Enterococcus faecalis strains are resident intestinal bacteria associated with invasive infections, inflammatory bowel diseases, and colon cancer. Although factors promoting E. faecalis colonization of intestines are not fully known, one implicated pathway is a phosphotransferase system (PTS) in E. faecalis strain OG1RF that phosphorylates gluconate and contains the genes OG1RF_12399 to OG1RF_12402 (OG1RF_12399-12402).
KEYWORDS: Enterococcus, colitis, gluconate, phosphotransferase system
ABSTRACT
Enterococcus faecalis strains are resident intestinal bacteria associated with invasive infections, inflammatory bowel diseases, and colon cancer. Although factors promoting E. faecalis colonization of intestines are not fully known, one implicated pathway is a phosphotransferase system (PTS) in E. faecalis strain OG1RF that phosphorylates gluconate and contains the genes OG1RF_12399 to OG1RF_12402 (OG1RF_12399-12402). We hypothesize that this PTS permits growth in gluconate, facilitates E. faecalis intestinal colonization, and exacerbates colitis. We generated E. faecalis strains containing deletions/point mutations in this PTS and measured bacterial growth and PTS gene expression in minimal medium supplemented with selected carbohydrates. We show that E. faecalis upregulates OG1RF_12399 transcription specifically in the presence of gluconate and that E. faecalis strains lacking, or harboring a single point mutation in, OG1RF_12399-12402 are unable to grow in minimal medium containing gluconate. We colonized germfree wild-type and colitis-prone interleukin-10-deficient mice with defined bacterial consortia containing the E. faecalis strains and measured inflammation and bacterial abundance in the colon. We infected macrophage and intestinal epithelial cell lines with the E. faecalis strains and measured intracellular bacterial survival and proinflammatory cytokine secretion. The presence of OG1RF_12399-12402 is not required for E. faecalis colonization of the mouse intestine but is associated with an accelerated onset of experimental colitis in interleukin-10-deficient mice, altered bacterial composition in the colon, enhanced E. faecalis survival within macrophages, and increased proinflammatory cytokine secretion by colon tissue and macrophages. Further studies of bacterial carbohydrate metabolism in general, and E. faecalis PTS-gluconate in particular, during inflammation may identify new mechanisms of disease pathogenesis.
INTRODUCTION
Enterococcus species are Gram-positive, facultative anaerobes that commonly colonize the human gastrointestinal tract early in life (1). Although enterococci are usually found in <1% of the total colonic bacterial population in adult humans, this percentage varies depending on age and diet (2). One species of enterococci, Enterococcus faecalis, has garnered particular attention as a human pathogen because it is a relatively common cause of endocarditis, septicemia, urinary tract infections, and persistent endodontic disease. Within the gastrointestinal tract, E. faecalis is associated with colorectal cancer and Crohn’s disease in humans (3, 4) and causes experimental colitis in mice (5–7). Understanding how E. faecalis colonizes the intestinal tract may provide clues to disease pathogenesis and treatment.
While relatively little is known about E. faecalis factors that facilitate colonization of the human intestine, more is known about E. faecalis colonization of the mouse intestine. For example, transcriptional analysis shows that E. faecalis strain OG1RF in the mouse colon upregulates the expression of several genes compared with E. faecalis OG1RF grown in laboratory culture media (8). Interestingly, seven phosphotransferase system (PTS) genes are among the most highly upregulated genes. The primary function of PTS proteins is to import and phosphorylate extracellular sugars that are used by the bacteria as an energy source or for biosynthetic processes. The E. faecalis OG1RF genome harbors 39 predicted PTS operons based on current NCBI genome annotation, but the sugar substrates for most of these have not been demonstrated experimentally. The OG1RF_12398 to OG1RF_12405 (OG1RF_12398-12405) operon is a putative E. faecalis PTS operon that is significantly upregulated in fecal bacteria from E. faecalis monocolonized mice and is predicted by current NCBI annotation to encode a mannose/fructose/sorbose family PTS (8).
Amino acid sequence alignment of OG1RF_12398-12405 with the corresponding genes in the well-studied E. faecalis strain V583 (EF3135-3142) demonstrates near-complete sequence similarity. Others have cloned EF3136 and solved its crystal structure (9). Moreover, Brockmeier et al. reported direct interactions between purified recombinant EF3136 and EF3137 (10). The interest in studying EF3136 and EF3137 was presumably based on the biochemical purification and characterization of homologous proteins in E. faecalis strain 26487 that were shown to phosphorylate gluconate, an oxidized form of glucose (9–11). However, evidence that this PTS is required for E. faecalis growth in gluconate-containing minimal medium is lacking.
In addition to being a product of glucose oxidation under oxidizing conditions, gluconate can also enter the intestinal tract from dietary sources (12). Others have shown that Escherichia coli upregulates gluconate utilization genes when cultured in the presence of gastrointestinal mucus and uses gluconate in mucus as a carbon source during early phases of colonization of the mouse gut (13, 14). However, the role of gluconate utilization by E. faecalis during colonization of the intestinal tract is unknown. We hypothesize that E. faecalis OG1RF_12398-12405 encode PTS proteins that import gluconate, are necessary for efficient colonization of the mouse gastrointestinal tract, and exacerbate colon inflammation in colitis-prone interleukin-10 (IL-10)-deficient (Il10−/−) mice.
RESULTS
OG1RF_12399-12402 encode gluconate metabolism proteins.
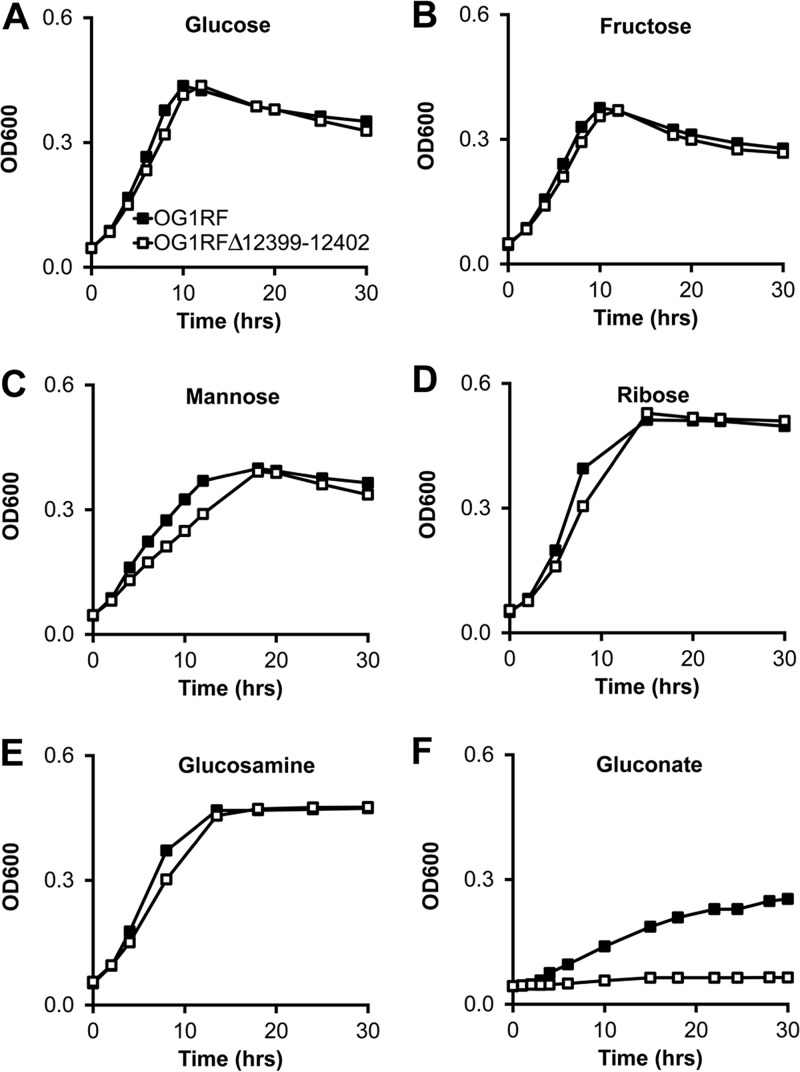
NCBI annotations of OG1RF_12398-12405 indicate that this putative operon might harbor genes important for the utilization of fructose, mannose, or sorbose. To experimentally test whether these predictions are correct, we first created an in-frame isogenic deletion mutant of OG1RF_12399-12402 (OG1RFΔ12399-12402), genes that are predicted to encode the core PTS components EIIA, EIIB, EIID, and EIIC of this operon. We then grew the parental and OG1RFΔ12399-12402 mutant strains in minimal media containing single sugars as carbon sources. While neither OG1RFΔ12399-12402 nor the parental strain grows in media containing sorbose (data not shown), both strains grow similarly well in media containing glucose, fructose, and mannose (Fig. 1). These data indicate that the OG1RF_12399-12402 operon does not harbor PTS genes important for utilizing glucose, fructose, mannose, or sorbose. In addition, we measured the growth of the two E. faecalis strains in minimal media containing ribose and glucosamine and detected no significant differences between the strains (Fig. 1).
FIG 1.
OG1RF_12399-12402 are required for E. faecalis growth in minimal medium containing gluconate. Shown are data for growth of OG1RF and OG1RFΔ12399-12402 in minimal media containing the indicated carbohydrates. Data are presented as means ± standard deviations (SD) (n = 6 cultures per bacterial strain). Differences in optical density at 600 nm (OD600) values between strains were statistically significant (P < 0.05 by Student’s t test) only for gluconate-containing medium (F).
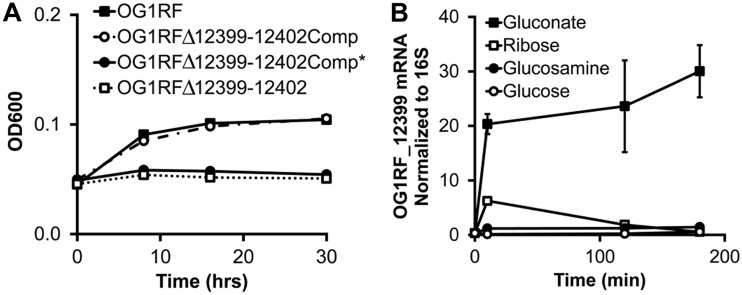
Examination of the NCBI annotation of genes adjacent to OG1RF_12399-12402 revealed two genes that are predicted to encode gluconate metabolism proteins, 2-dehydro-3-deoxyphosphogluconate aldolase (OG1RF_12397) and 6-phosphogluconate dehydrogenase (OG1RF_12405). In addition, others have shown using biochemical approaches that homologs of OG1RF_12399 and OG1RF_12400 in another E. faecalis strain phosphorylate gluconate (11). We therefore tested whether the OG1RF_12399-12402 operon is required for growth in gluconate-containing minimal medium by growing OG1RF and OG1RFΔ12399-12402 in minimal medium containing gluconate as the primary carbon source. OG1RF grows moderately well in gluconate-containing minimal medium, whereas OG1RFΔ12399-12402 fails to grow (Fig. 1 and 2). To confirm that the growth defect was not due to a spurious mutation elsewhere on the chromosome of OG1RFΔ12399-12402, we reintroduced the native OG1RF_12399-12402 genes back into the chromosome of the deletion mutant (OG1RFΔ12399-12402Comp) and found that the growth phenotype in gluconate-containing medium reverts to that of the parental strain (Fig. 2).
FIG 2.
OG1RF_12399 Asp66 is critical for protein function, and OG1RF_12399 mRNA is upregulated in the presence of gluconate. (A) Growth of the indicated bacterial strains in minimal medium containing gluconate. OG1RFΔ12399-12402Comp and OG1RFΔ12399-12402Comp* are strains in which OG1RF_12399-12402 and OG1RF_12399-12402 containing the D66H mutation in OG1RF_12399 have been introduced onto the chromosome of OG1RFΔ12399-12402, respectively. Data are presented as means ± SD (n = 6 cultures per strain). Differences in OD600 values between OG1RF/OG1RFΔ12399-12402Comp and OG1RFΔ12399-12402/OG1RFΔ12399-12402Comp* are statistically significant (P < 0.05 by Student’s t test). (B) Relative OG1RF_12399 mRNA levels in OG1RF grown in minimal medium containing the indicated carbohydrates. Data are presented as means ± SD (n = 3 cultures under each condition per time point). Differences in OG1RF_12399 mRNA abundances in bacteria grown in gluconate-containing medium versus other media are statistically significant (P < 0.05 by Student’s t test).
To further verify that OG1RF_12399-12402 encode gluconate PTS proteins, we quantified transcripts of OG1RF_12399, a gene predicted to encode PTS EIIA, in E. faecalis OG1RF before and after the addition of gluconate to the minimal growth medium. If OG1RF_12399-12402 encode gluconate PTS proteins, we predicted that E. faecalis OG1RF would upregulate their transcription in the presence of gluconate. Indeed, we detected significantly higher levels of OG1RF_12399 mRNA in E. faecalis OG1RF grown in the presence of gluconate than in the presence of ribose, glucosamine, and glucose (Fig. 2). Together with the growth curve data, these data support that the OG1RF_12398-12405 operon encodes PTS proteins necessary for gluconate metabolism.
OG1RF_12399 aspartate 66 is critical for growth in gluconate.
In the course of generating a genetically complemented strain of OG1RFΔ12399-12402, we serendipitously found that a random mutation in OG1RF_12399 introduced by PCR error into an expression plasmid abolishes the ability of the plasmid to complement the growth defect of OG1RFΔ12399-12402 in gluconate-containing minimal medium (data not shown). DNA sequencing revealed that the mutation is a G-to-C transversion at nucleotide position 196 in OG1RF_12399, which causes an aspartate-to-histidine missense mutation in amino acid 66. As determined by others (9), the D66H mutation disrupts the conformation of the catalytic active site by eliminating critical hydrogen bonding between the Asp66 and His9 residues. To more clearly characterize the functional effects of this point mutation, we introduced OG1RF_12399-12402 harboring the mutation onto the OG1RFΔ12399-12402 chromosome using a shuttle vector as described above to generate OG1RFΔ12399-12402Comp*. As expected, OG1RFΔ12399-12402Comp* does not grow in gluconate-containing minimal medium (Fig. 2). These findings confirm the predictions by Reinelt et al. that Asp66 is critical for the physiological function of OG1RF_12399 (9).
OG1RF_12399-12402 are not required for colonization of the mouse gut.
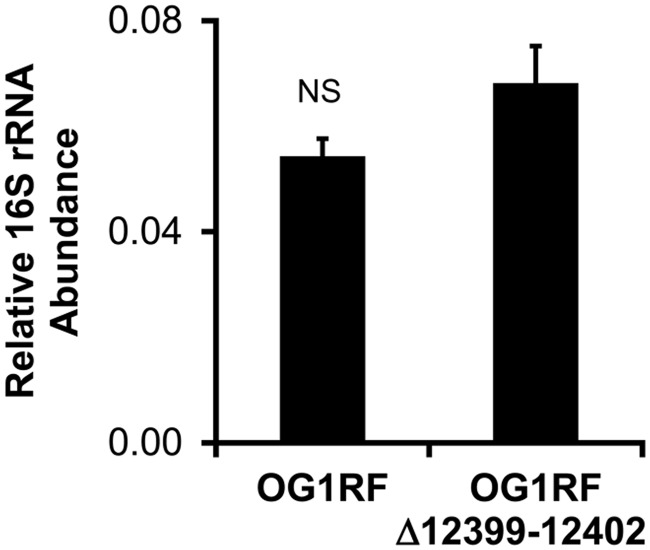
Since others have previously found that mRNA levels of OG1RF_12398-12405 in colonic E. faecalis from E. faecalis OG1RF monocolonized wild-type mice are elevated compared with laboratory-cultured E. faecalis OG1RF, we hypothesized that these genes are important for robust growth of E. faecalis in the mouse colon (8). To test this hypothesis, we colonized germfree wild-type mice (SvEv/129 background) for 5 weeks with a mixture of eight bacterial strains (Table 1) that represent each of the major bacterial phyla in the mammalian intestine, including E. faecalis OG1RF or OG1RFΔ12399-12402. We then quantified luminal bacterial numbers using 16S rRNA real-time PCR of bacterial DNA in cecal contents. We detected no difference in E. faecalis abundances in mice colonized with the mixture that included E. faecalis OG1RF versus OG1RFΔ12399-12402, indicating that OG1RF_12399-12402 do not affect the growth of E. faecalis in the mouse colon under these conditions (Fig. 3).
TABLE 1.
Bacteria used in this study
| Species | Strain | Source |
|---|---|---|
| Escherichia coli | NC101 | Mouse feces |
| Enterococcus faecalis | OG1RF | Human oral cavity |
| Bacteroides vulgatus | ATCC 8482 | Human feces |
| Bacteroides thetaiotaomicron | VPI-5482 | Human feces |
| Bifidobacterium longum | ATCC 15697 | Human feces |
| Lactobacillus rhamnosus | GG | Human feces |
| Ruminococcus gnavus | ATCC 29149 | Human feces |
| Faecalibacterium prausnitzii | A2-165 | Human feces |
FIG 3.
The absence of OG1RF_12399-12402 does not alter the abundance of E. faecalis OG1RF bacteria in gnotobiotic wild-type mice. Shown are E. faecalis 16S rRNA gene copy numbers relative to the total bacterial 16S rRNA gene copy number in cecal contents of wild-type mice colonized with a mixture of the eight bacterial strains listed in Table 1 for 5 weeks. Data are expressed as means ± standard errors of the means (SEM) (n = 6 mice per group). NS, not statistically significant.
OG1RF_12399-12402 exacerbate experimental colitis.
Others have previously shown that E. faecalis is associated with human inflammatory bowel disease and causes experimental colitis in genetically susceptible Il10−/− mice (4–7). Although OG1RF_12399-12402 do not affect colonization levels in healthy wild-type mice, we asked whether these genes affect colonization levels and colitis severity in Il10−/− mice.
To answer these questions, we colonized germfree wild-type and Il10−/− mice (SvEv/129 background) with the same eight bacterial strains as the ones described above (Table 1) for 5, 8, or 10 weeks and measured histological inflammation in colon segments and spontaneous secretion of the proinflammatory cytokine IL-12/23p40 by colon explant cultures in these mice. As expected, histological inflammation and IL-12/23p40 secretion in the colons of Il10−/− versus wild-type mice are increased at each time point (Fig. 4). Moreover, IL-12/23p40 secretion increases over time in Il10−/− mice with colitis, suggesting time-dependent worsening of inflammation.
FIG 4.
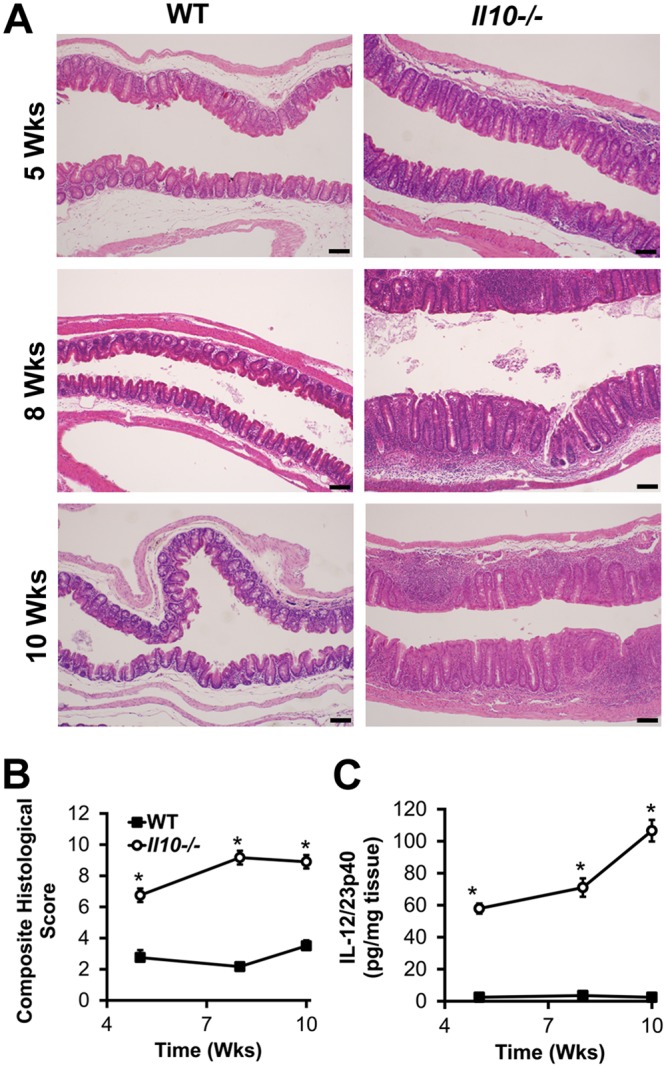
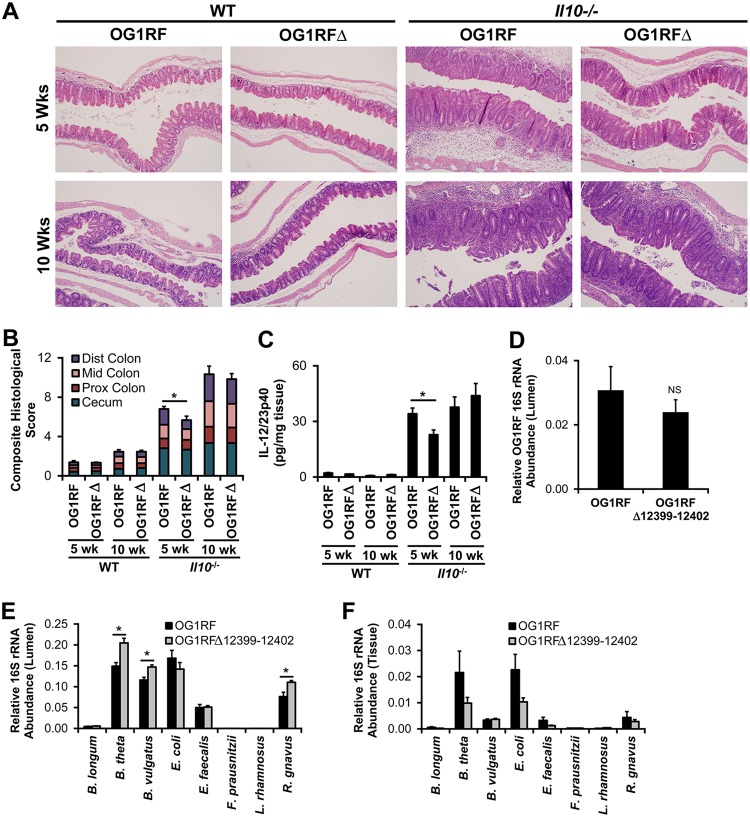
Gnotobiotic Il10−/− mice selectively colonized with eight bacterial strains, including E. faecalis OG1RF, develop colitis. (A) Representative photomicrographs of hematoxylin-and-eosin-stained cecal tissue from wild-type (WT) and Il10−/− mice colonized with the eight bacterial strains listed in Table 1 for the indicated times (bar = 100 μm). (B) Composite blind histological colitis scores (scale, 0 to 16) for mice referenced in panel A. (C) Spontaneous IL-12/23p40 secretion by colon explant cultures from mice referenced in panel A. Data in panels B and C are presented as means ± SEM (n = 5 to 6 mice/group/time point). *, P < 0.05 by Student’s t test.
We directly tested the effects of OG1RF_12399-12402 on colitis by colonizing wild-type and Il10−/− mice for 5 or 10 weeks with the mixture of eight bacterial strains (Table 1) containing either parental E. faecalis OG1RF or OG1RFΔ12399-12402 and then measured colitis severity using histological scoring and spontaneous secretion of IL-12/23p40 by colonic explant cultures as described above. As expected, Il10−/− mice have worse histological inflammation and higher levels of IL-12/23p40 secretion at 5 and 10 weeks than do wild-type mice (Fig. 5). The presence of OG1RF_12399-12402 does not affect histological inflammation or IL-12/23p40 secretion in wild-type mice or in Il10−/− mice at 10 weeks. However, we detected significantly less histological inflammation in the colon and spontaneous IL-12/23/p40 secretion by colonic explant cultures in mice colonized with E. faecalis OG1RFΔ12399-12402 than in mice colonized with E. faecalis OG1RF at 5 weeks (Fig. 5). These data indicate that OG1RF_12399-12402 accelerate the development of colitis in Il10−/− mice.
FIG 5.
OG1RF_12399-12402 accelerate the onset of colitis in gnotobiotic Il10−/− mice and alter luminal bacterial composition. (A) Representative photomicrographs of hematoxylin-and-eosin-stained cecal tissue from wild-type and Il10−/− mice colonized with the bacterial strains listed in Table 1 but including either E. faecalis OG1RF (OG1RF) or E. faecalis OG1RFΔ12399-12402 (OG1RFΔ). (B) Composite blind histological colitis scores (scale, 0 to 16) of mice referenced in panel A. (C) Spontaneous IL-12/23p40 secretion by colon explant cultures from mice referenced in panel A. (D) Relative 16S rRNA E. faecalis OG1RF gene abundance in midcolon contents of mice referenced in panel A, colonized for 5 weeks. (E) Relative 16S rRNA gene abundance of each bacterial strain in cecal contents of mice referenced in panel A, colonized for 5 weeks. (F) Relative 16S rRNA gene abundance of each bacterial strain in washed cecal tissue of mice referenced in panel A, colonized for 5 weeks. Data in panels D to F are expressed as a fraction of the total bacterial 16S rRNA. Data in panels B to F are presented as means ± SEM (n = 6 mice/group/time point). *, P < 0.05 by Student’s t test. NS, not statistically different.
We then hypothesized that the decreased colitis observed in Il10−/− mice colonized with OG1RFΔ12399-12402 for 5 weeks is associated with a decreased abundance of E. faecalis OG1RF in the colon. Similar to observations in wild-type mice, we detected no significant differences in E. faecalis numbers in the cecal or midcolon contents of Il10−/− mice colonized for 5 weeks with bacterial mixtures containing E. faecalis OG1RF versus OG1RFΔ12399-12402 (Fig. 5). However, we observed elevated levels of Bacteroides thetaiotaomicron, Bacteroides vulgatus, and Ruminococcus gnavus in cecal contents from Il10−/− mice colonized with OG1RFΔ12399-12402 versus OG1RF. The mechanisms by which OG1RF_12399-12402 affect the growth of these three bacterial strains and colitis severity in Il10−/− mice remain to be determined. Since mucosa-associated bacteria may impact host immune responses more than luminal bacteria, we tested whether OG1RF_12399-12402 affect the bacterial composition in washed cecal tissue from these mice. We detected no statistically significant differences in the abundances of any strain in washed cecal tissue from mice colonized with the two bacterial consortia (Fig. 5). However, the impact of OG1RF_12399-12402 on E. faecalis numbers in specific cell types within the intestinal mucosa is unknown.
OG1RF_12399-12402 increase survival of E. faecalis in macrophages in vitro and translocation to mesenteric lymph node cells in vivo.
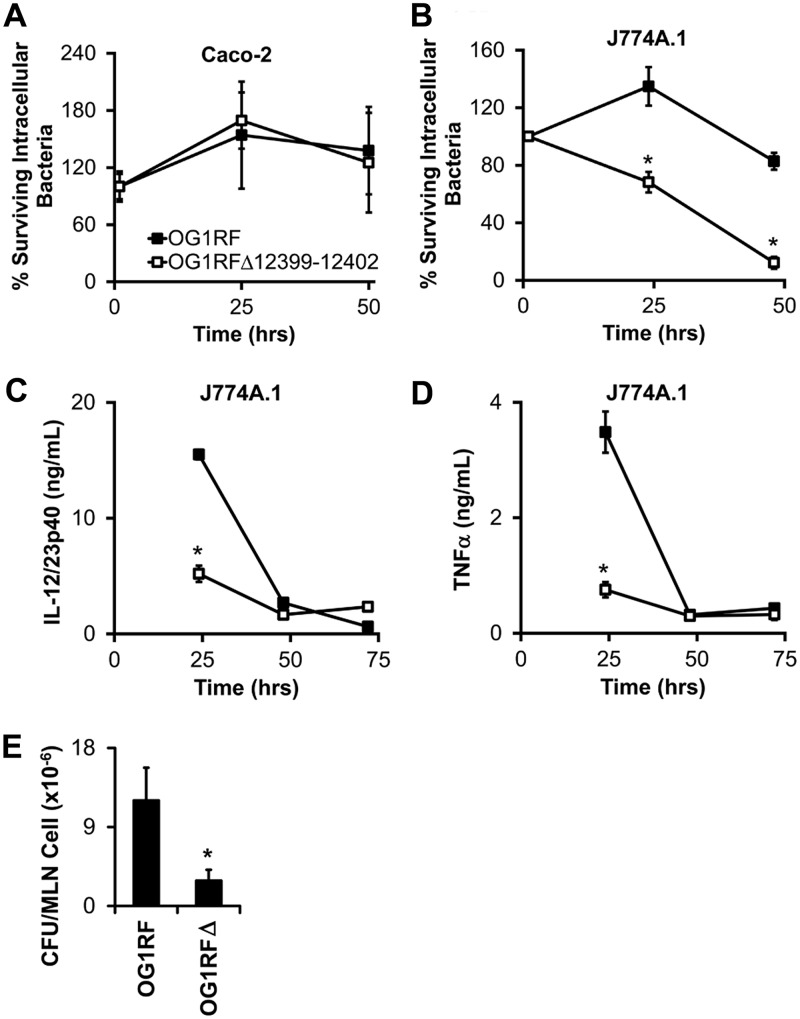
Invasion of resident intestinal bacteria through the intestinal epithelium and into the lamina propria is thought to contribute to the pathogenesis of inflammatory bowel diseases and experimental colitis (15–18). Therefore, we hypothesized that OG1RF_12399-12402 accelerate colitis by enhancing intracellular survival in intestinal epithelial cells and macrophages. To test this hypothesis, we infected Caco-2 intestinal epithelial cells or J774A.1 monocyte-macrophage cells with E. faecalis for 1 h, killed extracellular bacteria with gentamicin, and then plated epithelial/macrophage lysates on brain heart infusion (BHI) agar to enumerate viable intracellular bacteria. We detected similar survival rates of E. faecalis OG1RF and OG1RFΔ12399-12402 at 25 and 50 h postinfection in Caco-2 cells (Fig. 6). On the other hand, the survival rate of E. faecalis OG1RFΔ12399-12402 was significantly lower than that of OG1RF in J774A.1 macrophages at 25 and 48 h postinfection (Fig. 6), similar to previous reports (19). Moreover, the decreased survival of E. faecalis OG1RFΔ12399-12402 in macrophages corresponds with decreased secretion of the proinflammatory cytokines IL-12/23p40 and tumor necrosis factor (TNF) at 25 h postinfection (Fig. 6). To determine whether OG1RF_12399-12402 enhances bacterial translocation in vivo, we enumerated E. faecalis bacteria in mesenteric lymph node cell lysates from mice monocolonized with E. faecalis OG1RF or OG1RFΔ12399-12402. We detected significantly fewer E. faecalis bacteria in mesenteric lymph node cells from mice colonized with OG1RFΔ12399-12402 than in mice colonized with OG1RF (Fig. 6). Whether the OG1RF_12399-12402-associated increased intracellular bacterial survival and proinflammatory cytokine secretion in macrophages in vitro and increased bacterial translocation in vivo cause the accelerated colitis in Il10−/− mice is unknown.
FIG 6.
OG1RF_12399-12402 increase intramacrophage E. faecalis survival and proinflammatory cytokine secretion in vitro and bacterial translocation to mesenteric lymph nodes (MLN) in vivo. (A and B) Fraction of gentamicin-resistant (i.e., intracellular) E. faecalis bacteria in Caco-2 human intestinal epithelial cells (A) and J774A.1 mouse monocyte-macrophage cells (B). (C and D) IL-12/23p40 (C) and TNF-α (D) secretion by E. faecalis-infected J774A.1 cells referenced in panel B. (E) CFU per mesenteric lymph node cell in wild-type mice monocolonized for 2 weeks with either E. faecalis OG1RF or E. faecalis OG1RFΔ12399-12402. Data are presented as means ± SEM (n = 3 wells/bacterial strain/time point or 3 to 5 mice/group). Data in panels A and B are expressed as a percentage of intracellular bacteria at 1 h postinfection. *, P < 0.05 by Student’s t test.
DISCUSSION
In this work, we show that genes in the predicted E. faecalis operon, including OG1RF_12399, are expressed in response to gluconate, but not other sugars, and are necessary for growth in medium containing gluconate as the primary carbon source. Therefore, we propose that this operon, containing the genes OG1RF_12398-12405, be named PTS(Gnt), OG1RF_12399 be named EIIA(Gnt), OG1RF_12400 be named EIIB(Gnt), OG1RF_12401 be named EIID(Gnt), and OG1RF_12402 be named EIIC(Gnt), consistent with nomenclature proposed by Reinelt et al. (9). We also show that E. faecalis PTS(Gnt) does not affect the efficiency of colonization of the mouse colon but accelerates the onset of immune-mediated experimental colitis in Il10−/− mice. The mechanisms by which E. faecalis PTS(Gnt) accelerates colitis may involve its effects on other resident intestinal bacteria or its effects on intramacrophage bacterial survival and proinflammatory cytokine secretion. However, this remains to be shown experimentally. Also, it is important to note that other factors besides E. faecalis gluconate utilization more heavily contribute to the colitogenic potential of this bacterial consortium, since OG1RF PTS(Gnt) does not affect colitis severity by 10 weeks.
We found that the absence of OG1RF PTS(Gnt) increases the relative abundances of B. thetaiotaomicron, B. vulgatus, and R. gnavus in cecal contents of mice. One potential explanation for this finding is that E. faecalis competes with these three bacterial strains to use gluconate as a growth substrate in the cecum. A literature search did not reveal reported experimental evidence that these three strains metabolize gluconate. A survey of the NCBI annotated genomes of each of these strains identified gluconate metabolism genes in each strain. We found that the R. gnavus genome encodes a predicted gluconate transporter and permease, but neither Bacteroides species genome encodes a predicted gluconate transporter or permease. Data showing that these three strains can grow in minimal medium containing gluconate as the primary carbon source would support our hypothesis that these bacteria compete for gluconate in vivo.
Our current findings are significant because gluconate is found in common foodstuffs, including plants, fruits, dairy products, wine, honey, and vinegar, as well as antiseptics and calcium and iron supplements (12). Since gluconate is not known to be a structural component of intestinal mucus or secreted by intestinal epithelial cells, we hypothesize that the source of gluconate during inflammation may be oxidized dietary glucose because others have shown increased concentrations of tissue-reactive oxygen species and luminal glucose during intestinal inflammation (20–23). We plan to test this hypothesis by measuring gluconate concentrations in the lumen and at the mucosal interface in healthy and colitic mice. Based on the abundance of potential dietary and environmental sources of gluconate, further investigation of the effects of gluconate on E. faecalis PTS(Gnt) expression and E. faecalis virulence properties is warranted.
PTS components are associated with increased bacterial virulence in selected situations. For instance, phosphorylation of histidine residues on the PTS regulator protein HPr in Streptococcus pyogenes activates Mga, a transactivator of adhesion and antiphagocytic virulence genes (24). On the other hand, phosphorylation of serine residues on HPr activates CcpA, leading to enhanced activity of the E. faecalis virulence factor Ace (25).
PTS components have also previously been associated with reduced bacterial virulence. For example, HPr interacts with the CroSR two-component system to reduce cephalosporin resistance of E. faecalis (26). Also, the PTS-mannose system increases the sensitivity of E. faecalis to killing by bacteriocins (27).
There are several limitations of this work. We studied the gluconate transporter genes in only one strain of E. faecalis, OG1RF. Although the sequence similarity of this region is high across reported E. faecalis genomes, we cannot rule out strain-dependent differences in the functions of these genes. We show that PTS(Gnt) increases E. faecalis survival within macrophages in vitro, but the exact mechanisms by which this occurs or by which PTS(Gnt) might increase colitis in Il10−/− mice are unknown. Interestingly, others have shown that Salmonella enterica serovar Typhimurium upregulates the expression of its genes coding for gluconate transport and metabolism during intracellular growth within macrophages (28). Together with our findings, these data suggest that intramacrophage bacteria may use gluconate to sustain growth in the otherwise bacterium-toxic environment of the phagocytic vacuole. Finally, based on NCBI annotations of the E. faecalis OG1RF genome, there are 39 PTS operons, only 4 of which have been characterized. Whether they each affect experimental colitis and intramacrophage bacterial survival similarly to PTS(Gnt) is unknown. Further work is needed to investigate each of these questions.
The strengths of this work are that it provides, to the best of our knowledge, the first direct genetic evidence that OG1RF_12399-12402 encode proteins necessary for gluconate metabolism by E. faecalis, functional evidence that OG1RF_12399-12402 accelerate the onset of experimental colitis, and potential mechanistic explanations for how OG1RF_12399-12402 might alter E. faecalis virulence during colitis. These results lay the foundation for future investigations into the roles of gluconate metabolism by E. faecalis in health and disease.
MATERIALS AND METHODS
Mice.
Germfree wild-type and Il10−/− mice (both on the 129S6/SvEv background), originally derived under sterile conditions by hysterectomy at the Gnotobiotic Laboratory (University of Wisconsin—Madison), were purchased from the UNC National Gnotobiotic Rodent Resource Center. Gnotobiotic experiments were conducted in flexible-film isolators at the UNC National Gnotobiotic Rodent Resource Center. Male and female adult (8- to 12-week-old) germfree mice were colonized by oral gavage with a 200-μl equi-volume mixture of individual bacterial cultures grown for 48 h at 37°C in an anaerobic chamber. Oral gavage was repeated 3 days after the initial inoculation. After 5, 8, or 10 weeks of colonization, mice were sacrificed by CO2 asphyxiation and cervical dislocation, and tissues were harvested. Animal use protocols were approved by the UNC-Chapel Hill Institutional Animal Care and Use Committees.
Bacterial strains.
Bacterial strains (Table 1) were obtained from the following sources. E. coli NC101, Lactobacillus rhamnosus, Bifidobacterium longum, Faecalibacterium prausnitzii, and E. faecalis were kind gifts from R. Balfour Sartor, UNC-Chapel Hill. B. vulgatus and B. thetaiotaomicron were kind gifts from Jeffrey I. Gordon, Washington University, St. Louis, MO. R. gnavus was purchased from the ATCC (Manassas, VA). Prior to colonization of mice, bacteria were cultured anaerobically at 37°C for 48 h in the following media. E. coli NC101, E. faecalis OG1RF, B. thetaiotaomicron VPI-5482, and B. vulgatus ATCC 8482 were cultured in brain heart infusion (BHI) medium. R. gnavus ATCC 29149 was cultured in chopped-meat carbohydrate broth. F. prausnitzii was cultured in LY-BHI broth (BHI broth containing 0.05% l-cysteine, 0.1% cellobiose, 0.1% maltose, 5 g/liter yeast extract). L. rhamnosus and B. longum were cultured in MRS broth. All media were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ).
E. faecalis OG1RFΔ12399-12402 was generated by homologous recombination as previously described (29). Briefly, a 1-kb region of genomic DNA exactly upstream of the OG1RF_12399 open reading frame was PCR amplified using the primers 5′-TCCCGGGAATATAGGTCAACCCCATTGCTC-3′ and 5′-CTCTAGAGAATGTAGTAAAGGGTGAAAGCAATG-3′ and cloned into pJRS233 (a kind gift from Michael Caparon, Washington University, St. Louis, MO) using XmaI and XbaI restriction sites to create pJH191A. A 1-kb region of genomic DNA exactly downstream of the OG1RF_12402 open reading frame was PCR amplified using the primers 5′-CTCTAGAAGTTCCTCCTTCTTATCCTTGGTT-3′ and 5′-TCCGCGGGGTGCCGTCACTTTTCAAGGAA-3′ and cloned into pJH191A using XbaI and SacII restriction sites to create pJH191. Electrocompetent E. faecalis OG1RF bacteria were transformed with pJH191, and transformants were plated onto BHI agar containing 25 μg/ml erythromycin (Erm) and incubated at 30°C for 48 h. A single clone was then serially cultured in BHI agar containing Erm at 42°C for 96 h to force plasmid integration onto the chromosome, followed by BHI medium at 30°C for 96 h to select for bacteria lacking OG1RF_12399-12402. In-frame deletion of OG1RF_12399-12402 was confirmed by PCR amplification and Sanger sequencing of the entire operon.
E. faecalis OG1RFΔ12399-12402Comp was created by homologous recombination, similarly as described above. Briefly, OG1RF_12399-12402, including 1-kb upstream and downstream regions, were PCR amplified from OG1RF genomic DNA using the primers 5′-TCCGCGGGGTGCCGTCACTTTTCAAGGAA-3′ and 5′-TCCCGGGAATATAGGTCAACCCCATTGCTC-3′. The PCR product was cloned into pJRS233 using SacII and XmaI sites to produce pJH230. The OG1RF_12399-12402 genes were introduced onto the chromosome of OG1RFΔ12399-12402 by homologous recombination as described above and sequence confirmed by PCR amplification and Sanger sequencing of the operon.
E. faecalis OG1RFΔ12399-12402Comp* was generated by site-directed mutagenesis (G to C) of nucleotide 196 in OG1RF_12399 on pJH230 using the Q5 site-directed mutagenesis kit (NEB, Ipswich, MA) according to the manufacturer’s instructions and the PCR primers 5′-GGTCATGGTTCACTTATTAAGTGCG-3′ and 5′-AACACGCCATCCCCTTGT-3′ to create pJH231. The mutant was introduced onto the chromosome of OG1RFΔ12399-12402 by homologous recombination as described above and sequence confirmed by PCR amplification and Sanger sequencing of the operon.
Growth curves.
M9-based E. faecalis minimal medium was prepared to contain the following: 1× M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 2× minimal essential medium (MEM) nonessential amino acids (Gibco, Gaithersburg, MD), 1× MEM amino acids (Gibco, Gaithersburg, MD), and 2× MEM vitamin solution (Gibco, Gaithersburg, MD). The indicated carbohydrates were purchased from Sigma-Aldrich (St. Louis, MO) and added to a final concentration of 1% (wt/vol). Three milliliters of minimal medium was inoculated with 30 μl of a culture of the indicated E. faecalis strain grown overnight in BHI broth at 37°C. At the indicated time points, 100 μl of each culture was transferred to a 96-well plate, and the absorbance at 600 nm was measured using a BioTek (Winooski, VT) Synergy HTX plate reader.
Real-time PCR.
E. faecalis OG1RF was grown overnight at 37°C in BHI broth, diluted 1:100 in M9-based minimal medium containing 0.02% (wt/vol) glucose to a final volume of 2 ml, and incubated for 2 h at 37°C, and the indicated carbohydrate was then added to a final concentration of 0.2% (wt/vol). At the indicated time points after the addition of the carbohydrate, bacteria were rapidly pelleted, and pellets were resuspended in 0.5 ml bacterial RNAprotect (Qiagen, Germantown, MD) and frozen at −80°C for later use. Pellets were thawed and centrifuged at 14,000 × g for 2 min, and the supernatant was discarded. Bacterial RNA was isolated using a combination of phenol-chloroform extraction, mechanical bead beating, ethanol precipitation, and silica gel column purification as described previously, and cDNA was synthesized as described previously (30). The relative transcript abundance of OG1RF_12399 was determined by SYBR green real-time PCR using primers Efa16SF (5′-CGCGGTGCATTAGCTAGTTG-3′), Efa16SR (5′-TCACCCTCTCAGGTCGGCTAT-3′), Ef12399F (5′-TCCCCTTGTTGGACGTTCTC-3′), and Ef12399R (5′-GAAAGACGCAGCTACCGTGA-3′), and results are expressed relative to OG1RF 16S rRNA transcripts using the ΔCT method as described previously (30).
Bacterial DNA from cecal contents or phosphate-buffered saline (PBS)-washed pieces of cecal tissue was isolated, and 16S rRNA abundance was determined using SYBR green real-time PCR as described previously (18, 31) and the following primers: Blo16SF (5′-GCAACGCGAAGAACCTTACC-3′), Blo16SR (5′-CGCCCCGAAGGGAAAC-3′), Bth16SF (5′-CAGTGTCAGTTGCAGTCCAGTGA-3′), Bth16SR (5′-GTGTAGCGGTGAAATGCTTAGATATC-3′), Bvu16SF (5′-GGTCGCCATCGGCCTTA-3′), Bvu16SR (5′-CTAGCTACAGGCTTAACACATGCAA-3′), Efa16S (see above), Efa16R (see above), Eco16SF (5′-GAATGCCACGGTGAATACGTT-3′), Eco16SR (5′-ACCCACTCCCATGGTGTGA-3′), Fpr16SF (5′-CCAGCCGCAGGTTCTCCTA-3′), FPr16SR (5′-CGCCGCCGAAGGTAAAA-3′), Lrh16SF (5′-GAGAAGAATGGTCGGCAGAGTAA-3′), Lrh16SR (5′-CACGTAGTTAGCCGTGGCTTT-3′), Rgn16SF (5′-AACGTACTCCCCAGGTGGAAT-3′), Rgn16SR (5′-CGGGTGGCAAAGCCATT-3′), and universal bacterial 16S rRNA primers UniF (5′-GTGSTGCAYGGYTGTCGTCA-3′) and UniR (5′-ACGTCRTCCMCACCTTCCTC-3′). For cecal tissue, the cecum was opened longitudinally, and contents were gently removed by scraping with closed forceps. Tissue was then grasped with small forceps and shaken vigorously twice in 40 ml of sterile PBS for 5 s each, after which ∼20 mg was removed for bacterial DNA isolation. Results are presented relative to the universal bacterial 16S rRNA gene using the ΔCT method.
Quantification of colitis severity.
Histological inflammation was quantified by blind histological scoring of formalin-fixed, hematoxylin-and-eosin-stained sections of cecum, proximal colon, midcolon, and distal colon as described previously (30). Colon explant cultures were performed as described previously (30).
Gentamicin protection assays.
Gentamicin protection assays to assess intramacrophage bacterial survival were performed as described previously (30). Briefly, J774A.1 mouse monocytes-macrophages (purchased from the Lineberger Cancer Center Tissue Culture Facility, Chapel Hill, NC) were cultured in Dulbecco's modified Eagle's high glucose medium (DMEM-H) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. A total of 1 × 106 J774A.1 cells per well were plated in 12-well plates. The following day, 1 × 107 PBS-washed, mid-log-growth-phase E. faecalis bacteria were added to each well in DMEM-H and incubated at 37°C for 1 h (the end of which was designated time zero). Macrophages were washed three times with 1 ml PBS/well, after which DMEM-H plus 10% FBS containing 300 μg/ml gentamicin was added, and plates were incubated at 37°C for 1 h. Macrophages were washed three times with 1 ml PBS/well, and either the macrophages were then harvested (time 1 h) or DMEM-H plus 10% FBS containing 30 μg/ml gentamicin was added until the indicated time. At the indicated time, macrophages were washed three times with 1 ml PBS/well, and 0.5 ml of 1% Triton X-100 was then added to lyse mammalian cells. Lysates were diluted and plated onto BHI agar to enumerate CFU.
Gentamicin protection assays in human Caco-2 intestinal epithelial cells were performed similarly, with the following modifications. Only 5 × 105 epithelial cells and 5 × 106 bacterial cells per well were used. Growth medium was a 1:1 mixture of DMEM-H and F-12 medium containing 10% FBS and 1% penicillin-streptomycin.
Enzyme-linked immunosorbent assays.
Mouse TNF-α and IL-12/23p40 levels were measured in supernatants of cell cultures and colon explant cultures as described previously (30).
Bacterial translocation assay.
Wild-type germfree C57/B6 mice were transferred to sterilized, individually ventilated cages and colonized with E. faecalis cultures grown overnight in BHI broth by oral and rectal swabs in a laminar flow hood using sterile gloves and swabs. Two weeks later, mice were euthanized, mesenteric lymph nodes were aseptically removed and mechanically disrupted using a Teflon pestle in a 1.5-ml microcentrifuge tube, and viable lymph node cells were counted using an automated cell counter (Bio-Rad, Hercules, CA). The lymph node cells were lysed for 5 min in 1% Triton X-100, and lysates were immediately plated onto BHI agar to enumerate CFU.
ACKNOWLEDGMENTS
We acknowledge the Gnotobiotic, Histology, and Immunotechnology Cores at the UNC Center for Gastrointestinal Biology and Disease (supported by NIH grant P30DK34987, NIH grant P40OD 010995, and the Crohn’s and Colitis Foundation). This work was also funded by startup funds from the University of North Carolina at Chapel Hill.
We also thank Gary Port for technical advice during the construction of the E. faecalis deletion mutant.
REFERENCES
- 1.Fanaro S, Chierici R, Guerrini P, Vigi V. 2003. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 91:48–55. [DOI] [PubMed] [Google Scholar]
- 2.Tannock GW, Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p 101–132. In Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 3.Zhou Y, He H, Xu H, Li Y, Li Z, Du Y, He J, Zhou Y, Wang H, Nie Y. 2016. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget 7:80794–80802. doi: 10.18632/oncotarget.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y, Chen H, He H, Du Y, Hu J, Li Y, Li Y, Zhou Y, Wang H, Chen Y, Nie Y. 2016. Increased Enterococcus faecalis infection is associated with clinically active Crohn disease. Medicine (Baltimore) 95:e5019. doi: 10.1097/MD.0000000000005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. 2005. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. 2007. Dual-association of gnotobiotic IL-10−/− mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis 13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 7.Ocvirk S, Sava IG, Lengfelder I, Lagkouvardos I, Steck N, Roh JH, Tchaptchet S, Bao Y, Hansen JJ, Huebner J, Carroll I, Murray BE, Sartor RB, Haller D. 2015. Surface-associated lipoproteins link Enterococcus faecalis virulence to colitogenic activity in IL-10-deficient mice independent of their expression levels. PLoS Pathog 11:e1004911. doi: 10.1371/journal.ppat.1004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindenstrauss AG, Ehrmann MA, Behr J, Landstorfer R, Haller D, Sartor RB, Vogel RF. 2014. Transcriptome analysis of Enterococcus faecalis toward its adaption to surviving in the mouse intestinal tract. Arch Microbiol 196:423–433. doi: 10.1007/s00203-014-0982-2. [DOI] [PubMed] [Google Scholar]
- 9.Reinelt S, Koch B, Hothorn M, Hengstenberg W, Welti S, Scheffzek K. 2009. Structure of the Enterococcus faecalis EIIA(gnt) PTS component. Biochem Biophys Res Commun 388:626–629. doi: 10.1016/j.bbrc.2009.08.054. [DOI] [PubMed] [Google Scholar]
- 10.Brockmeier A, Skopnik M, Koch B, Herrmann C, Hengstenberg W, Welti S, Scheffzek K. 2009. Activity of the Enterococcus faecalis EIIA(gnt) PTS component and its strong interaction with EIIB(gnt). Biochem Biophys Res Commun 388:630–636. doi: 10.1016/j.bbrc.2009.08.100. [DOI] [PubMed] [Google Scholar]
- 11.Bernsmann P, Alpert CA, Muss P, Deutscher J, Hengstenberg W. 1982. The bacterial PEP-dependent phosphotransferase system mechanism of gluconate phosphorylation in Streptococcus faecalis. FEBS Lett 138:101–103. doi: 10.1016/0014-5793(82)80404-3. [DOI] [PubMed] [Google Scholar]
- 12.Ramachandran S, Fontanille P, Pandey A, Larroche C. 2006. Gluconic acid: properties, applications and microbial production. Food Technol Biotechnol 44:185–195. [Google Scholar]
- 13.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun 64:3504–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vrakas S, Mountzouris KC, Michalopoulos G, Karamanolis G, Papatheodoridis G, Tzathas C, Gazouli M. 2017. Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS One 12:e0170034. doi: 10.1371/journal.pone.0170034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, De Hertogh G, Valentino E, Braun E, Zheng J, Boisgard R, Neut C, Dubuquoy L, Chiappini F, Samuel D, Lepage P, Guerrieri F, Dore J, Brechot C, Moniaux N, Faivre J. 2018. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology 154:1009.e14–1023.e14. doi: 10.1053/j.gastro.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Dheer R, Santaolalla R, Davies JM, Lang JK, Phillips MC, Pastorini C, Vazquez-Pertejo MT, Abreu MT. 2016. Intestinal epithelial Toll-like receptor 4 signaling affects epithelial function and colonic microbiota and promotes a risk for transmissible colitis. Infect Immun 84:798–810. doi: 10.1128/IAI.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchaptchet S, Fan TJ, Goeser L, Schoenborn A, Gulati AS, Sartor RB, Hansen JJ. 2013. Inflammation-induced acid tolerance genes gadAB in luminal commensal Escherichia coli attenuate experimental colitis. Infect Immun 81:3662–3671. doi: 10.1128/IAI.00355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Z, Ehrmann MA, Waldhuber A, Niemeyer C, Miethke T, Frick JS, Xiong T, Vogel RF. 2017. Phosphotransferase systems in Enterococcus faecalis OG1RF enhance anti-stress capacity in vitro and in vivo. Res Microbiol 168:558–566. doi: 10.1016/j.resmic.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. 2011. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 21.Keshavarzian A, Banan A, Farhadi A, Komanduri S, Mutlu E, Zhang Y, Fields JZ. 2003. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut 52:720–728. doi: 10.1136/gut.52.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshavarzian A, Sedghi S, Kanofsky J, List T, Robinson C, Ibrahim C, Winship D. 1992. Excessive production of reactive oxygen metabolites by inflamed colon: analysis by chemiluminescence probe. Gastroenterology 103:177–185. doi: 10.1016/0016-5085(92)91111-G. [DOI] [PubMed] [Google Scholar]
- 23.Simmonds NJ, Allen RE, Stevens TR, Van Someren RN, Blake DR, Rampton DS. 1992. Chemiluminescence assay of mucosal reactive oxygen metabolites in inflammatory bowel disease. Gastroenterology 103:186–196. doi: 10.1016/0016-5085(92)91112-H. [DOI] [PubMed] [Google Scholar]
- 24.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol 88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao P, Pinkston KL, Bourgogne A, Cruz MR, Garsin DA, Murray BE, Harvey BR. 2013. Library screen identifies Enterococcus faecalis CcpA, the catabolite control protein A, as an effector of Ace, a collagen adhesion protein linked to virulence. J Bacteriol 195:4761–4768. doi: 10.1128/JB.00706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder H, Kellogg SL, Skarda LM, Little JL, Kristich CJ. 2014. Nutritional control of antibiotic resistance via an interface between the phosphotransferase system and a two-component signaling system. Antimicrob Agents Chemother 58:957–965. doi: 10.1128/AAC.01919-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hechard Y, Pelletier C, Cenatiempo Y, Frere J. 2001. Analysis of sigma(54)-dependent genes in Enterococcus faecalis: a mannose PTS permease (EII(Man)) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575–1580. doi: 10.1099/00221287-147-6-1575. [DOI] [PubMed] [Google Scholar]
- 28.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Casal J, Price JA, Maguin E, Scott JR. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol 8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 30.Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, Hansen JJ. 2011. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology 141:1842.e10–1851.e10. doi: 10.1053/j.gastro.2011.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan TJ, Tchaptchet SY, Arsene D, Mishima Y, Liu B, Sartor RB, Carroll IM, Miao EA, Fodor AA, Hansen JJ. 2018. Environmental factors modify the severity of acute DSS colitis in caspase-11-deficient mice. Inflamm Bowel Dis 24:2394–2403. doi: 10.1093/ibd/izy244. [DOI] [PMC free article] [PubMed] [Google Scholar]