Half of all humans harbor Helicobacter pylori in their stomachs. Helical cell shape is thought to facilitate H. pylori’s ability to bore into the protective mucus layer in a corkscrew-like motion, thereby enhancing colonization of the stomach.
KEYWORDS: Helicobacter pylori, chronic infection, gastric colonization, helical cell shape, pathology
ABSTRACT
Half of all humans harbor Helicobacter pylori in their stomachs. Helical cell shape is thought to facilitate H. pylori’s ability to bore into the protective mucus layer in a corkscrew-like motion, thereby enhancing colonization of the stomach. H. pylori cell shape mutants show impaired colonization of the mouse stomach, highlighting the importance of cell shape in infection. To gain a deeper understanding of how helical cell morphology promotes host colonization by H. pylori, we used three-dimensional confocal microscopy to visualize the clinical isolate PMSS1 and an isogenic straight-rod mutant (Δcsd6) within thick longitudinal mouse stomach sections. We also performed volumetric image analysis to quantify the number of bacteria residing within corpus and antral glands in addition to measuring total CFU. We found that straight rods show attenuation during acute colonization of the stomach (1 day or 1 week postinfection) as measured by total CFU. Our quantitative imaging revealed that wild-type bacteria extensively colonized antral glands at 1 week postinfection, while csd6 mutants showed variable colonization of the antrum at this time point. During chronic infection (1 or 3 months postinfection), total CFU were highly variable but similar for wild-type and straight rods. Both wild-type and straight rods persisted and expanded in corpus glands during chronic infection. However, the straight rods showed reduced inflammation and disease progression. Thus, helical cell shape contributes to tissue interactions that promote inflammation during chronic infection, in addition to facilitating niche acquisition during acute infection.
INTRODUCTION
Helicobacter pylori is a Gram-negative, helical bacterium that has evolved to survive in the human stomach. H. pylori chronically colonizes the gastric mucosa of approximately 20% of the population in developed countries and greater than 70% of the population in the developing world (1). Most H. pylori infections are asymptomatic; however, chronic infection increases the risk of developing chronic active gastritis, peptic ulcer disease, duodenal ulcers, gastric adenocarcinoma, and gastric extranodal marginal-zone lymphoma of mucosa-associated lymphoid tissue type (MALT lymphoma) (2). The stomach is an unfavorable environment for bacteria due to its acidity, active digestive enzymes, and low partial oxygen pressure. As a neutrophile, H. pylori can only survive for minutes in the stomach lumen and overcomes the acidic environment using urease, an enzyme that hydrolyzes urea to produce NH3, locally elevating the pH to near neutral. Successful colonization of the stomach by H. pylori requires both urease (3–6) and flagellum-mediated, chemosensory-directed motility to swim out of the lumen and through the mucus layer (7–10). Helical cell shape is thought to facilitate H. pylori’s ability to bore into the mucus layer in a corkscrew-like motion, further enhancing its motility through the highly viscous gastric mucus layer that overlies the gastric epithelium. Upon penetrating this thick (∼300-μm) mucus layer, H. pylori preferentially colonizes a narrow band (∼25 μm thick) of mucus immediately overlying the gastric epithelial cell surface (11). While H. pylori actively adheres to gastric epithelial cells, it remains extracellular and is only rarely observed within cells (12–14). Upon attachment, H. pylori disrupts the tight junctions of epithelial cells to exploit them as a site for replication, where the bacteria can grow as cell-associated microcolonies (15). H. pylori also penetrates gastric pits and grows in microcolonies deep in the gastric glands (8).
The helical cell shape of H. pylori is generated and maintained by the peptidoglycan (PG) cell wall (16, 17). PG-modifying enzymes (Csd1, Csd2, Csd3/HdpA, Csd4, and Csd6) are required to maintain helical cell shape in H. pylori (16–22). We and others have shown that H. pylori cell shape mutants (Δcsd1 curved-rod and Δcsd3 variably c-shaped-rod mutants) are attenuated in stomach colonization (16, 19). Straight-rod mutants (Δcsd4) also show reduced stomach colonization loads and are outcompeted by wild-type bacteria during coinfection (17). The cell shape mutants do not show an impaired ability to infect human gastric adenocarcinoma (AGS) cells in vitro, and infected cells produce a proinflammatory interleukin-8 (IL-8) response similar to that of wild-type H. pylori infection (17). We previously showed that variation in both cell body helical parameters (helical pitch and radius) and flagellum number among different H. pylori clinical isolates (LSH100, PMSS1, and B128) leads to distinct and broad swimming speed distributions that reflect both temporal variation in the swimming speed of individual bacterial cells and morphologic variation within the population (23). Furthermore, isogenic mutants with straight-rod morphology (Δcsd6) showed reduced swimming speeds and a higher fraction of immobilized bacteria in purified gastric mucin gels (23). Whether altered motility behavior in mucus fully accounts for the altered stomach colonization potential of cell shape mutants remains an open question.
Different H. pylori strains preferentially colonize the corpus or antrum of the human stomach. The inner lining of the stomach consists of four layers, the serosa, muscularis, submucosa, and mucosa. The mucosa is densely packed with branched tubular gastric glands. Corpus glands are mostly comprised of chief cells, which secrete pepsin, and parietal cells, which secrete hydrochloric acid. The antrum, which comprises about one-fourth of the stomach, is lined by glands mostly containing mucus-secreting cells and endocrine cells. Chronic infection with H. pylori triggers inflammation in the corpus or antrum, further resulting in distinct disease outcomes. Antrum-predominant gastritis is associated with increased acid production, a risk factor for duodenal ulcers (24, 25). Corpus-predominant gastritis leads to loss of parietal cells and eventual reduced acid secretion, increasing the risk for gastric cancer (24, 26, 27). As in the human stomach, H. pylori can colonize both the corpus and antrum regions of the mouse gastric mucosa (28). The mouse stomach contains two grossly distinct stomach regions, a nonglandular (forestomach) region and a glandular region (corpus and antrum). The forestomach, which does not become colonized by H. pylori, is lined with keratinized squamous epithelium and is separated from the glandular corpus region by a raised mucosal fold referred to as the limiting ridge. Several mouse-adapted H. pylori isolates induce gastritis and gland atrophy in C57BL/6 mice but do not induce neoplasia (29, 30). However, chronic infection with PMSS1, a strain shown to be more virulent than other clinical isolates in mice, triggers inflammation, gland hyperplasia, gastric atrophy, and early signs of metaplasia (31, 32).
Here, we used a mouse model of infection to investigate how helical cell shape helps H. pylori establish infection and acquire a replicative niche within the stomach. Both Δcsd4 and Δcsd6 straight-rod mutants display shape-dependent motility phenotypes in vitro (17, 23). For the purposes of this study, we used a Δcsd6 mutant in the PMSS1 strain to test whether this mutation confers colonization defects similar to those previously described for the Δcsd4 mutant and to extend these results to the commonly used PMSS1 strain background (our previous colonization assays utilized derivatives of strain G27). In addition to enumerating CFU of H. pylori in homogenized stomach tissue, we used three-dimensional (3D) confocal microscopy and volumetric image analysis to localize and quantify the number of bacteria present within corpus and antral glands. We discovered that Δcsd6 straight rods are attenuated at 1 day and 1 week postinfection, both in CFU load and localization within gastric glands, yet nonetheless can establish chronic infection. At 1 week postinfection, straight rods show reduced localization within antral glands, while at 1 month postinfection, localization within both antral and corpus glands is similar to or greater than that of wild-type H. pylori. In spite of their ability to localize within the glands, Δcsd6 straight rods elicited less inflammation and hyperplasia in the antrum and the transition zone between corpus and antrum at 1 and 3 months postinfection. Our study supports a role for helical cell shape in promoting efficient stomach colonization during acute infection and in driving gastric pathology during chronic infection.
RESULTS
Csd6-dependent helical cell shape of H. pylori confers an advantage during initial colonization of the stomach.
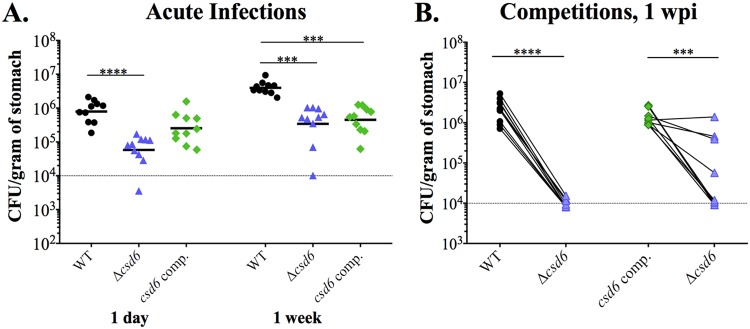
To investigate how helical cell morphology contributes to H. pylori stomach colonization and persistence, we conducted single-strain infections and competitions with wild-type H. pylori PMSS1 and an isogenic Δcsd6 straight-rod mutant (Table 1). We harvested a third of the stomach to assess bacterial load by CFU per gram of stomach tissue, a third to fix in paraformaldehyde (PFA) for immunohistochemistry for enumeration of bacteria within gastric glands, and a third for pathological evaluation (Fig. 1). As expected, we did not recover H. pylori from the mock-infected groups at any time point (data not shown). The Δcsd6 straight-rod mutant showed significantly attenuated gastric loads at 1 day, with a 2-log difference in CFU/g of stomach tissue from that of the wild type (P < 0.0001, unpaired nonparametric two-tailed Mann Whitney U test) (Fig. 2A). At 1 week, the mutant had a 1-log difference in recovery compared to that of the wild type (P < 0.0001), as had been previously reported for another straight-rod mutant (Δcsd4) generated in a different H. pylori strain background (LSH100) (17). In a competition experiment, wild-type bacteria strongly outcompeted the Δcsd6 mutant at 1 week postinfection (P = 0.0005, paired t test) (Fig. 2B). Complementation of the Δcsd6 mutant by expressing the csd6 gene at a distal intragenic locus (22, 33) restored helical cell shape, and no significant differences in side curvature distributions were observed between the wild-type and csd6 complemented strains (P = 0.6, Kolmogorov-Smirnov statistics) (see Fig. S1 in the supplemental material). In mice, the csd6 complemented strain showed colonization loads comparable to those of the wild-type strain at 1 day postinfection, but interestingly it was comparable to the Δcsd6 mutant at 1 week postinfection (Fig. 2A). However, in competitive infection, the Δcsd6 mutant was outcompeted by the csd6 complemented strain in 6 of 10 mice at 1 week in two independent experiments (Fig. 2B). The less than complete complementation may result from altered expression of Csd6 at the locus used for complementation during infection compared to that at the native locus; our previous work showed that cell morphology can be altered by either increased or decreased expression of Csd6 (22). Nonetheless, we tested both the deletion strain and the complemented strain in subsequent experiments.
TABLE 1.
Bacterial strains used in this study
| Strain name | Genotype or description | Shape phenotype | Reference or source |
|---|---|---|---|
| LSH100 | Wild-type H. pylori; mouse-adapted derivative from H. pylori G27 | Helical | 48–50 |
| PMSS1 | Wild-type H. pylori | Helical | 31, 46 |
| LMH6 | Δcsd6::cat in PMSS1 | Straight | This study |
| TSH17 | Δcsd6::cat in LSH100 | Straight | 22 |
| TSH31 | Δcsd6::cat McGee:csd6:aphA3 in LSH100 | Helical and straight morphologies | 22 |
| EPH1 | csd6 complemented strain, Δcsd6::cat McGee:csd6:aphA3 in PMSS1 | Helical | This study |
| LMH12 | csd6 complemented strain, EPH1 backcrossed in PMSS1 | Helical | This study |
| CLH1 | ΔcagE::cat in PMSS1 | Helical | This study |
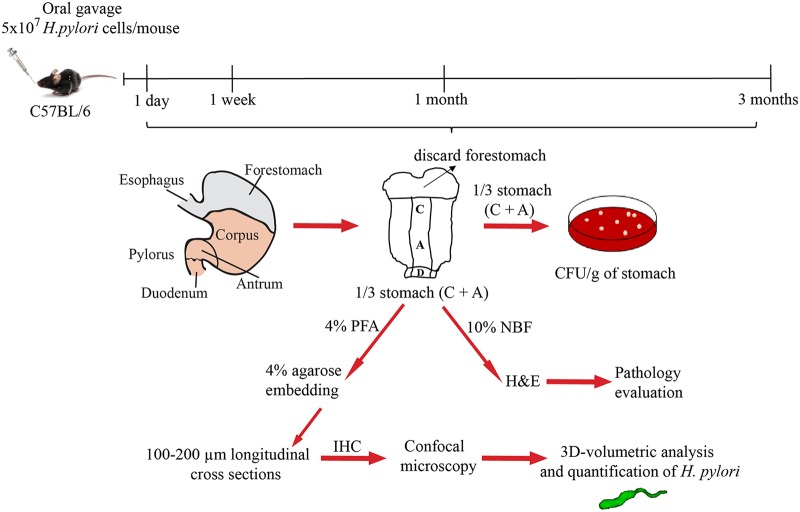
FIG 1.
Experimental outline. C57BL6/J mice were infected by oral gavage with wild-type (PMSS1), straight-rod mutant (Δcsd6), or csd6 complemented H. pylori bacteria or mock infected with broth. At the indicated time points, the stomach was removed and one-third used to determine bacterial load, one-third for pathology evaluation, and one-third for bacterial localization within glands. C, corpus; A, antrum; PFA, paraformaldehyde; NBF, neutral buffered formalin; H&E, hematoxylin and eosin; IHC, immunohistochemistry.
FIG 2.
H. pylori Δcsd6 straight-rod mutant shows early colonization defects compared to wild-type bacteria. Single or competitive infections were performed with the wild-type strain, Δcsd6 mutant, csd6 complemented strain, or broth (mock infection control). (A) Stomach loads at 1 day and 1 week of infection. P < 0.001 (***) and P < 0.0001 (****) by Kruskal-Wallis test with Dunn’s multiple-test correction. (B) Competitive infections between wild-type and Δcsd6 mutant or csd6 complemented strain and Δcsd6 mutant, with lines connecting the bacterial load values for each genotype from the same mouse. P < 0.001 (***) and P < 0.0001 (****) by Mann-Whitney U test. Dotted line indicates the average limit of detection. Data provided in panels A and B are from two independent experiments with a total of n = 10 mice per group. WT, wild type; comp, complemented; wpi, week postinfection.
3D volumetric image analysis of H. pylori indicates an antral gland preference for both wild-type and straight-rod mutant bacteria during acute infection.
Others have shown that during experimental infection in C57BL/6 mice, H. pylori bacteria reside in the mucus layer that overlies the stomach epithelium and a subpopulation of bacteria penetrate deep in the gastric glands, where they can adhere to gastric epithelial cells comprising the mid-glandular proliferative zone (8, 32). We questioned whether the Δcsd6 mutant could occupy the same niches. In thin (4- to 5-μm) sections, H. pylori is difficult to quantify, because the gastric gland lumen is rarely captured. Thus, we followed a recently established method for 3D confocal microscopy to visualize and quantify the number of H. pylori bacteria colonizing corpus and antral glands in thick (100- to 200-μm) sections using volumetric image analysis of individual bacterial cells and bacteria within microcolonies (8, 32). Similar to prior studies, the mean volume for individual cells analyzed was 9.5 μm3 (n = 203 cells analyzed), and clusters of two bacteria showed a proportional increase in volume (Fig. S2).
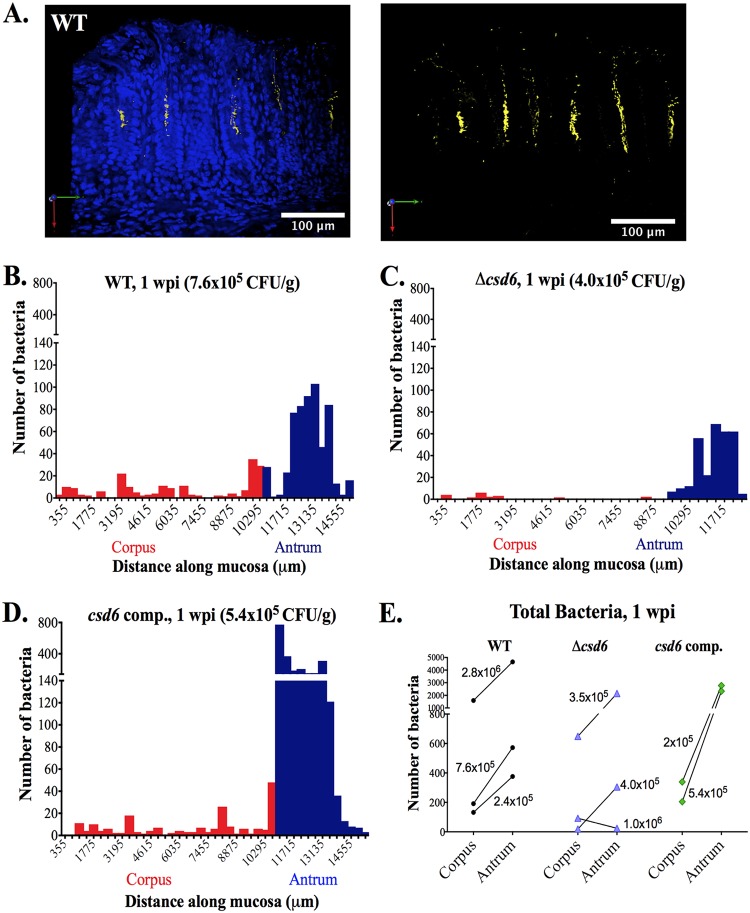
At 1 day postinfection, both the wild type and the Δcsd6 mutant were detected in corpus and antral glands at very low densities (data not shown). At 1 week, 3D visualization of the antrum showed wild-type bacteria associated with gastric epithelial cells near the luminal surface, as well as deeper in the glands, where they form dense microcolonies (Fig. 3A). To explore bacterial localization differences among strains, we quantified the number of bacteria in each field of view along the length of the stomach for one mouse from each genotype with similar colonization loads (wild type, 7.6 × 105 CFU/g; Δcsd6 mutant, 4.0 × 105 CFU/g; csd6 complemented, 5.4 × 105 CFU/g) (Fig. 3B to D). Wild-type bacteria were easily detected in antral glands and the transition zone between the corpus and antrum (C/A) junction (Fig. 3B). Fewer bacteria were observed in corpus glands (<40 bacteria per field of view). The Δcsd6 mutant bacteria were detectable but at lower numbers in both corpus and antral glands (fewer bacteria per gland as well as fewer glands colonized) (Fig. 3C and Fig. S3A). Like the wild type, csd6 complemented bacteria were observed predominantly in the antrum (Fig. 3D and Fig. S3B) and the C/A junction. We extended this analysis to additional animals from each genotype (Fig. 3E). In all animals infected with wild-type and csd6 complemented strains, as well as two out of three animals infected with the Δcsd6 mutant, the bacteria preferentially localized to the antrum instead of the corpus. However, the levels of bacteria detected in the glands did not correlate well with CFU loads. While there was a trend toward lower levels of gland localization for the Δcsd6 mutant than for the wild-type and csd6 complemented bacteria, the Δcsd6 mutant was able to penetrate and multiply within both corpus and antral glands in at least a subset of animals, despite exhibiting a significantly lower CFU load at this time point.
FIG 3.
Δcsd6 straight-rod mutant is attenuated in colonizing the corpus and antrum at 1 week postinfection. Thick stomach sections from the 1-week infections shown in Fig. 2A were stained for H. pylori, and the number of bacteria within the glands was quantified along the entire length of the section. (A) Representative images of the antrum of a mouse infected with wild-type H. pylori for 1 week. Shown are maximum intensity projections of Z stacks, with blue (DAPI, left) staining nuclei and yellow staining H. pylori. Scale bar, 100 μm. (B to D) Gland analysis for wild-type H. pylori (B, same mouse as that used for panel A), Δcsd6 mutant (C), and csd6 complemented strain (D), showing the number of bacteria detected by immunofluorescence within glands along the length of the stomach in microns. Red bars indicate the corpus, and blue bars indicate the antrum. Data presented in panels B to D are from one mouse per group and are representative of two independent experiments. (E) The total number of bacteria in the corpus and antral glands is shown for n = 2 to 3 mice per strain and is representative of two independent experiments, with the CFU per gram of stomach for each mouse indicated on the graph. WT, wild type; comp, complemented; wpi, week postinfection.
Both helical H. pylori and straight rods show expansion into the corpus after 1 month.
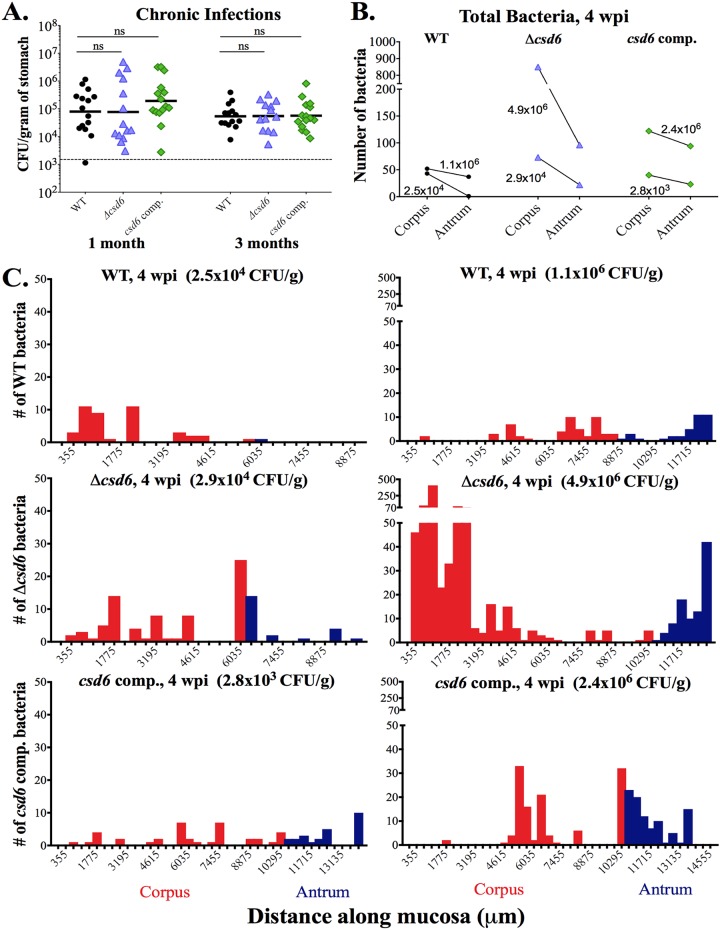
At 1 month postinfection, we observed a wide distribution of stomach loads from all strains tested, but in each case the geometric mean was around 105 CFU/g (Fig. 4A). A subset of mice showed stomach loads of 106 CFU/g or more, while other mice showed low loads near or below the limit of detection (103 CFU/g). At 3 months, stomach loads from each infected group were more tightly clustered, although the geometric mean was still around 105 CFU/g (Fig. 4A). Thus, Csd6-mediated helical cell shape is necessary for robust stomach colonization during the acute stages of gastric infection in mice but not for maintenance of chronic infection.
FIG 4.
Both wild-type H. pylori and the Δcsd6 straight-rod mutant can expand into the corpus by 1 month postinfection. Single infections were performed with the wild-type strain, Δcsd6 mutant, csd6 complemented strain, or broth (mock infection control). (A) Stomach loads at 1 month and 3 months postinfection. ns, not significant by Kruskal-Wallis test with Dunn’s multiple-test correction. Data are from two independent experiments with n = 15 mice per group; the limit of detection is shown with a dotted line. (B and C) Thick stomach sections from the 1-month infections shown in panel A were stained for H. pylori, and the number of bacteria within the glands was quantified along the entire length of the section. For each bacterial strain, a mouse with a low CFU load (left) and a high CFU load (right) was analyzed. (B) The total number of bacteria in the corpus and antrum is shown for n = 2 mice per bacterial strain and is from one experiment, with the CFU per gram of stomach for each mouse indicated on the graph. (C) Gland analysis for wild-type H. pylori, Δcsd6 mutant, and csd6 complemented strains, showing the number of bacteria within corpus and antral glands along the length of the stomach in microns. Two mice were analyzed per group from one experiment. Red bars indicate the corpus and blue indicates the antrum. WT, wild type; comp, complemented; wpi, week postinfection.
Given the variable stomach CFU at 1 month (Fig. 4A), we determined the localization of bacteria in gastric tissue samples with either low (∼104 CFU/g of stomach) or high (∼106 CFU/g of stomach) bacterial loads of the three strains (Fig. 4B). We observed two key differences from the 1-week analysis (compare to Fig. 3E): first, the total number of bacteria quantified at 1 month was generally lower than that at 1 week, and second, at 1 month there were more bacteria in the corpus than the antrum, the reverse of what was observed at 1 week. For mice with a low CFU load (Fig. 4C, left), the numbers of bacteria detected in corpus glands were similar among bacterial genotypes (fewer than 75 total bacteria). However, both the Δcsd6 mutant and csd6 complemented strains had more bacteria in the antrum (22 and 23 total bacteria, respectively) than the wild-type strain did (one bacterium). For mice with a high CFU load (Fig. 4C, right), the Δcsd6 mutant was different from the other two strains, with more bacteria detected in the corpus (849) than the antrum (96). The wild-type and csd6 complemented strains had more similar numbers of bacteria in the corpus (52 and 122, respectively) and antrum (37 and 94, respectively). While we did detect more total bacteria in the high-CFU mice than their low-CFU counterparts, CFU load did not correlate well with the number of bacteria detected in the glands.
Chronic infection with the straight-rod mutant results in reduced inflammation.
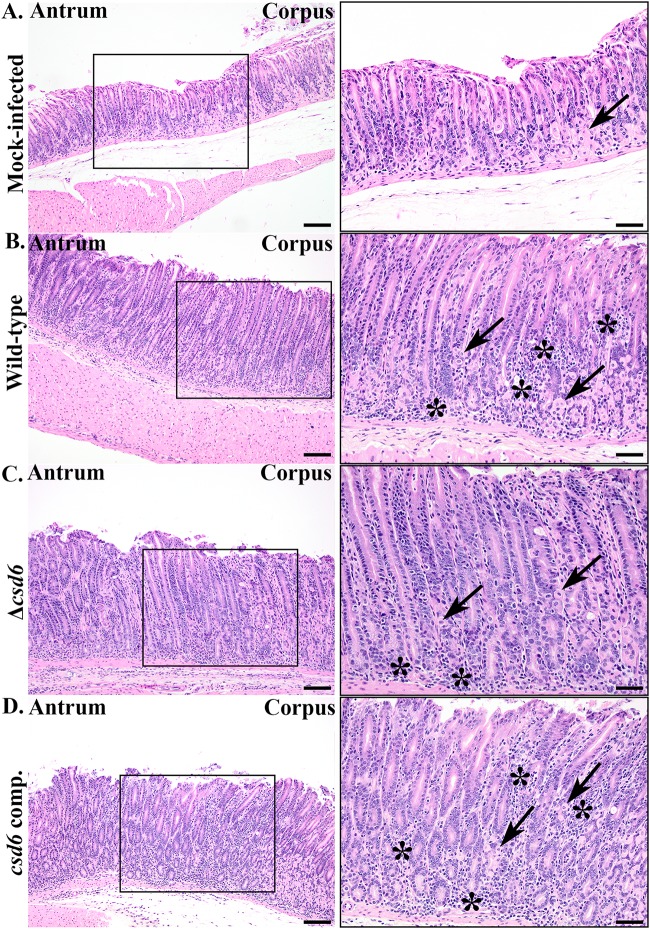
We next assessed pathological responses in mice infected with the different genotypes. The most severe lesions in each animal were scored according to previously developed pathology scoring criteria for inflammation, epithelial defects, oxyntic atrophy, hyperplasia, and metaplasia in the corpus and inflammation and hyperplasia in the antrum (see Table S2) (34). Individual scores (0 to 4) for each criterion were summed to generate a histological activity index (HAI) score. At 1 and 3 months postinfection, animals showed evidence of both inflammation and hyperplasia throughout the stomach. As shown in Fig. 5, which shows representative images of animals with the median HAI score for each genotype, pathological changes were most pronounced at the corpus/antrum (C/A) junction. Oxyntic atrophy (loss of parietal cells) and metaplasia were also observed within the distal corpus near the C/A junction (Fig. 5B).
FIG 5.
Δcsd6 straight-rod mutants elicit less immunopathology than wild-type and csd6 complemented bacteria. Images of hematoxylin- and eosin-stained sections from the 3-month infection, showing corpus, corpus/antral junction (box), and antral regions of the median histopathologic changes in each group. Panels on the right show higher-magnification images (20×) within the enclosed black boxes of 10× images on the left. Arrows point to remaining parietal cells in corpus glands, and the asterisks denote sites of infiltrating inflammatory cells. Images are from mock-infected (A), wild-type-infected (HAI = 13; 3.6 × 104 CFU/g stomach) (B), Δcsd6-infected (HAI = 8; 2.1 × 105 CFU/g stomach) (C), and csd6 complemented strain-infected (HAI = 14; 1.2 × 105 CFU/g of stomach) (D) mice. H&E images are representative of two independent experiments. WT, wild type; comp, complemented; HAI, histological activity index. Scale bar, 100 μm (left) and 50 μm (right).
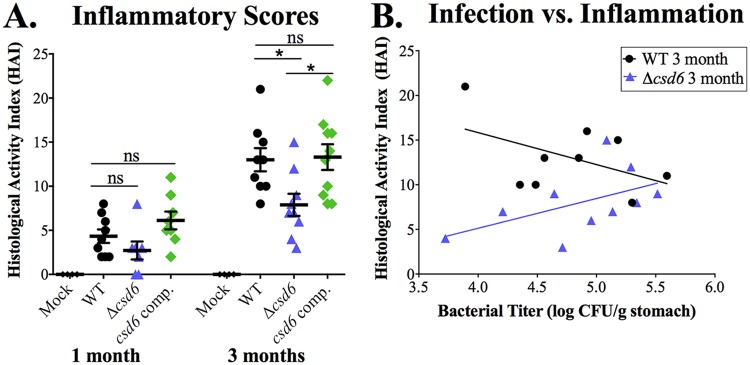
At 1 month postinfection, wild-type and csd6 complemented bacteria induced inflammation characterized by lymphocytic and neutrophilic infiltrates and low-level oxyntic atrophy at the antrum and C/A junction. In contrast, mice infected with the Δcsd6 mutant had little to no gastric inflammation and reduced hyperplasia (Fig. 6A and Fig. S4A and B), even though about half the mice had fairly high bacterial loads and bacteria were detected within gastric glands at this time point (Fig. 4A). The differences in pathology among bacterial genotypes became more pronounced at 3 months (Fig. 6A and Fig. S4C and D), by which time the Δcsd6 mutant-infected animals showed significantly reduced HAI scores and individual scores for inflammation, oxyntic atrophy, and hyperplasia in the C/A junction (Fig. S4C and D). We tested whether the Δcsd6 mutant had a general defect in inflammatory cytokine induction during coculture with gastric epithelial cells but found levels of IL-8 induction similar to those of the wild type, unlike the ΔcagE mutant, which cannot assemble the Cag type IV secretion system (35) (Fig. S5). Animals infected with wild-type bacteria showed evidence of an inverse relationship between stomach CFU load and HAI (Spearman correlation coefficient [r] of −0.22, two-tailed P value of 0.5722) (Fig. 6B), which is consistent with prior studies showing lower loads in animals with more severe gastritis (36). In contrast, animals infected with the Δcsd6 mutant showed the opposite trend (r = 0.518, two-tailed P value of 0.1286) (Fig. 6B). Thus, although bacterial loads are not statistically significantly different at 3 months postinfection (Fig. 4A), the Δcsd6 mutant elicits significantly less inflammation and hyperplasia than the wild-type or the csd6 complemented strain at this time point.
FIG 6.
Chronic Δcsd6 mutant infections show significantly less histological activity than wild-type and csd6 complemented infections. Thin stomach sections from the mice shown in Fig. 4A were used for an analysis conducted in a blind manner of stomach inflammation and pathology. (A) The total histological activity index (HAI) is provided for mock-infected and wild-type-, Δcsd6 mutant-, and csd6 complemented strain-infected mice at 1 and 3 months of infection. Nine to 11 mice were analyzed for pathology from each group, and data are representative of two independent experiments. Means ± standard deviations are shown. *, P < 0.05 by Kruskal-Wallis test with Dunn’s multiple-test correction. (B) Plot showing the correlation between wild-type and Δcsd6 stomach colonization loads and total HAI.
DISCUSSION
It has long been proposed that helical cell shape facilitates H. pylori’s ability to penetrate the thick gastric mucus layer by enhancing cell body propulsion, thereby promoting colonization of the stomach (37). Our study confirms and extends prior results that helical cell shape, while not required for stomach colonization, confers a significant advantage to H. pylori during acute infection (1 day and 1 week) (16, 17). Unlike our previously published work with csd1 and csd4 complemented mutants, the csd6 complemented strain did not restore colonization loads to wild-type levels at 1 week (although colonization levels were restored at 1 day). While shape appeared to be fully restored during growth in broth, we cannot rule out aberrant expression of Csd6 during infection or an unrelated genetic change during strain construction. Our previous studies revealed that cell shape is exquisitely sensitive to Csd6 expression levels (22). During chronic infection, the colonization levels of the wild-type strain and the Δcsd6 mutant were comparable, suggesting that the mutant can nonetheless chronically colonize glands of the stomach. Thus, straight rods may occupy a specific niche within the stomach, allowing them to persist long term. By performing 3D image analysis of H. pylori in thick stomach sections, we found that wild-type and csd6 complemented H. pylori had reduced localization within corpus and antral glands at 1 month postinfection relative to the number of bacteria observed in the glands at 1 week, which may be attributed to adaptive immune responses clearing the infection (36). While Δcsd6 mutant-infected tissues appeared to have somewhat lower levels of bacteria within the glands at 1 week, the contraction of the gland population at 1 month appeared less dramatic, particularly in the corpus.
Our 3D visualization and volumetric analysis provided better understanding of H. pylori’s localization early after infection and during chronic infection. However, our volumetric analysis has its limitations, as it only quantifies the total number of bacteria within gastric glands from each field of view analyzed (355-μm width by 355-μm height), and it only captures bacteria 40 to 50 μm deep into thick gastric tissue sections. In addition, our fixation conditions do not preserve the mucus layer overlying the epithelium, which may host a significant fraction of bacteria in both mutant and wild-type infections. For this reason, discrepancies exist between the bacterial loads quantified by our volumetric analysis and the CFU counts acquired per gram of stomach tissue sample, which includes both the gland and surface mucus bacterial populations. Future studies will characterize the fraction of H. pylori within the surface mucus layer, which may be essential to the establishment of chronic infection as bacteria may act as a reservoir of infection for the underlying gastric epithelium.
Finally, we found that the Δcsd6 mutant elicited less inflammation than wild-type or complemented bacteria at 1 and 3 months postinfection, despite having comparable bacterial CFU loads and similar or greater localization within the gastric glands. Thus, our study is the first to suggest a bacterial load-independent link between H. pylori’s helical cell shape and chronic gastritis.
During chronic infection with H. pylori, the degree of inflammation found at the corpus/antrum junction is often greater than that observed in the adjacent mucosa of either the corpus or antrum and may promote glandular atrophy, loss of parietal cells in the corpus, and eventually mucous cell gland hyperplasia (38). In the present study, we found highly variable colonization loads in mice infected with either the wild-type, Δcsd6 mutant, or csd6 complemented strain for 1 month, which may be related to differential host responses among mice. Chronic infection with wild-type PMSS1 or the csd6 complemented strain induced pathology in the antrum and the C/A junction. However, straight rods elicited less inflammation and gland hyperplasia. Chronic infection with H. pylori is controlled by innate and adaptive immune responses, regulated by CD4+ T-helper 1 (Th1) and/or Th17-polarized T-effector cell subsets, B cells, and their secreted cytokines (36, 39). Evaluating the adaptive immune response to infection with the Δcsd6 mutant and carrying out the infection to even longer time periods will determine if its differential localization in corpus glands at 1 month influenced its ability to induce inflammation or cause gastric disease. In addition, some H. pylori strains, including PMSS1 used in this study, express VacA, a virulence factor that has been shown to suppress T-cell responses to mediate longevity of infection (40–42). It will also be important to determine whether H. pylori cell shape mutants have unexpected effects on the expression of other factors that could mediate or suppress inflammation, such as VacA, to maintain a favorable niche in the stomach.
Our work demonstrates that the helical cell shape of H. pylori is important for acute colonization of the stomach and enhances pathology during chronic infection. Helical cell shape is also important for a bacterial pathogen that colonizes the human intestinal tract, Campylobacter jejuni. Isogenic straight-rod mutants of C. jejuni pgp1 and pgp2 (homologs of H. pylori csd4 and csd6, respectively) show attenuated motility in soft agar (43, 44) similar to that of H. pylori. However, C. jejuni pgp1 and pgp2 mutants completely fail to colonize intestinal crypts or to induce inflammatory responses in the mouse model tested (45).
In summary, our study provides insight into how helical cell shape impacts H. pylori’s niche acquisition and inflammation in the stomach. Loss of helical cell shape alters H. pylori’s ability to utilize some of the available niches within the stomach and its ability to promote inflammation and tissue hyperplasia when present in the glands. As morphological diversity exists between different clinical isolates of H. pylori in cell body and helical cell parameters (23), H. pylori clinical isolates with decreased helical pitch and twist may differ in their ability to colonize certain gastric niches and in the trajectories of pathogenesis. Thus, diversity in cell shape parameters may contribute to the diversity of pathogenic outcomes observed in infected individuals.
MATERIALS AND METHODS
H. pylori strains and growth conditions.
Strains used in this study are described in Table 1. Briefly, wild-type H. pylori strain PMSS1, also called 10700 (31, 46), and derivatives were cultured on horse blood plates or in liquid medium containing 90% (vol/vol) Brucella broth (BD Biosciences) and 10% fetal bovine serum (FBS; GIBCO) (BB10) in the absence of antimicrobials, as previously described (16). Cells were maintained at 37°C under microaerobic conditions in a tri-gas incubator equilibrated to 10% CO2 and 10% O2. Plates were incubated for 24 to 72 h, and liquid cultures were incubated for 12 to 16 h under constant agitation at 200 rpm. For resistance marker selection, horse blood plates were supplemented with chloramphenicol (15 μg/ml) or kanamycin (25 μg/ml).
Strain construction.
An isogenic mutant of csd6 (HPG27_477) in the PMSS1 strain background was generated by transfer of the mutation constructed in the G27/LSH100 strain background (16, 22) using natural transformation (47). LSH100 is a mouse-adapted derivative of the wild-type H. pylori G27 clinical strain (48–50). An isogenic mutant of cagE (HPG27_503) in the PMSS1 strain background was likewise generated by transfer of the mutation constructed in the G27 strain background using natural transformation. Transformants were confirmed by PCR using primers homologous to upstream and downstream flanking regions of the gene using the primers listed in Table S1 in the supplemental material. The mutations were then backcrossed into PMSS1 once by isolating genomic DNA from the resulting strain for natural transformation of PMSS1 to minimize any contribution of strain G27 polymorphisms beyond the targeted locus. The resulting backcrossed clones were evaluated by PCR to confirm replacement of the wild-type allele with the null allele. Clones were checked for urease activity and motility, and single clones were used for quantitative morphology analysis and for oral gavage of mice (in the case of the csd6 mutant) or coculture experiments (in the case of the cagE mutant).
The csd6 complemented strain was constructed by natural transformation of the PMSS1 Δcsd6 strain with genomic DNA from a csd6 complemented strain generated in the H. pylori G27 strain background, TSH31 (Δcsd6::cat McGee:csd6:aphA3), where a wild-type copy of csd6 (HPG27_477) was introduced at a neutral intergenic chromosomal site (between strain 26695 genes HP0203 and HP0204) (33, 55). In the H. pylori G27 strain, this region (204322 to 204784) is located between genes HPG27_186 (204127 to 204321; 195 bp in length) and HPG27_187 (204785 to 205168; 384 bp in length) (51). The TSH31 strain carries a kanamycin cassette gene (aphA3) (22). Genomic DNA from TSH31 was used for natural transformation of the PMSS1 Δcsd6 strain. Transformants were selected on horse blood plates supplemented with kanamycin (25 μg/ml). The recipient strain (EPH1) was PCR confirmed using primers homologous to upstream and downstream flanking regions of HPG27_186 and HPG27_187 using the primers listed in Table S1. The csd6 complemented strain was then backcrossed once by isolating genomic DNA from EPH1 for natural transformation of the PMSS1 Δcsd6 strain. The resulting backcrossed clones were evaluated by PCR to confirm integration of csd6 and were renamed LMH12 clones 1 to 3. The clones were then checked for urease activity and motility and were used for quantitative morphology analysis. Figure S1 shows morphology analysis of LMH12 clone 3 (csd6 complemented strain), which was the strain used for single-strain infections in mice and for bacterial localization studies summarized in the manuscript.
Morphology analysis.
Wild-type H. pylori PMSS1, Δcsd6 mutant, and csd6 complemented bacteria were grown in liquid culture to an optical density at 600 nm (OD600) of 0.4 to 0.6. Bacteria were fixed in 4% paraformaldehyde with 25% glycerol in 1× phosphate-buffered saline (PBS) and added to 0.1% poly-l-lysine (Sigma-Aldrich)-coated coverslips, which were then placed on a precleaned microscope slide and sealed with VaLP (1:1:1 vaseline-lanolin-paraffin). Single focal plane images were collected using a 100× ELWD Plan APO (numeric aperture, 1.40) oil lens objective mounted on a Nikon TE 200 microscope, equipped with a Nikon CoolSNAP HQ charge-coupled device camera controlled by MetaMorph software (MDS Analytical Technologies). Quantitative morphology analysis of manually thresholded phase-contrast images was performed as described by Sycuro et al. using the CellTool software program (16, 52).
Coculture experiments for type IV secretion system activity.
Cocultures were performed as previously described (53). Briefly, AGS cells (from a human gastric adenocarcinoma cell line; ATCC CRL-1739) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 10% heat-inactivated FBS (Gemini-Benchmark). Sixteen hours before infection, 1 × 105 AGS cells were seeded per well of a 24-well plate. On the day of infection, the cell culture medium was removed and replaced with H. pylori cells from mid-log phase (multiplicity of infection of 10:1) in 80% DMEM-FBS–20% BB10. Supernatants from three wells per strain were collected after 6 or 24 h of coculture, and IL-8 was detected using a human IL-8 enzyme-linked immunosorbent assay (ELISA) kit from BioLegend.
Parameter optimization for H. pylori 3D image analysis.
Wild-type H. pylori PMSS1 bacteria were grown in liquid culture to an OD600 of 0.4 to 0.6. A volume of 1 ml of bacterial culture was harvested in a 1.5-ml microcentrifuge tube and centrifuged at 5,000 rpm for 5 min. The cell pellet was resuspended in 100 to 200 μl of 4% PFA in 100 mM phosphate buffer (pH 7.4) and fixed for at least 30 min at room temperature. Bacteria were then embedded in 4% agarose (ultrapure low-melting-point agarose; Invitrogen) prepared in 1× phosphate-buffered saline (pH 7.4) (Gibco). The agarose solution was first cooled down to ∼55 to 65°C and then aliquoted into 1.5-ml microcentrifuge tubes. Aliquots of fixed bacteria were immediately added and gently resuspended into the solution before it solidified. The solidified slabs were gently removed by insertion of a metal spatula on the side of the tube. The slabs were then sectioned using a vibratome (Leica VT 1200 S fully automated vibrating blade microtome; Leica Biosystems, Germany) to generate 100- to 200-μm-thick sections. Sections were permeabilized in blocking buffer (3% bovine serum albumin [Sigma-Aldrich], 1% Saponin [Sigma-Aldrich], and 1% Triton X-100 [Sigma-Aldrich] in 1× PBS) and immunostained with primary anti-H. pylori rabbit polyclonal antibody (1:1,000 dilution) (gifted by Manuel Amieva at Stanford University) overnight at 4°C. A goat anti-rabbit Alexa Fluor-488 conjugated antibody (1:2,000) (Molecular Probes) was used to visualize H. pylori. Samples were incubated in the secondary antibody for 2 h at room temperature. Sections were then washed 5× with blocking buffer and mounted onto standard glass microscope slides with secure imaging spacers (9-mm diameter by 0.12-mm depth; Electron Microscopy Sciences). ProLong diamond antifade medium was added (Molecular Probes) before mounting on coverslips.
Ethics statement.
All procedures involving animals were done under practices and procedures of animal biosafety level 2 and carried out with strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (54). The facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and complies with the United States Department of Agriculture, Public Health Service, Washington State, and local area animal welfare regulations. All activities were approved by the FHCRC Institutional Animal Care and Use Committee (IACUC; protocol number 1531). Animals were euthanized by CO2 asphyxiation followed by cervical dislocation.
Mouse infections.
Four- to 6-week-old female C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were certified free of endogenous Helicobacter infection by the vendor. All animals were maintained in autoclaved microisolator cages (1 to 5 mice per cage) and provided with standard chow and water ad libitum. Mice were infected with a single dose of 5 × 107 H. pylori cells/strain (0.1 ml) via oral gavage. Mock-infected controls were gavaged with 0.1 ml of liquid culture medium containing 90% (vol/vol) Brucella broth and 10% fetal bovine serum (BB10); H. pylori was not recovered from mock-infected mice. Mice were euthanized by inhalation of CO2, and stomachs were harvested at 1 day, 1 week, and 1 or 3 months postinfection. Most of the nonglandular region (forestomach) was discarded, since this region of the stomach is lined with squamous rather than glandular epithelium. H. pylori has not been shown to colonize this region of the stomach. However, H. pylori may colonize the interface between the squamous forestomach and glandular stomach, where the corpus begins (the squamocolumnar junction). Regions of interest for H. pylori colonization include the corpus, antrum, and pyloric junction with the duodenum. Therefore, part of the forestomach was maintained and the glandular stomach (corpus and antrum) was opened along the lesser curvature from the esophagus through the proximal duodenum. For 1-day and 1-week harvests, half the stomach was used for plating for CFU enumeration and the other half was fixed in 4% PFA in 100 mM phosphate buffer (pH 7.4) for 1 to 2 h. For chronic infection time points (1 and 3 months), the stomach was divided into thirds. A third of the stomach was collected to measure CFU/gram of stomach load, a third was fixed in 4% PFA for immunofluorescence, and a third was fixed in 10% neutral buffered formalin (NBF) solution (Thermo Fisher Scientific) for histology. In each case, food was carefully removed and the stomach was laid flat on an index card and placed in a cassette with a sponge at the top, closed, and fixed in its respective solution. For CFU counts, one-half or one-third of stomachs were manually homogenized using a pestle in 0.5 ml of BB10. Serial 10-fold dilutions of stomach homogenate were plated on solid horse blood agar plates containing 4% Columbia agar base (Oxoid, Hampshire, UK), 5% defibrinated horse blood (HemoStat Labs, Dixon, CA), 0.2% β-cyclodextrin (Sigma, St. Louis, MO), 10 μg/ml vancomycin (Sigma), 5 μg/ml cefsulodin (Sigma), 2.5 U/ml polymyxin B (Sigma), 5 μg/ml trimethoprim (Sigma), 8 μg/ml amphotericin B (Sigma), and bacitracin (200 μg/ml) to eliminate normal mouse microbiota growth. Plates were incubated at 37°C using a tri-gas incubator (10% CO2, 10% O2; Thermo Fisher Scientific) for 4 to 5 days.
Competition experiments.
Mice were infected with an approximately 1:1 ratio of 107 CFU of wild-type H. pylori and the Δcsd6 isogenic straight-rod mutant or the csd6 complemented strain. Two independent experiments were conducted using 5 mice per group for each competition experiment. After 1 week, stomachs were removed, divided in half, and plated to determine bacterial loads as CFU/gram of stomach. Wild-type bacterial output was plated on horse blood plates containing the antibiotics described above. The Δcsd6 mutant was selected on horse blood plates with chloramphenicol (15 μg/ml), and the csd6 complemented strain was selected on horse blood plates with kanamycin (25 μg/ml).
Immunofluorescence of thick longitudinal mouse stomach sections.
Tissues from mouse stomachs were processed for confocal immunofluorescence microscopy as described previously (8, 32), with minor modifications. Gastric tissue was fixed in 4% PFA for 1 to 2 h at room temperature. Tissue was embedded in 4% agarose in 1× PBS (pH 7.4) (Gibco) and sectioned using a vibratome to generate 100- to 200-μm-thick longitudinal sections that include the limiting ridge at the forestomach/glandular junction to the pyloric junction with the duodenum. Tissue sections were then permeabilized in blocking buffer (3% bovine serum albumin [Sigma-Aldrich], 1% saponin [Sigma-Aldrich], 1% Triton X-100 [Sigma-Aldrich]) in 1× PBS (pH 7.4) (Gibco). Anti-H. pylori rabbit polyclonal antibody (1:1,000 dilution) was used to immunostain H. pylori in the tissue overnight at 4°C. The sections were then washed 5× with blocking buffer and incubated with a goat anti-rabbit Alexa Fluor 647-conjugated antibody (1:2,000) to visualize bacteria in tissue (Molecular Probes) and 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml) to stain nuclei for 2 h at room temperature. The sections were then washed 5× with blocking buffer and mounted onto standard glass microscope slides with secure imaging spacers (20-mm diameter by 0.12-mm thickness; Electron Microscopy Sciences) or hand-made imaging spacers using Parafilm (0.12 mm thick). ProLong diamond antifade medium (Molecular Probes) was added before mounting on coverslips.
Confocal microscopy.
Tissue samples were imaged with a Zeiss LSM 780 NLO confocal and multiphoton microscope with a 40× oil immersion lens objective (1.30 numeric aperture; EC Plan-Neofluar), and Z-stacks (355 μm by 355 μm) were generated using the ZEN acquisition software program. Images were acquired at a frame size of 1,024 by 1,024 with 8-bit depth and at a frame rate speed of 8 frames per second. Z-stacks were generated with a slice interval of 0.5 μm and penetrated 40 to 50 μm into the section. For each tissue section, multiple Z-stacks (ranging from 25 to 30) were acquired to capture the full length of longitudinal sections that include the limiting ridge of the forestomach to the glandular junction to the pyloric junction with the duodenum. For all Z-stacks, a collection of nonoverlapping images was acquired by manual translation of the microscope stage.
Volumetric image analysis and quantitation of H. pylori in the stomach.
Quantitation of H. pylori within individual gastric glands was performed using the Volocity 3D image analysis software program, as described previously (8, 32), with minor modifications. 3D reconstructed images were imported onto Volocity, and the total volume for individual bacterial cells was determined. The mean volume for a bacterium (9.5 μm3) was used to calculate the total number of bacteria near or at the surface epithelium and within gastric glands. The same measurement protocol was applied across all tissue samples analyzed for wild-type H. pylori, the Δcsd6 mutant, and the csd6 complemented bacteria. Analysis of 3 sections (>500 μm apart) provided consistent results in bacterial number counts. Our bacterial localization studies included analysis of one tissue section per infected mouse (n = 2 to 3 mice per genotype group). Therefore, 2 to 3 different tissue sections were analyzed per genotype at 1 week postinfection, and 2 tissue sections from 2 different mice were examined for each genotype at 1 month postinfection.
Histologic evaluation of H. pylori-infected stomachs.
Longitudinal gastric strips from the lesser curvature that include the squamocolumnar junction through the proximal duodenum were fixed in 10% NBF. Samples were paraffin embedded, sectioned (5 μm thick), and stained with hematoxylin and eosin (H&E) by the Experimental Histopathology Core at the Fred Hutchinson Cancer Research Center. Slides were interpreted and scored using the scoring criteria adapted from reference 34, shown in Table S2, by a veterinary pathologist (S. E. Knoblaugh) who was blind to the experimental details. The individual lesion scores of every mouse in each group were evaluated and compared for inflammation, epithelial defects, oxyntic atrophy, hyperplasia, and metaplasia in the corpus and inflammation and hyperplasia in the antrum. Individual scores (0 to 4) for each criterion were summed to generate a histological activity index (HAI) score. Nine to 11 tissue sections from each group (mock infected or infected with the wild-type, Δcsd6 mutant, or csd6 complemented strain) were evaluated for pathology at 1 and 3 months postinfection.
Statistical analyses.
We used the Kolmogorov-Smirnov statistics tool in CellTool to assay the differences in cell shape morphology, including cell length and side curvature distributions, as described previously (16, 17). For CFU data and histopathology scores, comparisons of three groups were performed using a Kruskal-Wallis one-way analysis of variance followed by Dunn’s multiple-test corrections, and pairwise comparisons (competition experiments) were performed with the Mann-Whitney U test using GraphPad Prism 7 (GraphPad Software, La Jolla, CA).
A P value of <0.05 was considered statistically significant. For the HAI, because mucous metaplasia and hyalinosis may develop spontaneously in mice, these subscores were excluded from the calculation of the total HAI.
Supplementary Material
ACKNOWLEDGMENTS
We thank Manuel Amieva for the wild-type H. pylori PMSS1 strain and antibodies, Julie Huang for advice in preparing gastric tissue for 3D confocal microscopy, and Simran Handa (supported by a generous donation from the AT&T Foundation to the FHCRC Summer High School Internship Program) and Ericka Pegues for assistance in generating the csd6 complemented strain.
This work was supported by the U.S. National Institutes of Health grant R01 AI136946 to N.R.S. and the NIH Ruth L. Kirschstein National Research Service Award (NRSA) F31 AI098424 from the NIAID to L.E.M. This research was supported by the Comparative Medicine, Electron Microscopy, Experimental Histopathology and Scientific Imaging Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704).
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute and the National Institute of Allergy and Infectious Diseases.
L.E.M. and N.R.S. designed the study. L.E.M., V.P.O., and C.K.L. performed experiments. L.E.M., V.P.O., C.K.L., and N.R.S. performed data analysis and interpretation. S.E.K. evaluated samples for pathology. L.E.M., V.P.O., S.E.K., and N.R.S. wrote the manuscript.
We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00904-18.
REFERENCES
- 1.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaton KA, Brooks CL, Morgan DR, Krakowka S. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun 59:2470–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun 66:4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuda M, Karita M, Mizote T, Morshed MG, Okita K, Nakazawa T. 1994. Essential role of Helicobacter pylori urease in gastric colonization: definite proof using a urease-negative mutant constructed by gene replacement. Eur J Gastroenterol Hepatol 6(Suppl 1):S49–S52. [PubMed] [Google Scholar]
- 6.Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun 62:3586–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton KA, Suerbaum S, Josenhans C, Krakowka S. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun 64:2445–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. 2011. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11. doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolig AS, Shanks J, Carter JE, Ottemann KM. 2012. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect Immun 80:3713–3720. doi: 10.1128/IAI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amieva MR, Salama NR, Tompkins LS, Falkow S. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol 4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 13.Chu YT, Wang YH, Wu JJ, Lei HY. 2010. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect Immun 78:4157–4165. doi: 10.1128/IAI.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok T, Backert S, Schwarz H, Berger J, Meyer TF. 2002. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect Immun 70:2108–2120. doi: 10.1128/IAI.70.4.2108-2120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S, Tompkins LS, Amieva MR. 2009. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog 5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. 2012. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog 8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An DR, Kim HS, Kim J, Im HN, Yoon HJ, Yoon JY, Jang JY, Hesek D, Lee M, Mobashery S, Kim SJ, Lee BI, Suh SW. 2015. Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase. Acta Crystallogr D Biol Crystallogr 71:675–686. doi: 10.1107/S1399004715000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonis M, Ecobichon C, Guadagnini S, Prevost MC, Boneca IG. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, Im HN, An DR, Yoon JY, Jang JY, Mobashery S, Hesek D, Lee M, Yoo J, Cui M, Choi S, Kim C, Lee NK, Kim SJ, Kim JY, Bang G, Han BW, Lee BI, Yoon HJ, Suh SW. 2015. The cell shape-determining Csd6 protein from Helicobacter pylori constitutes a new family of L,D-carboxypeptidase. J Biol Chem 290:25103–25117. doi: 10.1074/jbc.M115.658781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HS, Kim J, Im HN, An DR, Lee M, Hesek D, Mobashery S, Kim JY, Cho K, Yoon HJ, Han BW, Lee BI, Suh SW. 2014. Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape. Acta Crystallogr D Biol Crystallogr 70:2800–2812. doi: 10.1107/S1399004714018732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sycuro LK, Rule CS, Petersen TW, Wyckoff TJ, Sessler T, Nagarkar DB, Khalid F, Pincus Z, Biboy J, Vollmer W, Salama NR. 2013. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol 90:869–883. doi: 10.1111/mmi.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez LE, Hardcastle JM, Wang J, Pincus Z, Tsang J, Hoover TR, Bansil R, Salama NR. 2016. Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments. Mol Microbiol 99:88–110. doi: 10.1111/mmi.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atherton JC. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol 1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 25.Wroblewski LE, Peek RM Jr, 2013. Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterol Clin North Am 42:285–298. doi: 10.1016/j.gtc.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaser MJ, Atherton JC. 2004. Helicobacter pylori persistence: biology and disease. J Clin Investig 113:321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 28.Akada JK, Ogura K, Dailidiene D, Dailide G, Cheverud JM, Berg DE. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149:1901–1909. doi: 10.1099/mic.0.26129-0. [DOI] [PubMed] [Google Scholar]
- 29.Ferrero RL, Thiberge JM, Huerre M, Labigne A. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun 66:1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakagami T, Dixon M, O'Rourke J, Howlett R, Alderuccio F, Vella J, Shimoyama T, Lee A. 1996. Atrophic gastric changes in both Helicobacter felis and Helicobacter pylori infected mice are host dependent and separate from antral gastritis. Gut 39:639–648. doi: 10.1136/gut.39.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, Muller A. 2011. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, Nusse R, Torres J, Amieva MR. 2015. Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology 148:1392–1404. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 33.Langford ML, Zabaleta J, Ochoa AC, Testerman TL, McGee DJ. 2006. In vitro and in vivo complementation of the Helicobacter pylori arginase mutant using an intergenic chromosomal site. Helicobacter 11:477–493. doi: 10.1111/j.1523-5378.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers AB. 2012. Histologic scoring of gastritis and gastric cancer in mouse models. Methods Mol Biol 921:189–203. doi: 10.1007/978-1-62703-005-2_22. [DOI] [PubMed] [Google Scholar]
- 35.Algood HM, Cover TL. 2006. Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses. Clin Microbiol Rev 19:597–613. doi: 10.1128/CMR.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayi A, Kohler E, Hitzler I, Arnold I, Schwendener R, Rehrauer H, Muller A. 2009. The CD4+ T cell-mediated IFN-gamma response to Helicobacter infection is essential for clearance and determines gastric cancer risk. J Immunol 182:7085–7101. doi: 10.4049/jimmunol.0803293. [DOI] [PubMed] [Google Scholar]
- 37.Montecucco C, Rappuoli R. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol 2:457–466. doi: 10.1038/35073084. [DOI] [PubMed] [Google Scholar]
- 38.Goldenring JR, Nomura S. 2006. Differentiation of the gastric mucosa III. Animal models of oxyntic atrophy and metaplasia. Am J Physiol Gastrointest Liver Physiol 291:G999–G1004. doi: 10.1152/ajpgi.00187.2006. [DOI] [PubMed] [Google Scholar]
- 39.Velin D, Favre L, Bernasconi E, Bachmann D, Pythoud C, Saiji E, Bouzourene H, Michetti P. 2009. Interleukin-17 is a critical mediator of vaccine-induced reduction of Helicobacter infection in the mouse model. Gastroenterology 136:2237–2246. doi: 10.1053/j.gastro.2009.02.077. [DOI] [PubMed] [Google Scholar]
- 40.Boncristiano M, Paccani SR, Barone S, Ulivieri C, Patrussi L, Ilver D, Amedei A, D'Elios MM, Telford JL, Baldari CT. 2003. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J Exp Med 198:1887–1897. doi: 10.1084/jem.20030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 42.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. 2004. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci U S A 101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. 2012. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog 8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frirdich E, Vermeulen J, Biboy J, Soares F, Taveirne ME, Johnson JG, DiRita VJ, Girardin SE, Vollmer W, Gaynor EC. 2014. Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J Biol Chem 289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl M, Frirdich E, Vermeulen J, Badayeva Y, Li X, Vallance BA, Gaynor EC. 2016. The helical shape of Campylobacter jejuni promotes in vivo pathogenesis by aiding transit through intestinal mucus and colonization of crypts. Infect Immun 84:3399–3407. doi: 10.1128/IAI.00751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397. doi: 10.1016/S0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Roos KP, Taylor DE. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol 139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 48.Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR. 2007. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun 75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. 2013. Natural competence promotes Helicobacter pylori chronic infection. Infect Immun 81:209–215. doi: 10.1128/IAI.01042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191:7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, Mogilner A, Theriot JA. 2007. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol 5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gall A, Gaudet RG, Gray-Owen SD, Salama NR. 2017. TIFA signaling in gastric epithelial cells initiates the cag type 4 secretion system-dependent innate immune response to Helicobacter pylori infection. mBio 8:e01168-17. doi: 10.1128/mBio.01168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 55.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.