1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 26.Stojanova ZP, Kwan T, Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development. 2015;142:3529–3536. doi: 10.1242/dev.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yizhar-Barnea, O. et al. DNA methylation dynamics during embryonic development and postnatal maturation of the mouse auditory organ of Corti. bioRxiv (2018). [DOI] [PMC free article] [PubMed]
- 28.Galvez H, Tena JJ, Giraldez F, Abello G. The Repression of Atoh1 by Neurogenin1 during Inner Ear. Development. Frontiers in molecular neuroscience. 2017;10:321. doi: 10.3389/fnmol.2017.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol, Suppl. 1967;220:221–244. [PubMed] [Google Scholar]
- 30.Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 31.Slowik AD, Bermingham-McDonogh O. Notch signaling in mammalian hair cell regeneration. Trends in developmental biology. 2013;7:73–89. [PMC free article] [PubMed] [Google Scholar]
- 32.Cotanche DA, Kaiser CL. Hair cell fate decisions in cochlear development and regeneration. Hear Res. 2010;266:18–25. doi: 10.1016/j.heares.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Driver EC, Kelley MW. Specification of cell fate in the mammalian cochlea. Birth defects research. Part C. Embryo today: reviews. 2009;87:212–221. doi: 10.1002/bdrc.20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 Is Required for Sensory Epithelial Specification in the Developing Cochlea. The Journal of Neuroscience. 2008;28:5991–5999. doi: 10.1523/jneurosci.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci USA. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 37.Ohyama T, et al. BMP Signaling Is Necessary for Patterning the Sensory and Nonsensory Regions of the Developing Mammalian Cochlea. The Journal of Neuroscience. 2010;30:15044–15051. doi: 10.1523/jneurosci.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiernan A. E., Ahituv N., Fuchs H., Balling R., Avraham K. B., Steel K. P., Hrabe de Angelis M. The Notch ligand Jagged1 is required for inner ear sensory development. Proceedings of the National Academy of Sciences. 2001;98(7):3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiernan AE, Xu J, Gridley T. The Notch Ligand JAG1 Is Required for Sensory Progenitor Development in the Mammalian Inner Ear. PLoS Genetics. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci. 2011;31:8046–8058. doi: 10.1523/jneurosci.6671-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto N, Chang W, Kelley MW. Rbpj regulates development of prosensory cells in the mammalian inner ear. Developmental Biology. 2011;353:367–379. doi: 10.1016/j.ydbio.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng AG. Sensory hair cell development and regeneration: similarities and differences. Development. 2015;142:1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munnamalai V, Fekete DM. Wnt signaling during cochlear development. Seminars in Cell & Developmental Biology. 2013;24:480–489. doi: 10.1016/j.semcdb.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munnamalai V, Fekete DM. Notch-Wnt-Bmp crosstalk regulates radial patterning in the mouse cochlea in a spatiotemporal manner. Development. 2016;143:4003. doi: 10.1242/dev.139469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo XJ, et al. GATA3 controls the specification of prosensory domain and neuronal survival in the mouse cochlea. Hum Mol Genet. 2013;22:3609–3623. doi: 10.1093/hmg/ddt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiernan AE, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 48.Zheng W, et al. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dabdoub A, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong EY, Ahmed M, Xu PX. EYA1-SIX1 complex in neurosensory cell fate induction in the mammalian inner ear. Hear Res. 2013;297:13–19. doi: 10.1016/j.heares.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schimmang T. Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming. Hearing Research. 2013;297:84–90. doi: 10.1016/j.heares.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Costa A, Powell LM, Lowell S, Jarman AP. Atoh1 in sensory hair cell development: constraints and cofactors. Seminars in Cell & Developmental Biology. 2017;65:60–68. doi: 10.1016/j.semcdb.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Żak M, Klis SFL, Grolman W. The Wnt and Notch signalling pathways in the developing cochlea: Formation of hair cells and induction of regenerative potential. International Journal of Developmental Neuroscience. 2015;47:247–258. doi: 10.1016/j.ijdevneu.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Brooker R. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133(7):1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 55.Kiernan A. E. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132(19):4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- 56.Bermingham NA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 57.Żak, M. et al. LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea. Frontiers in Cellular Neuroscience10, 10.3389/fncel.2016.00186 (2016). [DOI] [PMC free article] [PubMed]
- 58.Erkman L, et al. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 59.Arnold K, et al. Sox2+ Adult Stem and Progenitor Cells Are Important for Tissue Regeneration and Survival of Mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Research. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M. D., McCarthy D. J., Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radde-Gallwitz K, et al. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. The Journal of comparative neurology. 2004;477:412–421. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rickheit G, et al. Endocochlear potential depends on Cl- channels: mechanism underlying deafness in Bartter syndrome IV. Embo j. 2008;27:2907–2917. doi: 10.1038/emboj.2008.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yizhar-Barnea O, et al. DNA methylation dynamics during embryonic development and postnatal maturation of the mouse auditory sensory epithelium. Scientific reports. 2018;8:17348. doi: 10.1038/s41598-018-35587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilken MS, et al. DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics & chromatin. 2015;8:8. doi: 10.1186/1756-8935-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zine Azel, Aubert Alexandre, Qiu Jiping, Therianos Stavros, Guillemot Francois, Kageyama Ryoichiro, de Ribaupierre Francois. Hes1andHes5Activities Are Required for the Normal Development of the Hair Cells in the Mammalian Inner Ear. The Journal of Neuroscience. 2001;21(13):4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murata J, et al. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. Journal of Neuroscience Research. 2009;87:3521–3534. doi: 10.1002/jnr.22169. [DOI] [PubMed] [Google Scholar]
- 70.Zheng, J. L., Shou, J., Guillemot, F., Kageyama, R. & Gao, W. Q. Hes1 is a negative regulator of inner ear hair cell differentiation. Development127 (2000). [DOI] [PubMed]
- 71.Chen C.-M. A. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development. 2002;129(23):5363–5375. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- 72.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu R, et al. Lineage tracing of Sox2-expressing progenitor cells in the mouse inner ear reveals a broad contribution to non-sensory tissues and insights into the origin of the organ of Corti. Dev Biol. 2016;414:72–84. doi: 10.1016/j.ydbio.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes & Development. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casey BH, Kollipara RK, Pozo K, Johnson JE, Intrinsic DNA. binding properties demonstrated for lineage-specifying basic helix-loop-helix transcription factors. Genome Res. 2018;28:484–496. doi: 10.1101/gr.224360.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong-Oh S, et al. CTCF Regulates Otic Neurogenesis via Histone Modification in the Neurog1 Locus. Mol. Cells. 2018;41:695–702. doi: 10.14348/molcells.2018.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gnedeva K, Jacobo A, Salvi JD, Petelski AA, Hudspeth AJ. Elastic force restricts growth of the murine utricle. eLife. 2017;6:e25681. doi: 10.7554/eLife.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loh SL, et al. Zebrafish yap1 plays a role in differentiation of hair cells in posterior lateral line. Scientific reports. 2014;4:4289. doi: 10.1038/srep04289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sleven H, et al. De Novo Mutations in EBF3 Cause a Neurodevelopmental Syndrome. The American Journal of Human Genetics. 2017;100:138–150. doi: 10.1016/j.ajhg.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garel S, Marin F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- 81.Jin K, Jiang H, Mo Z, Xiang M. Early B-cell factors are required for specifying multiple retinal cell types and subtypes from postmitotic precursors. J Neurosci. 2010;30:11902–11916. doi: 10.1523/jneurosci.2187-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steele-Perkins G, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/mcb.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell CE, et al. The transcription factor Nfix is essential for normal brain development. BMC developmental biology. 2008;8:52. doi: 10.1186/1471-213x-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao A, et al. An intragenic deletion of the NFIA gene in a patient with a hypoplastic corpus callosum, craniofacial abnormalities and urinary tract defects. European journal of medical genetics. 2014;57:65–70. doi: 10.1016/j.ejmg.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Malan V, et al. Distinct Effects of Allelic NFIX Mutations on Nonsense-Mediated mRNA Decay Engender Either a Sotos-like or a Marshall-Smith Syndrome. The American Journal of Human Genetics. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin S, Liu M, Niu W, Zhang CL. Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc Natl Acad Sci USA. 2011;108:21117–21121. doi: 10.1073/pnas.1112351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 89.Cusanovich DA, et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell. 2018;174:1309–1324.e1318. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 92.Cai T, et al. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J Neurosci. 2015;35:5870–5883. doi: 10.1523/jneurosci.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scheffer DI, Shen J, Corey DP, Chen Z-Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. The Journal of Neuroscience. 2015;35:6366–6380. doi: 10.1523/jneurosci.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maass JC, et al. Transcriptomic Analysis of Mouse Cochlear Supporting Cell Maturation Reveals Large-Scale Changes in Notch Responsiveness Prior to the Onset of Hearing. PLoS One. 2016;11:e0167286. doi: 10.1371/journal.pone.0167286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shibata S, Miwa T, Wu H-H, Levitt P, Ohyama T. Hepatocyte Growth Factor–c-MET Signaling Mediates the Development of Nonsensory Structures of the Mammalian Cochlea and Hearing. The Journal of Neuroscience. 2016;36:8200–8209. doi: 10.1523/jneurosci.4410-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trowe MO, Maier H, Schweizer M, Kispert A. Deafness in mice lacking the T-box transcription factor Tbx18 in otic fibrocytes. Development. 2008;135:1725–1734. doi: 10.1242/dev.014043. [DOI] [PubMed] [Google Scholar]
- 97.Freyer L, Aggarwal V, Morrow BE. Dual embryonic origin of the mammalian otic vesicle forming the inner ear. Development. 2011;138:5403–5414. doi: 10.1242/dev.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu J, et al. Identification of mouse cochlear progenitors that develop hair and supporting cells in the organ of Corti. Nature communications. 2017;8:15046. doi: 10.1038/ncomms15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crowson MG, Hertzano R, Tucci DL. Emerging Therapies for Sensorineural Hearing Loss. Otology & neurotology: official publication of the American Otological. Society, American Neurotology Society [and] European Academy of Otology and Neurotology. 2017;38:792–803. doi: 10.1097/mao.0000000000001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walters BJ, et al. In Vivo Interplay between p27(Kip1), GATA3, ATOH1, and POU4F3 Converts Non-sensory Cells to Hair Cells in Adult Mice. Cell reports. 2017;19:307–320. doi: 10.1016/j.celrep.2017.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atkinson PJ, Wise AK, Flynn BO, Nayagam BA, Richardson RT. Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PloS one. 2014;9:e102077–e102077. doi: 10.1371/journal.pone.0102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Izumikawa M, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 103.Shi F, et al. β-Catenin is required for hair-cell differentiation in the cochlea. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pan W, et al. Ectopic expression of activated notch or SOX2 reveals similar and unique roles in the development of the sensory cell progenitors in the mammalian inner ear. J Neurosci. 2013;33:16146–16157. doi: 10.1523/jneurosci.3150-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahmed M, et al. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Developmental cell. 2012;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng JL, Gao W-Q. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nature neuroscience. 2000;3:580. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 107.Atkinson PJ, et al. Sox2 haploinsufficiency primes regeneration and Wnt responsiveness in the mouse cochlea. J Clin Invest. 2018;128:1641–1656. doi: 10.1172/jci97248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ronaghi M, et al. Inner ear hair cell-like cells from human embryonic stem cells. Stem cells and development. 2014;23:1275–1284. doi: 10.1089/scd.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hartman, B. H., Durruthy-Durruthy, R., Laske, R. D., Losorelli, S. & Heller, S. Identification and characterization of mouse otic sensory lineage genes. Frontiers in Cellular Neuroscience9, 10.3389/fncel.2015.00079 (2015). [DOI] [PMC free article] [PubMed]
- 111.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Current protocols in molecular biology. 2015;109:21.29.21–29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krueger, F. Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files (2015).
- 113.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 114.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25–R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The, E. P. C An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Haeussler Maximilian, Zweig Ann S, Tyner Cath, Speir Matthew L, Rosenbloom Kate R, Raney Brian J, Lee Christopher M, Lee Brian T, Hinrichs Angie S, Gonzalez Jairo Navarro, Gibson David, Diekhans Mark, Clawson Hiram, Casper Jonathan, Barber Galt P, Haussler David, Kuhn Robert M, Kent W James. The UCSC Genome Browser database: 2019 update. Nucleic Acids Research. 2018;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Müllner D. Fastcluster: Fast Hierarchical, Agglomerative Clustering Routines for R and Python. 2013. 2013;53:18. doi: 10.18637/jss.v053.i09. [DOI] [Google Scholar]
- 124.Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics. 2017;18:287. doi: 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sergushichev, A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv, 060012, 10.1101/060012 (2016).
- 127.Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Briefings in Bioinformatics. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lawrence M, et al. Software for Computing and Annotating Genomic Ranges. PLoS computational biology. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer Publishing Company, Incorporated, 2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.