Abstract
Background
Patients with unprotected left main coronary artery disease (uLMCAD) have high mortality rate due to sudden heart failure and acute myocardial infarction, for which reliable diagnostic biomarkers to detect this disease at an early stage are in urgent need. Circulating microRNAs (miRNAs) have emerged as a class of novel biomarkers for cardiovascular diseases. The purpose of this study was to investigate utility of miRNAs as biomarkers for early detection of uLMCAD.
Methods
High-throughput sequencing (NGS) was initially employed to compare circulating miRNA expression profiles in uLMCAD patients to that in patients without coronary artery disease (CAD) to identify candidate miRNA biomarkers. We further validated the expression of candidate miRNAs by quantitative polymerase chain reaction (qPCR) in a larger cohort. Receiver operating characteristic (ROC) analysis with multivariate logistic regression was used to evaluate the diagnostic power of candidate miRNAs individually and combined.
Results
MiR-182-5p, miR-199a-5p and miR-5187-5p were found significantly differentially expressed through NGS (fold changes =1.35, 1.65, 0.5, P values =0.018, 0.046, 0.030, respectively, n=5 for both uLMCAD group and non-CAD control group). In a larger cohort (n=27 for uLMCAD patient and n=38 for non-CAD controls), qPCR confirmed that expression of miR-182-5p was up-regulated (2.57-fold, P=0.011) and expression of miR-5187-5p was down-regulated (0.47-fold, P=0.018) in the plasma of uLMCAD patients. ROC analysis with multivariate logistic regression show that miR-182 and miR-5187 have an AUC score of 0.97 and 0.94 respectively, indicating high diagnostic power as biomarkers for uLMCAD. Interestingly, correlation analysis suggests that the expression of two miRNAs were independent to each other.
Conclusions
These results suggested that circulating miR-182-5p and miR-5187-5p were suitable diagnostic biomarkers for uLMCAD, both potentially providing diagnostic information for discriminating uLMCAD patients from non-CAD population prior to invasive diagnostic coronary angiography (CAG).
Keywords: Unprotected left main coronary artery disease (uLMCAD), miR-182-5p, miR-5187-5p, diagnostic biomarker
Introduction
Unprotected left main coronary artery disease (uLMCAD) is a severe cardiovascular disease, which is defined as greater than or equal to 50% stenosis (≥50%) in left main coronary artery without bypass grafts to the left anterior descending or left circumflex coronary arteries. Significant stenosis in left main coronary artery is not rare and previously reported around 4–9% patients undergoing coronary angiography (CAG) with uLMCAD combining myocardial infarction (1). Patients with uLMCAD have high mortality without surgical management (2,3). Meanwhile these patients frequently suffer from cardiogenic shock or cardiac arrest and are at high risk for in-hospital major cardiac adverse events (4,5). Although coronary arteriography is an efficient exclusive method to diagnose uLMCAD, it still has various drawbacks including expensive cost, unavoidable limitation in early detection and diagnosis, operational complexity and potentially security risk. Therefore, effective non-invasive approaches and feasible biomarkers to detect uLMCAD, which allows early detection and intervention for uLMCAD, will greatly improve clinical management of patients with uLMCAD. Only a few studies examined the correlation between pretreatment plasma level of C-reactive protein or leukocyte count with the risk of death and myocardial infarction after percutaneous coronary intervention or coronary artery bypass graft surgery (6,7), but none provides diagnostic information for uLMCAD. Therefore, discovery of novel non-invasive markers to predict uLMCAD effectively in an early stage is pressing.
Increasing evidence indicates miRNAs are involved in many pathological cardiovascular conditions (8). For example, up-regulation of miR-130a mediates abnormal proliferation of vascular smooth muscle cells in hypertension (9). Elevated expression of miR-1 correlates with the progression of acute myocardial infarction (10). Recent research indicates that some circulating miRNAs plays critical roles in pathogenesis of acute coronary syndrome (11-13) The evidence predicts that miRNAs have broad prospects in early diagnosis of cardiovascular diseases. It is proposed using circulating miR-122-5p and miR-133b as a specific biomarker to identify patients with higher risk of an adverse prognosis after ST segment elevation myocardial infarction (14). However, there is no correlative research on seeking significant miRNA biomarkers for early diagnosis of uLMCAD.
In this study, we aimed to identify circulating miRNAs that may provide new diagnostic value for uLMCAD. We found significantly differentiated expression of circulating miR-182-5p, miR-199a-5p and miR-5187-5p between patients with uLMCAD and non-CAD controls in our screening cohort by high throughput sequencing (NGS). We further validated remarkably higher miR-182-5p expression and lower miR-5187-5p expression in patients with uLMCAD in a larger cohort. Interestingly, receiver operating characteristic (ROC) analysis manifested that miR-182-5p and miR-5187-5p had remarkable clinical diagnostic power for uLMCAD at early stage and may provide additional mechanistic insights into uLMCAD.
Methods
Patient selection
Patients with suspicious symptoms of coronary artery disease (CAD) underwent CAG as routine clinical diagnostic procedure. From March 2014 to July 2015 coronary blood plasma were collected from randomly selected patients for the purpose of examining biomarkers that may permit earlier diagnosis of unprotected LMCAD (ethics approval ID: 201502042). Inclusion criteria were defined as patients with de novo unprotected LMCAD confirmed by angiographic evidence of ≥50% stenosis of left main coronary artery. Non-CAD control group is consisted of patients without angiographic evidence of coronary artery stenosis (i.e., <50% stenosis in both left main coronary artery and triple vessel). The exclusion criteria was that patients could not have diagnosed high plasma glucose concentration, concomitant inflammation, infectious diseases and neoplastic diseases at enrollment. Risk factors related to unprotected LMCAD, age, gender, plasma high-density lipoprotein and cholesterol concentration, hypertension, coronary heart disease, myocardial infarction, diabetes mellitus, smoking and calcium channel blocker treatment were documented. Twenty-seven unprotected LMCAD patients and 38 non-CAD control subjects were enrolled in this study. This clinical study was approved by the Xiangya Hospital of Central South University Ethics Committee. Informed written consent was obtained from all participants.
Overall study design
In general, coronary blood (10 mL) was collected from the arterial catheter after angiographic imaging into EDTA collection tube. Plasma was immediately separated via centrifugation at 2,000× g for 15 min, and was transferred to a new tube and stored at –80 °C until subsequent RNA extraction. The subjects were randomly selected to a subset cohort (n=5 for uLMCAD patients and n=5 for non-CAD controls) for screening candidate miRNA biomarkers with high-throughput sequencing and the entire cohort was used for validating screened miRNA biomarkers.
miRNAs high-throughput sequencing for screening cohort
The expression of circulating miRNAs was determined in the screening cohort using high-throughput sequencing. The circulating RNA extraction and sequencing were performed at a commercial sequencing company (KangChen Bio-tech, Shanghai, China). Briefly, total circulating RNA in plasma was extracted using the miRNeasy Serum/Plasma kit (Qiagen, Germany). Total RNA was used to prepare miRNA sequencing libraries by using NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, USA) and RiboZero Magnetic Gold Kit (Human/Mouse/Rat) (Epicentre, Illumina, USA). The procedure included the following steps: (I) 3'-adaptor ligation with truncated T4 RNA ligase; (II) 5'-adaptor with T4 RNA ligase; (III) cDNA synthesis; (IV) cDNA PCR amplification; and (V) size selection of PCR-amplified fragments of ~120–140 bp (corresponding to ~15–25 nt small RNAs) from polyacrylamide gels. The libraries were quantified using an Agilent 2100 Bioanalyzer (Angilent, USA) and then were diluted to a final concentration of 8 pM, and pooled in equal volumes. Pooled libraries were denatured to single-stranded DNA molecules, which were then captured on Illumina flow cells (Illumina, CA, USA). In situ cluster generation was performed on an automated system cBot (Illumina) and finally libraries were sequenced (TruSeq Rapid SBS Kits) for 36 cycles on a HiSeq 2000 sequencer (Illumina) according to the manufacturer’s instructions.
Reverse transcription quantitative PCR (RT-qPCR)
RT-qPCR was used to confirm differential expression of three miRNA in 27 uLMCAD and 38 non-CAD patients. Circulating RNA was isolated from plasma using mirVana miRNA isolation kit (ThermoFisher Scientific, USA). Synthetic cel-miR-238-3p of 100 pM was spiked into plasma RNA and then RNAs were reverse-transcribed into cDNA using PrimeScript RT reagent kit (Takara, Japan) according to the manufacturer’s protocol. qPCR was performed using SYBR Premix Ex Taq kit (Takara) on an ABI 7500 real-time PCR system (Applied Biosystems, USA). The proprietary qPCR primers were used, which were designed and validated by a commercial company (RiboBio, Guangzhou, China). qPCR was performed in triplicates for each sample. The miRNA expression levels were analyzed using the comparative Ct method, normalized to the spiked-in exogenous cel-miR-238-3p.
GO and pathway analysis and protein-protein interaction (PPI) network establishment
The GO analysis and pathway analysis were carried out by Gene-Cloud of Biotechnology Information, Shanghai, China. Moreover, the PPI network was built online by DIANA TOOLS (http://diana.imis.athena-innovation.gr/).
Statistical analyses
Values were expressed as mean ± standard deviation (SD). Data were analyzed using the ‘heatmap.3’ function, SPSS and PrismGraphPad Version 5, whereby a P value <0.05 was considered statistically significant. The validation experiments generated values that were derived from individual RT-qPCR reactions and each of the miRNAs were transcribed from different genes and the miRNA data was log-transformed. Student’s t-tests and ANOVA were used to determine statistical differences when the data were normally distributed. Wilcoxon tests were used to detect statistical differences when the data were not normally distributed. Multivariate logistic regression models were constructed with stepwise addition of uLMCAD risk factors (age, gender, smoking, hypertension and HDL, total cholesterol, myocardial infarction history and calcium channel blockers) for each miRNA. Receiver operator characteristic (ROC) curves were analyzed to assess sensitivity and specificity of individual miRNAs and their combination using multiple logistic regression analysis. The optimal diagnostic point of the signature was assessed at cut-off values with the largest Youden’s index (sensitivity + specificity-1). ROC curves were analyzed to assess the sensitivity and specificity of each miRNA. This statistical test indicates the sensitivity and specificity of variable or individual miRNAs to discriminate between patients with uLMCAD and non-CAD controls. To consider multiple parameters, a multivariate logistic regression was performed with each individual miRNA together with age, gender, HDL and smoking to exclude their effect. The predicted probabilities from the multivariate logistic regression model were used to generate ROC curves.
Results
Identifying miRNAs differentially expressed in coronary plasma of uLMCAD patients using multiplex next-generation sequencing
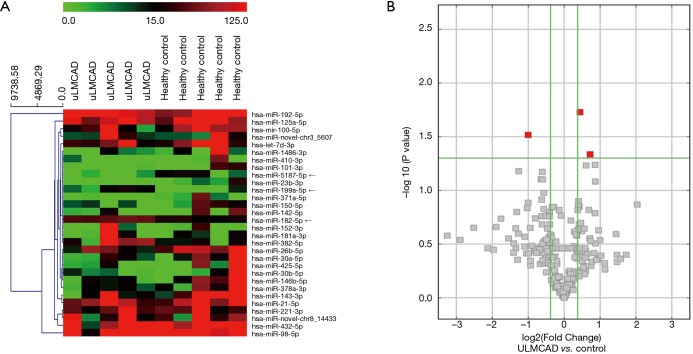
To identify miRNAs that are differentially expressed in the coronary plasma between uLMCAD patients and non-CAD control, multiplex miRNA sequencing was utilized to profiling total 1,457 miRNAs from the coronary plasma of the screening cohort (5 uLMCAD and 5 non-CAD controls; Table 1 for patient characteristics). A total of 1,457 circulating miRNAs were identified in coronary plasma and compared between uLMCAD patients and non-CAD controls. A heat map displays the top 30 dysregulated miRNAs (fold change >1.3) (Figure 1A). Among the candidate miRNAs, significantly up-regulated expression of hsa-miR-182-5p and hsa-miR-199a-5p and down-regulated expression of hsa-miR-5187-5p were exhibited in the plasma from patients with uLMCAD (fold changes =1.35, 1.65, 0.5, P values =0.018, 0.046, 0.030, respectively) (Figure 1B).
Table 1. Patient characteristics for sequencing.
| Characteristic | uLMCAD (n=5) | Non-CAD (n=5) | P value |
|---|---|---|---|
| Age(years), mean ± SD | 56.00±5.43 | 57.00±6.89 | 0.805 |
| Male gender, n (%) | 4 [80] | 2 [40] | 0.197 |
| Hypertension, n (%) | 2 [40] | 1 [20] | 0.490 |
| DM, n (%) | 0 [0] | 0 [0] | 1.000 |
| History of MI, n (%) | 1 [20] | 0 [0] | 0.292 |
| HDL (mmol/L), mean ± SD | 0.96±0.13 | 1.27±0.05 | 0.003* |
| LDL (mmol/L), mean ± SD | 3.11±2.36 | 2.84±0.51 | 0.863 |
| TC (mmol/L), mean ± SD | 1.65±0.56 | 1.61±0.70 | 0.931 |
| Past smoker, n (%) | 3 [60] | 0 [0] | 0.038* |
| Medication | |||
| CCB, n (%) | 4 [80] | 1 [20] | 0.058 |
*, P<0.05 for comparison between two groups. Patient characteristics were compared in patients with uLMCAD and non-CAD controls. DM, diabetes mellitus; CHD, coronary heart disease; MI, myocardial infarction; HDL, high density lipoprotein; LDL, low density lipoprotein; TC, total cholesterol; CCB, calcium channel blockers.
Figure 1.
Overview of miRNAs expression profiling in coronary plasma. (A) Heat map and unsupervised hierarchical clustering analysis of the top 30 miRNAs differential expression level between 5 uLMCAD cases and 5 non-CAD controls; (B) volcano plot of detectable genome-wide miRNA profiles in screening cohorts. The X-axis shows the log2 fold change in miRNAs’ expression levels in uLMCAD patients compared to non-CAD controls, while the Y-axis shows the –log10 of P value. The highlighted red square symbols mark the three miRNAs with the lowest P values found in the plasma from uLMCAD patients compared with the non-CAD controls: miR-182-5p, miR-5187-5p and miR-199a-5p.
Validation of candidate miRNAs by quantitative RT-Qpcr
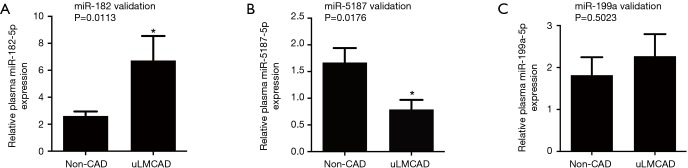
According to the miRNA high-throughput sequencing data, we further validated the expression levels of identified candidate miRNAs, (i.e., miR-182-5p, miR-199a-5p and miR-5187-5p) in the entire cohort of 27 patients with uLMCAD and 38 non-CAD controls (see Table 2 for patient characteristics) by RT-qPCR (Figure 2). We confirmed that patients with uLMCAD had significantly elevated expression of miR-182-5p (Figure 2A, fold change =2.57, P=0.011) and reduced expression of miR-5187-5p expression (Figure 2B, fold change =0.47, P=0.018) compared to non-CAD controls. While circulating miR-199a-5p (Figure 2C, P=0.502) were not differentially expressed between uLMCAD patients and non-CAD controls.
Table 2. Patient characteristics for validation.
| Characteristic | uLMCAD (n=27) | Non-CAD (n=38) | P value |
|---|---|---|---|
| Age (years), mean ± SD | 65.67±11.85 | 56.47±10.04 | 0.001* |
| Male gender, n (%) | 24 [89] | 18 [47] | 0.001* |
| Hypertension, n (%) | 17 [65] | 12 [32] | 0.008* |
| DM, n (%) | 1 [4] | 2 [5] | 1.000 |
| History of MI, n (%) | 4 [15] | 0 [0] | 0.026* |
| HDL (mmol/L), mean ± SD | 1.13±0.24 | 1.36±0.38 | 0.006* |
| LDL (mmol/L), mean ± SD | 2.32±0.86 | 2.77±0.91 | 0.055 |
| TC (mmol/L), mean ± SD | 3.95±1.16 | 4.55±1.06 | 0.043* |
| Past smoker, n (%) | 18 [69] | 16 [42] | 0.033* |
| Medication, n (%) | |||
| CCB | 12 [50] | 8 [21] | 0.018* |
| Anti-Plt | 27 [100] | 36 [94] | 0.226 |
*, P<0.05 for comparison between two groups. Patient characteristics were compared in patients with uLMCAD and non-CAD controls. DM, diabetes mellitus; CHD, coronary heart disease; MI, myocardial infarction; HDL, high density lipoprotein; LDL, low density lipoprotein; TC, total cholesterol; CCB, calcium channel blockers.
Figure 2.
Validation of 3 differentially expressed candidate miRNAs in coronary plasma between uLMCAD patients and non-CAD controls. Relative expression of miR-182-5p (A), miR-5187-5p (B) and miR-199a-5p (C) between 27 uLMCAD patients and 38 non-CAD controls by RT-qPCR, respectively. *, P<0.05. RT-qPCR, reverse transcription quantitative polymerase chain reaction; CAD, coronary artery disease; uLMCAD, unprotected left main coronary artery disease.
Diagnostic accuracy of candidate miRNAs
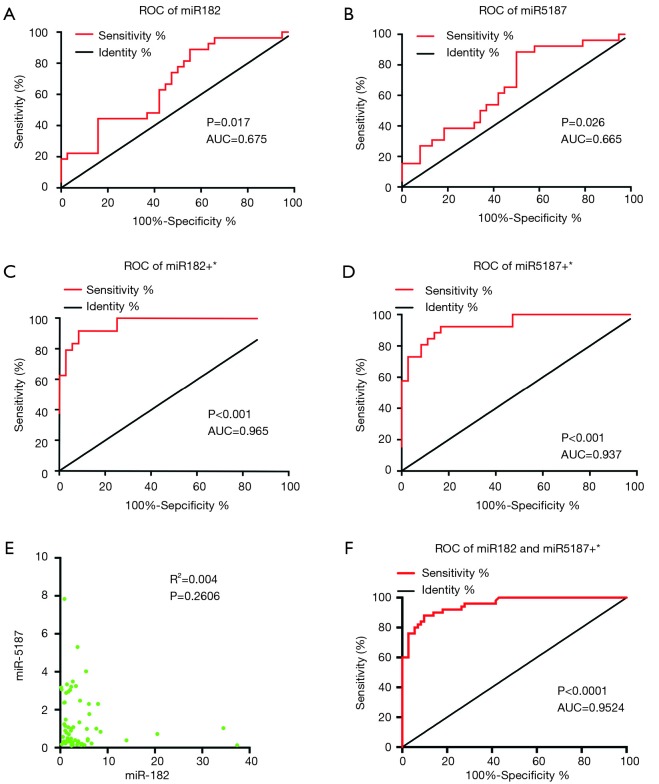
To further evaluate utility of the two differentially expressed miR-182-5p or miR-5187-5p as diagnostic markers for uLMCAD, ROC curve analysis was performed, without and with adjusting potential confounding factors. Multivariate logistic regression analysis was utilized to adjust effects of confounding factors age, HDL, gender, hypertension and smoking. The area under the curves (AUC) ranged 0.65–0.66 for individual miRNA (0.68 for miR-182-5p, cut-off value is 1.573, Figure 3A; 0.67 for miR-5187-5p, cut-off value is 1.115, Figure 3B) and reached more than 0.9 after adjusting those confounding factors (0.97 for miR182, Figure 3C; 0.94 for miR-5187, Figure 3D). We further examined correlation of expression of miR-182-5p and miR-5187-5p, and confirmed that regulation of the two miRNAs is independent to each other (P=0.2606, R2=0.004) (Figure 3E). As shown in Figure 3F, coordinated miR-182-5p and miR-5187-5p also showed powerful diagnosis ability with adjusting confounding factors (AUC =0.95).
Figure 3.
Investigation of miR-182 and miR-5187 acting as potential biomarkers for uLMCAD. ROC analysis of miR-182-5p (A) and miR-5187-5p (B); ROC analysis of miR182 (C) and miR-5187 (D) with adjusting the confounding influences of age, gender, Hypertension, MI, HDL, smoker and CCB; (E) correlation analysis between miR-182 and miR-5187; (F) ROC analysis of combined miR-182-5p and miR-5187-5p with adjusting the confounding factors. *, indicates adjust of age, gender, hypertension, MI, HDL, smoker and CCB. uLMCAD, unprotected left main coronary artery disease; ROC, receiver operating characteristic; MI, myocardial infarction; HDL, high density lipoprotein; CCB, calcium channel blockers.
Discussion
In our study, we are the first time to identified 2 novel circulating miRNAs, up-regulated miR-182-5p and down-regulated miR-5187-5p expression in patients with uLMCAD, validated with NGS and RT-qPCR. ROC curve analyses with multivariate logistic regression indicated that miR-182-5p and miR-5187-5p had significant diagnostic power in uLMCAD compared with non-CAD controls.
Previous studies have supported that percutaneous coronary intervention (PCI) and coronary artery bypass grafting can work as effective therapy to decrease mortality and improve the life quality for uLMCAD (15). However, besides CAG, there is no suitable diagnostic measurement for uLMCAD. Interestingly, increasing evidences suggest circulating miRNAs could function as biomarkers for diagnosis or prognosis of cardiovascular diseases (16). For example, circulating miR-499-5p was associated with risk of death in elderly patients after non-ST elevation myocardial infarction (17). In our study, we initially defined circulating miR-182-5p, miR-199a-5p and miR-5187-5p as candidate miRNAs of biomarkers to detect uLMCAD after the high-throughput sequencing in a subset screening cohort. Since several co-existing parameters might affect the diagnostic power of plasma miRNAs, we excluded the patients with diagnosed high plasma glucose concentration, acute myocardial infarction, infection and other influence factors. We further performed the validation by quantitative real-time PCR in a larger cohort and then found remarkably elevated plasma miR-182-5p and down-regulated miR-5187-5p in uLMCAD patients compared with non-CAD control. ROC analysis with multivariate logistic regression analysis for the diagnostic power of miR-182-5p presented an AUC of 0.97 in differentiating uLMCAD patients from non-CAD controls, and ROC of miR-5187-5p presented AUC of 0.94. These results suggested that circulating miR-182-5p and miR-5187-5p may be the most valuable biomarker among these 3 miRNAs regarding to its most significant correlation with the disease. However, the case size was relatively small, which may cause some statistical issues on logistic regression and the c-statistic. Therefore, a larger, multi-institutional, controlled trial will be beneficial to further validate utility of miR-182-5p and miR5187-5p as diagnostic biomarkers for uLMCAD. Several researches have demonstrated that multiplex miRNA biomarkers constitute a more accurate approach for diagnosing cardiovascular diseases compared to using any of miRNA biomarkers individually (18). However, no study has examined the combination of 2 or more miRNAs as biomarkers for the diagnosis of uLMCAD. After confirming that the gene expression is independent between miR-182-5p and miR-5187-5p, we examined and found that combined miR-182-5p and miR-5187-5p had high predictive power as well, and the AUC values were 0.95, suggesting that combination of two candidate miRNAs may be powerful in diagnosis as well.
Recently, increasing number of researches on functions of miR-182-5p and miR-199a reported, while there were few studies on miR-5187-5p. MiR-182-5p, first identified and cloned from the mouse eyes, was reported to be of great importance in several cancers (19). Regulating miR-182-5p could be considered as a potential therapeutic strategy for renal cell carcinoma (20). It could also work as an oncogene in human bladder cancer (21) and could act as a potential biomarker in breast cancer (22). Moreover, miR-182-5p could also be used as early sensitive cardiotoxicity biomarkers to screen potential drugs and environmental cardiotoxicants (23). For miR-199a, one study reported that downregulation of miR-199a associates with inhibition of hypoxia-inducible factor-1a in cardiac myocytes (24). There was a research reported that miR-199a could affect NF-κB activity in ovarian cancer cells through regulating IKKβ (25). We believe that a more thorough and systematic study of miR-182-5p, miR-199a and miR-5187-5p may help elucidate their biological roles and underlying mechanisms in cardiovascular diseases and other diseases.
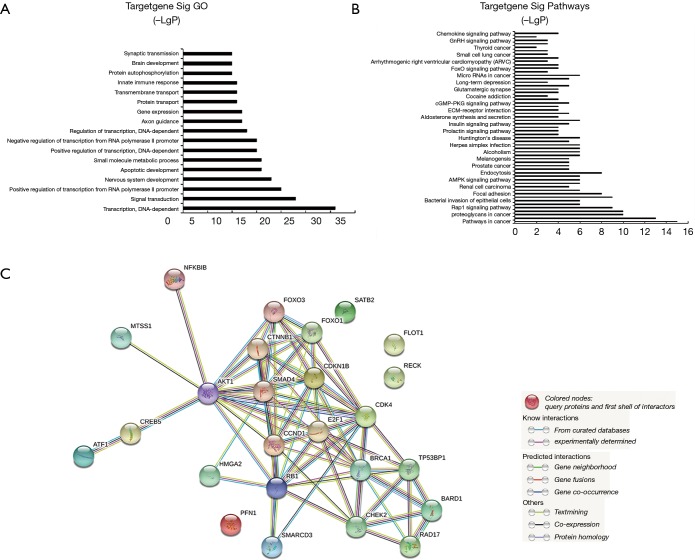
To explore the probable functions and downstream networks of the circulating miRNAs, gene ontology analysis predicted that the functions of miR-182-5p and miR-5187-5p were potentially mostly associated with DNA-dependent transcription (Figure 4A,B). Pathway analysis indicated that pathway in cancer signaling might be the downstream pathway of the above-mentioned miRNAs involved in uLMCAD. In order to further explore most important downstream targets of miR-182-5p, we constructed a PPI network of 26 miR-182-5p targets, which are validated by western blot and luciferase reporter gene assays (http://diana.imis.athena-innovation.gr/) (Figure 4C). Worthy of notice, most of them are network hubs, such as Activating transcription factor 1 (AKT1) (42 interactions), Catenin Beta 1 (CTNNB1) (20 interactions), SMAD family member 4 (SMAD4) (25 interactions), E2F family member 1 (E2F1) (30 interactions). Few validated targets of miR-5187 are available for constructing a PPI network. Therefore, pathway analyses predicted that pathway in cancer signaling was the most likely to be the downstream pathway target by the miRNAs, whose expression is dysregulated in uLMCAD. This prediction analysis hints that pathway in cancer signaling may involve in pathology of uLMCAD. Pathway in cancer signaling includes some subunit pathways, like the Notch signaling pathway and the VEGF signaling pathway. Both of them are very correlated to angiogenesis and arteriogenesis (26-28). As far as we know, arteriogenesis plays a very crucial part in CAD, and the patient who has good collateral circulation will avoid uLMCAD. Additionally, we found, AKT1, a validated target of miR-182-5p, possessed the highest degree of interaction among 26 validated targets of miR182-5p. Numerous studies showed that AKT1 played a very important role in cardiovascular disease. Loss of AKT1 will lead to severe atherosclerosis and CAD (29). And absence of AKT1 will induce cardiac dysfunction and reduce vascular smooth cell migration and survival (30). Given all that, in our future work, AKT1 is worthy of much focus and miR-182-5p might interact with it to function in uLMCAD .
Figure 4.
Prediction of target genes of miR-182-5p. GO (A) and signaling pathway analysis (B) of down-regulated genes by miR-182-5p and up-regulated downstream target genes by miR-5187 in uLMCAD patients compared with non-CAD; (C) protein-protein interaction network of 26 validated targets of miR-182-5p. CAD, coronary artery disease; uLMCAD, unprotected left main coronary artery disease.
Limitation
For the first time, to our knowledge, we identified that 2 circulating miRNAs, miR-182-5p and miR-5187-5p, all of them have potential to be biomarkers for diagnosis of uLMCAD. MiR-182-5p is the most promising one among them to be a suitable biomarker. However, the sample size is insufficient to make a conclusion. A large-scaled study is in need to assure our results and further validate the candidate biomarkers’ specificity and sensitivity. Finally, an experimental model should be established to further verify the targets of miR-182-5p and its role in uLMCAD process.
Acknowledgments
Funding: This study was supported in part by grants from the National Natural Science Foundation of China (No. 81873509, 81570239, 81770301, and 81670267).
Ethical Statement: This clinical study was approved by the Xiangya Hospital of Central South University Ethics Committee. Informed written consent was obtained from all participants.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Lenzen MJ, Boersma E, Bertrand ME, et al. Management and outcome of patients with established coronary artery disease: the Euro Heart Survey on coronary revascularization. Eur Heart J 2005;26:1169-79. 10.1093/eurheartj/ehi238 [DOI] [PubMed] [Google Scholar]
- 2.Taylor HA, Deumite NJ, Chaitman BR, et al. Asymptomatic left main coronary artery disease in the Coronary Artery Surgery Study (CASS) registry. Circulation 1989;79:1171-9. 10.1161/01.CIR.79.6.1171 [DOI] [PubMed] [Google Scholar]
- 3.Lee WC, Tsai TH, Chen YL, et al. Safety and feasibility of coronary stenting in unprotected left main coronary artery disease in the real world clinical practice—a single center experience. PloS One 2014;9:e109281. 10.1371/journal.pone.0109281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montalescot G, Brieger D, Eagle KA, et al. Unprotected left main revascularization in patients with acute coronary syndromes. Eur Heart J 2009;30:2308-17. 10.1093/eurheartj/ehp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdolrahimi S, Sanati H, Fatahian A, et al. Evaluation of Percutaneous Coronary Intervention and stenting of Left Main Coronary Artery Stenosis in Tehran’s Rajaie and Lavasani Hospitals from 2010 to 2011. Res Cardiovasc Med 2013;2:181-4. 10.5812/cardiovascmed.12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry TE, Muehlschlegel JD, Liu KY, et al. Preoperative C-reactive protein predicts long-term mortality and hospital length of stay after primary, nonemergent coronary artery bypass grafting. Anesthesiology 2010;112:607-13. 10.1097/ALN.0b013e3181cea3b5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmerini T, Marzocchi A, Marrozzini C, et al. Preprocedural levels of C-reactive protein and leukocyte counts predict 9-month mortality after coronary angioplasty for the treatment of unprotected left main coronary artery stenosis. Circulation 2005;112:2332-8. 10.1161/CIRCULATIONAHA.105.551648 [DOI] [PubMed] [Google Scholar]
- 8.Cui RR, Li SJ, Liu LJ, et al. MicroRNA-204 regulates vascular smooth muscle cell calcification in vitro and in vivo. Cardiovascular research 2012:96:320-9. 10.1093/cvr/cvs258 [DOI] [PubMed] [Google Scholar]
- 9.Wu WH, Hu CP, Chen XP, et al. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens 2011;24:1087-93. 10.1038/ajh.2011.116 [DOI] [PubMed] [Google Scholar]
- 10.Li LM, Cai WB, Ye Q, et al. Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J Emerg Med 2014;5:182-6. 10.5847/wjem.j.issn.1920-8642.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakob P, Kacprowski T, Briand-Schumacher S, et al. Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur Heart J 2017;38:511-5. [DOI] [PubMed] [Google Scholar]
- 12.Sayed AS, Xia K, Yang TL, et al. Circulating microRNAs: a potential role in diagnosis and prognosis of acute myocardial infarction. Dis Markers 2013;35:561-6. 10.1155/2013/217948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Xu J, Huang F. Recent players in the field of acute myocardial infarction biomarkers: circulating cell-free DNA or microRNAs? Int J Cardiol 2013;168:2956-7. 10.1016/j.ijcard.2013.03.118 [DOI] [PubMed] [Google Scholar]
- 14.Cortez-Dias N, Costa MC, Carrilho-Ferreira P, et al. Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ J 2016;80:2183-91. 10.1253/circj.CJ-16-0568 [DOI] [PubMed] [Google Scholar]
- 15.Vis MM, Beijk MA, Grundeken MJ, et al. A systematic review and meta-analysis on primary percutaneous coronary intervention of an unprotected left main coronary artery culprit lesion in the setting of acute myocardial infarction. JACC Cardiovasc Interv 2013;6:317-24. 10.1016/j.jcin.2012.10.020 [DOI] [PubMed] [Google Scholar]
- 16.Jung HJ, Suh Y. Circulating miRNAs in ageing and ageing-related diseases. J Genet Genomics 2014;41:465-72. 10.1016/j.jgg.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olivieri F, Antonicelli R, Spazzafumo L, et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int J Cardiol 2014;172:e276-8. 10.1016/j.ijcard.2013.12.203 [DOI] [PubMed] [Google Scholar]
- 18.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol 2011;80:193-208. 10.1016/j.critrevonc.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 19.Krishnan K, Steptoe AL, Martin HC, et al. MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 2013;19:230-42. 10.1261/rna.034926.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Wu J, Li S, et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol Cancer 2014;13:109. 10.1186/1476-4598-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21. 10.1016/j.ccell.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang PY, Gong HT, Li BF, et al. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol Lett 2013;6:1681-6. 10.3892/ol.2013.1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhari U, Nemade H, Gaspar JA, et al. MicroRNAs as early toxicity signatures of doxorubicin in human-induced pluripotent stem cell-derived cardiomyocytes. Arch Toxicol 2016;90:3087-98. 10.1007/s00204-016-1668-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rane S, He M, Sayed D, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res 2009;104:879-86. 10.1161/CIRCRESAHA.108.193102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R, Alvero AB, Silasi DA, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 2008;27:4712-23. 10.1038/onc.2008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Büchler P, Gazdhar A, Schubert M, et al. The Notch signaling pathway is related to neurovascular progression of pancreatic cancer. Ann Surg 2005;242:791-800. 10.1097/01.sla.0000189115.94847.f1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacker SA, Achen MG. The VEGF signaling pathway in cancer: the road ahead. Chin J Cancer 2013;32:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye QF, Zhang YC, Peng XQ, et al. Silencing Notch-1 induces apoptosis and increases the chemosensitivity of prostate cancer cells to docetaxel through Bcl-2 and Bax. Oncol Lett 2012;3:879-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Hernando C, Ackah E, Yu J, et al. Loss of Akt1 leads to severe atherosclerosis and occlusive coronary artery disease. Cell Metab 2007;6:446-57. 10.1016/j.cmet.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Hernando C, József L, Jenkins D, et al. Absence of Akt1 reduces vascular smooth muscle cell migration and survival and induces features of plaque vulnerability and cardiac dysfunction during atherosclerosis. Arterioscler Thromb Vasc Biol 2009;29:2033-40. 10.1161/ATVBAHA.109.196394 [DOI] [PMC free article] [PubMed] [Google Scholar]