Dear Editor,
Renal cell carcinoma (RCC) is among the most common human cancers in the United States, with approximately 63,990 new patients and 14,400 deaths annually [1]. However, RCC is not among the top 10 malignancies in China in terms of incidence and mortality [2]. The clinical and molecular features of RCC differ among distinct pathological types, mainly clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (PRCC), and chromophobe renal cell carcinoma (ChRCC). The most common subtype of RCC is ccRCC worldwide. According to The Cancer Genome Atlas (TCGA), the somatic mutation landscape of RCC has been revealed by whole-exome sequencing (WES) or whole-genome sequencing (WGS). In our previous WES study, we validated most of the significantly mutated genes reported by the TCGA and identified several novel somatically altered genes [3]. The TCGA study showed that only somatic mutations in BRCA1-associated protein 1 (BAP1) were associated with patients’ poor survival outcomes among all significantly mutated genes [4]. In our previous WES study, BAP1 was somatically mutated in 2 of 15 ccRCC samples [3]. Nevertheless, all of these RCC patients lacked follow-up information. Hence, further analysis is needed to determine whether there are any somatically mutated genes associated with the prognosis of Chinese patients with RCC. However, WES or WGS is time-consuming and costly. Furthermore, compared with targeted sequencing, WES was more likely to generate false positives and false negatives due to insufficient base coverage [5].
In recent years, immunotherapy has played an increasingly important role in the treatment of advanced RCC and other malignancies. Based on the current understanding, programmed death-1 (PD-1) can combine with programmed death-ligand 1 (PD-L1) to confine T cell activity in the tumor microenvironment, and inhibition of the PD-1/PD-L1 pathway can increase the anti-tumor immune response [6]. Nivolumab, a PD-1 immune checkpoint inhibitor, has been validated for the treatment of advanced RCC based on the overall survival (OS) benefit [7]. A recent study has shown that PD-L1 expression was a predictive factor in terms of response and OS benefit from nivolumab plus ipilimumab combination therapy or nivolumab monotherapy as a second-line treatment for advanced RCC [8]. In our previous study, we identified several somatically mutated genes associated with PD-L1 expression in RCC tumor cells, including CSPG4, DNAH11, INADL, and TMPRSS13 [3]. However, the sample size in the previous study was only 26 specimens, which was a little bit small. In the present study, we aimed to validate these discoveries with a larger sample size and investigate the association between somatic mutations and PD-L1 expression in RCC tumor cells.
In the present study, formalin-fixed paraffin-embedded (FFPE) RCC specimens from 40 patients were investigated using immunohistochemistry (IHC) and targeted sequencing. We designed a gene panel comprising of 173 genes, which contained the newly identified somatically mutated genes, the genes somatically mutated in at least two samples in our previous WES study, and the recurrently mutated genes reported in the TCGA and Catalogue of Somatic Mutations in Cancer (COSMIC) database. The sequencing depth was set to 500×. All the identified somatic mutations were annotated using Annovar [9]. The functional significance of missense mutations was predicted via several algorithms, including SIFT, PolyPhen2 HDIV, PolyPhen2 HVAR, LRT, MutationTaster, MutationAssessor, and FATHMM. The somatic mutations scored with at least two algorithms as deleterious were deemed as deleterious variants. Other variants, including nonsense, frameshift, and canonical ± 1 or ± 2 splice site mutations, were considered to be pathogenic according to the guidelines of the American College of Medical Genetics (ACMG) [10]. Among these 40 RCC patients, 27 were males and 13 were females, with a median age of 57 years (range 22–76 years). The median follow-up for these 40 patients was 74 months (range 15–86 months). Details of their clinicopathological information are listed in Table 1.
Table 1.
The clinicopathological information of 40 RCC patients
| Sample ID | Gender | Age | Subtype | Tumor grade | TNM stage | AJCC stagea | OS (months) | DFS (months) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 76 | ccRCC | G2 | T3aN0M0 | III | 34 | 34 | Death |
| 2 | Male | 74 | ccRCC | G2 | T1aN0M0 | I | 32 | 32 | Death |
| 3 | Male | 31 | ccRCC | G3 | T3aN0M0 | III | 63 | 63 | Death |
| 4 | Male | 74 | ccRCC | G2 | T1aN0M0 | I | 62 | 62 | Death |
| 5 | Male | 54 | ccRCC | G2 | T1aN0M0 | I | 57 | 57 | Death |
| 6 | Male | 62 | ccRCC | G2 | T1aN0M0 | I | 76 | 64 | Survival (metastasis) |
| 7 | Female | 40 | ccRCC | G3 | T3bN0M0 | III | 74 | 24 | Survival (metastasis) |
| 8 | Female | 57 | ccRCC | G2 | T1aN0M0 | I | 74 | 24 | Survival (metastasis) |
| 9 | Male | 56 | ccRCC | G2 | T3aN0M0 | III | 72 | 9 | Survival (metastasis) |
| 10 | Male | 59 | ccRCC | G3 | T1bN0M0 | I | 71 | 12 | Survival (metastasis) |
| 11 | Male | 55 | ccRCC | G1 | T1bN0M0 | I | 74 | 74 | Survival |
| 12 | Male | 62 | ccRCC | G2 | T1bN0M0 | I | 74 | 74 | Survival |
| 13 | Male | 54 | ccRCC | G2 | T1aN0M0 | I | 74 | 74 | Survival |
| 14 | Male | 60 | ccRCC | G2 | T1bN0M0 | I | 74 | 74 | Survival |
| 15 | Male | 48 | ccRCC | G1 | T1aN0M0 | I | 74 | 74 | Survival |
| 16 | Male | 68 | ccRCC | G2 | T1bN0M0 | I | 73 | 73 | Survival |
| 17 | Male | 48 | ccRCC | G2 | T1aN0M0 | I | 73 | 73 | Survival |
| 18 | Male | 73 | ccRCC | G1 | T1aN0M0 | I | 73 | 73 | Survival |
| 19 | Female | 58 | ccRCC | G2 | T1aN0M0 | I | 72 | 72 | Survival |
| 20 | Male | 48 | ccRCC | G2 | T1aN0M0 | I | 72 | 72 | Survival |
| 21 | Male | 49 | PRCC | G2 | T2N0M0 | II | 15 | 15 | Death |
| 22 | Female | 66 | PRCC | G2 | T3aN1M0 | III | 37 | 37 | Death |
| 23 | Male | 70 | PRCC | G2 | T3aN0M0 | III | 66 | 66 | Death |
| 24 | Male | 63 | PRCC | G2 | T3bN0M0 | III | 29 | 29 | Death |
| 25 | Male | 65 | PRCC | G2 | T1aN0M0 | I | 49 | 49 | Death |
| 26 | Female | 22 | PRCC | G1 | T1aN0M0 | I | 76 | 76 | Survival |
| 27 | Female | 60 | PRCC | G2 | T1aN0M0 | I | 74 | 74 | Survival |
| 28 | Male | 69 | PRCC | G2 | T1aN0M0 | I | 73 | 73 | Survival |
| 29 | Male | 59 | PRCC | G2 | T1aN0M0 | I | 71 | 71 | Survival |
| 30 | Male | 58 | PRCC | G2 | T1aN0M0 | I | 69 | 69 | Survival |
| 31 | Female | 49 | ChRCC | NA | T2N0M0 | II | 86 | 86 | Survival |
| 32 | Male | 64 | ChRCC | NA | T1aN0M0 | I | 85 | 85 | Survival |
| 33 | Female | 37 | ChRCC | NA | T1aN0M0 | I | 85 | 85 | Survival |
| 34 | Male | 36 | ChRCC | NA | T1bN0M0 | I | 84 | 84 | Survival |
| 35 | Female | 54 | ChRCC | NA | T1aN0M0 | I | 84 | 84 | Survival |
| 36 | Female | 52 | ChRCC | NA | T1aN0M0 | I | 82 | 82 | Survival |
| 37 | Female | 75 | ChRCC | NA | T1aN0M0 | I | 78 | 78 | Survival |
| 38 | Male | 40 | ChRCC | NA | T1bN0M0 | I | 77 | 77 | Survival |
| 39 | Female | 36 | ChRCC | NA | T1bN0M0 | I | 77 | 77 | Survival |
| 40 | Female | 49 | ChRCC | NA | T1bN0M0 | I | 76 | 76 | Survival |
RCC renal cell carcinoma, TNM tumor-node metastasis stage, AJCC American Joint Committee on Cancer, OS overall survival, DFS disease-free survival, ccRCC clear cell renal cell carcinoma, PRCC papillary renal cell carcinoma, ChRCC chromophobe renal cell carcinoma, NA not available
aThe 7th edition of the AJCC Cancer Staging Manual was used
Among all the significantly mutated genes in ccRCC from the TCGA database, VHL, PBRM1, SETD2, KDM5C, PTEN, BAP1, MTOR, and TP53 were the eight most significantly mutated genes [4]. All the eight genes were validated in the present study, whereas only six were validated in our previous WES study [3]. In the present study, VHL was somatically mutated in 10 ccRCC specimens, including five frameshift mutations, namely, p. K159fs, p. L135fs, p. P2fs, p. S183fs, and p. R58fs, all of which had not been reported previously and were deemed to be very strong evidence of pathogenicity. PBRM1 was somatically mutated in 7 ccRCC specimens, 5 PRCC specimens, and 3 ChRCC specimens. Most of the mutations in PBRM1 were frameshift mutations, which had not been reported previously and were predicted to be deleterious. The tumor mutation burden (TMB) of the 40 RCC specimens was calculated based on the custom-designed 173-gene panel. The TMB was significantly higher in RCC specimens with somatically mutated PBRM1 than in those without somatically mutated PBRM1 (P = 0.020). The sequencing depth in the present study was higher than that in our previous WES study. Consequently, more somatic mutations in each single specimen were revealed in the present study than in the TCGA data. There was usually more than one type of mutation identified in a single gene in multiple specimens. For instance, BAP1 was somatically mutated in 3 ccRCC specimens in the present study, namely a frameshift deletion (p. S432fs) and insertion (p. P462fs) in sample 9, a deletion–insertion mutation [p. E642_I643delins (39)] in sample 13, and a frameshift insertion (p. P339fs) and deletion–insertion mutation [p. I191_D192delins (18)] in sample 20. Mutated BAP1 or loss of BAP1 expression was reported to be associated with poor outcome in ccRCC [4, 11]. However, no significant association between BAP1 and prognosis was found in the present study.
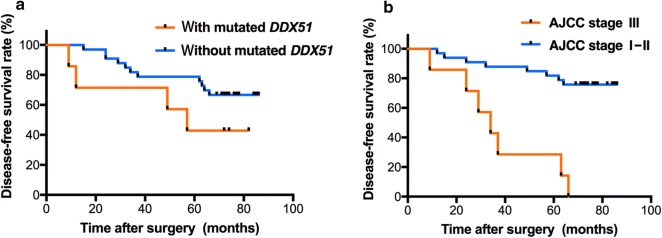
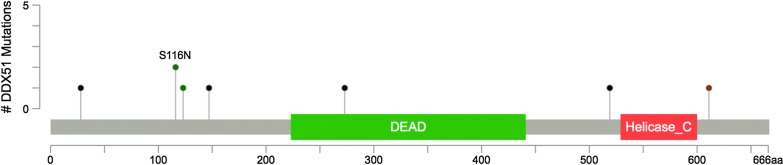
In our previous WES study, we identified several newly somatically mutated genes, including HGC6.3, DDX51, NWD2, CDC42EP1, NPIPB5, HSCB, HMCN2, and PCDHB9 in ccRCC; DEPDC4, PNLIP, SARDH, and ZAN in PRCC; and KRTAP4-8 in ChRCC [3]. All of these genes were enrolled in our custom-designed gene panel for further investigation with a larger sample size. As such, most of these newly identified somatically mutated genes were validated in the present study, except for HMCN2 and PCDHB9 in ccRCC and DEPDC4 and ZAN in PRCC. Three somatic mutations in DEPDC4 were identified in ccRCC specimens, namely 2 frameshift deletions (p. F150fs and p. R21fs) and 1 deletion–insertion [p. E147_L148delins (8)], all of which were predicted to be deleterious. Among the 40 RCC patients with complete follow-up information, univariate survival analysis with log-rank tests revealed that the disease-free survival (DFS) was shorter in patients with the maximum diameter of tumor > 7 cm than in patients with the maximum diameter of tumor ≤ 7 cm (P = 0.003) and shorter in patients with American Joint Committee on Cancer (AJCC) stage III than in patients with AJCC stage I–II (P < 0.001). In addition, we found a slight trend towards an association between DFS and somatically mutated DDX51 (P = 0.144). The three variables with P < 0.15 were all enrolled in the multivariate Cox regression survival analysis, which showed that somatically mutated DDX51 (P = 0.017) and AJCC stage III (P = 0.006) were independent risk factors for DFS among RCC patients (Table 2 and Fig. 1). However, no significant association between somatically mutated genes and OS was found in the present study. Among the 20 ccRCC specimens in the present study, DDX51 was somatically mutated in 5 specimens with six mutations, namely, a deletion–insertion mutation [p. K611_V612delins (35)] in 1 specimen, a missense mutation (p. S116N) in 2 specimens, two frameshift insertion mutations (p. G147fs and p. H28fs) in 1 specimen, and a frameshift deletion (p.A273fs) in 1 specimen. Notably, the frameshift deletion (p. A273fs) was located in the DEAD protein domain of DDX51 (Fig. 2). The missense mutation in DDX51 in both specimens was predicted to be benign or neutral, whereas the deletion–insertion mutation and three frameshift mutations were most likely to be deleterious according to the ACMG guidelines. Furthermore, somatic mutations in DDX51 were also identified in two other RCC subtypes, including a frameshift deletion (p. R519fs) in PRCC predicted to be deleterious and a missense mutation (p. P123R) in ChRCC predicted to be benign or neutral.
Table 2.
Multivariate Cox regression analysis for the DFS of RCC patients
| Variables | B | Wald | P value | Hazard ratio | 95% CI |
|---|---|---|---|---|---|
| Mutated DDX51 | 1.629 | 5.696 | 0.017 | 5.099 | 1.338–19.432 |
| AJCC stage III | 1.903 | 7.639 | 0.006 | 6.703 | 1.739–25.833 |
| Maximum diameter of tumor > 7 cm | 1.165 | 2.532 | 0.112 | 3.207 | 0.763–13.475 |
DFS disease-free survival, RCC renal cell carcinoma, CI confidence interval, DDX51 DEAD-box helicase 51, AJCC American Joint Committee on Cancer
Fig. 1.
The Kaplan–Meier disease-free survival (DFS) curves of 40 renal cell carcinoma (RCC) patients. a Survival curves of patients with or without nutated DEAD-box helicase 51 (DDX51); b survival curves of patients with American Joint Committee on Cancer (AJCC) stages I–II or III
Fig. 2.
Mutation Mapper interprets mutations with protein domains of DEAD-box helicase 51 (DDX51). The mutations are presented by circles and colors: green (missense), black (frameshift), brown (stop-gain)
In our previous study, PD-L1 expression in tumor cells was detected in 6 (23%) of 26 RCC specimens: 3 ccRCC specimens, 2 PRCC specimens, and 1 ChRCC specimen [3]. In the present study, PD-L1 expression in tumor cells was detected in 6 (15%) of the 40 RCC samples: 1 ccRCC sample, 4 PRCC samples, and 1 ChRCC sample (Fig. 3). Combined with the 26 RCC specimens investigated in our previous study, PD-L1 expression in tumor cells was positive in 4 (11%) of 35 ccRCC specimens. We identified 6 genes, VHL, INADL, MUC4, RAD21, CSPG4, and BAP1, that were somatically mutated in 3 of the 4 PD-L1-positive ccRCC specimens. Nevertheless, only mutated RAD21 and BAP1 were associated with PD-L1 expression in tumor cells. Among the 35 ccRCC specimens (15 from our previous WES study [3] and 20 in the present study), Fisher's exact test revealed that the PD-L1-positive rate in tumor cells was higher in specimens with somatically mutated RAD21 (P = 0.002) and BAP1 (P = 0.006) than in specimens without those mutated genes. The somatic mutations in BAP1 (p. P352fs, p. H193Q, p. S432fs, and p. P462fs) and RAD21 (p. F2 L, p. F304S, p. R402fs, and p. L515fs) detected in the 3 PD-L1-positive ccRCC samples were all predicted to be deleterious.
Fig. 3.
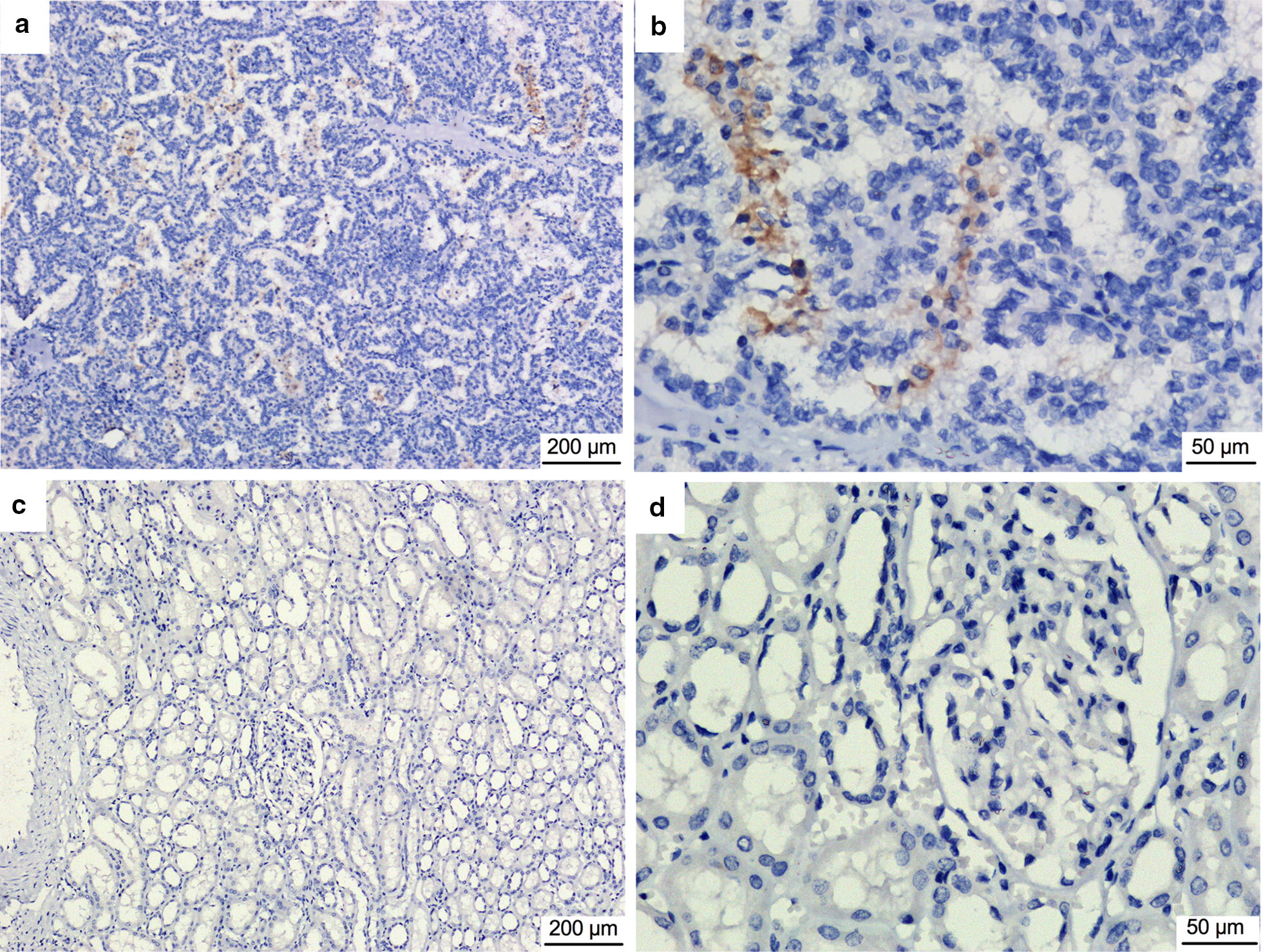
Immunohistochemical staining of programmed death-ligand 1 (PD-L1) in renal cell carcinoma specimens. a, b Yellowish-brown PD-L1-positive staining on cell membrane in a G1 tumor; c, d PD-L1-negative staining in adjacent normal tissue
In conclusion, RCC patients with somatically mutated PBRM1 tend to have higher TMB than those without it. The somatically mutated DDX51 is an independent risk factor for DFS among RCC patients and could be a new candidate gene for predicting the prognosis of RCC. The somatically mutated RAD21 and BAP1 are associated with PD-L1 expression in ccRCC tumor cells and might serve as a potential predictor of the response to immunotherapy with PD-1/PD-L1 inhibitors in ccRCC patients.
Acknowledgements
We thank Yongchen Ma’s contribution for performing the immunohistochemistry staining.
Abbreviations
- RCC
renal cell carcinoma
- ccRCC
clear cell renal cell carcinoma
- PRCC
papillary renal cell carcinoma
- ChRCC
chromophobe renal cell carcinoma
- TCGA
The Cancer Genome Atlas
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
- PD-1
programmed death-1
- PD-L1
programmed death-ligand 1
- OS
overall survival
- FFPE
formalin-fixed paraffin-embedded
- AJCC
American Joint Committee on Cancer
- IHC
immunohistochemistry
- COSMIC
Catalogue of Somatic Mutations in Cancer
- ACMG
American College of Medical Genetics
- TMB
tumor mutation burden
- DFS
disease-free survival
- BAP1
BRCA1-associated protein 1
- DDX51
DEAD-box helicase 51
Authors' contributions
YX and JL contributed to the acquisition of whole data independently and presented the same results. JW, HZ, and JL were responsible for DNA extraction, library preparation and immunohistochemical assays. JW and ZX were involved in the diagnosis and the recruitment of the patients in our affiliated hospitals and follow-up study. YX, HZ, and JW contributed to statistical analysis and data interpretation. ZX and RX contributed to revising it critically for important intellectual content. ZX, JX, and RX designed and organized the study. JW drafted the manuscript. All authors read and approved the final manuscript.
Funding
The study was funded by the National Natural Science Foundation of China (Grant No. 81272829).
Availability of data and materials
All the raw data generated and analyzed during this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study protocol conformed to the 1975 Declaration of Helsinki and all experiments involving human tissues and clinical data were performed in accordance with relevant guidelines. The present study was approved by the biomedical research ethics committee of Peking University First Hospital. Written informed contents were acquired from all the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Jie Wang, Email: wwyysswwjj@sina.com.
Jianzhong Xi, Email: jzxi@pku.edu.cn.
Hanshuo Zhang, Email: shzzhshuo@sina.com.
Juan Li, Email: jinyujin_0@163.com.
Yuchao Xia, Email: xiayuchao@pku.edu.cn.
Ruibin Xi, Email: ruibinxi@math.pku.edu.cn.
Zhijun Xi, Phone: +8601066175710, Email: xizhijun@hsc.pku.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Li H, Sun KX, Zheng RS, Zhang SW, Zeng HM, et al. Report of cancer incidence and mortality in China, 2014. Zhonghua Zhong Liu Za Zhi. 2018;40(1):5–13. doi: 10.3760/cma.j.issn.0253-3766.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Xi Z, Xi J, Zhang H, Li J, Xia Y, et al. Somatic mutations in renal cell carcinomas from Chinese patients revealed by whole exome sequencing. Cancer Cell Int. 2018;18:159. doi: 10.1186/s12935-018-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miya F, Kato M, Shiohama T, Okamoto N, Saitoh S, Yamasaki M, et al. A combination of targeted enrichment methodologies for whole-exome sequencing reveals novel pathogenic mutations. Sci Rep. 2015;5:9331. doi: 10.1038/srep09331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka S, Inoshita N, Miura Y, Oki R, Miyama Y, Nagamoto S, et al. The loss of BAP1 protein expression predicts poor prognosis in patients with nonmetastatic clear cell renal cell carcinoma with inferior vena cava tumor thrombosis. Urol Oncol. 2018;36(8):365. doi: 10.1016/j.urolonc.2018.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the raw data generated and analyzed during this study are available from the corresponding author on reasonable request.