Abstract
The initial binding of tropomyosin onto actin filaments and then its polymerization into continuous cables on the filament surface must be precisely tuned to overall thin-filament structure, function, and performance. Low-affinity interaction of tropomyosin with actin has to be sufficiently strong to localize the tropomyosin on actin, yet not so tight that regulatory movement on filaments is curtailed. Likewise, head-to-tail association of tropomyosin molecules must be favorable enough to promote tropomyosin cable formation but not so tenacious that polymerization precedes filament binding. Arguably, little molecular detail on early tropomyosin binding steps has been revealed since Wegner’s seminal studies on filament assembly almost 40 years ago. Thus, interpretation of mutation-based actin-tropomyosin binding anomalies leading to cardiomyopathies cannot be described fully. In vitro, tropomyosin binding is masked by explosive tropomyosin polymerization once cable formation is initiated on actin filaments. In contrast, in silico analysis, characterizing molecular dynamics simulations of single wild-type and mutant tropomyosin molecules on F-actin, is not complicated by tropomyosin polymerization at all. In fact, molecular dynamics performed here demonstrates that a midpiece tropomyosin domain is essential for normal actin-tropomyosin interaction and that this interaction is strictly conserved in a number of tropomyosin mutant species. Elsewhere along these mutant molecules, twisting and bending corrupts the tropomyosin superhelices as they “lose their grip” on F-actin. We propose that residual interactions displayed by these mutant tropomyosin structures with actin mimic ones that occur in early stages of thin-filament generation, as if the mutants are recapitulating the assembly process but in reverse. We conclude therefore that an initial binding step in tropomyosin assembly onto actin involves interaction of the essential centrally located domain.
Significance
Contacts between filamentous actin and tropomyosin facilitate thin-filament assembly, but plausible assembly mechanisms remain uncertain. We demonstrate that a single, centrally located tropomyosin domain is essential for normal actin-tropomyosin interaction and that this interaction is strictly conserved. Our premise is that this localized association occurs at the earliest stages of thin-filament generation. We expect that recognition of our work will lead to a more fundamental understanding of thin-filament assembly processes, ones that may fail in various cardiomyopathies and skeletal muscle diseases. Thus, we also expect that further consideration of our analysis will support new mechanistic paradigms for interpreting the role of tropomyosin in actin filament function and dysfunction.
Introduction
Coiled-coil tropomyosin is a modular protein consisting of seven pseudorepeats that contact seven successive actin subunits along thin filaments (1, 2). Reversible azimuthal repositioning of tropomyosin on actin under the influence of troponin, myosin, and Ca2+ is well documented and known to be instrumental in regulating muscle contraction and relaxation (3, 4, 5, 6, 7, 8, 9, 10). Inherent to this process is a tradeoff between tropomyosin’s binding specificity and its binding strength once linked to actin (11). In fact, single tropomyosin coiled-coil dimers interact with actin filaments extremely poorly, with dissociation constants (Kds) estimated to be in the millimolar range (12). It follows that tropomyosin only associates effectively on actin filaments once trapped head to tail with neighboring dimers as part of a polymerizing chain-like cable (2, 11). The growing cable then becomes confined topologically over the succeeding actin subunit substrate and biased to one azimuthal position or another by electrostatic interactions (13, 14).
Structural analysis combined with proteomic and/or bioinformatics approaches have provided very plausible atomic models of tropomyosin bound to actin (8, 15, 16, 17, 18, 19, 20, 21, 22). However, steps in the assembly of the filament, including initial contacts made between actin and tropomyosin, remain unclear. Most likely, the apparent binding constants frequently reported for tropomyosin and F-actin—based, for example, on cosedimentation—largely reflect nucleation and polymerization of tropomyosin into nascent cables on the actin filament substrate and not a true Kd of the tropomyosin molecule to F-actin. In turn, fluorescence light microscopy provides a means of following the polymerization process temporally but not at high resolution (23, 24, 25). Thus, characterizing the association of single tropomyosin molecules onto F-actin at a residue-residue level is challenging.
In principle, computationally directed steered docking of tropomyosin onto the F-actin surface can offer clues into the actin-tropomyosin assembly process (26). However, the number of choices involved in steering conformationally variable tropomyosin onto an actin model, to say nothing of scoring respective pathways, seemingly is limitless. Because a simple seven-actin-one-tropomyosin unit surrounded by associated water molecules contains up to 900,000 atoms, a thorough search of conformational space for an initial contact complex would be computationally impractical. Here, we describe a different and less daunting approach to computing likely early steps in the assembly of tropomyosin on actin filaments.
In the short story “All Rivers” by Amos Oz (27), the narrator suggests that “Memory distorts everything. Memory doesn’t move forward; it moves backward, from the end to the beginning.” Can our own experiments designed to move forward, from manipulation to effect and ideally to logical interpretation, also reveal a process lost in time that at present cannot be proven directly? In experiments performed in silico, we evaluate effects of selected point mutations on tropomyosin interaction with F-actin filaments. After observing the progressive destabilization of mutant tropomyosins bound to actin in molecular dynamics (MD) simulations, we conclude that this interaction process in effect recounts “lost” time by moving backward through the assembly pathway, memorializing initial events. The binding intermediates thus revealed provide a window to define likely initial events in the mechanism of thin-filament assembly.
We have previously carried out MD simulations on actin-tropomyosin complexes consisting of single dimeric αα-tropomyosin positioned on an 18-subunit actin filament segment and modeled as in Li et al. (19, 28). Periodic local electrostatic interactions were determined to be sufficiently well arranged to maintain tropomyosin on the filament at a ∼40 Å radius to the central filament axis. During MD, characteristic modulation of tropomyosin twist optimizes side-chain interactions with F-actin (29, 30, 31). In particular, this twisting aligns tropomyosin glutamate residues 139 and 142 very closely with oppositely charged Lys326, Lys328, and Arg147 on actin (29, 30). Coiled-coil twisting, midway along tropomyosin, relies on the presence of a strictly conserved and anomalously charged Asp137, located on tropomyosin in a position normally occupied by a nonpolar amino acid along an otherwise core hydrophobic stripe (29, 30) (Fig. 1).
Figure 1.
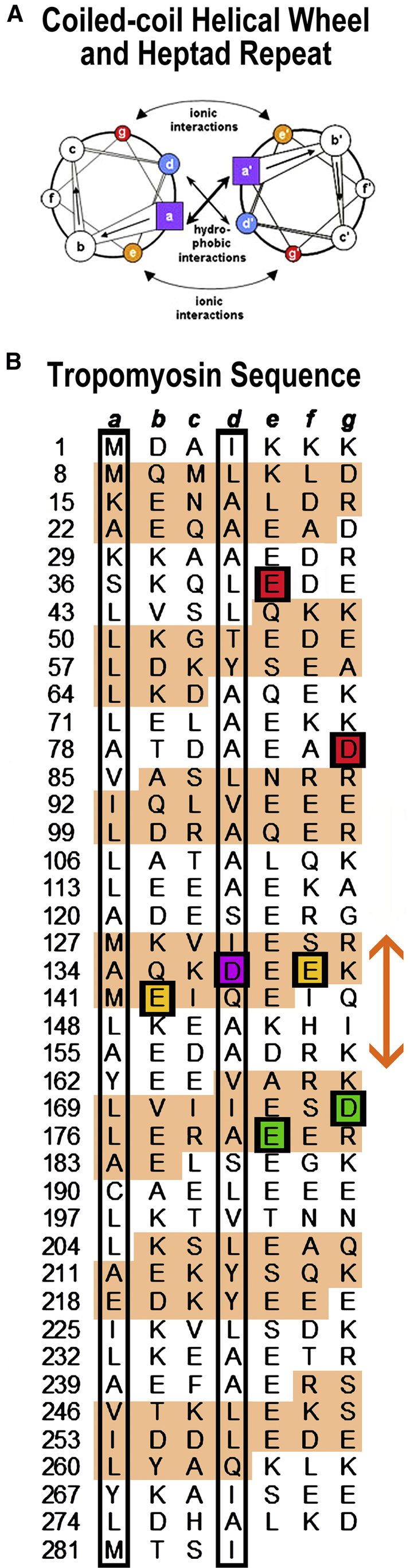
(A) Helical wheel diagram of a canonical dimeric coiled coil. Heptad positions are labeled a to g and a′ to g′ for the respective helices of the dimer; figure adapted from Hagemann et al. (75). (B) Striated muscle tropomyosin sequence annotation is shown; figure adapted and modified from Brown and Cohen (17) and Lehman et al. (29) to emphasize residues within heptad repeats (a, b, c, d, e, f, g) in each chain of the tropomyosin coiled-coil homodimer, with the first residue in each heptad repeat numbered. Residues in “α-zones” of each tropomyosin pseudorepeat, shaded beige, typically locate close to actin subdomains 1 and 3 on intact thin filaments, whereas those in “β-zones” (white background) bridge over the shallow aspect of actin subdomains 2 and 4 at radial distances to the filament surface that preclude tropomyosin-actin contact (17, 19). Generally, hydrophobic residues are localized at a and d residue positions (found within black borders in the diagram). Glutamate residues 139 and 142, which make unusually close contact with actin, are highlighted (gold) and are contained within the boundaries of the centrally located pseudorepeat 4 of tropomyosin (orange double arrow) (also see (18)). The anomalous d-position aspartate at residue 137 is highlighted in magenta. Residues 175 and 180 (green), when mutated, can result in HCM, and mutant residues at positions 40 and 84 (red) can lead to DCM. To see this figure in color, go online.
During MD, the association and position of tropomyosin on F-actin equilibrates quickly and is remarkably stable. No evidence suggests any local or global dissociation of any part of tropomyosin from actin as might, over time, be expected in vitro for a single tropomyosin coiled coil on F-actin (19). In these simulations, only the N- and C-terminal tips of tropomyosin lose intimate association with the actin surface, variably flailing around as if in search of a partner to make head-to-tail contact and participate in cable formation (19). Thus, over the 30–100 ns timescale of MD performed, no obvious information can be gleaned directly about any reversion to initial binding interactions. Hence, as we describe, no meaningful inferences emerge about the assembly process itself. However, in marked contrast, we show here that MD simulations of tropomyosin mutants linked to F-actin show strikingly different behavior. In many cases, the models progress from a canonically docked tropomyosin conformation on F-actin to converge on a poorly interacting actin-tropomyosin configuration, still distinguished by a common residual “core” subset of interacting residues on F-actin and tropomyosin. Barring significant hysteresis between the assembly and disassembly of tropomyosin on actin, we propose that these mutant tropomyosin structures mimic ones in early stages of thin-filament generation, as if the mutants are recapitulating the assembly process but in reverse. We regard the interactions that persist akin to intermediates assumed before the tropomyosin polymerization process occurred.
Materials and Methods
MD simulations were initiated on models built from atomic-resolution structures (19, 28). The MD performed here were designed to equilibrate single tropomyosin coiled-coil dimers on F-actin and thus emulate the behavior of the first tropomyosin that binds actin during thin filament assembly (19). We therefore did not model tropomyosin as a continuous cable, using periodic boundaries or other approaches, as in our most recent work (32, 33). We placed the Holmes-Lorenz canonical dimer model of tropomyosin (34), with side-chain conformations defined as previously described (28, 35), onto the surface of an F-actin model, locating the single tropomyosin over its energy-minimized position on F-actin, i.e., close to the blocked, B-state position, as defined in Li et al. (19). Amino-acid substitutions to characterize mutant tropomyosins were made in VMD (36), and the new homology structures were energy minimized, as previously (19). MD was run on these structures in explicit water for a minimum of 30 ns at 300 K and 1 atm using the program NAMD (37), as previously detailed (19). The average structures throughout MD were calculated in CHARMM (38). Local coiled-coil twisting and twist deviations from canonical models were measured using the program TWISTER (39) and VMD (36), as described in Lehman et al. (29). The observed twisting pattern for each particular tropomyosin variant equilibrated within 5–10 ns, but MD was performed at least 20–25 ns longer to validate this. To measure the tropomyosin position and its variance on the F-actin surface, we translated the Cartesian coordinates of the center of mass of each tropomyosin pseudorepeat to cylindrical coordinates. An average over the center-of-mass coordinates of seven tropomyosin pseudorepeats gives the average position of tropomyosin on F-actin in each MD frame. Azimuthal positioning and its variance stabilized within 10 ns. Molecular graphics was performed using VMD (36) and CHIMERA (40). Quantitative analyses of interaction energies during atomistic simulations were computed in CHARMM (38).
Results
Partial dissociation of D137L tropomyosin from F-actin
Coiled-coil dimers, like tropomyosin, are built from two amphipathic α-helices. In textbook examples, large nonpolar amino acids (leucine, isoleucine, and valine) occupy every first (“a-position”) and fourth (“d-position”) residue in a “heptad” pattern of seven residues repeating over and over along the two helical chains (Fig. 1, A and B). Hydrophobic bands formed by complementary a- and d-position residues in each of the chains zipper the two helices together into a coiled coil. However, coiled-coiled tropomyosins contain an anomalous polar aspartate-137 at a d-position and glutamate-218 at an a-position present along their otherwise hydrophobic stripe. These residues are conserved in tropomyosin variants across the animal kingdom and therefore are not phylogenetic flukes and, by definition, not sequence aberrations. On the contrary, numerous studies of D137 show its effects on thin-filament activity (41, 42, 43, 44, 45), as well as its role in localized tropomyosin twisting and superhelical bending for tropomyosin to fit properly on the actin filament surface (29, 30).
In this study, carried out in silico by MD, the effect of substituting leucine for tropomyosin’s d-position Asp137 on tropomyosin-F-actin interaction was examined. Homology models containing Leu137 tropomyosin but otherwise “canonically” configured actin-tropomyosin were first built as previously described (see Materials and Methods). Thus, D137L and wild-type tropomyosin were wrapped around F-actin as perfect superhelices tracing the F-actin helix at the very start of MD, as in Li et al. (19). Configurations of D137L and wild-type tropomyosin dimers on actin were then compared during the simulations (Fig. 2, A and B). Fitting averaged structures of wild-type and mutant filaments to each other throughout MD by simply matching the actin subunits of the trajectories shows that the tropomyosin pseudorepeat 4 region from each tropomyosin construct is directly superimposable on the other (Fig. 2, C and F). This is particularly true for the helical chain facing actin (Fig. 2 C). Not only does the pseudorepeat 4 helix match, but individual side chains of residues Glu139 and Glu142 along this domain contact oppositely charged partners on the actin surface with virtually identical average orientations (Fig. 2, G–I). However, this equivalency is not an invariant feature for the rest of the coiled coil. Notably, the configurations of the wild-type and the mutant tropomyosin chains diverge both at local and global levels distal to pseudorepeat 4, and these deviations become increasingly obvious toward the tropomyosin C-terminus and in the direction of the N-terminus (Fig. 2, A–F). Hence, except for the region near to E139 and E142, including D137L, the mutation causes tropomyosin to reconfigure, resulting in diminished electrostatic interaction between tropomyosin and actin (Table 1). It follows that pseudorepeat 4 may be largely responsible for maintaining residual F-actin-tropomyosin interaction in the mutant and preventing dissociation of the dimer from F-actin during MD. Quantification of the azimuthal positioning of D137L and wild-type tropomyosin on actin shows little mutation-induced flexural variance in the tropomyosin superhelices (Table 1). However, corresponding comparison indicates aberrant twisting behavior of the mutant on actin in regions abutting residue 137 (Fig. 3). Thus, normal interaction between the D137L tropomyosin and F-actin is highly localized despite or at the expense of mutation-induced twisting deformation of the remainder of the coiled coil. If indeed the ends of tropomyosin, even in the wild type, bind weakest to actin, then a consequence of structural disruptions observed in the mutant exaggerates this deficit, leading to deficient assembly.
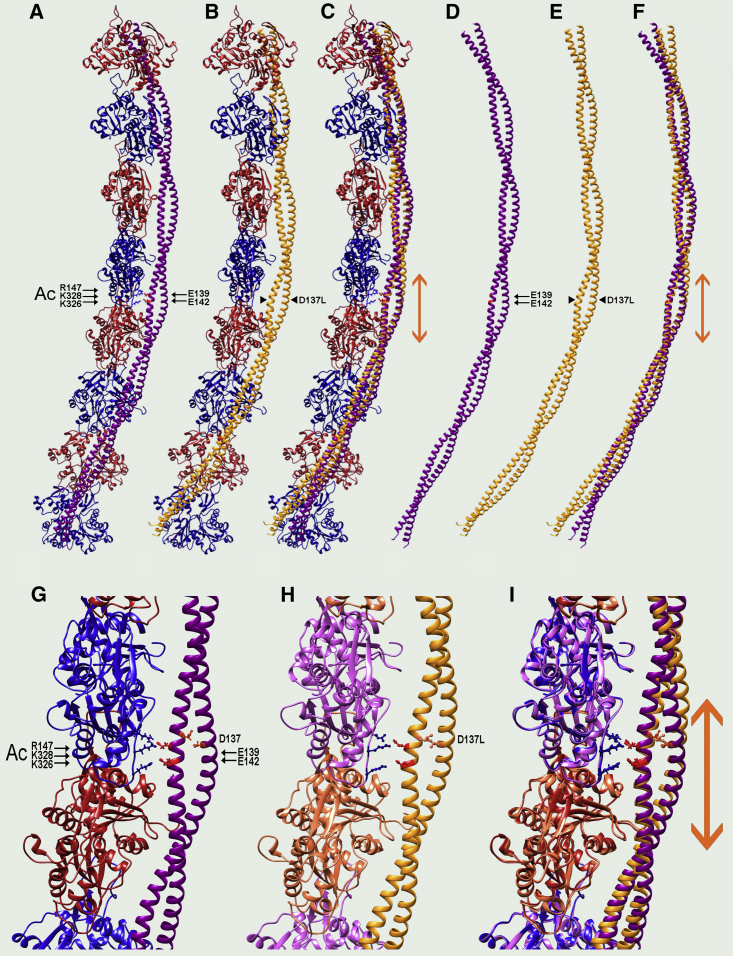
Figure 2.
Tropomyosin pseudorepeat 4 makes intimate contact with F-actin. (A–C) Ribbon structure showing the average position of wild-type tropomyosin (A) and D137L (B) on F-actin during MD is given. Only one strand of actin (blue, red actin subunits) and one tropomyosin chain (magenta for wild type and gold for D137L) are rendered for simplicity (filament pointed ends facing up, N-termini of tropomyosin pointed up). Double arrows (orange) indicate the pseudorepeat 4 region of tropomyosin that contacts F-actin; small black arrows show positions of actin residues 326, 328, and 147, which make electrostatic interactions with tropomyosin residues 139 and 142; and the arrowhead indicates the position of mutated residue Leu137 (D137L) in (B). In (C), the structures in (A) and (B) are superposed, showing conservation of the structure in pseudorepeat 4 between the mutant and the wild type. (D–F) Ribbon diagrams of the structures in (A)–(C), in which the actin has been removed to better illustrate and compare tropomyosin contributions, are shown in (D)–(F), respectively. The superposition of mutant and wild-type tropomyosin shown in (F) highlights the global divergence of the D137L tropomyosin backbone from the wild type, progressing from pseudodomain 4 to the C- and N-tropomyosin termini. Arrows are the same as in (A)–(C). (G–I) Magnified view of the central parts of (A)–(C) are shown in (G)–(I), respectively, to more clearly show the side chains of Glu142 and Glu139 (red), within tropomyosin pseudorepeat 4, that contact side chains of oppositely charged residues Lys326, Lys328, and Arg147 (blue) on actin (arrows same as in upper panels); side chains of tropomyosin’s core D137 are also shown (orange-red). Note that side chains on tropomyosin residues 139 and 142 linked to actin residues 326, 328, and 147 are almost perfectly aligned, as are the respective coiled-coil backbones in pseudorepeat 4, but the backbone chains already diverge in pseudorepeats 3 and 5; wild-type and mutant side chains D137 and L137 are also compared (orange-red and gold). To see this figure in color, go online.
Table 1.
Azimuthal Position and Interaction Energy between F-Actin and Wild-Type or Mutant Tropomyosins during MD
| Tropomyosin Sample | Average Tropomyosin Position and Its Azimuthal Deviation on F-Actin (Relative to Wild Type) (Å) | Average Coulombic Interaction Energy between Actin and Tropomyosin (kcal/mol) | Average vdW Interaction Energy between Actin and Tropomyosin (kcal/mol) |
|---|---|---|---|
| Wild type | 0 ± 0.77 | −3160 | −52 |
| D137L | −1.8 ± 0.50 | −2727 | −76 |
| E180G | +1.1 ± 0.75 | −2724 | −58 |
| D175N | +2.1 ± 0.37 | −2410 | −48 |
| D84N | +0.8 ± 0.90 | −2260 | −40 |
| E40K | −0.9 ± 0.50 | −3450 | −107 |
Note the small change in azimuthal position of tropomyosin and its variance in each case, suggesting limited tropomyosin flexural flexibility once linked to F-actin. Compared to the wild type, the diminished negative interaction energies of D137L, E180G, D175N, and D84N are consistent with lower actin-tropomyosin affinity, whereas the more negative value for E40K suggests higher affinity. vdW, van der Waals.
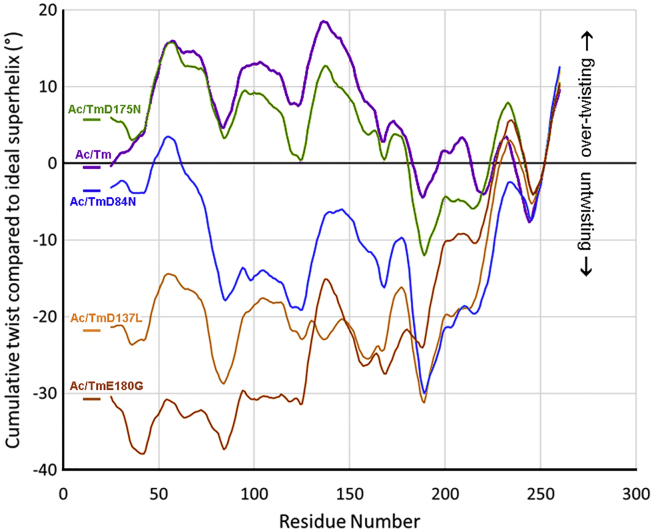
Figure 3.
Comparison of mutant and wild-type tropomyosin twisting on F-actin during MD. Plots show the cumulative twist angle deviation from an idealized tropomyosin superhelical model (0° along the x axis), generated as in Lorenz et al. (34), as described earlier in Lehman et al. (30) and below. The twist behavior of actin-bound wild-type tropomyosin (magenta tracing) during MD is compared to the twisting of D137L tropomyosin (gold), as done earlier in Lehman et al. (30), and now also compared to the twisting of D175N (green), D84N (blue), and E180G (orange) mutant tropomyosins. Note that periodic twist undulations in the various plots reflect the presence of alanine clusters that are partly responsible for the characteristic superhelical bending of tropomyosin (29, 30). All mutants show a variable degree of untwisting relative to the wild-type pattern. D175N tropomyosin displays the least twist divergence, which largely is confined to the C-terminal terminal region of the coiled coil near to residue 175. Plots of the D137L, D84N, and E180G mutants indicate substantially more tropomyosin untwisting on F-actin, and this is reflected by conformational mismatching to the helical curvature of F-actin (see Figs. 2 and 4). The plots shown in this figure record the average cumulative twist deviation relative to that of an idealized, “canonical” tropomyosin coiled coil. The canonical coiled-coil model has been bent to match the surface of F-actin’s long-pitch helix at a 39 Å radius and to traverse seven-actin subunits, as in Lorenz et al. (34). Because canonical tropomyosin is modeled so that coiled-coil twist is constant over the entire molecule, the residue to residue twist anywhere over its length is constant and the cumulative twist set to 0° on the plot. During MD, the average bending and coiled-coil twist pattern of the C-terminal half of wild-type tropomyosin most closely matches that of the canonical superhelical model. To reflect this correspondence in plots of cumulative twist deviation and to be consistent with plots in previous publications (29, 30), the twist over residue 252, near to the C-terminal end of tropomyosin, was set as a 0° reference point in each of the subsequent plots, and cumulative twist deviation from a canonical model was then measured proceeding from this point to N- and C termini. Thus, the plots measure both end-to-end and local overtwisting and untwisting of wild-type and mutant tropomyosins relative to each other and to the canonical twisted model (i.e., the normalized 0° values in the plot). (Please note, coiled-coil twist itself was measured during MD and not superhelical twist). To see this figure in color, go online.
Partial dissociation of tropomyosin with HCM-linked mutations from F-actin
The distinctive patterns attributable to D137L tropomyosin-actin binding noted above may be traits specific to the widely studied but non-native tropomyosin construct and therefore of relatively narrow academic interest. However, this does not appear to be the case, given the description below of two well-characterized, naturally occurring tropomyosin mutations, E180G and D175N (Fig. 1 B). The mutants have been linked to the development of HCMs (46) and display functional behavior in vitro similar to that caused by D137L (47, 48, 49, 50). These mutations are located on tropomyosin next to f-position E181 and b-position E177, residues that normally are involved in electrostatic interactions between F-actin and pseudorepeat 5 of tropomyosin. It is not surprising, therefore, that the earlier work showed that the mutants exhibit appreciably reduced apparent affinity for actin and atypical sigmoidal binding isotherms (48).
MD simulations show that the overall superhelical bending outlines of E180G and D175N chains in F-actin-tropomyosin complexes become obviously distorted (Fig. 4, B and C). Overlaying averaged MD trajectories demonstrate that the mutant tropomyosins diverge from the wild type as well as from each other and D137L (Fig. 4, E and F). However, once again, such superposition shows that tropomyosin pseudodomain 4 remains almost perfectly matched to the cognate wild-type molecule in the corresponding actin-tropomyosin complex. In addition, identical Glu139- and Glu142-actin salt-bridge orientations are maintained by the hypertrophic cardiomyopathy (HCM) mutants during MD (Fig. 4, J, K, M, and N). Nevertheless, aberrant bending and twisting over the remainder of the molecules is apparent, particularly for the E180G mutant at the level of the mutation (Fig. 4, B and E), whereas the D175N deviates from the control largely in its N-terminal half (Fig. 4, C and F). Again, quantitation of coiled-coil twisting on F-actin corroborates the molecular graphics (Fig. 3), whereas azimuthal variance is not obviously affected (Table 1). Still, the global alterations in tropomyosin twist patterning likely disrupt overall interactions between actin and mutant tropomyosin in the B-state, resulting in increased Ca2+ activation at low Ca2+ levels (48) and presumably prompting the development of HCM.
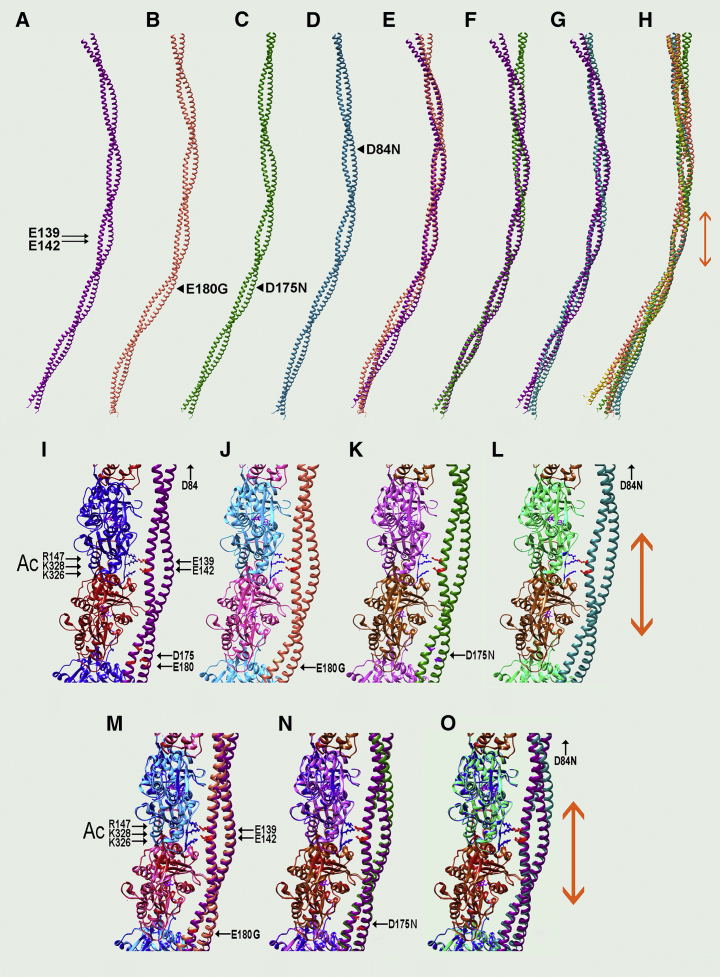
Figure 4.
Conformational adjustment of mutant tropomyosins on F-actin. Ribbon diagrams showing the average tropomyosin structures on F-actin during MD are given. (A–H) The actin component of the structures is hidden to better illustrate and compare tropomyosin conformations; tropomyosin N-termini pointed up. (A) Wild-type tropomyosin (magenta) and (B) E180G (orange), (C) D175N (green), and (D) D84N (blue) mutant tropomyosins are shown; positions of key residues noted. (E) Superposing (B) on (A), (F) superposing (C) on (A), and (G) superposing (D) on (A) highlights the near-perfect alignment of pseudorepeat 4 of the mutant and wild-type tropomyosin structures and also the global divergence of mutant and wild-type tropomyosin backbones progressing away from pseudodomain 4. (H) Superposition of the three mutant tropomyosins as well as D137L (gold) is shown; note that only the region within pseudodomain 4 is consistently well-matched in all examples. Double arrows (orange) indicate the pseudorepeat 4 region of tropomyosin; small black arrows show positions of residues 142 (lower arrow) and 139 (upper arrow) as in Fig. 2. Black arrowheads indicate the positions of the mutated residues in (B)–(D). (I–O) Magnified view of the central part of the above structures is given, now showing corresponding actin subunits from the MD as well as side chains of Glu142 and Glu139 (red) within tropomyosin pseudorepeat 4 that contact oppositely charged residues Lys326, Lys328, and Arg147 (blue) on actin as in Fig. 2. (I) Wild-type actin-tropomyosin, (J) actin-E180G tropomyosin, (K) actin-D175N tropomyosin, and (L) actin-D84N tropomyosin (blue) are shown. (M) Superposing (J) on (I), (N) superposing (K) on (I), and (O) superposing (L) on (I) are shown; note that side chains of tropomyosin residues 139 and 142, which link to actin residues 326, 328, and 147, are all well-matched to each other, as are the respective coiled-coil backbones within pseudorepeat 4. N.B., use zoom feature to visualize side chains on small-dimension monitors. The distance of tropomyosin residue 84 from pseudorepeat 4 precludes including the mutated residue in the magnified panels, but here again conformational constancy over pseudorepeat 4 is evident. To see this figure in color, go online.
Behavior of DCM-linked tropomyosin mutations on F-actin
In principle, an enhanced bias of tropomyosin for the thin filament blocking state should lead to decreased cardiac thin-filament activation, in this case potentially predisposing heart muscle to dilated cardiomyopathy (DCM) (46). For example, the interaction energetics of DCM-linked E40K tropomyosin for F-actin is considerably stronger than that for wild-type tropomyosin (Table 1; also see (51)), whereas the Ca2+ sensitivity of reconstituted filaments containing E40K is lower (32, 52, 53, 54, 55, 56, 57). As expected, the E40K-F-actin complex shows stable interactions during MD and no evidence of localized detachment from F-actin. In fact, MD suggests that this mutant makes as close or even closer side-chain contacts with actin subunits than the wild type does (32) and enhances overtwisting (data not shown).
Contrary to expectation, the DCM-linked D84N mutant tropomyosin (also associated with severe left ventricular noncompaction cardiomyopathy (58, 59)) shows a lower than normal actin affinity and weaker than control interaction energetics with actin (Table 1). The mutation, located on the N-terminal side of the above HCM mutation sites, maps to a tropomyosin β-zone, several heptads removed from the nearest residues likely to bind to actin subunits (Fig. 1 B). Consistent with the energetics calibration, the behavior of the D84N DCM-linked mutant during MD is much like the actin-associated HCM mutants described above. Again, the overall superhelical bending of the mutant on F-actin diverges from controls (as well as from E180G, D175N, and D137L tropomyosins) (Fig. 4, D, G, and H). Still, superposition of averaged D84N trajectories to wild-type filaments shows excellent alignment over pseudodomain 4 (Fig. 4 G). And, once again, Glu139- and Glu142-actin salt bridges are retained in the mutant tropomyosins (Fig. 4 O). Quantifying the mutant’s twisting also demonstrates localized divergence from the control pattern (Fig. 3).
Discussion
The center must hold
Extensive MD simulations have been carried out to characterize and compare actin-tropomyosin filaments containing wild-type and various mutant tropomyosins (19, 22, 29, 30, 43, 60, 61, 62). Our own MD shows that previously unrecognized local twisting of the wild-type tropomyosin optimizes interactions between residues Glu139 and Glu142 in the central pseudorepeat 4 of tropomyosin and residues Lys326, Lys328, and Arg147 on neighboring actin subunits of the thin filament (29, 30). Acidic residues located in the peripheral tropomyosin pseudorepeats, including Asp20, Asp58, Glu62, Glu97, Asp100, Glu104, Glu177, Glu181, Asp219, Glu223, and Asp258 on periods 1–3 and 5–7, also contact oppositely charged Lys326, Lys328, and Arg147 on actin, but often less intimately (29, 30). We now have additionally found that although all the mutant tropomyosins examined in this study maintain close contacts between residues 139 and 142 and actin partners, this appears to occur at the expense of the remaining potential links noted above. In turn, corresponding side-chain realignment in the mutants is accompanied by less favorable overall interaction energy (Table 1), seemingly a product of aberrant coiled-coil twisting and bending in the mutants. (Note the diminished negative interaction energies for the mutants in Table 1.) Still, mutant tropomyosins remain tethered to F-actin during MD by linkages facilitated by pseudorepeat 4 and, to a variable extent, by ones from repeats 5 to 7 flanking the central domain. At the same time, the superhelical topological bias of tropomyosin also favors residual interfacial complementarity.
It is noteworthy that the midregion of tropomyosin is strictly conserved among sarcomeric and somatic cell tropomyosin isoforms. Given that actin isoforms and their tropomyosin binding interface also are largely invariant, it makes sense that the actin-tropomyosin interactions described here are likely to be universally applied by all tropomyosins but not responsible for differentiation between isoform types. In fact, tropomyosin isoform sorting on actin filaments, targeting distinct cellular domains, is defined by unique C- and N-terminal sequences and by not pseudorepeat 4 specialization involving the tropomyosin midpiece (reviewed in (30, 63)).
Early life on the filament aufbau
As mentioned, pathways possible for building low-affinity, conformationally variable, and multivalent tropomyosin onto actin filaments seem limitless. Yet, buried in the protein’s “blueprint,” sequence specificity is designed to overcome this entropic challenge, thereby discounting numerous other initial “choices” during thin-filament assembly.
It appears that tropomyosin preferentially assembles near to the pointed ends of filaments, which contain actin subunits associated with bound ADP (64). (In fact, the F-actin model used in this study includes only ADP-actin subunits.) However, the preference for ADP-actin is not specifically or necessarily restricted to the filament pointed end itself; instead, just close to that end (64). (Nor is the Z-disk a likely nucleation center for tropomyosin binding in striated muscle (65), as might be imagined.) We also note that it is doubtful that major mammalian-type formins fulfill a nucleation function in higher organisms. Unlike such formin involvement in yeast (66), mDia1 and mDia3 formins do not recruit or nucleate mammalian tropomyosin onto thin filaments (67). Therefore, yeast tropomyosin may not be a good model for characterization of more “advanced” tropomyosins. Unlike vertebrate and invertebrate tropomyosins, the yeast variants contain possible coiled-coil sequence interruptions and lack proper alanine clusters (68, 69), whereas tropomyosins found elsewhere are defined by uninterrupted coiled coils and alanine cluster-based curvature (28). Therefore, these studies still call for a guiding principle that describes tropomyosin assembly on actin, one that possibly is intrinsic to filament mechanochemistry per se.
As raised in the work’s Introduction, we contend that tropomyosin favors replicating steps detected in mutation-sponsored filament destabilization taking place during MD but here progressing in reverse. Our premise is that the residual association noted for mutant D137L, E180G, D175N, and D84N mirrors intermediates in tropomyosin assembly on F-actin. Thus, we propose that during the normal assembly process, favorable contacts are initiated between pseudorepeat 4 and F-actin, mimicking those retained by the mutants. These initial, and presumably transient, contacts limit diffusion of the coiled coil away from actin while setting tropomyosin polarity relative to the actin filament axis. We cannot exclude the prospect that differences observed between wild-type and mutant tropomyosins in silico may have an even larger impact in vitro or in cells or, alternatively, a damped effect.
Based on previous work, we know that on average, the superhelical outlines of the C-terminal half of the actin-free tropomyosin molecule approximate those of the F-actin helix (29, 70). Thus, once contact is made by pseudorepeat 4 and F-actin, it is likely that semirigid C-terminal domains of wild-type tropomyosin zipper onto the thin filament surface. This leaves the N-terminal region of tropomyosin poised to capture unbound tropomyosin dimers or similarly linked neighbors on actin, annealing them to the tropomyosin and hence sponsoring growth of a nascent tropomyosin cable on F-actin.
Although tropomyosin end-to-end bonds form readily in solution as dimers and short oligomers (71, 72), actin-free tropomyosin cables are rarely observed in solution (72). Their instability suggests behavior analogous to that of long single-stranded “Einstein polymers” (73). In contrast, interactions between tropomyosin and actin transform the thin-filament complex into a long-range stabilized copolymer. Thus, extended tropomyosin cables form readily by the end-to-end association once present on F-actin while the cables wrap around their thin filament actin partner. Subsequently, the tropomyosin cable then protects the core actin component of thin filaments from depolymerization (74). Finally, our results also indicate that the dynamics of the tropomyosin cable on F-actin cannot be easily characterized by thermal reptation but rather is governed by a taut twist of the superhelix. We propose that our description of thin-filament behavior, based on realistic full atomistic models of actin-tropomyosin, reasonably approximates events occurring in vitro. We expect that our work will support new mechanistic paradigms for interpreting the role of tropomyosin in actin filament function and dysfunction.
Author Contributions
W.L. thought of the general approach taken and formulated the principal concepts presented. M.J.R. carried out the MD simulation and data computation. J.R.M. and S.G.C. contributed to ongoing discussion of the data and the manuscript together with W.L. and M.J.R. W.L. and M.J.R analyzed the data. W.L. wrote the manuscript. W.L. prepared the molecular graphics and M.J.R. the tables and graphs.
Acknowledgments
This work was funded by National Institutes of Health grants R01HL036153 (to W.L.), R01HL123774 (to J.R.M. and W.L.), and R01HL136590 (to S.G.C.). Computational work was carried out on the computational resources provided by the Massachusetts Green High-Performance Computing Center. S.G.C. has equity ownership in Propria LLC, a company that develops technology for producing engineered heart tissue from induced pluripotent stem-cell-derived cardiomyocytes.
Editor: Michael Ostap.
References
- 1.McLachlan A.D., Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J. Mol. Biol. 1976;103:271–298. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]; McLachlan, A. D., and M. Stewart. 1976. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J. Mol. Biol. 103:271-298. [DOI] [PubMed]
- 2.Hitchcock-DeGregori S.E. Tropomyosin: function follows structure. Adv. Exp. Med. Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]; Hitchcock-DeGregori, S. E. 2008. Tropomyosin: function follows structure. Adv. Exp. Med. Biol. 644:60-72. [DOI] [PubMed]
- 3.Haselgrove J.C. X-ray evidence for a conformational change in the actin-containing filaments of vertebrate striated muscle. Cold Spring Harb. Symp. Quant. Biol. 1973;37:341–352. [Google Scholar]; Haselgrove, J. C. 1973. X-ray evidence for a conformational change in the actin-containing filaments of vertebrate striated muscle. Cold Spring Harb. Symp. Quant. Biol. 37:341-352.
- 4.Huxley H.E. Structural changes in the actin- and myosin-eontaining filaments during contraction. Cold Spring Harb. Symp. Quant. Biol. 1973;37:361–376. [Google Scholar]; Huxley, H. E. 1973. Structural changes in the actin- and myosin-eontaining filaments during contraction. Cold Spring Harb. Symp. Quant. Biol. 37:361-376.
- 5.Parry D.A., Squire J.M. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J. Mol. Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]; Parry, D. A., and J. M. Squire. 1973. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J. Mol. Biol. 75:33-55. [DOI] [PubMed]
- 6.Lehman W., Craig R., Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]; Lehman, W., R. Craig, and P. Vibert. 1994. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 368:65-67. [DOI] [PubMed]
- 7.Vibert P., Craig R., Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]; Vibert, P., R. Craig, and W. Lehman. 1997. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 266:8-14. [DOI] [PubMed]
- 8.Poole K.J., Lorenz M., Holmes K.C. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]; Poole, K. J., M. Lorenz, …, K. C. Holmes. 2006. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 155:273-284. [DOI] [PubMed]
- 9.Galińska-Rakoczy A., Engel P., Lehman W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J. Mol. Biol. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]; Galińska-Rakoczy, A., P. Engel, …, W. Lehman. 2008. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J. Mol. Biol. 379:929-935. [DOI] [PMC free article] [PubMed]
- 10.Yang S., Barbu-Tudoran L., Lehman W. Three-dimensional organization of troponin on cardiac muscle thin filaments in the relaxed state. Biophys. J. 2014;106:855–864. doi: 10.1016/j.bpj.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, S., L. Barbu-Tudoran, …, W. Lehman. 2014. Three-dimensional organization of troponin on cardiac muscle thin filaments in the relaxed state. Biophys. J. 106:855-864. [DOI] [PMC free article] [PubMed]
- 11.Holmes K.C., Lehman W. Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]; Holmes, K. C., and W. Lehman. 2008. Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 29:213-219. [DOI] [PubMed]
- 12.Wegner A. The interaction of α, α- and α, β-tropomyosin with actin filaments. FEBS Lett. 1980;119:245–248. doi: 10.1016/0014-5793(80)80263-8. [DOI] [PubMed] [Google Scholar]; Wegner, A. 1980. The interaction of α, α- and α, β-tropomyosin with actin filaments. FEBS Lett. 119:245-248. [DOI] [PubMed]
- 13.Lehman W. Thin filament structure and the steric blocking model. Compr. Physiol. 2016;6:1043–1069. doi: 10.1002/cphy.c150030. [DOI] [PubMed] [Google Scholar]; Lehman, W. 2016. Thin filament structure and the steric blocking model. Compr. Physiol. 6:1043-1069. [DOI] [PubMed]
- 14.Moore J.R., Campbell S.G., Lehman W. Structural determinants of muscle thin filament cooperativity. Arch. Biochem. Biophys. 2016;594:8–17. doi: 10.1016/j.abb.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moore, J. R., S. G. Campbell, and W. Lehman. 2016. Structural determinants of muscle thin filament cooperativity. Arch. Biochem. Biophys. 594:8-17. [DOI] [PMC free article] [PubMed]
- 15.Barua B., Winkelmann D.A., Hitchcock-DeGregori S.E. Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc. Natl. Acad. Sci. USA. 2012;109:18425–18430. doi: 10.1073/pnas.1212754109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barua, B., D. A. Winkelmann, …, S. E. Hitchcock-DeGregori. 2012. Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc. Natl. Acad. Sci. USA. 109:18425-18430. [DOI] [PMC free article] [PubMed]
- 16.Barua B., Fagnant P.M., Hitchcock-DeGregori S.E. A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J. Biol. Chem. 2013;288:9602–9609. doi: 10.1074/jbc.M113.451161. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barua, B., P. M. Fagnant, …, S. E. Hitchcock-DeGregori. 2013. A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J. Biol. Chem. 288:9602-9609. [DOI] [PMC free article] [PubMed]
- 17.Brown J.H., Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv. Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]; Brown, J. H., and C. Cohen. 2005. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv. Protein Chem. 71:121-159. [DOI] [PubMed]
- 18.Dominguez R. Tropomyosin: the gatekeeper’s view of the actin filament revealed. Biophys. J. 2011;100:797–798. doi: 10.1016/j.bpj.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dominguez, R. 2011. Tropomyosin: the gatekeeper’s view of the actin filament revealed. Biophys. J. 100:797-798. [DOI] [PMC free article] [PubMed]
- 19.Li X.E., Tobacman L.S., Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, X. E., L. S. Tobacman, …, W. Lehman. 2011. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 100:1005-1013. [DOI] [PMC free article] [PubMed]
- 20.Orzechowski M., Li X.E., Lehman W. An atomic model of the tropomyosin cable on F-actin. Biophys. J. 2014;107:694–699. doi: 10.1016/j.bpj.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Orzechowski, M., X. E. Li, …, W. Lehman. 2014a. An atomic model of the tropomyosin cable on F-actin. Biophys. J. 107:694-699. [DOI] [PMC free article] [PubMed]
- 21.von der Ecken J., Müller M., Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]; von der Ecken, J., M. Muller, …, S. Raunser. 2015. Structure of the F-actin-tropomyosin complex. Nature. 519:114-117. [DOI] [PMC free article] [PubMed]
- 22.Williams M.R., Lehman S.J., Schwartz S.D. Atomic resolution probe for allostery in the regulatory thin filament. Proc. Natl. Acad. Sci. USA. 2016;113:3257–3262. doi: 10.1073/pnas.1519541113. [DOI] [PMC free article] [PubMed] [Google Scholar]; Williams, M. R., S. J. Lehman, …, S. D. Schwartz. 2016. Atomic resolution probe for allostery in the regulatory thin filament. Proc. Natl. Acad. Sci. USA. 113:3257-3262. [DOI] [PMC free article] [PubMed]
- 23.Desai R., Geeves M.A., Kad N.M. Using fluorescent myosin to directly visualize cooperative activation of thin filaments. J. Biol. Chem. 2015;290:1915–1925. doi: 10.1074/jbc.M114.609743. [DOI] [PMC free article] [PubMed] [Google Scholar]; Desai, R., M. A. Geeves, and N. M. Kad. 2015. Using fluorescent myosin to directly visualize cooperative activation of thin filaments. J. Biol. Chem. 290:1915-1925. [DOI] [PMC free article] [PubMed]
- 24.Schmidt W.M., Lehman W., Moore J.R. Direct observation of tropomyosin binding to actin filaments. Cytoskeleton (Hoboken) 2015;72:292–303. doi: 10.1002/cm.21225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schmidt, W. M., W. Lehman, and J. R. Moore. 2015. Direct observation of tropomyosin binding to actin filaments. Cytoskeleton (Hoboken). 72:292-303. [DOI] [PMC free article] [PubMed]
- 25.Nicovich P.R., Janco M., Böcking T. Effect of surface chemistry on tropomyosin binding to actin filaments on surfaces. Cytoskeleton (Hoboken) 2016;73:729–738. doi: 10.1002/cm.21342. [DOI] [PubMed] [Google Scholar]; Nicovich, P. R., M. Janco, …, T. Bocking. 2016. Effect of surface chemistry on tropomyosin binding to actin filaments on surfaces. Cytoskeleton (Hoboken). 73:729-738. [DOI] [PubMed]
- 26.Williams M.R., Tardiff J.C., Schwartz S.D. Mechanism of cardiac tropomyosin transitions on filamentous actin as revealed by all-atom steered molecular dynamics simulations. J. Phys. Chem. Lett. 2018;9:3301–3306. doi: 10.1021/acs.jpclett.8b00958. [DOI] [PMC free article] [PubMed] [Google Scholar]; Williams, M. R., J. C. Tardiff, and S. D. Schwartz. 2018. Mechanism of cardiac tropomyosin transitions on filamentous actin as revealed by all-atom steered molecular dynamics simulations. J. Phys. Chem. Lett. 9:3301-3306. [DOI] [PMC free article] [PubMed]
- 27.Oz A. All Rivers. New Yorker. 2019 https://www.newyorker.com/magazine/2019/01/14/all-rivers [Google Scholar]; Oz, A. 2019. All Rivers. New Yorker. https://www.newyorker.com/magazine/2019/01/14/all-rivers.
- 28.Li X.E., Holmes K.C., Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J. Mol. Biol. 2010;395:327–339. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]; Li, X. E., K. C. Holmes, …, S. Fischer. 2010. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J. Mol. Biol. 395:327-339. [DOI] [PubMed]
- 29.Lehman W., Li X., Rynkiewicz M.J. Precise binding of tropomyosin on actin involves sequence-dependent variance in coiled-coil twisting. Biophys. J. 2018;115:1082–1092. doi: 10.1016/j.bpj.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lehman, W., X. Li, …, M. J. Rynkiewicz. 2018. Precise binding of tropomyosin on actin involves sequence-dependent variance in coiled-coil twisting. Biophys. J. 115:1082-1092. [DOI] [PMC free article] [PubMed]
- 30.Lehman W., Rynkiewicz M.J., Moore J.R. A new twist on tropomyosin binding to actin filaments: perspectives on thin filament function, assembly and biomechanics. J. Muscle Res. Cell Motil. 2019 doi: 10.1007/s10974-019-09501-5. Published online February 15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lehman, W., M. J. Rynkiewicz, and J. R. Moore. 2019. A new twist on tropomyosin binding to actin filaments: perspectives on thin filament function, assembly and biomechanics. J. Muscle Res. Cell Motil. Published online February 15, 2019. [DOI] [PMC free article] [PubMed]
- 31.Rynkiewicz M.J., Fischer S., Lehman W. The propensity for tropomyosin twisting in the presence and absence of F-actin. Arch. Biochem. Biophys. 2016;609:51–58. doi: 10.1016/j.abb.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rynkiewicz, M. J., S. Fischer, and W. Lehman. 2016. The propensity for tropomyosin twisting in the presence and absence of F-actin. Arch. Biochem. Biophys. 609:51-58. [DOI] [PMC free article] [PubMed]
- 32.Farman G.P., Rynkiewicz M.J., Moore J.R. HCM and DCM cardiomyopathy-linked α-tropomyosin mutations influence off-state stability and crossbridge interaction on thin filaments. Arch. Biochem. Biophys. 2018;647:84–92. doi: 10.1016/j.abb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Farman, G. P., M. J. Rynkiewicz, …, J. R. Moore. 2018. HCM and DCM cardiomyopathy-linked α-tropomyosin mutations influence off-state stability and crossbridge interaction on thin filaments. Arch. Biochem. Biophys. 647:84-92. [DOI] [PMC free article] [PubMed]
- 33.Kiani F.A., Lehman W., Rynkiewicz M.J. Spontaneous transitions of actin-bound tropomyosin toward blocked and closed states. J. Gen. Physiol. 2019;151:4–8. doi: 10.1085/jgp.201812188. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kiani, F. A., W. Lehman, …, M. J. Rynkiewicz. 2019. Spontaneous transitions of actin-bound tropomyosin toward blocked and closed states. J. Gen. Physiol. 151:4-8. [DOI] [PMC free article] [PubMed]
- 34.Lorenz M., Poole K.J.V., Holmes K.C. An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol. 1995;246:108–119. doi: 10.1006/jmbi.1994.0070. [DOI] [PubMed] [Google Scholar]; Lorenz, M., K. J. V. Poole, …, K. C. Holmes. 1995. An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol. 246:108-119. [DOI] [PubMed]
- 35.Rynkiewicz M.J., Schott V., Fischer S. Electrostatic interaction map reveals a new binding position for tropomyosin on F-actin. J. Muscle Res. Cell Motil. 2015;36:525–533. doi: 10.1007/s10974-015-9419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rynkiewicz, M. J., V. Schott, …, S. Fischer. 2015. Electrostatic interaction map reveals a new binding position for tropomyosin on F-actin. J. Muscle Res. Cell Motil. 36:525-533. [DOI] [PMC free article] [PubMed]
- 36.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28.. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]; Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38, 27-28. [DOI] [PubMed]
- 37.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phillips, J. C., R. Braun, …, K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781-1802. [DOI] [PMC free article] [PubMed]
- 38.Brooks B.R., Brooks C.L., III, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brooks, B. R., C. L. Brooks III, …, M. Karplus. 2009. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30:1545-1614. [DOI] [PMC free article] [PubMed]
- 39.Strelkov S.V., Burkhard P. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]; Strelkov, S. V., and P. Burkhard. 2002. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137:54-64. [DOI] [PubMed]
- 40.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]; Pettersen, E. F., T. D. Goddard, …, T. E. Ferrin. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed]
- 41.Sumida J.P., Wu E., Lehrer S.S. Conserved Asp-137 imparts flexibility to tropomyosin and affects function. J. Biol. Chem. 2008;283:6728–6734. doi: 10.1074/jbc.M707485200. [DOI] [PubMed] [Google Scholar]; Sumida, J. P., E. Wu, and S. S. Lehrer. 2008. Conserved Asp-137 imparts flexibility to tropomyosin and affects function. J. Biol. Chem. 283:6728-6734. [DOI] [PubMed]
- 42.Shchepkin D.V., Matyushenko A.M., Levitsky D.I. Stabilization of the central part of tropomyosin molecule alters the Ca2+-sensitivity of actin-myosin interaction. Acta Naturae. 2013;5:126–129. [PMC free article] [PubMed] [Google Scholar]; Shchepkin, D. V., A. M. Matyushenko, …, D. I. Levitsky. 2013. Stabilization of the central part of tropomyosin molecule alters the Ca2+-sensitivity of actin-myosin interaction. Acta Naturae. 5:126-129. [PMC free article] [PubMed]
- 43.Matyushenko A.M., Artemova N.V., Levitsky D.I. Structural and functional effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin molecule. FEBS J. 2014;281:2004–2016. doi: 10.1111/febs.12756. [DOI] [PubMed] [Google Scholar]; Matyushenko, A. M., N. V. Artemova, …, D. I. Levitsky. 2014. Structural and functional effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin molecule. FEBS J. 281:2004-2016. [DOI] [PubMed]
- 44.Scellini B., Piroddi N., Tesi C. The relaxation properties of myofibrils are compromised by amino acids that stabilize α-tropomyosin. Biophys. J. 2017;112:376–387. doi: 10.1016/j.bpj.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scellini, B., N. Piroddi, …, C. Tesi. 2017. The relaxation properties of myofibrils are compromised by amino acids that stabilize α-tropomyosin. Biophys. J. 112:376-387. [DOI] [PMC free article] [PubMed]
- 45.Matyushenko A.M., Shchepkin D.V., Levitsky D.I. Functional role of the core gap in the middle part of tropomyosin. FEBS J. 2018;285:871–886. doi: 10.1111/febs.14369. [DOI] [PubMed] [Google Scholar]; Matyushenko, A. M., D. V. Shchepkin, …, D. I. Levitsky. 2018. Functional role of the core gap in the middle part of tropomyosin. FEBS J. 285:871-886. [DOI] [PubMed]
- 46.Redwood C., Robinson P. Alpha-tropomyosin mutations in inherited cardiomyopathies. J. Muscle Res. Cell Motil. 2013;34:285–294. doi: 10.1007/s10974-013-9358-5. [DOI] [PubMed] [Google Scholar]; Redwood, C., and P. Robinson. 2013. Alpha-tropomyosin mutations in inherited cardiomyopathies. J. Muscle Res. Cell Motil. 34:285-294. [DOI] [PubMed]
- 47.Li X.E., Suphamungmee W., Lehman W. The flexibility of two tropomyosin mutants, D175N and E180G, that cause hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2012;424:493–496. doi: 10.1016/j.bbrc.2012.06.141. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, X. E., W. Suphamungmee, …, W. Lehman. 2012. The flexibility of two tropomyosin mutants, D175N and E180G, that cause hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 424:493-496. [DOI] [PMC free article] [PubMed]
- 48.Kremneva E., Boussouf S., Levitsky D.I. Effects of two familial hypertrophic cardiomyopathy mutations in alpha-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin-bound tropomyosin. Biophys. J. 2004;87:3922–3933. doi: 10.1529/biophysj.104.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kremneva, E., S. Boussouf, …, D. I. Levitsky. 2004. Effects of two familial hypertrophic cardiomyopathy mutations in alpha-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin-bound tropomyosin. Biophys. J. 87:3922-3933. [DOI] [PMC free article] [PubMed]
- 49.Janco M., Kalyva A., Geeves M.A. α-Tropomyosin with a D175N or E180G mutation in only one chain differs from tropomyosin with mutations in both chains. Biochemistry. 2012;51:9880–9890. doi: 10.1021/bi301323n. [DOI] [PMC free article] [PubMed] [Google Scholar]; Janco, M., A. Kalyva, …, M. A. Geeves. 2012. α-Tropomyosin with a D175N or E180G mutation in only one chain differs from tropomyosin with mutations in both chains. Biochemistry. 51:9880-9890. [DOI] [PMC free article] [PubMed]
- 50.Janco M., Suphamungmee W., Geeves M.A. Polymorphism in tropomyosin structure and function. J. Muscle Res. Cell Motil. 2013;34:177–187. doi: 10.1007/s10974-013-9353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Janco, M., W. Suphamungmee, …, M. A. Geeves. 2013. Polymorphism in tropomyosin structure and function. J. Muscle Res. Cell Motil. 34:177-187. [DOI] [PMC free article] [PubMed]
- 51.Orzechowski M., Fischer S., Farman G.P. Energy landscapes reveal the myopathic effects of tropomyosin mutations. Arch. Biochem. Biophys. 2014;564:89–99. doi: 10.1016/j.abb.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Orzechowski, M., S. Fischer, …, G. P. Farman. 2014. Energy landscapes reveal the myopathic effects of tropomyosin mutations. Arch. Biochem. Biophys. 564:89-99. [DOI] [PMC free article] [PubMed]
- 52.Chang A.N., Harada K., Potter J.D. Functional consequences of hypertrophic and dilated cardiomyopathy-causing mutations in alpha-tropomyosin. J. Biol. Chem. 2005;280:34343–34349. doi: 10.1074/jbc.M505014200. [DOI] [PubMed] [Google Scholar]; Chang, A. N., K. Harada, …, J. D. Potter. 2005. Functional consequences of hypertrophic and dilated cardiomyopathy-causing mutations in alpha-tropomyosin. J. Biol. Chem. 280:34343-34349. [DOI] [PubMed]
- 53.Chang A.N., Greenfield N.J., Pinto J.R. Structural and protein interaction effects of hypertrophic and dilated cardiomyopathic mutations in alpha-tropomyosin. Front. Physiol. 2014;5:460. doi: 10.3389/fphys.2014.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang, A. N., N. J. Greenfield, …, J. R. Pinto. 2014. Structural and protein interaction effects of hypertrophic and dilated cardiomyopathic mutations in alpha-tropomyosin. Front. Physiol. 5:460. [DOI] [PMC free article] [PubMed]
- 54.Mirza M., Robinson P., Marston S. The effect of mutations in alpha-tropomyosin (E40K and E54K) that cause familial dilated cardiomyopathy on the regulatory mechanism of cardiac muscle thin filaments. J. Biol. Chem. 2007;282:13487–13497. doi: 10.1074/jbc.M701071200. [DOI] [PubMed] [Google Scholar]; Mirza, M., P. Robinson, …, S. Marston. 2007. The effect of mutations in alpha-tropomyosin (E40K and E54K) that cause familial dilated cardiomyopathy on the regulatory mechanism of cardiac muscle thin filaments. J. Biol. Chem. 282:13487-13497. [DOI] [PubMed]
- 55.Bai F., Wang L., Kawai M. A study of tropomyosin’s role in cardiac function and disease using thin-filament reconstituted myocardium. J. Muscle Res. Cell Motil. 2013;34:295–310. doi: 10.1007/s10974-013-9343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bai, F., L. Wang, and M. Kawai. 2013. A study of tropomyosin’s role in cardiac function and disease using thin-filament reconstituted myocardium. J. Muscle Res. Cell Motil. 34:295-310. [DOI] [PMC free article] [PubMed]
- 56.Kopylova G.V., Shchepkin D.V., Matyushenko A.M. Effect of cardiomyopathic mutations in tropomyosin on calcium regulation of the actin-myosin interaction in skeletal muscle. Bull. Exp. Biol. Med. 2016;162:42–44. doi: 10.1007/s10517-016-3540-x. [DOI] [PubMed] [Google Scholar]; Kopylova, G. V., D. V. Shchepkin, …, A. M. Matyushenko. 2016. Effect of cardiomyopathic mutations in tropomyosin on calcium regulation of the actin-myosin interaction in skeletal muscle. Bull. Exp. Biol. Med. 162:42-44. [DOI] [PubMed]
- 57.Deranek A.E., Klass M.M., Tardiff J.C. Moving beyond simple answers to complex disorders in sarcomeric cardiomyopathies: the role of integrated systems. Pflugers Arch. 2019;471:661–671. doi: 10.1007/s00424-019-02269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deranek, A. E., M. M. Klass, and J. C. Tardiff. 2019. Moving beyond simple answers to complex disorders in sarcomeric cardiomyopathies: the role of integrated systems. Pflugers Arch. 471:661-671. [DOI] [PMC free article] [PubMed]
- 58.van de Meerakker J.B., Christiaans I., Postma A.V. A novel alpha-tropomyosin mutation associates with dilated and non-compaction cardiomyopathy and diminishes actin binding. Biochim. Biophys. Acta. 2013;1833:833–839. doi: 10.1016/j.bbamcr.2012.11.003. [DOI] [PubMed] [Google Scholar]; van de Meerakker, J. B., I. Christiaans, …, A. V. Postma. 2013. A novel alpha-tropomyosin mutation associates with dilated and non-compaction cardiomyopathy and diminishes actin binding. Biochim. Biophys. Acta. 1833:833-839. [DOI] [PubMed]
- 59.Gupte T.M., Haque F., Mercer J.A. Mechanistic heterogeneity in contractile properties of α-tropomyosin (TPM1) mutants associated with inherited cardiomyopathies. J. Biol. Chem. 2015;290:7003–7015. doi: 10.1074/jbc.M114.596676. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gupte, T. M., F. Haque, …, J. A. Mercer. 2015. Mechanistic heterogeneity in contractile properties of α-tropomyosin (TPM1) mutants associated with inherited cardiomyopathies. J. Biol. Chem. 290:7003-7015. [DOI] [PMC free article] [PubMed]
- 60.Moore J.R., Li X., Lehman W. Structural implications of conserved aspartate residues located in tropomyosin’s coiled-coil core. Bioarchitecture. 2011;1:250–255. doi: 10.4161/bioa.18117. [DOI] [PMC free article] [PubMed] [Google Scholar]; Moore, J. R., X. Li, …, W. Lehman. 2011. Structural implications of conserved aspartate residues located in tropomyosin’s coiled-coil core. Bioarchitecture. 1:250-255. [DOI] [PMC free article] [PubMed]
- 61.Zheng W., Barua B., Hitchcock-DeGregori S.E. Probing the flexibility of tropomyosin and its binding to filamentous actin using molecular dynamics simulations. Biophys. J. 2013;105:1882–1892. doi: 10.1016/j.bpj.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng, W., B. Barua, and S. E. Hitchcock-DeGregori. 2013. Probing the flexibility of tropomyosin and its binding to filamentous actin using molecular dynamics simulations. Biophys. J. 105:1882-1892. [DOI] [PMC free article] [PubMed]
- 62.Zheng W., Hitchcock-DeGregori S.E., Barua B. Investigating the effects of tropomyosin mutations on its flexibility and interactions with filamentous actin using molecular dynamics simulation. J. Muscle Res. Cell Motil. 2016;37:131–147. doi: 10.1007/s10974-016-9447-3. [DOI] [PubMed] [Google Scholar]; Zheng, W., S. E. Hitchcock-DeGregori, and B. Barua. 2016. Investigating the effects of tropomyosin mutations on its flexibility and interactions with filamentous actin using molecular dynamics simulation. J. Muscle Res. Cell Motil. 37:131-147. [DOI] [PubMed]
- 63.Manstein D.J., Meiring J.C.M., Gunning P.W. Actin-tropomyosin distribution in non-muscle cells. J. Muscle Res. Cell Motil. 2019 doi: 10.1007/s10974-019-09514-0. Published online May 4, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; Manstein, D. J., J. C. M. Meiring, …, P. W. Gunning. 2019. Actin-tropomyosin distribution in non-muscle cells. J. Muscle Res. Cell Motil. Published online May 4, 2019. [DOI] [PMC free article] [PubMed]
- 64.Hsiao J.Y., Goins L.M., Mullins R.D. Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 2015;25:1573–1582. doi: 10.1016/j.cub.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hsiao, J. Y., L. M. Goins, …, R. D. Mullins. 2015. Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 25:1573-1582. [DOI] [PMC free article] [PubMed]
- 65.Luther P.K. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Luther, P. K. 2009. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 30:171-185. [DOI] [PMC free article] [PubMed]
- 66.Johnson M., East D.A., Mulvihill D.P. Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 2014;24:1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]; Johnson, M., D. A. East, and D. P. Mulvihill. 2014. Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 24:1525-1530. [DOI] [PubMed]
- 67.Meiring J.C.M., Bryce N.S., Gunning P.W. Tropomyosin concentration but not formin nucleators mDia1 and mDia3 determines the level of tropomyosin incorporation into actin filaments. Sci. Rep. 2019;9:6504. doi: 10.1038/s41598-019-42977-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meiring, J. C. M., N. S. Bryce, …, P. W. Gunning. 2019. Tropomyosin concentration but not formin nucleators mDia1 and mDia3 determines the level of tropomyosin incorporation into actin filaments. Sci. Rep. 9:6504. [DOI] [PMC free article] [PubMed]
- 68.Liu H.P., Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]; Liu, H. P., and A. Bretscher. 1989. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 57:233-242. [DOI] [PubMed]
- 69.Hitchcock-DeGregori S.E., Singh A. What makes tropomyosin an actin binding protein? A perspective. J. Struct. Biol. 2010;170:319–324. doi: 10.1016/j.jsb.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hitchcock-DeGregori, S. E., and A. Singh. 2010. What makes tropomyosin an actin binding protein? A perspective. J. Struct. Biol. 170:319-324. [DOI] [PMC free article] [PubMed]
- 70.Fischer S., Rynkiewicz M.J., Lehman W. Tropomyosin diffusion over actin subunits facilitates thin filament assembly. Struct. Dyn. 2016;3:012002. doi: 10.1063/1.4940223. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fischer, S., M. J. Rynkiewicz, …, W. Lehman. 2016. Tropomyosin diffusion over actin subunits facilitates thin filament assembly. Struct. Dyn. 3:012002. [DOI] [PMC free article] [PubMed]
- 71.Flicker P.F., Phillips G.N., Jr., Cohen C. Troponin and its interactions with tropomyosin. An electron microscope study. J. Mol. Biol. 1982;162:495–501. doi: 10.1016/0022-2836(82)90540-x. [DOI] [PubMed] [Google Scholar]; Flicker, P. F., G. N. Phillips, Jr., and C. Cohen. 1982. Troponin and its interactions with tropomyosin. An electron microscope study. J. Mol. Biol. 162:495-501. [DOI] [PubMed]
- 72.Sousa D., Cammarato A., Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys. J. 2010;99:862–868. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sousa, D., A. Cammarato, …, W. Lehman. 2010. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys. J. 99:862-868. [DOI] [PMC free article] [PubMed]
- 73.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Inc.; 2001. Chapter 9. Polymerization of cytoskeletal filaments; pp. 151–164. [Google Scholar]; Howard, J. 2001. Chapter 9. Polymerization of cytoskeletal filaments. In Mechanics of Motor Proteins and the Cytoskeleton, (Sinauer Associates, Inc.), pp.151-164.
- 74.Broschat K.O., Weber A., Burgess D.R. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28:8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]; Broschat, K. O., A. Weber, and D. R. Burgess. 1989. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 28:8501-8506. [DOI] [PubMed]
- 75.Hagemann U.B., Mason J.M., Arndt K.M. Selectional and mutational scope of peptides sequestering the Jun-Fos coiled-coil domain. J. Mol. Biol. 2008;381:73–88. doi: 10.1016/j.jmb.2008.04.030. [DOI] [PubMed] [Google Scholar]; Hagemann, U. B., J. M. Mason, …, K. M. Arndt. 2008. Selectional and mutational scope of peptides sequestering the Jun-Fos coiled-coil domain. J. Mol. Biol. 381:73-88. [DOI] [PubMed]