Abstract
Molecular recognition is critical for the fidelity of signal transduction in biology. Conversely, the disruption of protein-protein interactions can lead to disease. Thus, comprehension of the molecular determinants of specificity is essential for understanding normal biological signaling processes and for the development of precise therapeutics. Although high-resolution structures have provided atomic details of molecular interactions, much less is known about the influence of cooperativity and conformational dynamics. Here, we used the Tiam2 PSD-95/Dlg/ZO-1 (PDZ) domain and a quadruple mutant (QM), engineered by swapping the identity of four residues important for specificity in the Tiam1 PDZ into the Tiam2 PDZ domain, as a model system to investigate the role of cooperativity and dynamics in PDZ ligand specificity. Surprisingly, equilibrium binding experiments found that the ligand specificity of the Tiam2 QM was switched to that of the Tiam1 PDZ. NMR-based studies indicated that Tiam2 QM PDZ, but not other mutants, had extensive microsecond to millisecond motions distributed throughout the entire domain suggesting structural cooperativity between the mutated residues. Thermodynamic analyses revealed energetic cooperativity between residues in distinct specificity subpockets that was dependent upon the identity of the ligand, indicating a context-dependent binding mechanism. Finally, isothermal titration calorimetry experiments showed distinct entropic signatures along the mutational trajectory from the Tiam2 wild-type to the QM PDZ domain. Collectively, our studies provide unique insights into how structure, conformational dynamics, and thermodynamics combine to modulate ligand-binding specificity and have implications for the evolution, regulation, and design of protein-ligand interactions.
Significance
High-resolution structures have provided the atomic details of countless molecular interactions. However, the influence of cooperativity and conformational dynamics on the specificity of molecular interactions has received little attention. Here, we show that energetic cooperativity between distal sites and conformational dynamics in a model PDZ ligand system play a critical role in dictating molecular specificity.
Introduction
Molecular recognition is critical for signal transduction in cells. Over the years, structural studies have provided a wealth of information concerning the direct interactions that lead to molecular recognition, but far less is known about the thermodynamics and cooperativity of the residues involved in these interactions. Furthermore, we know little about the role of protein dynamics in molecular recognition and its relationship to binding specificity. PSD-95/Dlg/ZO-1 (PDZ) domains have become model systems for interrogating the fundamental principles of protein-protein interactions (1, 2, 3). They are small modular domains that generally bind to the C-terminus of their partner proteins to assemble signaling complexes at the cell membrane (3). Here, we focus on the T cell lymphoma invasion and metastasis 1 and 2 proteins (Tiam1 and Tiam2), which are guanine nucleotide-exchange factors specific for the Rho-family GTPase member Rac1. Both Tiam1 and Tiam2 play crucial roles in neuronal migration, dendritic spine formation, and neuronal polarization (4, 5, 6). These multidomain signaling proteins contain a catalytic domain composed of a Dbl and Pleckstrin homology region, a Pleckstrin homology-coiled coil-extension region, a Ras-binding domain, and a single PDZ domain.
Tiam1 and Tiam2 PDZ domains share only ∼28% identity in amino acid sequence and have distinct ligand specificities (7). The Tiam1 PDZ domain is known to bind the C-terminus of several adhesion proteins, whereas data for the Tiam2 PDZ domain are limited, and only two adhesion proteins have been identified as interaction partners (7). The Tiam1 PDZ domain binds the cell adhesion receptor syndecan (SDC) 1, and this interaction is important for the regulation of cell-cell adhesion and cell migration (8). The dissociation constants (Kds) of PDZ domain-mediated interactions have been shown to vary over a wide range (Kd ∼μM to mM) while still being biologically significant (9, 10). Relative to known PDZ/ligand complexes, quantification of the Tiam1 PDZ domain and the SDC1 peptide binding showed the interaction is of average strength (Kd ∼20 μM), whereas the interaction of the SDC1 peptide with the Tiam2 PDZ domain is weaker (Kd ∼200 μM). We also have found that Caspr4 and neurexin 1 (NRXN1), members of the neuronal presynaptic protein neurexin superfamily, have differential specificity for Tiam PDZ domains. The Tiam2 PDZ domain bound both Caspr4 and NRXN1 C-terminal peptides tightly (Kds of 3.4 and 5.0 μM, respectively), which was six-fold and 500-fold tighter than the interaction with the Tiam1 PDZ domain (7, 11, 12). These studies indicated that Tiam1 and Tiam2 PDZ domains have distinct ligand-binding specificities.
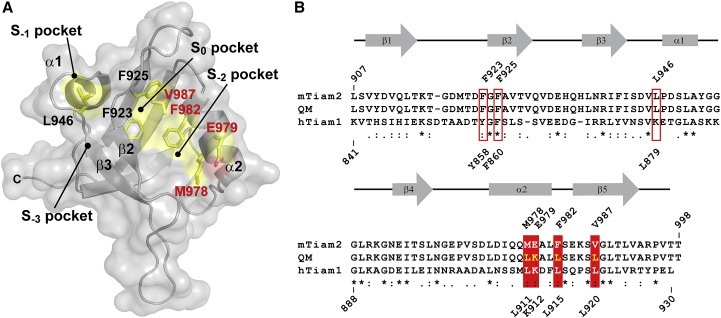
Structures of the Tiam1 PDZ domain have provided insight into determinants of ligand specificity. In general, ligands interact with the PDZ domain via backbone interactions, and several subpockets accommodate side chains in the ligand to provide specificity. In describing PDZ/ligand interactions, the convention is to number the peptide ligand from the C-terminus, which is defined as P0, and the remaining preceding residues are numbered as P-1, P-2, etc., whereas the PDZ binding subpockets are numbered as S0, S-1, S-2, etc. The crystal structure of the Tiam1 PDZ/SDC1 complex showed that residues in the S0 (L915 and L920) and S-2 (L911 and K912) subpockets are critical molecular determinants for ligand recognition (13). Sequence comparison between the Tiam1 and Tiam2 PDZ wild-type (WT) showed that these four residues are not conserved and, in the Tiam2 PDZ domain, correspond to F982 and V987 in S0 pocket and M978 and E979 in the S-2 pocket (Fig. 1). Previously, we reported that substituting these four residues in the Tiam1 PDZ domain with those in the Tiam2 PDZ domain (Tiam1 PDZ quadruple mutant (QM), L911M/K912E/L915F/L920V) switched the ligand-binding specificity. Specifically, the affinity of the Tiam1 QM PDZ was reduced fourfold for SDC1 and increased ∼50-fold for NRXN1 relative to the Tiam1 WT PDZ (7). In addition, structural analyses of Tiam1 QM PDZ/ligand complexes found that among these four residues, E912 and F915 played a significant role in switching ligand-binding specificity compared to M911 and V920 by shaping the ligand subpockets and maintaining electrostatic interactions with the specific ligands (14).
Figure 1.
Homology model of the Tiam2 WT PDZ domain and location of the four residues mutated in this study. (A) A space-filled representation of the Tiam2 WT PDZ homology model is shown. The homology model was created using the SWISS-MODEL server (28) and the Tiam1 WT PDZ structure (PDB: 4GVC) as a template. Mutated residues are labeled and colored in red. The S0 and S-2 specificity pockets are colored yellow. (B) Shown is the amino acid sequence alignment of human Tiam1, mouse Tiam2, and the mouse Tiam2 quadruple mutant (QM) PDZ domains. The secondary structure elements are indicated by rectangles (α-helix) and arrows (β-strand). The residues highlighted in (A) are boxed and labeled. Tiam1 and Tiam2 sequence numbering is indicated at above and below of the sequence, respectively. To see this figure in color, go online.
To gain insight into the molecular determinants of Tiam1/2 PDZ domain distinct ligand-binding specificity, we created the Tiam2 QM PDZ domain (M978L, E979K, F982L, and V987L mutant). Using a panel of C-terminal peptides to probe its binding profile, we found that the specificity of the Tiam2 QM PDZ was switched to that of the Tiam1 WT PDZ. Solution NMR-based analyses of the WT and QM PDZs showed that two loop regions in Tiam2 WT PDZ were dynamic, whereas the Tiam2 QM PDZ had dynamic motions detected in many regions across the entire domain. However, the dynamics in both WT and QM PDZ domains were largely quenched upon ligand binding. The difference in protein dynamics may be related to the distinct contributions of entropy during ligand-binding events. Further, thermodynamic cycle and cube analyses revealed cooperativity between the S0 and S-2 subpockets and highlighted the importance of the E979K mutation in the switch of ligand specificity. Taken together, the engineered Tiam2 QM PDZ domain provides unique insight into how structure, dynamics, and thermodynamics combine to modulate ligand-binding specificity.
Materials and Methods
Expression, mutagenesis, and purification of proteins
The Tiam2 WT PDZ domain expression vector has been described (7). Briefly, the DNA sequence for the mouse Tiam2 PDZ domain was ligated into a modified pET21 expression vector (Novagen, Madison, WI) that contains an N-terminal 6× His-tag and tobacco etch virus protease site. All Tiam2 PDZ domain mutants were produced using oligonucleotide-directed mutagenesis with the Tiam2 WT PDZ domain DNA as a template and verified by automated DNA sequencing (Genomics Division, Iowa Institute of Human Genetics, University of Iowa, Iowa City, IA). Protein expression was achieved using Escherichia coli Rosetta cells (Novagen) grown in Luria Bertani or M9 minimal media containing uniformly labeled 15NH4Cl (99%) and D-glucose (U-13C-99%) to produce 15N and 15N/13C isotopic-labeled proteins. Protein purification followed our previously published protocol and included nickel-chelate and size-exclusion (S-75) chromatography (GE Healthcare, Chicago, IL) (7). The N-terminal 6× His affinity tag was proteolytically removed by incubation of the His-tagged PDZ protein with recombinant tobacco etch virus protease overnight at room temperature. The undigested PDZ fusion protein, cleaved 6× His-tag, and 6×His-recombinant tobacco etch virus protein were separated from the digested PDZ domain by nickel-chelate chromatography.
Fluorescence anisotropy binding measurements
Fluorescence anisotropy binding assays were performed on a Fluorolog3 (Jobin Yvon, Horiba, Kyoto, Japan) spectrofluorometer. Binding experiments were performed at 25°C using 1 μM dansylated peptides in phosphate buffer (20 mM NaPO4 and 50 mM NaCl (pH 6.8)) and increasing amounts of Tiam2 PDZ domains. The fluorescence anisotropy signal was recorded using excitation and emission wavelengths of 340 and 550 nm, respectively. Typically, each measurement was made in triplicate, and their average and SE reported. The titration curves were analyzed by nonlinear regression fitting of binding isotherms to generate the dissociation constants (Kd) as previously described (15). Analyses of thermodynamic cycles and cubes used the propagated error obtained from binding experiments for each PDZ/ligand pair.
NMR spectroscopy
Tiam2 PDZ samples in phosphate buffer (20 mM NaPO4 and 50 mM NaCl (pH 6.8)) were used at a concentration of 0.5 mM for the titration experiments and 1 mM for collection of assignment and relaxation data. NMR spectra were collected on Varian Inova 600 MHz (room temperature probe), Bruker Avance II 500 MHz (room temperature probe), and Avance II 800 MHz (CryoProbe™) spectrometers. All spectra were collected at 25°C except where noted. Standard two-dimensional 1H-15N-HSQC, three-dimensional HNCACB, CBCA(CO)NH, and HNCA were acquired for backbone resonance assignments. Peptide NMR titration experiments were carried out by recording a series of two-dimensional 1H-15N-HSQC spectra of Tiam2 PDZ domains in the presence of Caspr4 or SDC1 ligand added in eight steps to saturation (with a final molar ratio of PDZ domains to ligand ranging from 1:5 to 1:10, depending on the complex). 15N relaxation-compensated CPMG experiments were performed at 500 and 800 MHz field-strength spectrometers (16, 17). The experiment was carried out with a reference experiment, 12 τcp values, and three duplicates using a total CPMG period of 60 ms recorded in an interleaved fashion. 1H-15N HSQC spectra were collected before and after each experiment to ensure sample integrity.
All NMR data were processed using NMRPipe (18), NMRView (19), and Sparky (20). The CPMG experiments were analyzed as previously described (14). Peak intensities from all resonances, except for those with significant overlapping or weak intensities, were extracted and analyzed. In all, 64/72 and 56/64 nonproline amides were analyzed for the WT and QM Tiam2 PDZ domains, respectively. R2,eff values from two fields were calculated and fitted simultaneously. Residues with significant chemical exchange (>2 Hz) were identified by fitting their intensities using both a model assuming no exchange and a simple two-state model, followed with F test (αcritical = 0.01). These residues were then fitted using the full Carver-Richards equation (21) to obtain the relaxation parameters using the program exrate2.0 (22). Monte Carlo simulations were used to determine the errors in the fitted parameters. Residue-dependent local fits were carried out before the global fits. The criteria for group fitting was χ2group/χ2local ≤ 2, where χ2local is the χ2 when a residue is locally fit, and χ2group is the χ2 of the same residue when fit in a group (23, 24). If this criterion was not met, then individual local fits were used.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed on a VP-ITC microcalorimeter (MicroCal, Northampton, MA). The Tiam2 PDZ domains were dialyzed extensively in phosphate buffer (20 mM NaPO4 and 50 mM NaCl (pH 6.8)) before all experiments. N-terminally acetylated peptides were used in these experiments and dissolved in the same dialysis buffer. The concentration of peptide and protein was determined using the bicinchoninic acid assay (Pierce Biotechnology, Thermo Fisher Scientific, Waltham, MA). Typically, ∼1.4 mL of PDZ domain (0.03–0.4 mM) was placed in the sample cell with 300 μL peptide (0.4–6.5 mM) loaded into the syringe equilibrated at 25°C. Heats of dilution were determined in separate experiments by titrating peptides into ITC buffer. Thermograms of PDZ/peptide interactions were corrected for heats of dilution using ORIGIN software (v7.0, MicroCal). The change in enthalpy (ΔH), association constant (Ka), and stoichiometry (n) were obtained by nonlinear least squares fitting of the data using a single-site binding model in ORIGIN software. Representative data are reported, and the average fitted parameters were determined from two to three replicate experiments.
Chemical denaturation
Chemical denaturation experiments were carried out by monitoring the circular dichroism (CD) signal while varying the concentration of guanidinium hydrochloride (Gdn-HCl). WT and mutant Tiam2 PDZ domains were used at the final concentration of 10 μM in the phosphate buffer (20 mM NaPO4 and 50 mM NaCl (pH 6.8)). The proteins were incubated with a given concentration of Gdn-HCl for 24 h before each CD measurement. The concentration of the stock Gdn-HCl solution was determined by measuring the refractive index (Abbe-3L; Thermo Fisher Scientific) (25, 26). A 1 mm optical length cuvette was used to measure the changes in ellipticity at the wavelength of 222 nm and at 25°C in a Jasco J-815 spectropolarimeter. The data were normalized to the maximal and minimal signals and fitted into a two-state model with a single transition between native and denatured protein (25, 27). The average free energy of folding (ΔGf) and Cm and their associated errors were derived from measurements obtained in triplicate.
Results
Mutations in the Tiam2 PDZ domain switch ligand specificity
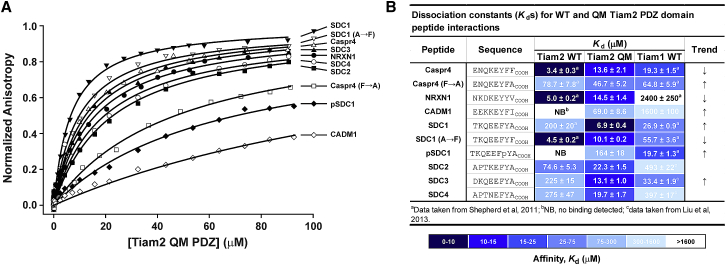
We previously designed a QM in the Tiam1 PDZ domain (Tiam1 QM) and showed that its binding specificity is switched to that of the Tiam2 PDZ domain (7, 14). To determine whether the analogous four residues in the Tiam2 WT PDZ are important for specificity, we produced the Tiam2 QM PDZ (M978L/E979K/F982L/V987L) (Fig. 1) and performed fluorescence-based binding assays using a panel of Tiam family ligands (C-terminal peptides derived from Caspr4, NRXN1, and SDC1–4 proteins). The dissociation constants (Kd) for the interaction between Caspr4, NRXN1, and SDC1 and the Tiam2 QM PDZ domain were switched, approaching those of the Tiam1 WT PDZ domain. For instance, the Tiam2 QM PDZ bound the Caspr4 peptide with a Kd of 13.6 μM (ΔGb = −6.64 kcal/mol), which was weaker than that for Tiam2 WT PDZ (Kd = 3.4 μM; ΔGb = −7.46 kcal/mol) and similar to the Tiam1 WT PDZ (Kd = 19.3 μM; ΔGb = −6.43 kcal/mol) (Fig. 2). The Kd of the Tiam2 QM PDZ/NRXN1 interaction was slightly weaker (14.5 μM; ΔGb = −6.60 kcal/mol) than that determined for Tiam2 WT PDZ/NRXN1 (5.0 μM; ΔGb = −7.22 kcal/mol)—trending toward the much weaker Tiam1 WT PDZ/NRXN1 affinity (Kd = 2400 μM; ΔGb = −3.57 kcal/mol).
Figure 2.
Ligand-binding specificity of the Tiam2 QM PDZ domain. (A) Representative binding curves of the Tiam2 QM PDZ. The Kd values reflect the mean and SD from at least three technical replicates. (B) Shown is a summary of the peptide-binding data for Tiam2 WT, Tiam2 QM, and Tiam1 WT PDZ domains. The color coding shows graphically the trend in affinity of the Tiam2 QM and Tiam1 WT compared to the Tiam2 WT PDZ. The column labeled “Trend” indicates with an arrow the trend in affinity (higher (↑) or lower (↓)). The scale of the color coding is defined at the bottom. To see this figure in color, go online.
The dissociation constant of Tiam2 QM PDZ/SDC1 complex was 6.9 μM, tighter than both WT Tiam1 and Tiam2 PDZ domains. Indeed, all the ligands from the SDC family bound to Tiam2 QM PDZ relatively tightly, with Kds ranging from 6.9 to 22.3 μM (ΔGb ranging from −7.04 to −6.34 kcal/mol). Among the SDC family members, Tiam2 QM PDZ bound SDC1 and SDC3 more tightly than SDC2 and SDC4. This result is in harmony with the binding specificity of the Tiam1 WT PDZ, which prefers SDC1 and SDC3 (13). In contrast, the Tiam2 WT PDZ had a binding preference for SDC2, whereas other SDC family members bound weaker (Kds > 200 μM; Fig. 2). Moreover, the affinity of the SDC1 A → F mutant was slightly weaker for the Tiam2 QM compared to the WT, whereas the affinity of the Caspr4 F → A mutant was increased for the Tiam2 QM compared to the WT. Overall, 8 of the 10 peptides bound the Tiam2 QM with a Kd trending toward that of Tiam1 WT, indicating a general specificity change in the Tiam2 QM PDZ domain (Fig. 2 B).
Intrinsic dynamics of the WT and QM Tiam2 PDZ domains
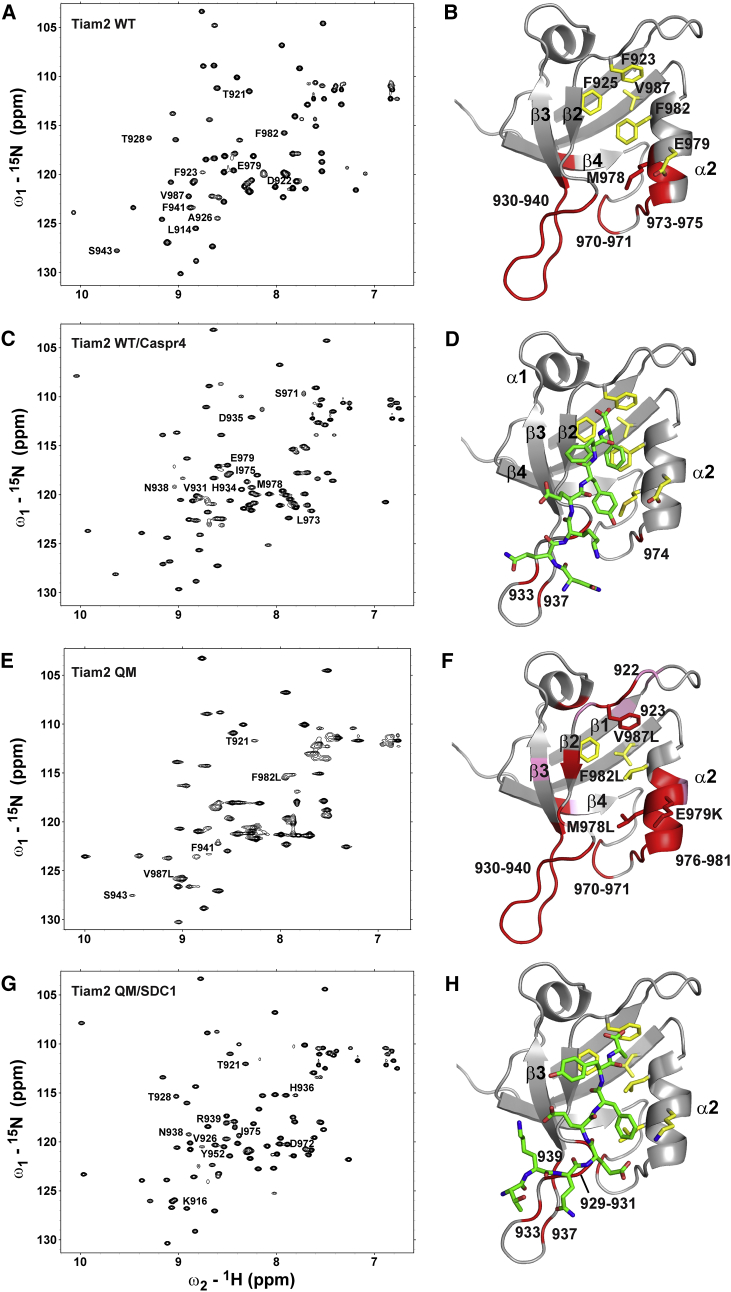
Nitrogen HSQC (1H-15N HSQC) spectra were collected to examine structural features in the WT and QM Tiam2 PDZ domains (Fig. 3). Overall, the Tiam2 WT PDZ domain had dispersed 1H and 15N chemical shifts, indicative of a folded protein. However, only 72 of 91 nonproline residues were observed in the 1H-15N HSQC spectrum of the Tiam2 WT PDZ, with the remaining peaks exchange broadened beyond detection (Fig. 3 A). Resonance assignment of amide peaks indicated that the 19 broadened resonances originate from residues 930–940, 952, 970–971, 973–975, to 977–978. Because there is currently no structure of the Tiam2 WT PDZ domain available, these residues were mapped onto a homology model of the Tiam2 WT PDZ generated using the SWISS-MODEL server (28) and the Tiam1 WT PDZ structure (Protein Data Bank [PDB]: 4GVC) as template. The exchange-broadened residues cluster together to form a continuous region comprised of the β2-β3 loop, the N-terminal region of the β3 strand, and the β4-α2 loop to the beginning of α2 helix (Fig. 3 B). From this data, we conclude that the β2-β3 loop, β4-α2 loop, and peripheral regions are highly dynamic on the μs-ms timescale in solution.
Figure 3.
1H-15N HSQC spectra and structural models of WT and QM Tiam2 PDZ domain free and bound to ligands. (A and B) WT PDZ Tiam2 PDZ in ligand-free state is shown. (C and D) Tiam2 WT PDZ bound to Caspr4 is shown. (E and F) Tiam2 QM PDZ in ligand-free state is shown. (G and H) Tiam2 QM PDZ bound to SDC1 is shown. Each panel represents an HSQC spectrum and the backbone resonance assignments mapped onto the structural model. Select residues are labeled to guide the comparison between spectra. Residues broadened beyond detection are colored in red, whereas residues with weak intensities are colored in pink. The Tiam2 QM homology model was generated by mutating the four residues in the WT homology model created in SWISS-MODEL (26). The PDZ/peptide complexes were generated starting with the apo Tiam2 WT PDZ. The Tiam2 WT/Caspr4 complex was generated by overlaying the Tiam2 WT model with the Tiam1 QM/Caspr4 structure (PDB: 4NXQ) and including the Caspr4 pose with the Tiam2 WT model. The Tiam2 QM/SDC1 complex was generated by overlaying the QM model with Tiam1 WT/SDC1 structure (PDB: 4GVD), keeping the SDC1 pose. These models are shown simply to indicate the location of the peptide-binding site. To see this figure in color, go online.
The four substituted residues in the Tiam2 QM PDZ had a dramatic effect on the 1H-15N-HSQC spectrum. Only 64 peaks appeared in the spectrum, and their intensities were not uniform, suggesting a significant increase in internal dynamics (Fig. 3 E). Assignment of these 64 resonances provided the identity of the 27 broadened resonances. Mapping these residues onto a homology model showed a similar pattern seen with the Tiam2 WT PDZ domain with the same 19 residues in the β2-β3 and β4-α2 loops being exchange broadened. The remaining broadened residues were found in the α2 helix (residues 976, 979, and 981), the β1-β2 loop (residues 922 and 923), the β2 strand (residues 926 and 928), and the α1 helix (residue 951) (Fig. 3 F). In addition, residues 914, 916, 921, 924, 943, 964, and 980 had significantly reduced intensities.
Enhanced dynamics of the QM PDZ were also observed in side chain methyl groups. We examined this phenomenon by acquiring methyl 1H-13C heteronuclear single quantum coherence (HSQC) for the WT and QM PDZ domains. 1H-13C-HSQC methyl side chain spectra resolved 59 of 64 methyl resonances (from Met, Ala, Thr, Val, Leu, and Ile residues) in the Tiam2 WT PDZ, whereas only 50 of 67 resonances were observed in the QM PDZ (Fig. S1). Of the 20 exchange-broadened amide resonances seen in the WT 1H-15N-HSQC spectrum, there were only eight methyl-bearing residues (containing a total of 15 methyl groups). Moreover, 12 of the 28 missing amide resonances in the QM were methyl-bearing residues (containing a total of 20 methyl groups). However, only 5 out of 64 methyl signals (8%) were found to be exchange broadened in the WT 1H-13C-HSQC spectrum, whereas 17 out of 67 methyl signals (∼25%) in the QM PDZ domain were exchange broadened (Fig. S1). Thus, a larger proportion of the methyl resonances were exchange broadened in the QM relative to the WT PDZ domain, which may be a critical parameter linked to changes in specificity seen in the Tiam2 QM PDZ. Combined, this data shows that mutating four residues in the Tiam2 PDZ domain dramatically increased the number of residues experiencing dynamic motions at both the backbone and side chain level.
Next, we used NMR spectroscopy to monitor the effect of ligand binding on the exchange-broadened peaks of the WT and QM Tiam2 PDZ domains. A series of 1H-15N-HSQC spectra were collected for 15N-labeled Tiam2 WT or QM PDZ as a function of an increasing peptide concentration. Typically, we found the complexes to be in slow exchange upon the titration of peptide when the dissociation constant was ∼10 μM or tighter, whereas weaker peptide binding with Kd > 20 μM generally resulted in a fast exchange on the NMR timescale. Titration with the SDC1 peptide (Kd ∼200 μM) was found to be in fast exchange and resulted in several peaks being shifted from the apo state, but no additional peaks became resolved in the spectrum (Fig. S2 A). In contrast, marked chemical shift perturbations were observed in the presence of the Caspr4 peptide (Kd ∼3 μM). Moreover, 15 additional resonances became resolved in the spectrum upon saturation with the Caspr4 peptide, giving 87/91 peaks in total (Fig. 3 C). The other four expected resonances (Q930, E933, L937, and D974), located in the β2-β3 loop, remained broadened (Fig. 3 D). Thus, peptide binding stabilized the Tiam2 WT PDZ domain, resulting in quenched dynamics.
The effect of ligand binding on the of 1H-15N-HSQC spectrum of the Tiam2 QM PDZ domain was also examined. In contrast to Tiam2 WT PDZ spectrum, titration of the QM PDZ with the Caspr4 peptide (Kd ∼14 μM) resulted in 78 resonances being resolved (Fig. S2 B). Moreover, titration with SDC1 (Kd ∼7 μM) resulted in resonances in the slow exchange regime and a spectrum with 84 resonances of uniform intensity (Fig. 3 G). The seven broadened resonances (929–931, 933, 937, and 939 in the β2-β3 loop and residue 970 in the β4-α2 loop) are mapped onto a homology model of Tiam2 QM PDZ domain (Fig. 3 H). These data show that the broadened resonances in the α2 helix, β1-β2 loop, and α1 helix are observable in the presence of SDC1 and Caspr4 ligands, indicating dampened dynamic motions in these regions.
Microsecond-millisecond timescale motions in WT and QM Tiam2 PDZ domains
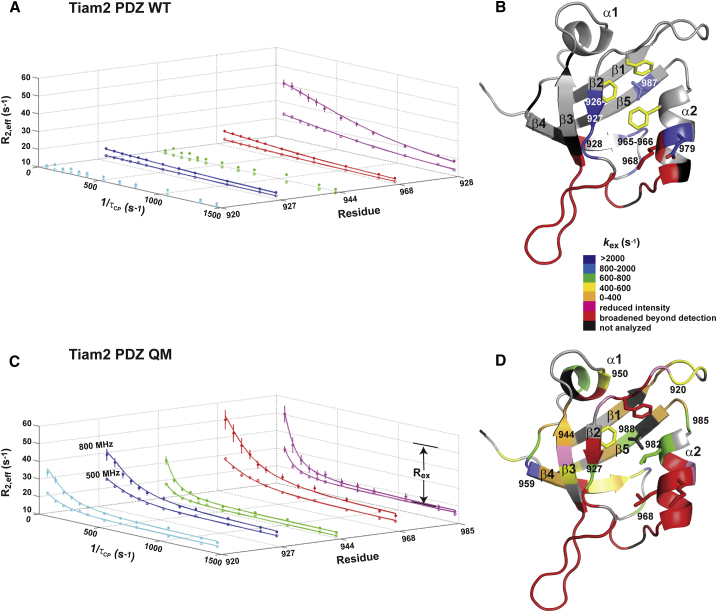
To quantitatively characterize the distinct microsecond to millisecond (μs-ms) timescale dynamic behavior in the WT and QM Tiam2 PDZ domains, we conducted 15N Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments (17, 24). This approach relies on using a CPMG pulsing scheme to measure transverse relaxation (R2) and its dependence on the effective field strength (νcpmg) to quantify chemical exchange (Rex) (from conformational dynamics). Representative relaxation dispersion curves for the Tiam2 WT PDZ domain are shown in Fig. 4 A. Eight residues exhibited chemical exchange on the μs-ms timescale, whereas the other 56 residues had no measurable Rex (Rex ≈ 0). The eight dispersion curves were fit to a global, two-state model, yielding an exchange rate constant (kex = 2355 s−1), the population of the major and minor states (pA and pB), and the difference in chemical shift between these states (Δω) (Fig. S3; Table 1). The eight residues are mapped onto the structural model of the Tiam2 WT PDZ domain (Fig. 4 B).
Figure 4.
Microsecond to millisecond timescale motions in the WT and QM Tiam2 PDZ domain. Representative relaxation dispersion curves from the Tiam2 WT (A) and QM (C) PDZ domains collected at 500 (open circle) and 800 (closed circle) MHz are shown. Curves for each residue with Rex are shown in Figs. S3 and S4, whereas their fitted parameters are indicated in Tables 1 and 2. Error bars were determined by the analysis of peak intensities from duplicate experiments. (B and D) Structural models of WT and QM Tiam2 PDZ with highlighted residues showing relaxation dispersion are shown. To see this figure in color, go online.
Table 1.
Fitted Parameters for the Tiam2 WT PDZ Domain 15N-CPMG Relaxation Dispersion Curves
| Residue | Δω (ppm) | R2,B0(s−1) (500 MHz) | R2,B0(s−1) (800 MHz) |
|---|---|---|---|
| 926 | 1.248 ± 0.099 | 11.34 ± 0.18 | 12.47 ± 0.42 |
| 927 | 0.567 ± 0.043 | 11.63 ± 0.06 | 12.91 ± 0.11 |
| 928 | 1.561 ± 0.123 | 11.60 ± 0.19 | 12.49 ± 0.40 |
| 965 | 1.334 ± 0.102 | 11.75 ± 0.17 | 13.81 ± 0.48 |
| 966 | 1.586 ± 0.123 | 11.95 ± 0.16 | 12.63 ± 0.39 |
| 968 | 0.719 ± 0.053 | 10.78 ± 0.05 | 11.59 ± 0.10 |
| 979 | 1.494 ± 0.117 | 12.09 ± 0.15 | 13.20 ± 0.36 |
| 987 | 1.129 ± 0.087 | 12.21 ± 0.09 | 12.95 ± 0.20 |
Results from global fitting of data with kex = 2355 ± 53 s−1 and pA = 0.864 ± 0.026. The error for each parameter was estimated from Monte Carlo simulations.
In contrast, the Tiam2 QM PDZ domain had enhanced μs-ms motions. Overall, 34 residues from Tiam2 QM PDZ domain had significant Rex values (Fig. S4; Table 2), and most of these residues did not show chemical exchange (Rex ≈ 0) in the WT PDZ domain. Only resonances from residues 927, 965, 966, and 968 were common to both PDZ domains. Furthermore, residues 926, 928, and 979 were broadened beyond detection. Representative dispersion curves for several residues and their location on the PDZ domain are shown in Fig. 4, C and D. The residues with significant Rex were distributed throughout the PDZ domain, including the α2 helix, β1-β2 loop, β2- β3 loop, and β3-α1 region, indicating widespread dynamic motions on the μs-ms timescale (Fig. 4 D). The relaxation dispersion data for these residues did not fit to a single, global process. Instead, the data was best represented by individual fits for each residue with kex values ranging from 170 to 1700 s−1 (Table 2), suggesting distinct motions in the Tiam2 QM PDZ domain compared to the WT. Overall, these experiments indicate that the QM PDZ domain had a pronounced increase in μs-ms dynamics compared to the WT protein.
Table 2.
Fitted Parameters for the QM PDZ Domain 15N-CPMG Relaxation Dispersion Curves
| Residue | kex (s−1) | Δω (ppm) | pA | R2,B0 (s−1) (500 MHz) | R2,B0 (s−1) (800 MHz) |
|---|---|---|---|---|---|
| 909 | 401 ± 31 | 1.012 ± 0.099 | 0.847 ± 0.032 | 13.37 ± 0.11 | 16.96 ± 0.28 |
| 910 | 321 ± 55 | 0.583 ± 0.121 | 0.705 ± 0.086 | 16.70 ± 0.20 | 20.07 ± 0.93 |
| 913 | 242 ± 53 | 0.625 ± 0.094 | 0.944 ± 0.013 | 14.88 ± 0.10 | 17.03 ± 0.26 |
| 915 | 220 ± 39 | 0.493 ± 0.053 | 0.708 ± 0.072 | 14.76 ± 0.17 | 15.82 ± 0.64 |
| 916 | 171 ± 70 | 0.494 ± 0.107 | 0.857 ± 0.069 | 16.68 ± 0.25 | 20.25 ± 0.63 |
| 917 | 552 ± 71 | 0.902 ± 0.176 | 0.922 ± 0.038 | 13.36 ± 0.17 | 14.38 ± 0.39 |
| 919 | 504 ± 277 | 0.597 ± 0.200 | 0.914 ± 0.055 | 21.48 ± 0.32 | 24.14 ± 0.72 |
| 920 | 517 ± 38 | 0.713 ± 0.092 | 0.855 ± 0.053 | 14.17 ± 0.12 | 16.55 ± 0.31 |
| 927 | 658 ± 49 | 1.024 ± 0.141 | 0.892 ± 0.037 | 15.58 ± 0.15 | 17.75 ± 0.39 |
| 942 | 496 ± 110 | 0.668 ± 0.162 | 0.908 ± 0.036 | 18.20 ± 0.20 | 20.58 ± 0.55 |
| 944 | 297 ± 29 | 0.498 ± 0.052 | 0.768 ± 0.061 | 11.52 ± 0.11 | 13.42 ± 0.29 |
| 945 | 233 ± 62 | 0.710 ± 0.152 | 0.935 ± 0.032 | 12.99 ± 0.16 | 14.27 ± 0.43 |
| 950 | 497 ± 105 | 0.700 ± 0.159 | 0.897 ± 0.047 | 18.12 ± 0.23 | 20.72 ± 0.66 |
| 953 | 709 ± 160 | 0.615 ± 0.125 | 0.936 ± 0.025 | 16.90 ± 0.15 | 18.25 ± 0.36 |
| 954 | 788 ± 173 | 0.485 ± 0.077 | 0.936 ± 0.023 | 15.29 ± 0.10 | 17.77 ± 0.21 |
| 957 | 337 ± 66 | 0.581 ± 0.090 | 0.941 ± 0.017 | 16.09 ± 0.10 | 18.22 ± 0.20 |
| 958 | 603 ± 119 | 0.592 ± 0.143 | 0.933 ± 0.036 | 16.57 ± 0.14 | 18.20 ± 0.35 |
| 959 | 1108 ± 668 | 0.484 ± 0.197 | 0.936 ± 0.065 | 17.10 ± 0.21 | 19.69 ± 0.50 |
| 960 | 261 ± 29 | 0.699 ± 0.089 | 0.883 ± 0.029 | 15.45 ± 0.09 | 18.75 ± 0.25 |
| 961 | 350 ± 66 | 0.726 ± 0.142 | 0.692 ± 0.104 | 17.28 ± 0.34 | 19.07 ± 1.12 |
| 963 | 528 ± 155 | 0.857 ± 0.209 | 0.933 ± 0.037 | 18.31 ± 0.30 | 20.73 ± 0.69 |
| 964 | 263 ± 47 | 0.479 ± 0.059 | 0.902 ± 0.031 | 16.77 ± 0.09 | 19.26 ± 0.20 |
| 965 | 474 ± 85 | 1.047 ± 0.204 | 0.941 ± 0.022 | 17.41 ± 0.17 | 20.60 ± 0.43 |
| 966 | 1712 ± 439 | 0.707 ± 0.121 | 0.929 ± 0.028 | 17.34 ± 0.22 | 19.14 ± 0.52 |
| 967 | 487 ± 123 | 0.469 ± 0.087 | 0.942 ± 0.026 | 15.40 ± 0.09 | 17.59 ± 0.22 |
| 968 | 791 ± 43 | 0.752 ± 0.093 | 0.666 ± 0.073 | 15.23 ± 0.16 | 18.68 ± 0.56 |
| 982 | 613 ± 177 | 2.179 ± 0.439 | 0.887 ± 0.046 | 17.42 ± 0.51 | 21.32 ± 1.29 |
| 984 | 638 ± 70 | 1.130 ± 0.160 | 0.926 ± 0.023 | 15.93 ± 0.18 | 18.25 ± 0.47 |
| 985 | 289 ± 45 | 0.662 ± 0.107 | 0.764 ± 0.074 | 15.84 ± 0.21 | 16.86 ± 0.64 |
| 986 | 372 ± 47 | 0.482 ± 0.079 | 0.930 ± 0.029 | 14.14 ± 0.05 | 16.61 ± 0.13 |
| 988 | 643 ± 120 | 0.900 ± 0.225 | 0.898 ± 0.055 | 16.34 ± 0.31 | 19.23 ± 0.88 |
| 989 | 375 ± 93 | 0.490 ± 0.084 | 0.869 ± 0.048 | 16.72 ± 0.19 | 18.69 ± 0.55 |
| 991 | 435 ± 81 | 0.531 ± 0.077 | 0.901 ± 0.036 | 17.47 ± 0.15 | 19.82 ± 0.35 |
| 994 | 493 ± 23 | 0.601 ± 0.056 | 0.900 ± 0.020 | 11.92 ± 0.05 | 13.52 ± 0.12 |
Results from fitting individual residues to the data. The error for each parameter was estimated from Monte Carlo simulations.
Structural consequences caused by individual mutations
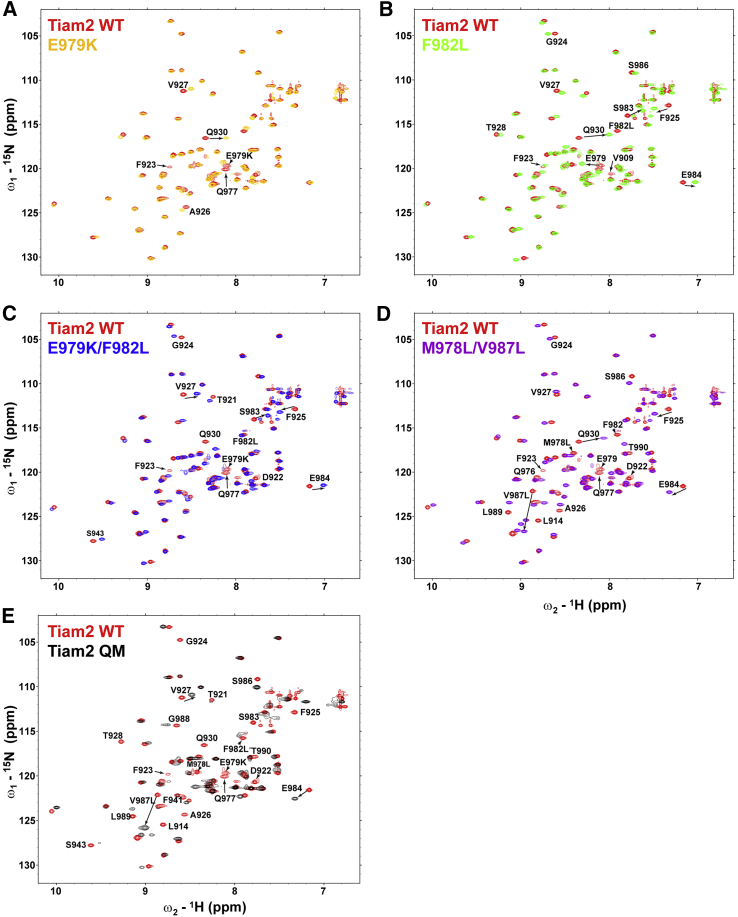
Having examined the structural and dynamic effects of mutating the four residues in the Tiam2 WT PDZ (i.e., the QM), we sought to investigate the contribution of individual and paired mutations. Fig. 5 shows the overlay of the 1H-15N-HSQC spectra of the WT and mutant PDZ proteins. Although chemical shift perturbations were evident in the spectra for the single and double mutant PDZ domains compared to Tiam2 WT PDZ spectrum, none approached the dramatic changes observed in the QM PDZ domain. The E979K mutant showed perturbed chemical shifts in residues E979 and Q977 and in the β1-β2 loop (F923, V927, and Q930) (Fig. 5 A). The F982L mutation induced additional changes, perturbing resonances in both the α2 helix (Q977, F982, S983, E984, and S986) and β1-β2 loop (residues 923–928) (Fig. 5 B). The E979K/F982L double mutant had an “additive effect” on the spectrum, in which the two mutations affected the chemical shifts of the same residues in the single mutants with no additional resonances being perturbed (Fig. 5 C). Likewise, the M978L/V987L double mutant affected both residues in the α2 helix and those in β1-β2 loop—resonances from Q976, Q977, M978, V987, L989, T990, D922, G924, F927, and Q930 were either shifted or broadened (Fig. 5 D). In contrast, the four mutations in the QM PDZ domain had a widespread effect on the HSQC spectrum (Fig. 5 E), including residues from the α2 helix through the β2 strand to β1-β2 loop and β3-α1 loop as stated above (Fig. 3, E and F). Importantly, none of the single or double mutant PDZ domain spectra studied here shared the significant line-broadening effects seen in the QM PDZ spectrum, suggesting that a cooperative effect of the four residues is required for altering the structure and dynamics. Here, we use “cooperative” to describe the structure and dynamics observed when multiple mutations do not add up to the effects observed in QM. However, we did not test all permutations of the four mutations, so it is possible that other combinations of double or triple mutations are cooperative.
Figure 5.
Structural changes of WT and mutants of the Tiam2 PDZ domain. Shown is an overlay of 1H-15N HSQC spectra of WT Tiam2 PDZ with Tiam2 PDZ mutants: Tiam2 PDZ E979K (A), Tiam2 PDZ F982L (B), Tiam2 PDZ E979K/F982L (C), Tiam2 PDZ M978L/V987L (D), and Tiam2 PDZ M978L/E979K/F982L/V987L (QM) (E). Select residues with perturbed chemical shifts are labeled. To see this figure in color, go online.
Distinct enthalpic and entropic contributions along the mutation trajectory
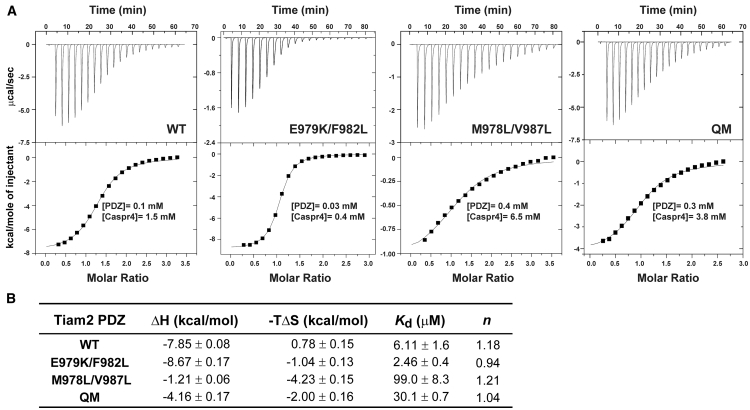
Ligand-binding thermodynamics of PDZ/ligand interactions were examined by ITC. The analyses of the interactions of the Caspr4 ligand with WT and QM Tiam2 PDZ revealed that the binding affinities (Kds) were approximately two-fold weaker than the values obtained from fluorescence anisotropy measurements (Figs. 2 and 6). This is consistent with previous studies that found similar differences in Tiam1 WT PDZ/ligand interactions in the presence and absence of a fluorophore (8, 13, 14). Caspr4 binding to the WT and QM Tiam2 PDZ gave similar binding free energies, −7.07 and −6.16 kcal/mol, respectively. However, the contribution from the enthalpy and entropy was distinct (Fig. 6 B). The Tiam2 WT PDZ/Caspr4 interaction was an enthalpically driven (ΔH = −7.85 kcal/mol) process coupled with unfavorable entropy. In contrast, the enthalpy in the Tiam2 QM PDZ/Caspr4 interaction was approximately half of the WT/Caspr4 value (ΔH = −4.16 kcal/mol), though with a favorable entropy change (−TΔS = −2.00 kcal/mol).
Figure 6.
Thermodynamic analysis of Tiam2 PDZ/Caspr4 peptide interactions determined by ITC. (A) Representative thermograms and integrated titration curves are shown for the Caspr4 ligand bound to Tiam2 WT, E979K/F982L, M978L/V987L, and QM PDZ domains. Protein and ligand concentrations used in the experiments are indicated. (B) Average thermodynamic parameters for Tiam2 PDZ/Caspr4 interactions at 25°C are shown. The change in enthalpy (ΔH), association constant (Ka), and stoichiometry (n) were fit by nonlinear least squares analysis using a single-site binding model in ORIGIN software. Each parameter represents the mean and SD from two to three technical replicates.
Next, we sought to dissect the molecular origin of the entropic contributions by analyzing the two double mutant PDZ domains. Caspr4 binding to the Tiam2 E979K/F982L PDZ remained enthalpically driven (ΔH = −8.67 kcal/mol), whereas Caspr4 binding to the M978L/V987L double mutant was entropically driven (−TΔS = −4.23 kcal/mol) (Fig. 6). These data imply that the Tiam2 QM PDZ utilizes a distinct thermodynamic mechanism compared to the WT in binding the Caspr4 ligand. These data also reveal that residues M978L and V987L play a significant role in redistributing the enthalpy/entropy balance in the QM/Caspr4 complex.
Modulating ligand binding through individual PDZ domain specificity pockets
Significantly, ligand-binding experiments revealed that the Tiam2 QM PDZ domain generally bound SDC family members much tighter than the WT Tiam1 and Tiam2 PDZ domains (Fig. 2). To further examine the molecular origins of this change in specificity, we examined the binding of single mutants to SDC family peptides, NRXN1 and Caspr4. Our recent structure of Tiam1 QM PDZ/ligand complexes showed that residues E912 and F915 play significant roles in ligand recognition (14). Therefore, these substitutions at the analogous sites in the Tiam2 PDZ domain (E979K and F982L) were examined. The two mutant PDZ domains had a significantly higher affinity for the SDC family peptides, whereas their affinity for NRXN1 and Caspr4 was only marginally weaker compared to the Tiam2 WT PDZ (Table 3). Specifically, the F982L PDZ mutant bound the SDC1–4 peptides with affinities ranging from 39 to 126 μM (1.8–3.5-fold tighter than WT), with the highest affinity being for SDC2. This trend mirrored the Tiam2 WT PDZ specificity for the SDC family. In contrast, the E979K PDZ mutant had switched specificity—having SDC1 and SDC3 as its tightest interaction partners. This is distinct from the SDC family specificity for the WT and F982L Tiam2 PDZ domains but in line with the Tiam2 QM and Tiam1 WT PDZ domains. Indeed, the E979K mutant bound to SDC1–4 with affinities ranging from ∼10–41 μM, which was ∼1.8–∼20-fold tighter than the Kds found in Tiam2 WT PDZ binding with SDC1 peptide binding the tightest. Comparison of the E979K and F982L affinity for the SDC family members shows that the E979K mutant had a larger effect on binding SDC1, 3, and 4. However, F982L played a larger role in altering the affinity for SDC2, NRXN1, and Caspr4, all of which have a lysine at position P-4. Together, these results suggest that both mutations play critical, but distinct, roles in switching the Tiam2 WT PDZ specificity. The F982L effect likely occurs through hydrophobic interactions, whereas the E979K effects are mediated by an electrostatic interaction with ligands.
Table 3.
Coupling Free Energies of Binding for Tiam2 PDZ Mutants as a Function of Binding to SDC Family, Caspr4, and NRXN1 Ligands
| Tiam2 PDZ Domain | Kd (μM) | ΔGb (kcal/mol)a | ΔGint (kcal/mol)b |
|---|---|---|---|
| SDC1 (TKQEEFYACOOH) | |||
| WT | 200 ± 20c | −5.05 ± 0.06 | |
| E979K | 10.2 ± 0.7 | −6.81 ± 0.04 | |
| F982L | 56.7 ± 6.9 | −5.77 ± 0.06 | |
| E979K/F982L | 5.4 ± 0.9 | −7.23 ± 0.04 | 0.30 ± 0.10 |
| QM | 6.9 ± 0.6 | −7.04 ± 0.05 | |
| SDC2 (APTKEFYACOOH) | |||
| WT | 74.6 ± 5.3 | −5.63 ± 0.04 | |
| E979K | 41.3 ± 5.8 | −5.98 ± 0.09 | |
| F982L | 38.7 ± 4.8 | −6.02 ± 0.10 | |
| E979K/F982L | 14.1 ± 0.6 | −6.62 ± 0.03 | −0.25 ± 0.14 |
| QM | 22.5 ± 1.5 | −6.34 ± 0.04 | |
| SDC3 (DKQEEFYACOOH) | |||
| WT | 225 ± 11 | −4.98 ± 0.03 | |
| E979K | 18.0 ± 0.6 | −6.47 ± 0.02 | |
| F982L | 112 ± 5 | −5.39 ± 0.03 | |
| E979K/F982L | 8.4 ± 0.4 | −6.92 ± 0.03 | −0.04 ± 0.05 |
| QM | 13.3 ± 1.0 | −6.65 ± 0.05 | |
| SDC4 (APTNEFYACOOH) | |||
| WT | 275 ± 47 | −4.86 ± 0.10 | |
| E979K | 29.8 ± 7.3 | −6.20 ± 0.21 | |
| F982L | 126 ± 3 | −5.32 ± 0.02 | |
| E979K/F982L | 18.6 ± 5.0 | −6.59 ± 0.23 | 0.07 ± 0.33 |
| QM | 19.7 ± 1.7 | −6.42 ± 0.05 | |
| NRXN1 (NKDKEYYVCOOH) | |||
| WT | 5.0 ± 0.2c | −7.22 ± 0.04 | |
| E979K | 6.7 ± 0.1 | −7.06 ± 0.01 | |
| F982L | 14.9 ± 3.6 | −6.59 ± 0.14 | |
| E979K/F982L | 5.0 ± 0.3 | −7.22 ± 0.04 | −0.80 ± 0.09 |
| QM | 14.5 ± 1.4 | −6.60 ± 0.06 | |
| Caspr4 (ENQKEYFFCOOH) | |||
| WT | 3.4 ± 0.3c | −7.46 ± 0.05 | |
| E979K | 4.9 ± 1.1 | −7.25 ± 0.14 | |
| F982L | 5.9 ± 1.6 | −7.15 ± 0.18 | |
| E979K/F982L | 1.9 ± 0.1 | −7.84 ± 0.04 | −0.90 ± 0.23 |
| QM | 13.6 ± 2.1 | −6.64 ± 0.09 | |
Abbreviations: b, binding; int, interaction (or coupling); QM, quadruple mutant (M978L/E979K/F982L/V987L).
ΔGint = [ΔGb (WT) − ΔGb (mutant1)] − [ ΔGb (mutant2) − ΔGb (mutant1,2)].
Data taken from Shepherd et al. (7). The error for each parameter was estimated from triplicate measurements. The error of individual measurements was propagated to obtain the error in coupling free energy.
Thermodynamic analysis of interpocket cooperativity
The Tiam2 PDZ E979K/F982L double mutant was produced to assess the energetic cooperativity between these two residues located in separate specificity pockets (S-2 and S0; Fig. 1) with respect to stability and ligand binding. The E979K/F982L double mutant bound SDC1–4 peptides more tightly than either the WT and QM Tiam2 PDZ domains, suggesting that the M978L and V987L mutations oppose the E979K and F982L mutations in determining ligand affinity (Table 3). These results also imply a synergistic role for the E979K and F982L residues in ligand binding. To test this idea, we constructed thermodynamic cycles (29, 30) using WT, E979K, F982L, and E979K/F982L Tiam2 PDZ domains and determined the coupling free energies for several ligands. We found that residues E979 and F982 were energetically coupled in a ligand-dependent fashion (Table 3). No significant cooperativity was found in the binding of SDC3 (ΔGint = −0.04 kcal/mol) and SDC4 (ΔGint = 0.07 kcal/mol), whereas residues E979 and F982 were coupled in response to binding SDC1 (ΔGint = 0.30 kcal/mol). In the case of SDC1, the E979K/F982L mutant had an increased affinity compared to the two individual mutants, and most of the energy change was due to the E979K mutation. The E979K and F982L residues were also coupled in SDC2 (ΔGint = −0.25 kcal/mol), NRXN1 (ΔGint = −0.80 kcal/mol), and Caspr4 (ΔGint = −0.90 kcal/mol) binding, such that the double mutant enhanced the binding compared to the individual mutations. Interestingly, although both single mutants (E979K and F982L) reduced binding to NRXN1 and Caspr4, together they worked cooperatively to enhance the affinity compared to both the WT and QM PDZ domains. Together, these data revealed ligand-dependent cooperativity between the E979K and F982L side chains that are located in distinct (S0 and S-2) specificity pockets.
Protein folding stability of WT and QM Tiam2 PDZ domains
Amino acid substitutions in proteins often affect their thermodynamic stability. We used the CD signal at 220 nm to monitor the unfolding transition of Tiam2 PDZ domains as a function of increasing concentrations of Gdn-HCl. The folding energies of the Tiam2 PDZ domains ranged from −3 to −5 kcal/mol (Fig. S5; Table 4). The Tiam2 WT PDZ was the least stable (ΔGf = −3.08 kcal/mol), whereas the E979K and F982L mutants were slightly more stable than the WT (ΔGf = −3.19 and −3.27, respectively). The E979K/F982L double mutant was significantly more stable (ΔGf = −3.91 kcal/mol). Double mutant cycle analysis revealed weak cooperativity (ΔGint = −0.53 kcal/mol) between the E979K and F982L residues. The M978L/V987L double mutant was even more stable with a folding energy of −4.46 kcal/mol, whereas the QM PDZ was the most stable PDZ variant (ΔGf = −5.04 kcal/mol). For reference, the Tiam1 WT and QM stabilities (ΔGf) were −2.6 and −0.82 kcal/mol, respectively (14). Interestingly, the mutations reported here generally had a stabilizing effect, unlike what is typically seen for most mutations in the literature. This was likely because the region surrounding the mutations is inherently dynamic in the WT (Fig. 4); therefore, the penalty for having defects in QM side chain packing is mitigated (31, 32). The substitutions introduced in QM may lead to new interactions (i.e., van der Waals or ionic) that are favorable for side chain packing, which has yet to be validated by structural studies. These data indicate that the Tiam2 QM is markedly more stable than the WT and that the E979K and F982L residues (and the S0 and S-2 specificity pockets) are coupled during the folding process.
Table 4.
Coupling Free Energies of Folding for Tiam2 PDZ Mutants
| Tiam2 PDZ Domain | Cm [mM]a | ΔGf (kcal/mol) | ΔGint (kcal/mol)b |
|---|---|---|---|
| WT | 1.26 ± 0.02 | −3.08 ± 0.18 | |
| E979K | 1.29 ± 0.01 | −3.19 ± 0.16 | |
| F982L | 1.33 ± 0.02 | −3.27 ± 0.15 | |
| E979K/F982L | 1.41 ± 0.02 | −3.91 ± 0.17 | −0.53 ± 0.33 |
| M978L/V987L | 1.60 ± 0.02 | −4.46 ± 0.17 | |
| QM | 1.78 ± 0.03 | −5.04 ± 0.17 |
Abbreviations: Cm, concentration of Gdn-HCl at ΔGf = 0; f, folding; int, interaction; QM, quadruple mutant (M978L/E979K/F982L/V987L).
ΔGint = [ΔGf (WT) − ΔGf (mutant1)] − [ΔGf (mutant2) − ΔGf (mutant1,2)]. The error of individual measurements was estimated from triplicate measurements and propagated to obtain the error in coupling free energy.
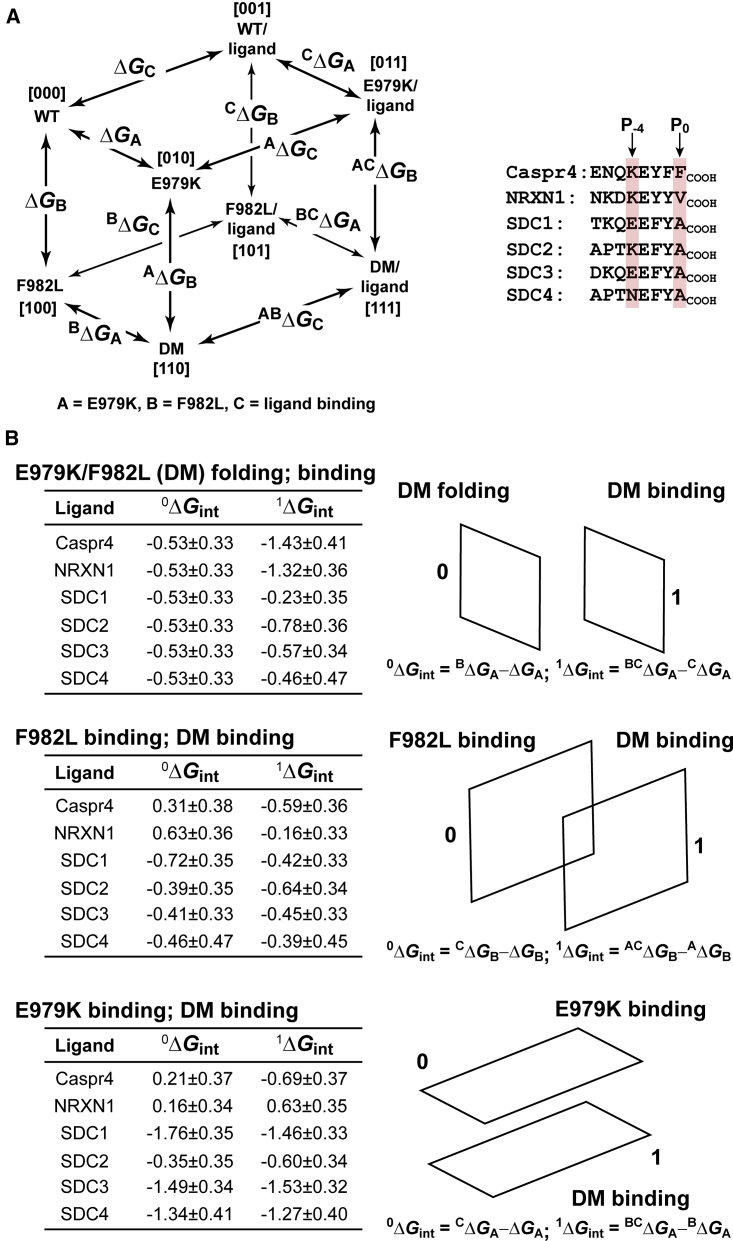
Coupling between the S0 and S-2 pockets from thermodynamic cube analyses
The binding and folding double mutant cycle analysis of Tiam2 PDZ E979K and F982L mutants provides evidence for the energetic coupling of these residues, indicating that the S0 and S-2 specificity pockets are energetically linked. Together, the two thermodynamic squares can be linked to form a thermodynamic cube, in which the folding and binding squares comprise two faces of the cube (29, 31, 32). Fig. 7 A shows a schematic for a general thermodynamic cube. In all, six cubes were constructed using the peptide ligands shown. Fig. 7 B depicts the transitions used to calculate the free energy couplings for each cube. The first transition (E979K/F982L (DM) folding; DM binding) showed a small coupling energy (0ΔGint) in folding between the side chains of E979K and F982L in the absence of any ligand. However, in the presence of the ligand, the two side chains (located in separate specificity pockets) were energetically coupled, depending on the identity of the ligand, with Caspr4 and NRXN1 having the largest couplings. The second transition (F982L binding and DM binding) describes the effect of the F982L mutation on ligand binding and how E979K modulates this interaction. Generally, F982L was weakly or not coupled to ligand binding and the E979K mutation had little or no influence on this process. The third transition (E979K binding and DM binding) describes the effect of the E979K mutant on ligand binding and how the F982L mutation affects this process. Both the single E979K mutation and the double mutant were highly energetically coupled to ligand binding for SDC1, 3, and 4. In contrast, the SDC2, Caspr4, and NRXN1 ligands were not coupled. These data conclusively show that residues E979K and F982L, located in separate specificity pockets, are energetically coupled to ligand binding.
Figure 7.
Pairwise free energy couplings for a three-dimensional cycle between Tiam2 E979K, F982L, and ligand binding. (A) Schematic of the three-dimensional thermodynamic cycle is shown. The peptide ligands used in the analysis are shown with two positions (P0 and P-4) highlighted. (B) Pairwise free energy couplings for three transitions described in the text are depicted schematically. The coupling energy (0 or 1ΔGint) is shown for the 0 and 1 planes, respectively. The error in free energy couplings was obtained by propagating the errors obtained from binding experiments for each PDZ/ligand pair. To see this figure in color, go online.
A third specificity pocket is required for the Tiam2 PDZ/SDC1 phosphorylation interaction
SDC-PDZ protein interactions and downstream signaling are regulated by SDC1 phosphorylation (pSDC1) at P-1 tyrosine (P-1 Tyr) through differential binding to PDZ proteins (i.e., Syntenin binds SDC1 but not pSDC1, whereas Tiam1 and CASK PDZs bind both SDC1 and pSDC1) (1, 13, 33, 34, 35). The Tiam1 PDZ domain interacts with the phosphate moiety of pSDC1 through an electrostatic interaction with the side chain of residue K879 (13). In contrast, the Tiam2 WT PDZ domain does not bind pSDC1 (Table 5). Based on the homology model of Tiam2 WT PDZ domain, residue L946 is located in the analogous position of K879 (S-1 pocket) in the Tiam1 PDZ domain (Fig. 1). Thus, we created the L946K mutation in the Tiam2 PDZ domain to test its ability to bind the pSDC1 peptide. The L946K mutant showed an improved affinity for pSDC1, suggesting a key role for the S-1 pocket in the pSDC1 recognition (Table 5). Although mutations at S0 and S-2 pockets (F982L and E979K) can switch SDC family ligand-binding specificity, only E979K was capable of binding the pSDC1 peptide (Table 5). However, the E979K/F982L double mutant had a Kd = 90 μM, indicating an enhanced effect when mutating F982L in the context of E979K. The QM also bound to pSDC1 but weaker than the E979K/F982L double mutant and similar to the affinity of E979K (Table 5). These results indicate that the three specificity pockets (S0, S-1, and S-2) play a role in supporting phosphorylated SDC family ligand interactions.
Table 5.
Tiam2 PDZ Domain Interactions with pSDC1
| Tiam2 PDZ Domain | Kd [μM] |
|---|---|
| WT | NBa,b |
| L946K | 104 ± 14 |
| E979K | 172 ± 26 |
| F982L | NB |
| E979K/F982L | 90 ± 17 |
| QM | 164 ± 18 |
Data taken from Shepherd et al. (7).
NB, no binding detected. The error for each parameter was estimated from triplicate measurements.
Discussion
Protein-protein and protein-peptide interactions are critical for all facets of signal transduction. The structure of hundreds of protein complexes provides structural detail of these interactions. However, far less is known about the thermodynamics of these complexes and how interactions throughout the binding site depend on each other—i.e., cooperativity. Although it is well recognized that proteins are dynamic, the impact of dynamics on molecular recognition has received much less attention. In this study, we used the Tiam2 PDZ domain as a model system to probe the relationship between dynamics, thermodynamics, and ligand-binding specificity. Here, we used the Tiam2 QM PDZ to gain insight into the dynamics, thermodynamics, and specificity of protein-ligand interactions. The central theme that emerges indicates that energetic cooperativity between residues in subpockets is fundamental for ligand specificity.
The P-4 ligand position governs SDC family specificity in Tiam family PDZ domains
We previously reported that the Tiam1 WT PDZ domain has specificity for a subset of SDC family members (13). The binding between the Tiam1 WT PDZ domain and SDC1 and SDC3 peptides was relatively high (Kd = 29 and 33 μM, respectively), ∼20-fold tighter than that with SDC2 and SDC4. The crystal structure of the Tiam1 WT PDZ/SDC1 complex revealed an electrostatic interaction between K912 in the PDZ domain and P-4 Glu of the ligand. This electrostatic interaction is also present with SDC3 but becomes repulsive for SDC2 and SDC4 interactions. Thus, this electrostatic interaction is proposed to be a significant molecular determinant for the SDC family specificity. This notion is further supported by the binding experiments with the Tiam1 QM PDZ, which contains the K912E mutation and showed a switched specificity for SDC family peptides in which the SDC2 peptide bound with the highest affinity (Kd = 100 μM) (14) similar to the Tiam2 WT PDZ domain (Kd = 74.6 μM). However, the specificity of the Tiam2 QM PDZ (with E979K) was switched such that SDC1 and SDC3 were preferred (Kd = 6.9 and 13.1 μM, respectively) compared to SDC2 (Kd = 22.3 μM). This specificity switch was also found in the E979K single mutant of Tiam2 PDZ but not the F982L Tiam2 PDZ (Table 3). Thus, the origin of the SDC family specificity switch in the Tiam2 QM PDZ domain is the E979K mutation. Combined with our previous results, the electrostatic attraction/repulsion between residue E979 in the Tiam2 PDZ (residue K912 in Tiam1 PDZ domain) and the identity of the residue at P-4 in SDC isoforms largely dictates ligand specificity. Like the interaction between the microtubule-associated serine-threonine kinases 2 and the rabies virus envelope glycoprotein (36), this example illustrates the capacity of a single residue change to modulate ligand specificity in a PDZ domain.
pSDC1 binding is regulated by three individual pockets
As expected, mutating L946 to Lys (L946K) resulted in significant improvement of pSDC1 binding. This result indicates that maintaining the electrostatic interaction between the S-1 pocket and the phosphoryl group is critical for pSDC1 binding. Moreover, the E979K mutant also had improved binding (Table 5). The enhanced binding of E979K suggests that the electrostatic interaction with Glu P-4 is also important. Our previous structural results with the Tiam1 QM PDZ/ligand complexes indicated that the volume of the S0 pocket is a stringent requirement for specificity. The increased S0 pocket size due to conformational changes surrounding the L915F Tiam1 PDZ mutation decreased the binding affinity with SDC members (14). Therefore, we tested the effect of the analogous mutation in the Tiam2 PDZ domain (F982L). The binding data showed that the F982L mutation alone did not improve the binding until it was coupled with E979K (Table 5). This finding suggests that reducing the S0 pocket size is necessary for optimized recognition of the pSDC1 ligand and implies cooperativity between these two residues. Interestingly, the Tiam2 QM (E979K/F982L/M978L/V987L) PDZ interacted with pSDC1 weaker than with the E979K/F982L mutant. Thus, in the context of the Tiam2 QM PDZ, the M978L/V987L double mutation opposes ligand binding. Together, these data provide evidence for the importance of S-1 (L946K), S-2 (E979K), and S0 (F982L) specificity subpockets in accommodating pSDC1 binding. More generally, these results indicate the need for cooperativity in binding pSDC1.
Interpocket cooperativity
Double mutant cycle analysis has been widely used for studying energetic cooperativity of sites in proteins, including PDZ domains (7, 15, 29, 37, 38). In a previous study with the Tiam1 WT PDZ domain, we showed that residues within a given specificity subpocket displayed cooperativity in binding peptide ligands (7). This “intrapocket” cooperativity occurred in both the S0 and S-2 subpockets and in a ligand-dependent manner. However, the cooperativity between these two subpockets (“interpocket cooperativity”) remains uncharacterized. Here we examined the thermodynamic coupling between residues from these subpockets (E979K in S-2 and F982L in S0) separated by ∼6 Å in the Tiam2 PDZ domain. Our pairwise analysis verified the nonadditivity (i.e., cooperativity) between the two side chains (and subpockets) with respect to ligand binding and folding. Moreover, the binding cooperativity was ligand dependent. In binding to SDC1, the free energy changes from WT to E979K/F982L came predominantly from E979K. The electrostatic interaction between SDC1 P-4 Glu and E979K mutant made the binding more favorable. The F982L mutant also preferred SDC1, likely because a reduced S0 pocket better accommodates the P0 Ala residue. Therefore, both the E979K and F982L mutations lead to a more favorable binding of SDC1 compared to Tiam2 WT PDZ domain.
NRXN1 and Caspr4 have a Lys at position P-4 (Fig. 2 B), and this residue predicts that the binding of these peptides to the Tiam2 E979K mutant would be less favorable than binding to the Tiam2 WT PDZ domain. Our results support this prediction, but the reduction in affinity was relatively small (Table 3). The F982L mutation is predicted to reduce the size of the S0 pocket, which would favor ligands with smaller P0 side chains (e.g., Ala) and disfavor ligands with larger P0 side chains (e.g., Phe and Val). Indeed, when compared to the Tiam2 WT PDZ, the Tiam2 F982L mutant bound all the SDC family members more favorably and bound NRXN1 and Caspr4 less favorably. The double mutant (E979K/F982L) bound all the ligands tighter than the Tiam2 WT PDZ. In addition, this double mutant had the largest energetic couplings when binding NRXN1 and Caspr4, despite these peptides having large side chains at P0. One possibility is that unfavorable changes in the S0 pocket and unfavorable electrostatics between Lys P-4 and E979K are compensated by a favorable electrostatic interaction between the Glu P-3 and E979K residue.
The results of the thermodynamic cube analyses reinforce and extend the observations made by the pairwise double mutant cycles. In contrast to the pairwise analyses, the cube analyses provide information about the energetic coupling between three perturbations: E979K, F982L, and ligand binding. Consistent with the pairwise analyses, the cube analyses found that the energetic coupling between E979K and F982L was dependent upon ligand binding and modulated by the identity of the ligand. Moreover, the degree of coupling was dependent on the identity of the residue in the ligand at the P0 and P-4 positions. This is best shown by the E979K binding and DM binding transitions (Fig. 7 B), in which the degree of energetic coupling was clearly ligand dependent and occurred in the presence of the E979K and E979K/F982L mutations. The E979K mutation and ligand binding were highly energetically coupled for SDC1, 3, and 4. SDC1 and 3 have a Glu at P-4, whereas SDC4 has an Asn at P-4. The E979K mutation provides a complementary charge-charge interaction for SDC1 and SDC3 and a potential hydrogen bonding partner with Asn in SDC4, all of which favor a gain in binding affinity. In contrast, the SDC2, Caspr4, and NRXN1 ligands were not coupled. Notably, they have Lys at P-4 that could lead to a charge-charge repulsion. Including the F982L mutation led to some degree of energetic coupling for all the ligands, with SDC1, 3, and 4 showing the largest couplings. This result is consistent with the idea that the F982L reshapes the S0 pocket to better accommodate the smaller P0 Ala side chain of SDC ligands. Together, the thermodynamic cube analyses clearly show that the S0 and S-2 specificity subpockets are energetically coupled in the context of ligand binding, despite being separated by ∼6 Å. The structural rationale for these results awaits the determination of high-resolution WT and mutant Tiam2 PDZ domain complexes.
Microsecond-millisecond timescale dynamics, ligand specificity, allostery and, statistical co-evolution
Exchange-broadened resonances in 1H-15N-HSQC spectra indicated that both the WT and QM Tiam2 PDZ domains have slow timescale dynamics. To quantify these dynamics, we carried out CPMG-based relaxation dispersion experiments. The results from these experiments revealed that several residues in the WT and QM PDZ domains had a chemical exchange (Rex). Importantly, the QM PDZ had a marked increase in the number of residues possessing Rex and a broad distribution of rates of exchange (kex). As indicated in Fig. 4, the residues with chemical exchange in the WT PDZ were limited to the α2 helix and part of the β2-β3 loop. Moreover, regions near the β4-α2 and most of the β2-β3 loops were exchange broadened. In contrast, the QM PDZ domain had a significant number of residues experiencing Rex beyond those regions observed for the WT PDZ. Specifically, regions in the α2 helix, the β1-β2 loop, β2 strand, and α1 helix had dynamic motions on a μs-ms timescale, which were not observed in the WT. Among these residues, F923 and F925 are in the β1-β2 loop and participate in forming the S0 pocket (Fig. 1). The change in protein dynamics at these positions is reminiscent of the analogous residues Y858 and F860 in the Tiam1 PDZ domain, which play crucial roles in propagating dynamics. Previously, we proposed that Y858 and F860 are “hub” residues that communicate/transmit dynamics within the Tiam1 WT PDZ (13). In the Tiam1 QM PDZ, F860 was found to have perturbed chemical shifts, further supporting a role of this residue in propagating dynamics distally. In the Tiam2 QM PDZ, F923 was completely exchange broadened, and F925 had a significant chemical shift perturbation relative to the WT. These results implicate a general role of these two hub residues in supporting the intradomain communication (i.e., allostery) between α2 helix and distal regions, including the β1-β2 loop, α1 helix, and β2 stand. Changes in dynamics in the β2 strand and β2-β3 loop could also be mediated by interactions between the side chains of mutant residues and the β4-α2 loop. Side chains from the β4-α2 loop help form the hydrophobic core of the protein, and they became dynamic after mutation (Fig. S1). Taken together, mutations in the α2 helix resulted in more dynamic motions that extend beyond the mutations to distal sites via the two hub residues and buried side chain interactions.
Interestingly, the mutation site and many of the dynamically perturbed residues constitute the ligand-binding groove. Analysis of the Tiam2 QM CPMG data highlighted the enhanced conformational exchange in these regions. Importantly, binding data also shows that the Tiam2 QM PDZ domain has a broader ligand-binding specificity—it interacted with Caspr4, NRXN1, and most of the SDC family members with relatively high affinity (Fig. 2). We can speculate that the enhanced dynamics in the QM PDZ domain might contribute to the broader specificity by promoting multiple conformations in the apo state, which could be selected by a variety of ligands (39). This result is similar to what was observed in the Tiam1 PDZ domain, in which the QM mutation induced a chemical exchange in the protein that otherwise had none (14). In that study, the Tiam1 QM PDZ domain was dynamic in both fast and slow timescales. The fast timescale side chain methyl order parameters were used as a proxy for the entropic contribution to ligand binding (14, 40, 41), whereas the relaxation dispersion experiments provided insight into conformational switching of the mutant. In this study, the Tiam2 QM PDZ is found more dynamic than the WT on the slow timescale, and the ITC data suggest that changes in entropy may also be important. Our results are also consistent with a correlation between conformational dynamics and specificity observed in other PDZ domains (14, 42, 43). Simply stated, re-engineering of PDZ ligand specificity is associated with manipulating protein conformational dynamics and specificity, which has been described as a key parameter for the evolution of proteins to gain new functionality (44).
PDZ domains have been the intense subject of amino acid co-conservation and bioinformatic studies since the initial studies by Ranganathan and colleagues (45, 46, 47, 48). These studies have sought to identify functionally important residues from multiple sequence alignments and statistical co-evolution. Indeed, Ranganathan and colleagues have proposed the sector concept whereby a network of co-evolved residues are energetically and functionally coupled (49). A more recent study using a novel “deep coupling scan” method that combines large-scale mutagenesis with a bacterial two-hybrid assay—a high throughput version of thermodynamic double mutant cycle—uncovered apparent energetic couplings between several positions along helix α2 in five members of the PDZ domain family (50). Specifically, it was shown that a network of residues, including those at analogous positions of F982, E979, and M978 in the Tiam2 QM PDZ domain studied here, shared a pattern of couplings across the binding interface. Our thermodynamic analyses on the Tiam2 PDZ domain are consistent with these couplings and provide an independent validation of the deep coupling scan methodology. Together, these data suggest that a common network of residues are inextricably linked to ligand binding and specificity. Previous NMR dynamics studies with PDZ domains showed that many of these sector residues have perturbed dynamics upon ligand binding, suggesting a link between sector communication and ligand binding—i.e., allostery (13, 51, 52, 53). Here, we show that mutating sector positions have important consequences for protein dynamics and binding specificity.
In passing, we note that our strategy for re-engineering new PDZ/ligand interactions may be general because it has been successful for both the Tiam1 and Tiam2 PDZ domains; however, further work will be required to confirm this. Future applications may include using our approach to re-engineering PDZ domains to probe the specificity of signal transduction pathways.
Conclusions
We present a comprehensive study concerning the role of thermodynamics and dynamics in ligand binding of the Tiam2 QM PDZ domain. This mutant was engineered by substituting four residues in two ligand specificity subpockets in the Tiam2 PDZ with their counterparts in the Tiam1 PDZ domain. The Tiam2 QM PDZ had switched ligand-binding specificity compared to that of the Tiam1 and Tiam2 PDZ domains. Importantly, the Tiam2 QM PDZ domain was significantly more dynamic than the WT. Thermodynamic binding analyses found distinct enthalpic and entropic contributions along the mutation trajectory. Further analyses revealed that the E979K/F982L mutations altered the ligand-binding specificity by modulating the size of the S0 subpocket and an electrostatic interaction in the S-2 subpocket. In addition, we found energetic coupling between the E979K and F982L side chains that was ligand dependent, providing insight into the energetics of specificity. Together, these data show that distinct thermodynamic and dynamic features in the Tiam2 QM PDZ work cooperatively to drive the altered ligand-binding specificity.
Author Contributions
X.L. and E.J.F. designed the research, analyzed the data, and wrote the manuscript. X.L., E.J.F., L.C.G., J.A.L., T.R.S., and L.Y performed the research.
Acknowledgments
The authors thank members of the Fuentes lab for helpful discussions and comments on the manuscript. We are grateful to C. Andrew Fowler in College of Medicine NMR Facility for assistance with NMR experiments.
X.L. and E.J.F. were supported by the American Heart Association (Predoctoral Fellowship (E155500) to X.L. and Scientist Development Grant (0835261N) to E.J.F.). This research was supported in part by National Institutes of Health grant (R21 AI135305) to E.J.F. and Predoctoral Fellowship (T32 GM008365) to T.R.S.
Editor: James Cole.
Footnotes
Xu Liu’s present address is Department of Biochemistry, Emory University, Atlanta, Georgia.
Lisa C. Golden’s present address is Department of Neurology, University of California Los Angeles, Los Angeles, California.
Josue A. Lopez’s present address is Department of Molecular Physiology and Biophysics, University of Iowa, Iowa City, Iowa.
Tyson R. Shepherd’s present address is Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.05.008.
Supporting Material
References
- 1.Liu X., Fuentes E.J. Emerging themes in PDZ domain signaling: structure, function, and inhibition. Int. Rev. Cell Mol. Biol. 2019;343:129–218. doi: 10.1016/bs.ircmb.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, X., and E. J. Fuentes. 2019. Emerging themes in PDZ domain signaling: structure, function, and inhibition. Int. Rev. Cell Mol. Biol. 343:129-218. [DOI] [PMC free article] [PubMed]
- 2.Ernst A., Appleton B.A., Sidhu S.S. A structural portrait of the PDZ domain family. J. Mol. Biol. 2014;426:3509–3519. doi: 10.1016/j.jmb.2014.08.012. [DOI] [PubMed] [Google Scholar]; Ernst, A., B. A. Appleton, …, S. S. Sidhu. 2014. A structural portrait of the PDZ domain family. J. Mol. Biol. 426:3509-3519. [DOI] [PubMed]
- 3.Lee H.J., Zheng J.J. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, H. J., and J. J. Zheng. 2010. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 8:8. [DOI] [PMC free article] [PubMed]
- 4.Ehler E., van Leeuwen F., Salinas P.C. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol. Cell. Neurosci. 1997;9:1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]; Ehler, E., F. van Leeuwen, …, P. C. Salinas. 1997. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol. Cell. Neurosci. 9:1-12. [DOI] [PubMed]
- 5.Matsuo N., Hoshino M., Nabeshima Y. Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J. Biol. Chem. 2002;277:2860–2868. doi: 10.1074/jbc.M106186200. [DOI] [PubMed] [Google Scholar]; Matsuo, N., M. Hoshino, …, Y. Nabeshima. 2002. Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J. Biol. Chem. 277:2860-2868. [DOI] [PubMed]
- 6.Tolias K.F., Bikoff J.B., Greenberg M.E. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc. Natl. Acad. Sci. USA. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tolias, K. F., J. B. Bikoff, …, M. E. Greenberg. 2007. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc. Natl. Acad. Sci. USA. 104:7265-7270. [DOI] [PMC free article] [PubMed]
- 7.Shepherd T.R., Hard R.L., Fuentes E.J. Distinct ligand specificity of the Tiam1 and Tiam2 PDZ domains. Biochemistry. 2011;50:1296–1308. doi: 10.1021/bi1013613. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shepherd, T. R., R. L. Hard, …, E. J. Fuentes. 2011. Distinct ligand specificity of the Tiam1 and Tiam2 PDZ domains. Biochemistry. 50:1296-1308. [DOI] [PMC free article] [PubMed]
- 8.Shepherd T.R., Klaus S.M., Fuentes E.J. The Tiam1 PDZ domain couples to Syndecan1 and promotes cell-matrix adhesion. J. Mol. Biol. 2010;398:730–746. doi: 10.1016/j.jmb.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shepherd, T. R., S. M. Klaus, …, E. J. Fuentes. 2010. The Tiam1 PDZ domain couples to Syndecan1 and promotes cell-matrix adhesion. J. Mol. Biol. 398:730-746. [DOI] [PMC free article] [PubMed]
- 9.Masuda M., Maruyama T., Murakami Y. CADM1 interacts with Tiam1 and promotes invasive phenotype of human T-cell leukemia virus type I-transformed cells and adult T-cell leukemia cells. J. Biol. Chem. 2010;285:15511–15522. doi: 10.1074/jbc.M109.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]; Masuda, M., T. Maruyama, …, Y. Murakami. 2010. CADM1 interacts with Tiam1 and promotes invasive phenotype of human T-cell leukemia virus type I-transformed cells and adult T-cell leukemia cells. J. Biol. Chem. 285:15511-15522. [DOI] [PMC free article] [PubMed]
- 10.Babault N., Cordier F., Wolff N. Peptides targeting the PDZ domain of PTPN4 are efficient inducers of glioblastoma cell death. Structure. 2011;19:1518–1524. doi: 10.1016/j.str.2011.07.007. [DOI] [PubMed] [Google Scholar]; Babault, N., F. Cordier, …, N. Wolff. 2011. Peptides targeting the PDZ domain of PTPN4 are efficient inducers of glioblastoma cell death. Structure. 19:1518-1524. [DOI] [PubMed]
- 11.Spiegel I., Salomon D., Peles E. Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol. Cell. Neurosci. 2002;20:283–297. doi: 10.1006/mcne.2002.1110. [DOI] [PubMed] [Google Scholar]; Spiegel, I., D. Salomon, …, E. Peles. 2002. Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol. Cell. Neurosci. 20:283-297. [DOI] [PubMed]
- 12.Südhof T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sudhof, T. C. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903-911. [DOI] [PMC free article] [PubMed]
- 13.Liu X., Shepherd T.R., Fuentes E.J. The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics. Structure. 2013;21:342–354. doi: 10.1016/j.str.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, X., T. R. Shepherd, …, E. J. Fuentes. 2013. The structure of the Tiam1 PDZ domain/ phospho-syndecan1 complex reveals a ligand conformation that modulates protein dynamics. Structure. 21:342-354. [DOI] [PMC free article] [PubMed]
- 14.Liu X., Speckhard D.C., Fuentes E.J. Distinct roles for conformational dynamics in protein-ligand interactions. Structure. 2016;24:2053–2066. doi: 10.1016/j.str.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, X., D. C. Speckhard, …, E. J. Fuentes. 2016. Distinct roles for conformational dynamics in protein-ligand interactions. Structure. 24:2053-2066. [DOI] [PMC free article] [PubMed]
- 15.Shepherd T.R., Fuentes E.J. Structural and thermodynamic analysis of PDZ-ligand interactions. Methods Enzymol. 2011;488:81–100. doi: 10.1016/B978-0-12-381268-1.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shepherd, T. R., and E. J. Fuentes. 2011. Structural and thermodynamic analysis of PDZ-ligand interactions. Methods Enzymol. 488:81-100. [DOI] [PMC free article] [PubMed]
- 16.Mulder F.A., Skrynnikov N.R., Kay L.E. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]; Mulder, F. A., N. R. Skrynnikov, …, L. E. Kay. 2001. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 123:967-975. [DOI] [PubMed]
- 17.Loria J.P., Rance M., Palmer A.G. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]; Loria, J. P., M. Rance, and A. G. Palmer. 1999. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121:2331-2332.
- 18.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]; Delaglio, F., S. Grzesiek, …, A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277-293. [DOI] [PubMed]
- 19.Johnson B.A., Blevins R.A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]; Johnson, B. A., and R. A. Blevins. 1994. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 4:603-614. [DOI] [PubMed]
- 20.Goddard T.D., Kneller D.G. Univeristy of California; San Fransico, CA: 2007. SPARKY 3. [Google Scholar]; Goddard, T. D., and D. G. Kneller. 2007. SPARKY 3: Univeristy of California, San Fransico, CA.
- 21.Carver J.P., Richards R.E. General two-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J. Magn. Reson. 1972;6:89–105. [Google Scholar]; Carver, J. P., and R. E. Richards. 1972. General two-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J. Magn. Reson. 6:89-105.
- 22.Mauldin R.V., Carroll M.J., Lee A.L. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mauldin, R. V., M. J. Carroll, and A. L. Lee. 2009. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 17:386-394. [DOI] [PMC free article] [PubMed]
- 23.McDonald L.R., Boyer J.A., Lee A.L. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure. 2012;20:1363–1373. doi: 10.1016/j.str.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; McDonald, L. R., J. A. Boyer, and A. L. Lee. 2012. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure. 20:1363-1373. [DOI] [PMC free article] [PubMed]
- 24.Mulder F.A., Mittermaier A., Kay L.E. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]; Mulder, F. A., A. Mittermaier, …, L. E. Kay. 2001. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 8:932-935. [DOI] [PubMed]
- 25.Pace C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]; Pace, C. N. 1986. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131:266-280. [DOI] [PubMed]
- 26.Nozaki Y. The preparation of guanidine hydrochloride. Methods Enzymol. 1972;26:43–50. doi: 10.1016/s0076-6879(72)26005-0. [DOI] [PubMed] [Google Scholar]; Nozaki, Y. 1972. The preparation of guanidine hydrochloride. Methods Enzymol. 26:43-50. [DOI] [PubMed]
- 27.Walters J., Milam S.L., Clark A.C. Practical approaches to protein folding and assembly: spectroscopic strategies in thermodynamics and kinetics. Methods Enzymol. 2009;455:1–39. doi: 10.1016/S0076-6879(08)04201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Walters, J., S. L. Milam, and A. C. Clark. 2009. Practical approaches to protein folding and assembly: spectroscopic strategies in thermodynamics and kinetics. Methods Enzymol. 455:1-39. [DOI] [PMC free article] [PubMed]
- 28.Arnold K., Bordoli L., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]; Arnold, K., L. Bordoli, …, T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22:195-201. [DOI] [PubMed]
- 29.Horovitz A., Fersht A.R. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J. Mol. Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]; Horovitz, A., and A. R. Fersht. 1990. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J. Mol. Biol. 214:613-617. [DOI] [PubMed]
- 30.Ackers G.K., Smith F.R. Effects of site-specific amino acid modification on protein interactions and biological function. Annu. Rev. Biochem. 1985;54:597–629. doi: 10.1146/annurev.bi.54.070185.003121. [DOI] [PubMed] [Google Scholar]; Ackers, G. K., and F. R. Smith. 1985. Effects of site-specific amino acid modification on protein interactions and biological function. Annu. Rev. Biochem. 54:597-629. [DOI] [PubMed]
- 31.Fersht A.R. W. H. Freeman and Company; New York: 1998. Structure and Mechanism in Protein Science. [Google Scholar]; Fersht, A. R. 1998. Structure and Mechanism in Protein Science: W. H. Freeman and Company, New York.
- 32.di Cera E. Site-specific analysis of mutational effects in proteins. Adv. Protein Chem. 1998;51:59–119. doi: 10.1016/s0065-3233(08)60651-8. [DOI] [PubMed] [Google Scholar]; di Cera, E. 1998. Site-specific analysis of mutational effects in proteins. Adv. Protein Chem. 51:59-119. [DOI] [PubMed]
- 33.Sulka B., Lortat-Jacob H., Rousselle P. Tyrosine dephosphorylation of the syndecan-1 PDZ binding domain regulates syntenin-1 recruitment. J. Biol. Chem. 2009;284:10659–10671. doi: 10.1074/jbc.M807643200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sulka, B., H. Lortat-Jacob, …, P. Rousselle. 2009. Tyrosine dephosphorylation of the syndecan-1 PDZ binding domain regulates syntenin-1 recruitment. J. Biol. Chem. 284:10659-10671. [DOI] [PMC free article] [PubMed]
- 34.Ott V.L., Rapraeger A.C. Tyrosine phosphorylation of syndecan-1 and -4 cytoplasmic domains in adherent B82 fibroblasts. J. Biol. Chem. 1998;273:35291–35298. doi: 10.1074/jbc.273.52.35291. [DOI] [PubMed] [Google Scholar]; Ott, V. L., and A. C. Rapraeger. 1998. Tyrosine phosphorylation of syndecan-1 and -4 cytoplasmic domains in adherent B82 fibroblasts. J. Biol. Chem. 273:35291-35298. [DOI] [PubMed]
- 35.Cheng B., Montmasson M., Rousselle P. Syndecans as cell surface receptors in cancer biology. A focus on their interaction with PDZ domain proteins. Front. Pharmacol. 2016;7:10. doi: 10.3389/fphar.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheng, B., M. Montmasson, …, P. Rousselle. 2016. Syndecans as cell surface receptors in cancer biology. A focus on their interaction with PDZ domain proteins. Front. Pharmacol. 7:10. [DOI] [PMC free article] [PubMed]
- 36.Préhaud C., Wolff N., Lafon M. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci. Signal. 2010;3:ra5. doi: 10.1126/scisignal.2000510. [DOI] [PubMed] [Google Scholar]; Prehaud, C., N. Wolff, …, M. Lafon. 2010. Attenuation of rabies virulence: takeover by the cytoplasmic domain of its envelope protein. Sci. Signal. 3:ra5. [DOI] [PubMed]
- 37.Saro D., Li T., Spaller M.R. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 2007;46:6340–6352. doi: 10.1021/bi062088k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Saro, D., T. Li, …, M. R. Spaller. 2007. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 46:6340-6352. [DOI] [PMC free article] [PubMed]
- 38.Carter P.J., Winter G., Fersht A.R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]; Carter, P. J., G. Winter, …, A. R. Fersht. 1984. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 38:835-840. [DOI] [PubMed]
- 39.Kastritis P.L., Bonvin A.M. Molecular origins of binding affinity: seeking the Archimedean point. Curr. Opin. Struct. Biol. 2013;23:868–877. doi: 10.1016/j.sbi.2013.07.001. [DOI] [PubMed] [Google Scholar]; Kastritis, P. L., and A. M. Bonvin. 2013. Molecular origins of binding affinity: seeking the Archimedean point. Curr. Opin. Struct. Biol. 23:868-877. [DOI] [PubMed]
- 40.Frederick K.K., Marlow M.S., Wand A.J. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]; Frederick, K. K., M. S. Marlow, …, A. J. Wand. 2007. Conformational entropy in molecular recognition by proteins. Nature. 448:325-329. [DOI] [PMC free article] [PubMed]
- 41.Kasinath V., Sharp K.A., Wand A.J. Microscopic insights into the NMR relaxation-based protein conformational entropy meter. J. Am. Chem. Soc. 2013;135:15092–15100. doi: 10.1021/ja405200u. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kasinath, V., K. A. Sharp, and A. J. Wand. 2013. Microscopic insights into the NMR relaxation-based protein conformational entropy meter. J. Am. Chem. Soc. 135:15092-15100. [DOI] [PMC free article] [PubMed]
- 42.Murciano-Calles J., McLaughlin M.E., Sidhu S.S. Alteration of the C-terminal ligand specificity of the erbin PDZ domain by allosteric mutational effects. J. Mol. Biol. 2014;426:3500–3508. doi: 10.1016/j.jmb.2014.05.003. [DOI] [PubMed] [Google Scholar]; Murciano-Calles, J., M. E. McLaughlin, …, S. S. Sidhu. 2014. Alteration of the C-terminal ligand specificity of the erbin PDZ domain by allosteric mutational effects. J. Mol. Biol. 426:3500-3508. [DOI] [PubMed]
- 43.Münz M., Hein J., Biggin P.C. The role of flexibility and conformational selection in the binding promiscuity of PDZ domains. PLoS Comput. Biol. 2012;8:e1002749. doi: 10.1371/journal.pcbi.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]; Munz, M., J. Hein, and P. C. Biggin. 2012. The role of flexibility and conformational selection in the binding promiscuity of PDZ domains. PLoS Comput. Biol. 8:e1002749. [DOI] [PMC free article] [PubMed]
- 44.Tokuriki N., Tawfik D.S. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]; Tokuriki, N., and D. S. Tawfik. 2009. Protein dynamism and evolvability. Science. 324:203-207. [DOI] [PubMed]
- 45.Gautier C., Laursen L., Gianni S. Seeking allosteric networks in PDZ domains. Protein Eng. Des. Sel. 2018;31:367–373. doi: 10.1093/protein/gzy033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gautier, C., L. Laursen, …, S. Gianni. 2018. Seeking allosteric networks in PDZ domains. Protein Eng. Des. Sel. 31:367-373. [DOI] [PMC free article] [PubMed]
- 46.Morcos F., Pagnani A., Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA. 2011;108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Morcos, F., A. Pagnani, …, M. Weigt. 2011. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA. 108:E1293-E1301. [DOI] [PMC free article] [PubMed]
- 47.Morcos F., Schafer N.P., Wolynes P.G. Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. Proc. Natl. Acad. Sci. USA. 2014;111:12408–12413. doi: 10.1073/pnas.1413575111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Morcos, F., N. P. Schafer, …, P. G. Wolynes. 2014. Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. Proc. Natl. Acad. Sci. USA. 111:12408-12413. [DOI] [PMC free article] [PubMed]
- 48.Lockless S.W., Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]; Lockless, S. W., and R. Ranganathan. 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 286:295-299. [DOI] [PubMed]
- 49.Halabi N., Rivoire O., Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; Halabi, N., O. Rivoire, …, R. Ranganathan. 2009. Protein sectors: evolutionary units of three-dimensional structure. Cell. 138:774-786. [DOI] [PMC free article] [PubMed]
- 50.Salinas V.H., Ranganathan R. Coevolution-based inference of amino acid interactions underlying protein function. Elife. 2018;7:e34300. doi: 10.7554/eLife.34300. [DOI] [PMC free article] [PubMed] [Google Scholar]; Salinas, V. H., and R. Ranganathan. 2018. Coevolution-based inference of amino acid interactions underlying protein function. Elife. 7:e34300. [DOI] [PMC free article] [PubMed]
- 51.Fuentes E.J., Der C.J., Lee A.L. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]; Fuentes, E. J., C. J. Der, and A. L. Lee. 2004. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 335:1105-1115. [DOI] [PubMed]
- 52.Fuentes E.J., Gilmore S.A., Lee A.L. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J. Mol. Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]; Fuentes, E. J., S. A. Gilmore, …, A. L. Lee. 2006. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J. Mol. Biol. 364:337-351. [DOI] [PubMed]
- 53.Law A.B., Fuentes E.J., Lee A.L. Conservation of side-chain dynamics within a protein family. J. Am. Chem. Soc. 2009;131:6322–6323. doi: 10.1021/ja809915a. [DOI] [PMC free article] [PubMed] [Google Scholar]; Law, A. B., E. J. Fuentes, and A. L. Lee. 2009. Conservation of side-chain dynamics within a protein family. J. Am. Chem. Soc. 131:6322-6323. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.