Abstract
Background
Non-small cell lung cancer (NSCLC) is the most common cancer and the pathogenesis remain unclear. According to the competing endogenous RNA (ceRNA) theory, long noncoding RNA (lncRNA) have a competition with mRNAs for the connecting with miRNAs that affecting the level of mRNA. In this work, the ceRNA network and the important genes to predict the survival prognosis were explored.
Methods
In the study, we recognized differently expressed genes (mRNAs, lncRNAs and miRNAs) between NSCLC and normal tissues from The Cancer Genome Atlas database (fold change >2, P<0.01) using edgeR. Then, the interaction between lncRNA and miRNA or mRNA and miRNA was explored by miRcode, miRDB, TargetScan, and miRanda. Furthermore, the functions and KEGG pathway were analyzed with DAVID and KOBAS. The connections of these mRNAs were explored by STRING online database. The relation between genes in the network and survival time were further explored by survival package in R.
Results
By bioinformatics tools, we explored 155 lncRNAs, 30 miRNAs and 68 mRNAs and constructed ceRNA network. The functions and KEGG pathway of 68 mRNAs were further analyzed. AQP2, EGF, SLC12A1, TRPV5 and AVPR2 was in the center of network and may play key roles in the development of NSCLC. And mRNA (CCNB1, COL1A1, E2F7, EGLN3, FOXG1 and PFKP), miRNA (miR-31, miR-144 and miR-192) and lncRNA (AC080129.1, AC100791.1, AL163952.1, AP000525.1, AP003064.2, C2orf48, C10orf91, FGF12-AS2, HOTAIR, LINC00518, LNX1-AS1, MED4-AS1, MIG31HG, MUC2, TTTY16 and UCA1) were closely related with overall survival (OS).
Conclusions
In summary, the present study provides a deeper understanding of the lncRNA-related ceRNA network in NSCLC and some genes may be new target to treat for NSCLC patients.
Keywords: Long noncoding RNAs (lncRNAs), non-small cell lung cancer (NSCLC), The Cancer Genome Atlas, competing endogenous RNAs network, STRING
Introduction
Lung cancer is one of the most common cancer worldwide with the highest morbidity and mortality (1). Eighty to eighty-five percent of lung cancer is non-small cell lung cancer (NSCLC) (2). Though there is a great progress in surgery, chemotherapy and radiation therapy, the 5-year survival rate of NSCLC was still 15% (3). Hence, it is significantly important to have a deeper understanding of the molecular mechanism underlying NSCLC tumorigenesis and seek new therapeutic strategies for NSCLC. Moreover, the regional or distant metastasis will lead to high percent of mortality (4). Therefore, to better study the disease and find the new targets to cure has become extremely importantly. Long noncoding RNAs (lncRNAs) is a type of non-coding RNAs (ncRNAs) with 200 nucleotides to 100 kb in length (5). LncRNAs are with the function of regulating the target gene expression in the process of transcriptional and post-transcriptional but no protein-coding function (6). The research showed the manifestations of lncRNAs in tumors and normal tissues was different. Some lncRNAs played important roles in cancer progression and may be biomarkers in diagnosis, treatment and conditions of prognosis for its stronger tissue specificity (7). However, the clear mechanism of lncRNA in cancer was still not clear. In 2011, the theory of Competing endogenous RNA (ceRNA) hypothesis which is proposed by Sardina et al. was appealing (8). The main theory was some lncRNAs could have an effect of sponge on miRNAs which weak the impact of miRNAs on mRNAs. Additionally, there are more and more research showed the network of lncRNA, miRNA and mRNA is important in the pathogenesis and progression of cancers (9,10). For example, Zhang investigated the lncRNA analysed and built a lncRNA-miRNA-mRNA network in hepatocellular carcinoma (11). Besides, a ceRNA network of esophageal cancer was constructed (12). In renal cell cancer, a ceRNA network was constructed likewise (13). Nevertheless, the study of large-scale samples in NSCLC is uncommon. And the nexuses between prognosis and lncRNAs are undetermined. Therefore, building ceRNA network is very important for prognosis prediction, therapeutic decision and improving the overall survival (OS) for NSCLC sufferers.
At present we studied the expression of genes (lncRNA, miRNA and mRNA) in NSCLC from the database of TCGA. In addition, we built a ceRNA network in NSCLC by bioinformatics means which can find the novel targets and pathway to improve survival for patients.
Methods
Explore these genes with differentially expressed
We obtained the data of RNA sequencing from TCGA. One thousand and thirty-seven NSCLC tumor and 108 normal tissues of mRNA and lncRNA, 999 NSCLC tumor and 91 normal tissues of miRNA were obtained. The edgeR was used to detect the remarkable differentially expressed miRNAs (DEmiRNAs), lncRNAs (DElncRNAs) and mRNAs (DEmRNAs) (14). To maximize the reliability of these data, these lncRNAs which were not recoded in the database of GENCODE were excluded (15). |Log2FC| >2 and FDR <0.01 was set as the cut-off value (FC, fold change; FDR, false discovery rate) (16).
Explore the ceRNA network
The miRcode (http://www.mircode.org/) was used to predict lncRNA and miRNA interaction. Then to predict the miRNA and mRNA interaction, the database of miRDB, TargetScan and miRanda were utilized. At last we used the Cytoscape software to explore the lncRNA-miRNA-mRNA network with the interactions of results (17).
Function and pathway enrichment
We used the Database for Annotation Visualization and Integrated Discovery 6.8 (DAVID) to explore the function (18). Then the KOBAS was used to enrich the pathway of Kyoto Encyclopedia of Genes and Genomes (KEGG) (19). The connection between them was explored by STRING (20).
Statistic analysis
We used the unpaired t-test to explore the DEmiRNAs, DElncRNAs and DEmRNAs. To further understand these genes, the clinical data of NSCLC patients were explored as well. The R package of survival was utilized to plot the survivorship curve.
Results
Differentially expressed lncRNAs, miRNAs and mRNAs
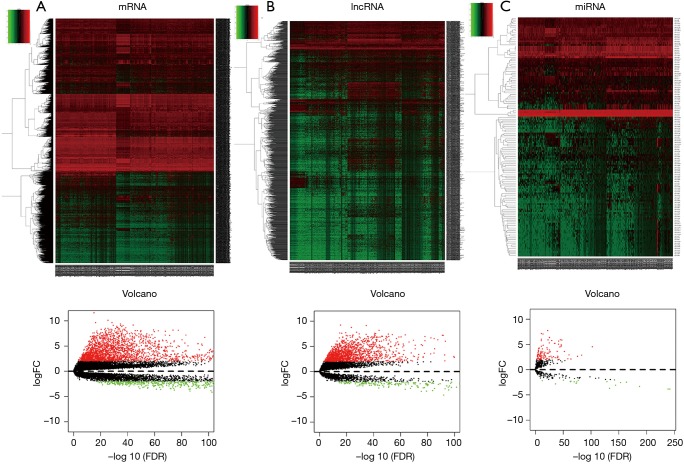
First, we explored the differentially expression of mRNA, miRNA and lncRNA. Two thousand and nine hundred sixty-five mRNAs (2,295 elevations and 670 downregulations, Figure 1A), 1,817 lncRNAs (1,605 elevations and 212 downregulations, Figure 1B) and 135 miRNAs between normal and cancerous tissues (117 elevated and 18 down-regulated, Figure 1C) were explored using the above standard filtration analysis (absolute fold change >2 and FDR value <0.01). The results indicate that these genes may distinguish between normal and cancerous tissues.
Figure 1.
Differentially expressed lncRNAs, miRNAs and mRNAs. (A) 2,965 mRNA (2,295 elevated and 670 downregulated); (B) 1,817 lncRNA (1,605 elevated and 212 downregulated); (C) 135 miRNA between normal tissues and cancer tissues.
The functions and KEGG pathway
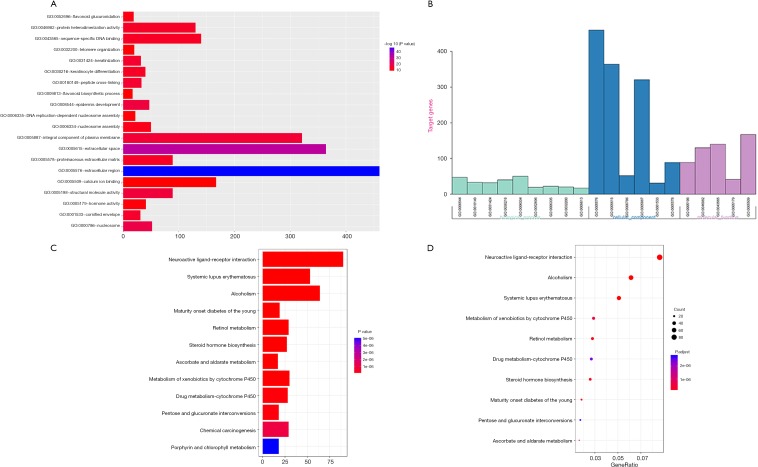
In addition, the function of DE mRNA and the KEGG pathway were explored by the DAVID and KOBAS online database. The top 20 GO function according to P value were extracellular region, extracellular space, nucleosome, epidermis development, structural molecules activity, integral component of plasma membrane and so on (Figure S1A,B). In the KEGG pathway analysis, the top 5 pathways according to P value was neuroactive ligand-receptor interaction, alcoholism, systemic lupus erythematosus, metabolism of xenobiotics by cytochrome P450 and steroid hormone biosynthesis (Figure S1C,D).
The network of DEmRNAs
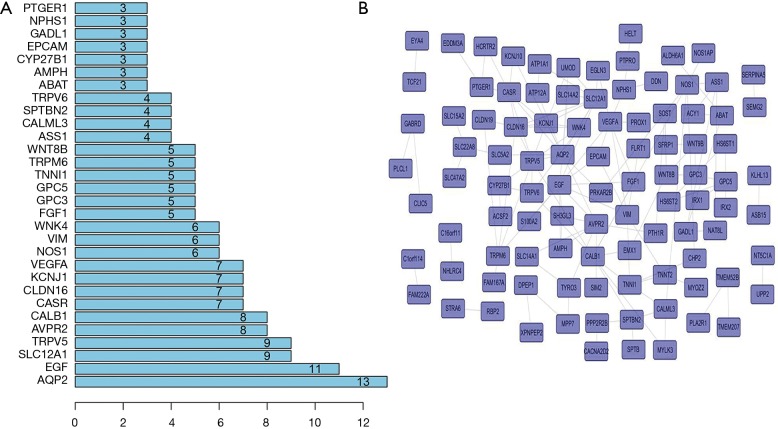
To explore the connections of these mRNAs whose expression is different between NSCLC and normal lung tissues, we used the STRING online database. The top 200 gene according to P value was explored by SRTING. The results showed AQP2, EGF, SLC12A1, TRPV5 and AVPR2 were in the center of network and may play key roles in the development of NSCLC (Figure 2).
Figure 2.
The network of DEmRNAs by STRING databases.
The construction of ceRNA network
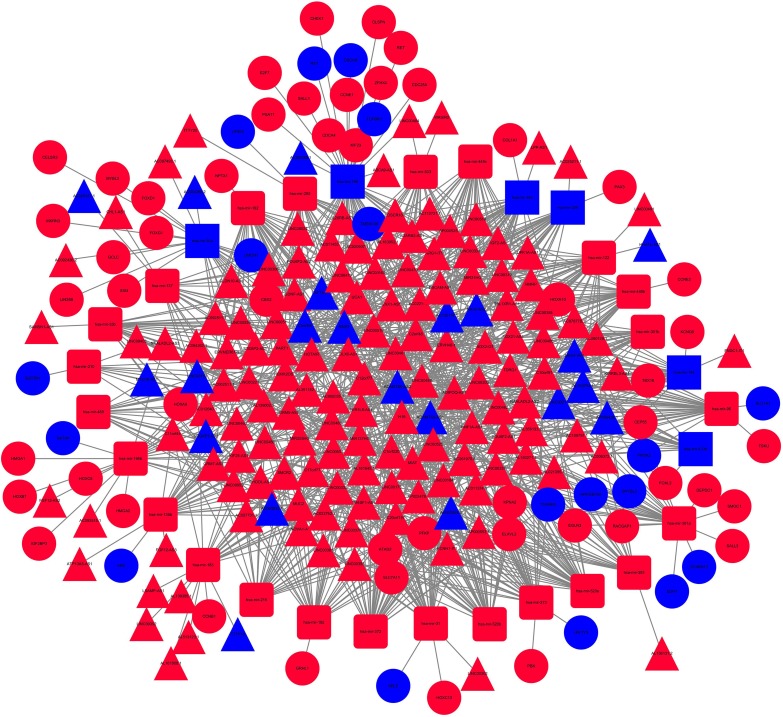
Then, potential interactions between these genes are predicted based on the ceRNA hypothesis. It is expected that the 155 DE1ncRNA can interact with 30 DEmiRNAs via the miRcode online tool. In addition, miRDB, miRanda and Targetscan were combined to predict 30 DEmRNA according to DEmiRNA. Furthermore, there are 68 target DEmRNAs involved in 2,897 different mRNAs were also enrolled in the network (Figure 3). Totally, there are 155 DElncRNAs, 30 DEmiRNAs and 68 DEmRNAs in the network. Besides, the interactions among DElncRNAs, DEmiRNAs and DEmRNAs were shown in Figure S2.
Figure 3.
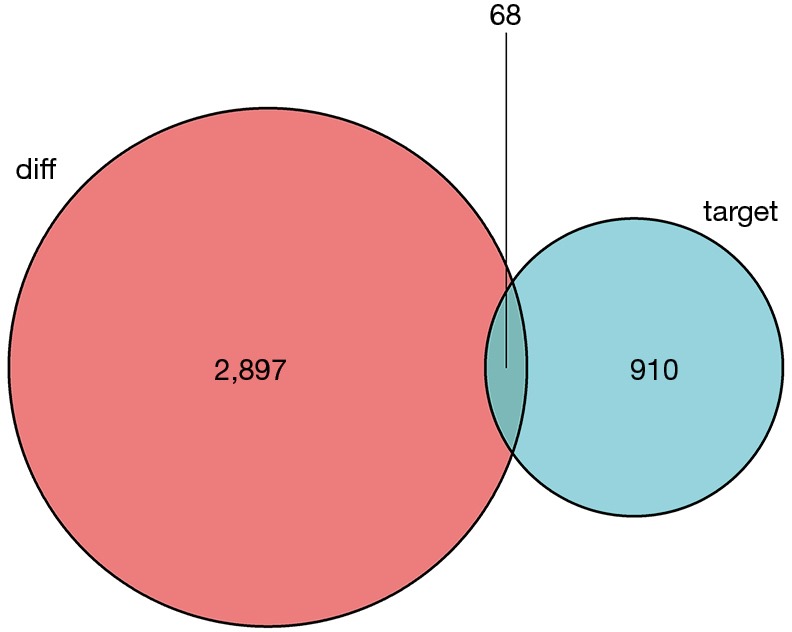
The 68 target DE mRNAs involved in 2,897 different mRNAs were involved in the ceRNA network.
Figure S2.
The interactions among DElncRNAs, DEmiRNAs and DEmRNAs.
The OS of ceRNA network
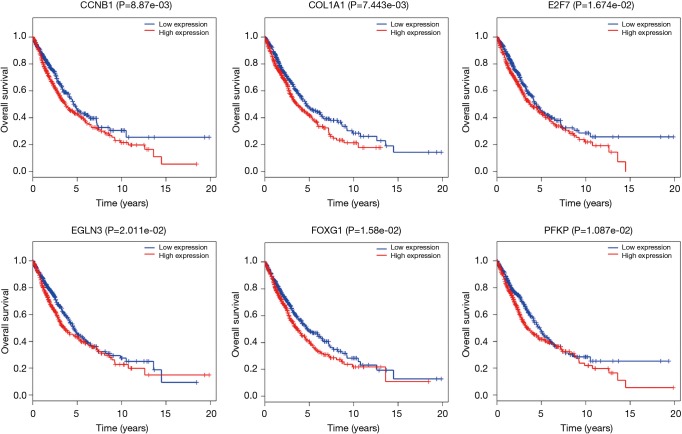
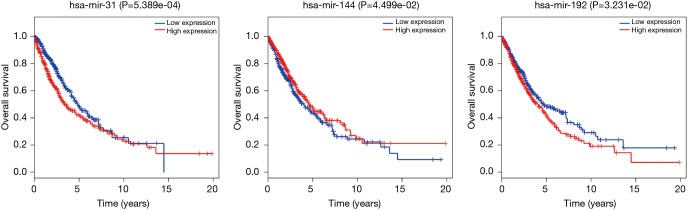
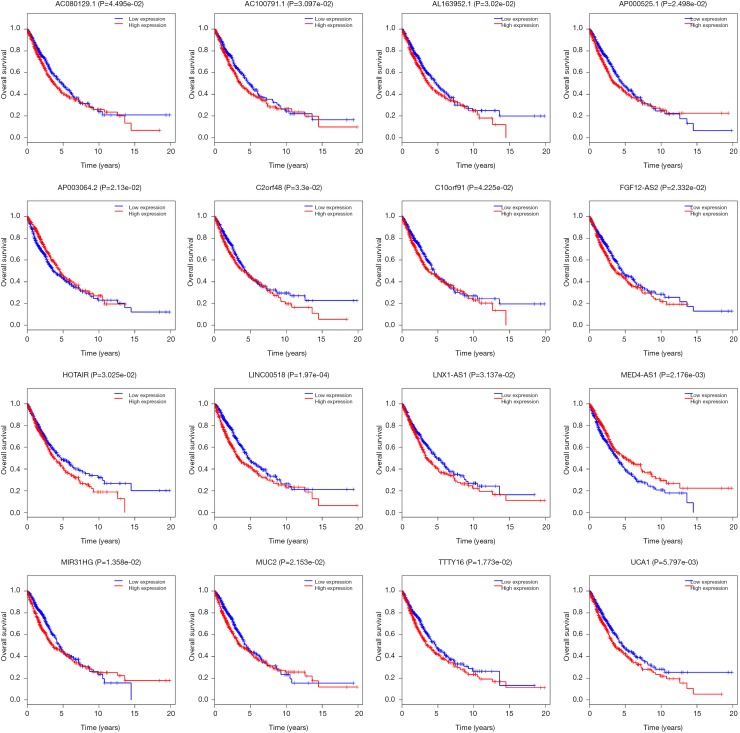
To further explore the function of the genes in the ceRNA network, we did the correlation between these genes and OS. There are 999 patients who have intact follow-up information enrolled in the study. The lncRNA, miRNA and mRNA in the ceRNA network were explored. The results showed the mRNA (CCNB1, COL1A1, E2F7, EGLN3, FOXG1 and PFKP) were closely related with OS (P<0.05, Figure 4). The miRNA (miR-31, miR-144 and miR-192, Figure 5) and lncRNA (AC080129.1, AC100791.1, AL163952.1, AP000525.1, AP003064.2, C2orf48, C10orf91, FGF12-AS2, HOTAIR, LINC00518, LNX1-AS1, MED4-AS1, MIG31HG, MUC2, TTTY16 and UCA1) were closely related with OS (P<0.05, Figure S3).
Figure 4.
The mRNA (CCNB1, COL1A1, E2F7, EGLN3, FOXG1 and PFKP) were closely related with overall survival.
Figure 5.
The miRNA (miR-31, miR-144 and miR-192, Figure 5) were closely related with overall survival.
Figure S3.
The lncRNA (AC080129.1, AC100791.1, AL163952.1, AP000525.1, AP003064.2, C2orf48, C10orf91, FGF12-AS2, HOTAIR, LINC00518, LNX1-AS1, MED4-AS1, MIG31HG, MUC2, TTTY16 and UCA1) were closely related with overall survival. lncRNA, long noncoding RNA.
Discussion
Lung cancer is the most common cause of cancer-related death in the world. Approximately 85% of lung cancer is NSCLC. Therefore, the mechanisms of this disease are urgently needed (21). LncRNAs play a significant role in the development of the disease according to many studies especially in cancer (22). In gastric cancer, lncRNA MT1JP could regulate FBXW7 expression through competitively binding to miR-92a (23). In ovarian cancer, lncRNA ABHD11-AS1 played a role in the tumorigenesis and progression by targeting Rhoc (24). In esophageal squamous cell carcinoma, lncRNA CASC9 promotes squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2 (25). In colorectal cancer, lncH19 acts oncogene in regulating cancer cell growth (26). In breast cancer, lncRNA-ATB promotes trastuzumab resistance and invasion (27). In prostate cancer, lncRNA HOXD-AS1 regulates proliferation and chemoresistance by recruiting WDR5 (28). Hence, lncRNAs play important biological roles by regulating gene expression and may be the biomarkers in diagnostics and target to treatment.
In our study, we found the differential expressed lncRNAs, miRNAs and mRNAs between NSCLC and normal lung tissues. Furthermore, we put the DEmRNAs into function and KEGG pathway analysis by using DAVID and KOBAS. The protein network was also constructed by String database. The connection between lncRNA and miRNA was conducted by miRcode (http://www.mircode.org/). The connection between miRNA and mRNA was conducted by miRDB database, TargetScan database and miRanda database. Next, the network was constructed by the Cytoscape software. Finally, the genes in the ceRNA network was also explored to detect whether they are related to OS. The results showed mRNA (CCNB1, COL1A1, E2F7, EGLN3, FOXG1 and PFKP) miRNA (miR-31, miR-144 and miR-192) and lncRNA (AC080129.1, AC100791.1, AL163952.1, AP000525.1, AP003064.2, C2orf48, C10orf91, FGF12-AS2, HOTAIR, LINC00518, LNX1-AS1, MED4-AS1, MIG31HG, MUC2, TTTY16 and UCA1) were closely related with OS (P<0.05).
But, a little of limitations still existed in this research. The major method of our research was bioinformatics technology, a useful tool for understanding the interactions, pathways and network. But, the networks and functions still need further verification.
Conclusions
In a word, the present study provides a deeper understanding of the lncRNA-related ceRNA network in NSCLC and provided new target for NSCLC patients.
Figure S1.
The functions and KEGG pathway. (A,B) Top 20 GO function of DEmRNAs according to P value; (C,D) the KEGG pathway.
Acknowledgments
Funding: This work was supported by the Fundamental Research Funds for the Provincial Universities in Heilongjiang (No. 2017LCZX94 to H Yin), Haiyan Foundation of Harbin Medical University Cancer Hospital (No. JJQN2017-10 to H Yin and No. JJQN2019-01 to X Wang), Chen Xiao-ping Foundation for the Development Science and Technology of Hubei Province (H Yin), by Hei Long Jiang Postdoctoral (X Wang) and China Postdoctoral Science Foundation (2018M641861 to X Wang), by Jieping Wu Foundation (No. 320.6799.15046 to J Ma) and Academy of Medical Sciences in Heilongjiang (No. 201606 to J Ma).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Barton MK. Encouraging long-term outcomes reported in patients with stage I non-small cell lung cancer treated with stereotactic ablative radiotherapy. CA Cancer J Clin 2017;67:349-50. 10.3322/caac.21375 [DOI] [PubMed] [Google Scholar]
- 3.Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2017: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin 2017;67:100-21. 10.3322/caac.21392 [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 5.Mendell JT. Targeting a Long Noncoding RNA in Breast Cancer. N Engl J Med 2016;374:2287-9. 10.1056/NEJMcibr1603785 [DOI] [PubMed] [Google Scholar]
- 6.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Gene regulation: Long RNAs wire up cancer growth. Nature 2013;500:536-7. 10.1038/nature12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sardina DS, Alaimo S, Ferro A, et al. A novel computational method for inferring competing endogenous interactions. Brief Bioinform 2017;18:1071-81. [DOI] [PubMed] [Google Scholar]
- 9.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Fan D, Jian Z, et al. Cancer Specific Long Noncoding RNAs Show Differential Expression Patterns and Competing Endogenous RNA Potential in Hepatocellular Carcinoma. PLoS One 2015;10:e0141042. 10.1371/journal.pone.0141042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue WH, Fan ZR, Li LF, et al. Construction of an oesophageal cancer-specific ceRNA network based on miRNA, lncRNA, and mRNA expression data. World J Gastroenterol 2018;24:23-34. 10.3748/wjg.v24.i1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Yuan N, Wu L, et al. An integrated analysis for long noncoding RNAs and microRNAs with the mediated competing endogenous RNA network in papillary renal cell carcinoma. Onco Targets Ther 2017;10:4037-50. 10.2147/OTT.S141951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760-74. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform 2008;6:9-15. 10.4137/CIN.S448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W , Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 19.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316-22. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fillon M. Immune checkpoint inhibitors are superior to docetaxel as second-line therapy for patients with non-small cell lung carcinoma. CA Cancer J Clin 2018;68:178-9. 10.3322/caac.21423 [DOI] [PubMed] [Google Scholar]
- 22.Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297-325. 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Li S, Lu J, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer 2018;17:87. 10.1186/s12943-018-0829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu DD, Chen X, Sun KX, et al. Role of the lncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol Cancer 2017;16:138. 10.1186/s12943-017-0709-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer 2017;16:150. 10.1186/s12943-017-0715-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. 10.18632/oncotarget.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget 2015;6:11652-63. 10.18632/oncotarget.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu P, Chen X, Xie R, et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol Ther 2017;25:1959-73. 10.1016/j.ymthe.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]