Abstract
Background
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have become important treatment options for non-small cell lung cancer (NSCLC) patients with EGFR sensitive mutation. However, the detection of EGFR driver mutation is impeded by the lack of adequate tumor tissues, histopathological type, long detection period, and the heterogeneity of a tumor. Therefore, it is necessary to develop a more convenient method to guide the clinical use of EGFR-TKI. Circular RNAs (circRNAs) are characterized as a closed structure with covalently joined ends resistant to exonucleases may be a potential biomarker. In the present study, we aimed to screen circRNAs that may be associated with the efficacy of EGFR-TKI.
Methods
The expression of circRNAs sequenced by circular microarray in plasma samples between gefitinib effective and ineffective groups were compared. RT-qPCR further validated the results in an independent cohort. Kaplan-Meier curves were used to analyze the association between circRNA and progression-free survival (PFS) of NSCLC patients treated with gefitinib.
Results
In total, 52 NSCLC patients treated with gefitinib were included for analysis. 1,377 circRNAs were differentially expressed in gefitinib effective and ineffective groups, among which 989 circRNAs were up-regulated, and 388 circRNAs were down-regulated in the effective group. Furthermore, two differentially expressed circRNAs, hsa_circ_0109320 and hsa_circ_0134501, were validated by RT-qPCR in an independent cohort of 38 gefitinib-treated NSCLC patients. Elevated hsa_circ_0109320 was associated with longer PFS in gefitinib-treated NSCLC patients.
Conclusions
Taken together, hsa_circ_0109320 may be a potential biomarker for the efficacy of EGFR-TKI in NSCLC patients. This provides a new molecular typing method for individualized precision treatment.
Keywords: Non-small cell lung cancer (NSCLC), circular RNA (circRNA), epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI), predictive biomarker
Introduction
Lung cancer is common, and it one of the leading causes of cancer-related deaths worldwide (1). There were more than 1.8 million new cases and almost 1.6 million deaths estimated in 2012 (2). Despite progress in its detection and treatment, the prognosis of lung cancer is still poor with a 5-year overall survival rate of only about 11% (3). Lung cancer has two main histological types: non-small cell lung cancer (NSCLC) and small cell lung cancer. NSCLC accounts for approximately 85% of all lung cancer cases and almost consists of squamous cell carcinoma, adenocarcinoma, and other types (3,4). Most patients with NSCLC have advanced local invasion and distant metastases at the time of diagnosis (3,4).
In recent years, with the development of molecular diagnostic techniques, new driver genes of lung cancer have been increasingly discovered. Consequently, new drugs designed to target these driver genes have emerged. Targeted drugs such as EGFR-TKIs including gefitinib, icotinib, and erlotinib have become important treatment options for patients with advanced NSCLC (5). The presence of EGFR mutations in tumors is a prerequisite for the administration of EGFR-TKIs (5). The detection rate of EGFR mutations in lung cancer is slightly low, especially in China, where it is only 50%. The most common causes for the low detection rate include a lack of adequate tumor tissues, pathological histological limitations (such as for squamous cell carcinoma), and long detection periods. Moreover, because of tumor heterogeneity, genetic testing of a single biopsy could not describe well the overall genetic changes in a tumor (5). Therefore, to achieve individualized risk stratification, a sensitive and convenient blood-based efficacy prediction system would be highly valuable.
Circular RNA (circRNA) is a newly identified class of non-coding RNA molecules that are formed by back splicing of more than one exon. CircRNAs are abundant in eukaryotic cells, exhibiting tissue, temporal, and disease specificities (6). Some circRNAs have a sponge effect on microRNAs (miRNAs), which releases the inhibitory effect of miRNAs on their target genes and up-regulates the expression of the target genes, playing an essential regulatory role in multiple biological processes and even some diseases (7). CircRNAs play essential roles in many diseases such as tumors by regulating gene transcription, and protein translation (8). Unlike linear RNAs, circRNAs are covalently closed continuous loops without 5’ to 3’ polarity or polyadenylated tails, making them less prone to be degraded by exonucleases and more stable than linear RNAs (7). Therefore, circRNAs can potentially become good molecular markers for they have these features.
Because of the non-invasive nature of plasma samples, circRNAs in plasma could be ideal biomarkers (9). To date, a few studies have reported the detection of circRNAs in cell-free plasma samples (9,10). However, plasma circRNAs that can be used to predict the therapeutic efficacy in cancer has never been reported.
In this study, we aimed to screen possible circRNA that were associated with the survival of NSCLC patients treated with gefitinib. We found that two circRNAs, hsa_circ_0109320 and hsa_circ_0134501, were highly expressed in gefitinib-effective NSCLC patients. Further, we demonstrated that hsa_circ_0109320 expression was associated with better progression-free survival (PFS). Taken together, hsa_circ_0109320 may be a useful biomarker for the prognosis in gefitinib-treated patients with NSCLC and further prospective studies are warranted.
Methods
Patients
Fifty-two NSCLC patients treated with gefitinib in the Cancer Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College between January 2010 and November 2014 were included in this study. All these patients had locally advanced or metastatic NSCLC, which was confirmed by a histological or cytological diagnosis. At the same time, the following patients were excluded incomplete electronic medical records and patients without evaluable target lesion. The circRNA microarray technology measured the expression of circRNAs in the plasma samples of 14 patients including 8 effective patients whose PFS were more than 10 months and 6 ineffective patients whose PFS were less than 3 months, and these samples were recorded as microarray samples. Also, treatment-effective patients (28 cases) and treatment-ineffective patients (10 cases) from an independent cohort were used to performing RT-qPCR validation of circRNA biomarker candidates. Moreover, these 38 validation samples were recorded as non-microarray samples. Detailed information of these 52 patients is summarized in Table S1. All patients provided informed consent, and this study was approved by the Clinical Research Ethics Committee of the Cancer Hospital of the Chinese Academy of Medical Sciences.
Table S1. Baseline characteristics of the patients.
| Characteristics | PFS ≥10 m, n=34 (%) | PFS ≤3 m, n=18 (%) |
|---|---|---|
| Age (years) | ||
| Median | 58.5 | 55.0 |
| Range | 35–74 | 36–74 |
| <60 | 17 (50.0) | 11 (61.1) |
| ≥60 | 17 (50.0) | 7 (38.9) |
| Sex | ||
| Male | 10 (29.4) | 10 (55.6) |
| Female | 24 (70.6) | 8 (44.4) |
| Smoking history | ||
| No | 29 (85.3) | 15 (83.3) |
| Yes | 5 (14.7) | 3 (16.7) |
| Prior chemo | ||
| No | 11 (32.4) | 7 (38.9) |
| 1–2 regimens | 23 (67.6) | 11 (61.1) |
m, month; PFS, progression-free survival.
Plasma sample preparation
Plasma samples were taken before the treatment of gefitinib. The samples were isolated, and fresh peripheral blood samples were collected in EDTA tubes and centrifuged at 820× g for 10 min. Aliquots of plasma from the upper layer were centrifuged at 14,000 rpm for 10 min. Plasma samples were transported frozen to the laboratory for storage at −80 °C.
RNA extraction and RNase R treatment
Four hundred µL of plasma was used to isolate the RNA in the total RNA using Qiagen miRNeasy MiniKit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The purity and concentration of RNA were determined with Qubit 3.0 (Thermo Fisher Scientific, USA). The integrity of the RNA was determined with the Bioanalyzer 2100 (Agilent Technologies, USA).
For RNase R treatment, total cfRNA was incubated with 0.5 µL RNase R (10 U) (Epicentre, Madison, WI, RNR07250) and a 2 µL RNase R 10× reaction buffer for 30 minutes at 37 °C in a 20 µL reaction. After the incubation, RNase R-treated RNA was purified with a Qiagen RNeasy MinElute Purification Kit (Qiagen, Hilden, Germany, 74204).
CircRNA microarray
Plasma RNAs extracted from the 14 subjects were used for microarray analysis. RNA amplification and labeling was performed according to the manufacturer’s protocol. Labeled RNAs were hybridized onto CapitalBio Technology Human CircRNA Array v2.0 (CapitalBio, Beijing, China). Each array contained probes representing approximately 170,340 human circRNAs.
RNA amplification, labeling, and hybridization to a microarray
CapitalBio circRNA Amplification and Labeling Kit (CapitalBio, Beijing, China) was used to amplify and transcribe the RNase R enriched circRNA into fluorescent cRNA with 100–500 ng purified circRNAs. The labeled cRNAs were hybridized onto the CapitalBio Technology Human CircRNA Array v2.0 (CapitalBio, Beijing, China). Array hybridization was performed in an Agilent Hybridization Oven overnight at a rotation speed of 20 rpm and a temperature of 42 °C and washed twice (0.2% SDS, 2× SSC at 42 °C for 5 min, followed by 0.2× SSC for 5 min at room temperature). Subsequently, the arrays were scanned using the Agilent Scanner G2565CA (Agilent, Santa Clara, CA, USA). Acquired array images were analyzed with the Agilent Feature Extraction (v10.7) software.
Data collection on circRNA expression
The raw data was extracted with the Agilent Feature Extraction (version 10.7) software. Data summarization, quality control, quantile normalization, and analysis of circRNAs differentially expressed in the effective vs. ineffective groups were performed with GeneSpring v13.0 (Agilent). A box plot was used to visualize the distribution of the intensities of all circRNAs in the sample after normalization. Unsupervised hierarchical clustering was performed to analyze circRNA expression in the samples with the R package “Pheatmap.” Scatterplot was used to assess the differences in circRNA expression between the two groups. Fold change (i.e., ratio of the group averages) for each circRNA and unpaired Student’s t-test was used to analyze the difference in circRNA expression between the two groups with the GeneSpring software. Individually, circRNAs with a fold change it 1.5 and Benjamini-Hochberg-corrected P<0.05 were considered significantly differentially expressed.
Quantitative real-time reverse transcription PCR (RT-qPCR) validation of circRNAs
RT-qPCR was performed to validate microarray data by evaluating the expression of the two up-regulated genes, hsa_circ_0109320 and hsa_circ_0134501, and the housekeeping gene β-actin. One, microarray sample set of 6 individuals in the effective group, compared with 8 individuals in the ineffective group. Two, an independent validation set of patients including 28 individuals in the effective group, compared with 10 individuals in the ineffective group. PCR validation was performed with first strand cDNA synthesis, standard curve preparation, and real-time PCR detection. Briefly, single strand cDNA was amplified from 10 ng RNA with FastQuant RT cDNA synthesis kit (Tiangen, Beijing, China) following the manufacturer’s instructions. Then, quantitative PCR was performed with a QuantStudio 7K Flex Real-Time PCR System (Thermo Fisher Scientific, Wilmington, DE, USA) following the manufacturer’s instructions. Rather than the commonly used convergent primers, divergent primers were designed for RT-qPCR of circRNAs. Sequences of forward and reverse primers used in this study are listed in Table S2. Each sample was analyzed with triplicate experiments for 40 cycles. Specificity of amplicons was determined by melt curve analysis. Data analysis was further performed using the ΔΔCT method. One point five percent agarose gel was used for electrophoresis to verify the PCR products.
Table S2. Primer of circRNA.
| Primer ID | Sequence |
|---|---|
| hsa_circ_0109320_Forward | ATGCTTTGCCACATTCTTCACA |
| hsa_circ_0109320_Reverse | TAAGATAATTCACACTGGAGAGA |
| hsa_circ_0134501_Forward | AGCTCTGCCACATTCTTCACATTTG |
| hsa_circ_0134501_Reverse | TACATAAAAGAATTCATACTGGAGA |
| beta Actin_Forward | CATGTACGTTGCTATCCAGGC |
| beta Actin_Reverse | CTCCTTAATGTCACGCACGAT |
Statistical analysis
All the analyses were performed by R software package or GraphPad PRISM 6.0c (GraphPad Software, La Jolla, CA, USA). PFS was defined as the time from the start of the first line therapy until the date of progression or last follow-up evaluation. Student’s t-test compared the difference of circRNA between groups. A high circRNAs expression was defined as the top 20% of expression. Receiver operating curve (ROC) was used to test the association between circRNA and survival. A sensitivity and specificity value were calculated when the probabilities >0.5 or >0.25 were predicted to be positive.
Results
Expression profiles of circRNAs in the effective and ineffective groups
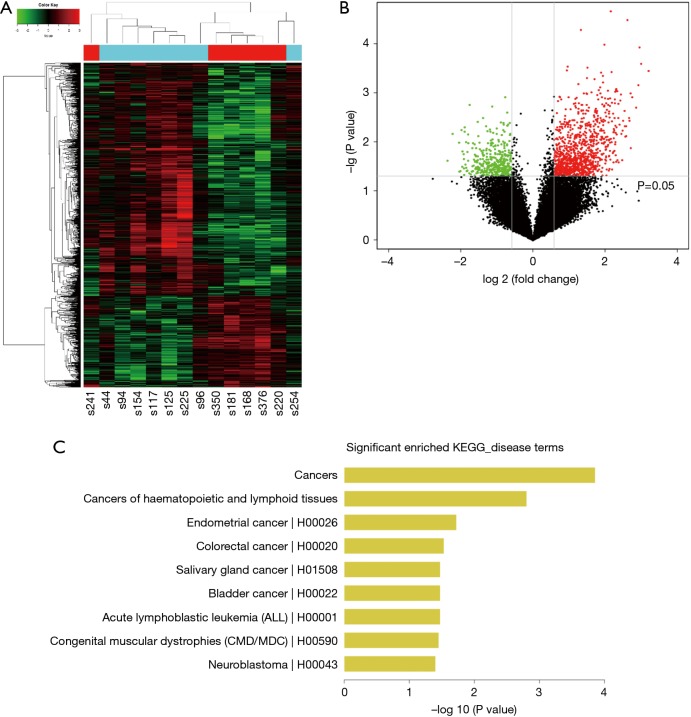
The normalized intensities of circRNA expression were similar across all the samples used in the circRNA microarray (Figure 1A). Unsupervised hierarchical cluster analysis indicated that the differentially expressed circRNAs could divide the samples in the effective vs. ineffective group into distinct expression patterns. The volcano plot showed clusters of circRNAs differentially expressed between the effective and ineffective groups (fold change ≥1.5, P value <0.05) (Figures 1B,S1,S2). In total, 1,377 circRNAs were differentially expressed between these two groups, among which 989 circRNAs were up-regulated, and 388 circRNAs were down-regulated in the effective group. Moreover, the distribution of the parent genes of these differentially expressed circRNAs in the human genome showed even distribution on all chromosomes. Also, a KEGG pathway enrichment analysis on the parent genes revealed that many of them are involved in the biological processes of various cancers, such as cancers of hematopoietic and lymphoid tissues, bladder cancer, colorectal cancer, and neuroblastoma (Figure 1C).
Figure 1.
circRNA expression profiles in effective and ineffective groups. (A) Unsupervised hierarchical clustering is showing distinct circRNA expression profiles among the 14 samples (light blue: the ineffective group; red: the effective group). Red represents the higher expression, and green represents the lower expression; (B) volcano plot showing clusters of circRNAs differentially expressed between the effective group and ineffective groups, with 989 and 388 up- and down-regulated circRNAs, respectively (fold change ≥1.5 and P<0.05). The vertical gray lines correspond to 1.5-fold up- and down-regulation and the horizontal gray line represents a P value of 0.05. Red and green points in the plot represent the significantly differentially expressed circRNAs; (C) significantly enriched pathways in genes regulated by differentially expressed cancer-related circRNAs were identified through the KEGG pathway enrichment analysis.
Figure S1.
Box plots are showing the distribution of circRNA expression in the 14 samples (red box: the effective group, blue box: the ineffective group). (A) Is showing the raw processed signal, and (B) is showing the normalized processed signal.
Figure S2.
Circos plots are showing the chromosomal distribution of the parent genes of the differentially expressed circRNAs.
Expression of circRNAs validated by RT-qPCR
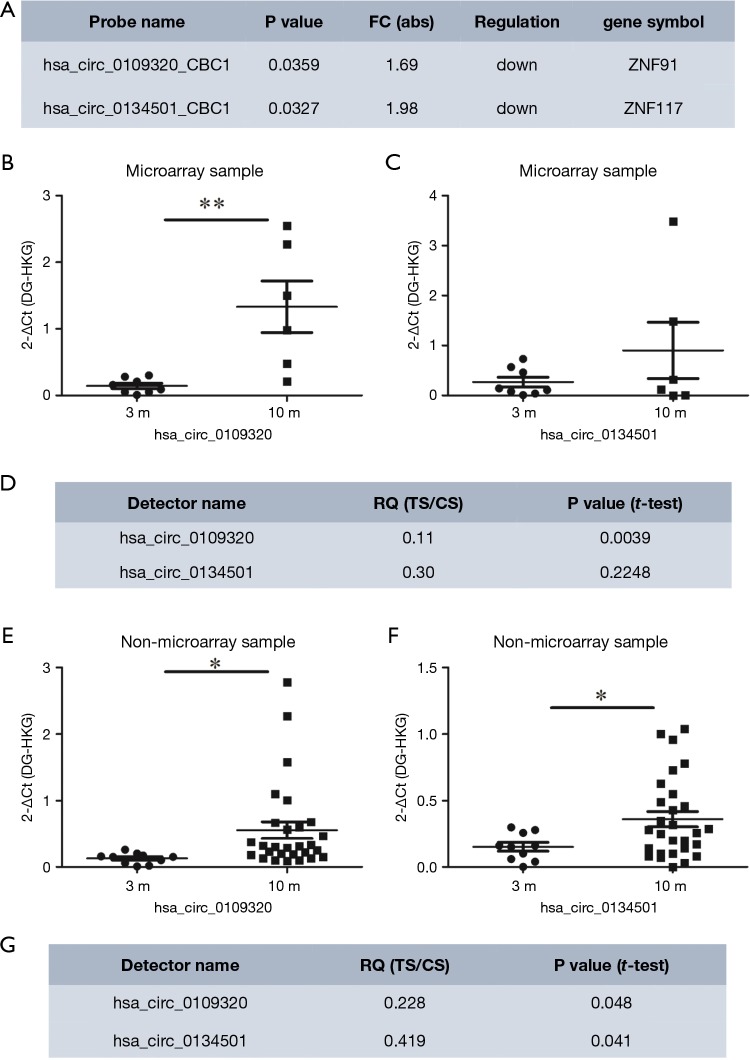
To identify circRNAs that can be used as biomarkers to estimate the therapeutic effect of gefitinib in NSCLC, 1,377 differentially expressed circRNAs were screened with the following criteria: (I) fold change ≥1.5 and P<0.05, and (II) the parent gene is associated with lung cancer. Numerous circRNAs met these standards including hsa_circ_0109320 and hsa_circ_0134501 (Figure 2A). They were ~2 folds down-regulated in the effective group by microarray. Also, ZNF117, the parent gene of hsa_circ_0134501, encodes a zinc finger protein that is involved in the process of lung cancer (11,12). The parent gene of hsa_circ_0109320 also encodes a zinc finger protein, ZNF91, that is associated with NSCLC (13).
Figure 2.
Expression levels of two selected circRNAs. (A) Two circRNAs selected as potential biomarkers to evaluate the effectiveness of gefitinib; (B-D) expression of hsa_circ_0109320 and hsa_circ_0134501 quantified by RT-qPCR in the cohort subjected to microarray profiling (*, P<0.05, **, P<0.01); (E-G) expression of hsa_circ_0109320 and hsa_circ_0134501 quantified by RT-qPCR in an independent cohort (*, P<0.05, **, P<0.01). The relative expression levels of circRNAs were normalized to levels of control (β-actin). 3M represents patients whose PFS is less than 3 months, and 10M represents patients whose PFS is longer than 10 months. RT-qPCR, real-time quantitative polymerase chain reaction; PFS, progression-free survival.
RT-qPCR assays were conducted in the subjects used in the microarray to verify the expression of these selected candidate circRNAs, (effective group, n=6; ineffective group, n=8) and an independent cohort (validation cohort, effective group, n=28; ineffective group, n=10). The specificity of the PCR primers was validated by the melt-curve analysis (Figure S3). hsa_circ_0109320 and hsa_circ_0134501 were detectable by RT-qPCR in these NSCLC samples. Consistent with the microarray result, the expression of hsa_circ_0134501 was higher in the effective group than in the ineffective group in both cohorts. The result of hsa_circ_0134501 modestly agreed with the result from microarray (Figure 2B,C,D,E,F,G). Thus, hsa_circ_0109320 and hsa_circ_0134501 are good biomarker candidates for predicting the therapeutic effect of gefitinib in NSCLC.
Figure S3.
Significantly enriched pathways in genes regulated by differentially expressed circRNAs were identified through the KEGG pathway enrichment analysis.
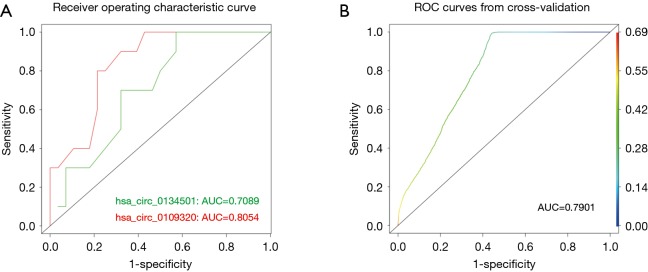
ROC curve analysis of circRNAs
To determine the diagnostic performance of hsa_circ_0109320 and hsa_circ_0134501 for estimating the therapeutic effect of gefitinib in NSCLC, we next performed ROC curve analysis for each circRNA. The analysis was conducted by calculating the sensitivities and specificities at different thresholds of circRNA expression in the validation cohort and plotting the resulting values in the ROC space (Figure 3). When the expression of hsa_circ_0109320 and hsa_circ_0134501 was analyzed for this purpose, the cutoff value was 0.18 and 0.17, respectively; based on this cutoff, the sensitivity and specificity were 80%/70% and 78.6%/67.8%, respectively. The values of AUC of hsa_circ_0109320 and hsa_circ_0134501 for estimating the therapeutic effect of gefitinib in NSCLC were 0.81 and 0.71, respectively. Furthermore, the combination of hsa_circ_0109320 and hsa_circ_0134501 gave a better prognostic value (AUC: 0.79) than hsa_circ_0134501 alone did (AUC: 0.71) (Figure 3B). Therefore, hsa_circ_0109320 alone or the combination of hsa_circ_0109320 and hsa_circ_0134501 can be used as a biomarker.
Figure 3.
The ROC curves of hsa_circ_0109320 and hsa_circ_0134501. (A) ROC curves of hsa_circ_0109320 and hsa_circ_0134501 were built for distinguishing plasma samples in the effective group from those in the ineffective group. The AUC of was 0.81 and 0.71, the threshold was 0.18 and 0.17, and the sensitivity and specificity were 80%/70% and 78.6%/67.8% for hsa_circ_0109320 and hsa_circ_0134501, respectively (P<0.001); (B) ROC curve analysis of hsa_circ_0134501 combined with hsa_circ_0109320 for distinguishing patients in the effective vs. ineffective group. The AUC was 0.79.
Collectively, the above results suggest that hsa_circ_0109320 and hsa_circ_0134501 could serve as potential indicators of the therapeutic effectiveness of gefitinib in NSCLC.
Potential prognostic values of hsa_circ_0109320 and hsa_circ_0134501 in NSCLC
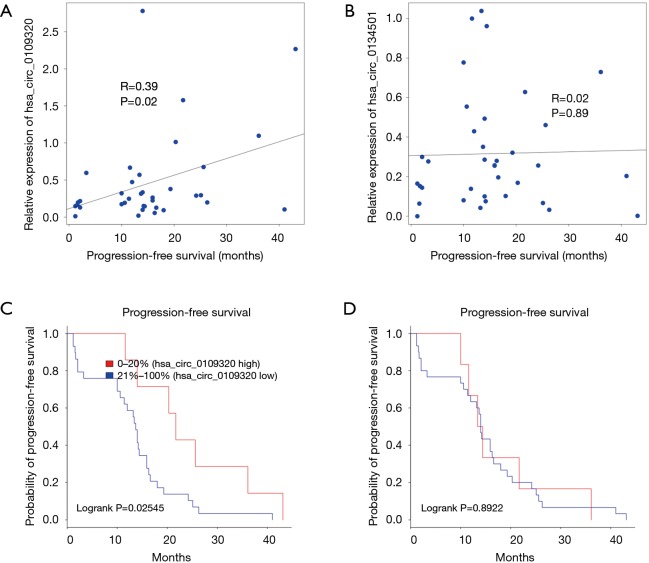
In clinical trials for drug development, PFS is often used as a primary endpoint or secondary endpoint to determine the efficacy of anti-cancer drugs. In this study, we examined the correlation between circRNA levels and PFS by Pearson’s correlation test and found that hsa_circ_0109320 was correlated with PFS, while hsa_circ_0134501 was not (Figure 4A,B).
Figure 4.
Correlation between hsa_circ_0109320 or hsa_circ_0134501 expression and PFS in NSCLC patients. (A,B) Pearson correlation analysis between hsa_circ_0109320 or hsa_circ_0134501 expression and PFS in NSCLC patients, R: Pearson correlation coefficient, P: a significant test of the correlation; (C,D) PFS curves of patients with NSCLC according to hsa_circ_0109320 or hsa_circ_0134501 expression. NSCLC, non-small cell lung cancer; PFS, progression-free survival.
We also tested whether hsa_circ_0109320 or hsa_circ_0134501 expression could predict the survival of NSCLC patients. In total, 38 patients were successfully followed up in this study. The PFS curves are shown in Figure 4. The Kaplan-Meier survival curve revealed that elevated hsa_circ_0109320 expression was associated with significantly better PFS in patients with NSCLC after gefitinib treatment (Figure 4C,D).
Collectively, our results suggested that hsa_circ_0109320 might be a useful biomarker for predicting the effectiveness of gefitinib in the treatment of NSCLC.
Correlation between the expression of hsa_circ_0109320 and hsa_circ_0134501 and clinical characteristics of NSCLC patients
Furthermore, to assess whether the ability of hsa_circ_0109320 and hsa_circ_0134501 to predict PFS is independent of other clinical or pathological factors, we evaluated the association of the high level of hsa_circ_0109320 and hsa_circ_0134501 with clinical characteristics in 38 patients with NSCLC. The patients were stratified according to hsa_circ_0109320 and hsa_circ_0134501 expression with the cut-off of 2−ΔCt(DG-HKG) =0.3. Then, the P value was computed by comparing different groups with a chi-square test. The expression of hsa_circ_0109320 and hsa_circ_0134501 showed no significant differences between groups when patients were stratified by gender (P=0.497 and 0.293, respectively), age (P=0.494, 1), smoking status (P=0.54 and 1, respectively), or PFS (P=0.101 and 0.11, respectively). These chi-square test results indicated that gender, age, smoking, and PFS did not affect the expression of these two circRNAs.
Discussion
In the present study, we found that two circRNAs, hsa_circ_0109320 and hsa_circ_0134501, were highly expressed in gefitinib-effective NSCLC patients. Further, hsa_circ_0109320 expression was associated with better PFS. Taken together, hsa_circ_0109320 may be a useful biomarker for the prognosis in gefitinib-treated patients with NSCLC and further prospective studies are warranted.
EGFR-TKI inhibitors including (5), Gefitinib, Icotinib, and Erlotinib, have become the critical treatment option for metastatic NSCLC (5). However, not all patients may respond to EGFR-TKI (14). Plasma samples can serve as non-invasive diagnostic carries in clinical application. circRNAs have the closed loop structure that makes them stable and easily cumulated in plasma (15). Indeed, recently, several reports have detected circRNAs in plasma (16,17), which give us an excellent opportunity to discover circRNA biomarkers and furthermore use them for the diagnosis, evaluation the treatment efficiency of inhibitors for cancer, and even to monitor cancer progression by non-invasively detecting these circRNAs in plasma.
In this study, we used circRNA expression microarray to measure circRNA expression in the plasma of NSCLC patients in EGFR-TKI-effective and -ineffective groups. The circRNA microarray used in this study covers most commonly used cricRNA databases including circBase and deepBase, enabling high-throughput, high-coverage detection of circRNAs in the plasma. This has dramatically improved the probability of discovering circRNA markers.
In this study, two circRNAs were found to be differentially expressed in gefitinib treatment-effective vs. ineffective groups. The elevated expression of hsa_circ_0109320 was associated with better PFS. Further studies are also needed to elucidate the regulatory mechanisms of hsa_circ_0109320 fully.
However, our study has several limitations. First, many other factors may influence the PFS of NSCLC patients treated with EGFR-TKI like mutations of oncogenic genes. Therefore, more parameters should be considered to identify the specific factors that may influence the predictive value of these two circRNAs. Second, we did not compare the predictive value of circRNA and EGFR driver mutation because for most patients the tissue sample was unavailable. At last, only 38 samples were used for the independent RT-qPCR validation. The cohort is indeed not large enough to get a definite conclusion. Therefore, more samples should be collected to confirm our conclusion.
In conclusion, hsa_circ_0109320 is highly expressed in EGFR-TKI effective patients and may be a useful biomarker for the prognosis in gefitinib-treated patients with NSCLC. Further prospective studies are warranted.
Acknowledgments
The authors thank all the patients who participated in this study and their families. We also thank the investigators, study coordinators, operation staff, and the whole project team who worked on this study.
Funding: This work is supported by grants CAMS Innovation Fund for Medical Sciences (CIFMS) [Grant number: 2016-I2M-1-001] and Chinese National Major Project for New Drug Innovation [Grant number: 2018ZX09201003].
Ethical Statement: All patients provided informed consent, and this study was approved by the Clinical Research Ethics Committee of the Cancer Hospital of the Chinese Academy of Medical Sciences.
Footnotes
Conflicts of Interest: ZH Yang is employee of Beijing CapitalBio Technology Company. The other authors have no conflicts of interest to declare.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- 2.World Cancer Report 2014, World Health Organization 2015.
- 3.Williams CD, Gajra A, Ganti AK, et al. Use and impact of adjuvant chemotherapy in patients with resected non-small cell lung cancer. Cancer 2014;120:1939-47. 10.1002/cncr.28679 [DOI] [PubMed] [Google Scholar]
- 4.Hsu CP, Chuang HC, Lee MC, et al. GLK/MAP4K3 overexpression associates with recurrence risk for non-small cell lung cancer. Oncotarget 2016;7:41748-57. 10.18632/oncotarget.9410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janne P, Sequist L, Lindeman N, et al. Long-term survival of NSCLC patients with EGFR mutations treated with EGFR TKIs gefitinib or erlotinib. Lung Cancer 2005;49:S109-S109. 10.1016/S0169-5002(05)80478-715950789 [DOI] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 7.Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol 2017;14:992-9. 10.1080/15476286.2016.1220473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao T, Chen Q, Fu L, et al. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res 2017;47:497-504. 10.1111/hepr.12871 [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform 2018. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 11.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19. 10.1101/gr.145144.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong D, Li G, Li K, et al. Exome sequencing identifies MXRA5 as a novel cancer gene frequently mutated in non-small cell lung carcinoma from Chinese patients. Carcinogenesis 2012;33:1797-805. 10.1093/carcin/bgs210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 2003;290:2149-58. 10.1001/jama.290.16.2149 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Xue W, Li X, et al. The Biogenesis of Nascent Circular RNAs. Cell Rep 2016;15:611-24. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- 16.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]