Abstract
Transcription, catalyzed by RNA polymerase II (Pol II) in eukaryotes, is the first step in gene expression. RNA Pol II is a 12-subunit enzyme complex regulated by many different transcription factors during transcription initiation, elongation, and termination. During elongation, Pol II encounters various types of obstacles that can cause transcriptional pausing and arrest. Through decades of research on transcriptional pausing, it is widely known that Pol II can distinguish between different types of obstacles by its active site. A major class of obstacles is DNA lesions. While some DNA lesions can cause transient transcriptional pausing, which can be bypassed by Pol II itself or with the help from other elongation factors, bulky DNA damage can cause prolonged transcriptional pausing and arrest, which signals for transcription coupled repair. Using biochemical and structural biology approaches, the outcomes of many different types of DNA lesions, DNA modifications, and DNA binding molecules to transcription were studied. In this mini review, we will describe the in vitro transcription assays with Pol II to investigate the impacts of various DNA lesions on transcriptional outcomes and the crystallization method of lesion-arrested Pol II complex. These methods can provide a general platform for the structural and biochemical analysis of Pol II transcriptional pausing and bypass mechanisms.
Keywords: RNA polymerase II, X-ray crystallography, DNA lesion, transcription
1. Introduction
RNA polymerase II (Pol II) is a key enzyme complex in eukaryotic gene expression that is responsible for synthesizing the precursors of mRNA, snRNA and microRNA [1, 2]. During transcription elongation, Pol II elongation complex translocates along the template DNA and frequently encounters numerous kinds of modifications and lesions that occurs on a daily basis [3]. While some DNA lesions can cause transient transcriptional pausing, bulky DNA damage can cause prolonged transcriptional pausing and arrest, which signals for transcription coupled repair. Defects of transcription coupled repair and prolonged transcription arrest can attribute a variety of diseases and aging [4].
Ever since the determination of the first structures of a 10-subunit Pol II core [5, 6] and a complete 12-subunit Pol II with subcomplex Rpb4/7 [7, 8], dozens of Pol II structures in complex with a wide range of transcription factors or DNA binding molecules, lesions, and modifications have been reported [9–30]. With these results, now we have a better understanding of how Pol II reads out template DNA strand and incorporates the correct nucleotide substrate during transcriptional elongation, and how Pol II recognizes and deals with different types of DNA modifications. While small DNA lesions can lead to transient pausing and error-prone transcriptional bypass, bulky DNA lesions can induce persistent transcriptional stalling and provide a strong signal for the recruitment of repair factors to initiate transcription coupled repair (TCR) or the degradation of stalled Pol II by ubiquitylation [31–35].
Combining structural and biochemical approaches in the past decade, we have gained important insights into understanding the mechanism of normal transcription process as well as transcriptional pausing/arrest and bypass. For example, the bridge helix, a conserved motif featuring a long α-helix, dictates the positioning of template strand DNA and separates the upstream DNA-RNA hybrid from the downstream DNA duplex. It is suggested that the bridge helix plays important roles in Pol II forward and backtracking translocation [19, 36, 37]. During forward translocation, the downstream template nucleobase needs to cross over the bridge helix to reach +1 position. During backtracking, the template nucleobase at +1 position also needs to cross over the bridge helix in the opposite direction to reach the downstream channel. We found that this crossing over of the bridge helix serves as an important check point for DNA lesion detection. Indeed, several DNA-lesion arrested Pol II complexes are trapped in the half-translocation intermediate state in which the DNA lesions are located above the bridge helix [10, 20, 26, 38].
In this mini-review, we will describe the methods of how in vitro transcription assays are carried out to investigate the effect of various DNA modifications on Pol II elongation. We will also describe the methods for the structural determination of Pol II with DNA modification or lesions. This mini-review is aimed to provide an overview for those who are interested in understanding the impact of DNA modifications or DNA lesions on Pol II transcription.
2. Biochemical analysis of the impacts of DNA lesions on RNA Pol II transcription pausing/arrest, bypass and transcriptional fidelity
2.1. Purification of RNA polymerase II
Yeast RNA polymerase II is purified as previously reported [9, 15, 24]. Briefly, cells are first resuspended in lysis buffer and then lysed by French press and followed by centrifugation. The cleared lysate is transferred to a new container and Pol II is precipitated by the addition of 50% saturated ammonium sulfate solution. The pellet is then dissolved in Ni buffer and purified by Ni-NTA affinity chromatography. After washing with high-salt buffer (Ni buffer with 1000 mM KCl and 7 mM imidazole) and Ni wash buffer, the protein is eluted with Ni wash buffer containing 100 mM imidazole and no protease inhibitors. The eluted protein is diluted with MonoQ buffer and then purified by anion exchange chromatography (MonoQ, GE healthcare) using a gradient ranging from 150 mM to 1500 mM KOAc. The last elution peak is collected and concentrated. For the strains containing recombinant protein A tag, Pol II is first purified using IgG sepharose resin (GE Healthcare)[16, 39, 40]. After cleavage of protein A tag by TEV protease, Pol II is further purified by Hi-Trap Heparin and Mono Q columns (GE Healthcare). For biochemical analysis, purified protein is stored in aliquots of elongation buffer and stored at −80 °C or liquid nitrogen. For crystallization study, Pol II is precipitated by adding saturated ammonium sulfate to 55% concentration at 4 °C. After centrifugation, the majority of supernatant is discarded, leaving a small remaining amount to protect the protein from air. The protein is then flash frozen and stored in −80 °C or liquid nitrogen.
In addition to direct purification [21, 27, 39, 40], 12-subunit RNA Pol II can also be reconstituted by mixing 10-subunit core RNA Pol II (strain name: CBC010 Δ4 Protein A-tagged) and Rpb 4/7 subcomplex [7, 8]. Rpb 4/7 is expressed in Rosetta 2 (Novagen) and purified using Ni-NTA affinity chromatography (Qiagen). Ten-subunit core RNA Pol II and Rpb 4/7 are mixed at a molar ratio of 1:4 in elongation buffer and purified by size exclusion chromatography to remove excess Rpb 4/7. Buffers used in this method are summarized in Table 1.
Table 1.
Buffers and reagents used in this study.
| Name | Components |
|---|---|
| Lysis buffer | 50 mM Tris-HCl [pH 7.9], 1 mM EDTA, 10 μM ZnCl2, 10% v/v glycerol, 1% v/v DMSO, 10 mM DTT and 1 × protease inhibitors |
| Ni buffer | 20 mM Tris-HCl [pH 7.9], 150 mM KCl, 10 μM ZnCl2, 10% v/v glycerol, 10 mM DTT and 1 × protease inhibitors |
| Ni wash buffer | 20 mM Tris-HCl [pH 7.9], 150 mM KC1, 7 mM imidazole, 10 μM ZnCl2, 10 mM DTT and 1 × protease inhibitors |
| MonoQ buffer | 20 mM Tris-acetate [pH 7.9], 0.5 mM EDTA, 10 μM ZnCl2, 10% v/v glycerol and 10 mM DTT |
| Elongation buffer | 20 mM Tris-HCl [pH 8.0], 40 mM KC1, 5 mM DTT and 5 mM MgCl2 |
| Quench-loading buffer | 90% (v/v) formamide, 50 mM EDTA, 0.05% (w/v) xylene cyanol and 0.05% (w/v) bromophenol blue |
| 10X TBE buffer | 1 M Tris base, 1 M Boric acid and 20 mM EDTA |
| TBE-Urea/PAGE | 1 × TBE, 7 M urea and 8–16 % (v/v) bis-aerylamide 19:1 |
| Exchange buffer | 25 mM Tris [pH 7.5], 20 mM NaCl, 5 mM DTT, 1 μM Zn(OAc)2 and 100 μM EDTA |
| Crystallization buffer | 390 mM Ammonium phosphate [pH 6.0], 5 mM DTT, 5 mM Dioxane and 9–13 % (w/v) PEG 6,000 |
| Cryo buffer | 100 mM MES [pH 6.0], 350 mM NaCl, 5mM DTT, 5mM Dioxane, 16 % PEG 6,000 and 17 % PEG400 |
2.2. Purification of site-specific DNA lesion-containing DNA template
There are two conventional methods for obtaining site-specific DNA lesion-containing DNA template. If lesion-modified nucleoside phosphoramidites are available, the DNA-lesion containing oligonucleotides can be synthesized using a solid-phase DNA synthesizer via phosphoramidite chemistry [20, 26]. Otherwise, DNA lesion can be formed in a post-synthesis manner [11, 16]. The short oligonucleotides are directly reacted with chemical agents. Extensive purification steps are often required to separate DNA-lesion containing oligonucleotides from undamaged oligonucleotides using HPLC (ion exchange and reverse phase) and PAGE purification [11, 16]. The identities and purities of these lesion-containing oligonucleotides can be confirmed by electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) analyses. For the long DNA-lesion containing DNA template (for full-bubble scaffold), additional preparation steps are required starting from a short DNA-lesion containing oligos via DNA ligation or primer extension [20, 26].
2.3. Preparation of Transcription Scaffold
The promoter-dependent transcription initiation is very inefficient in vitro and requires general transcription factors. To overcome this technical barrier, a synthetic transcription RNA:DNA scaffold is used to assemble Pol II elongation complex. This system allows us to achieve 100% transcription elongation efficiency in vitro and quantitatively measure the kinetic impact on transcription by site-specific DNA lesions at base resolution.
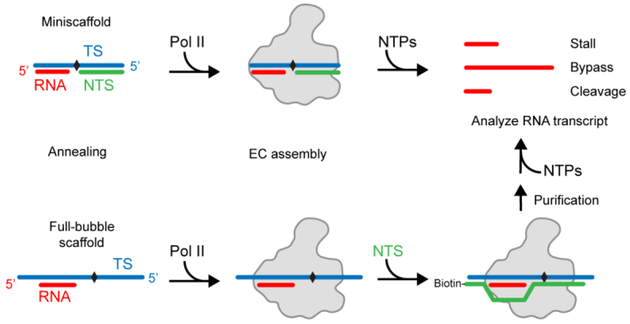
The scaffold can be further divided into two classes: mini-scaffold and full-bubble scaffold (Fig. 1). The mini-scaffold represents a part of the transcription bubble that harbors 8–9 bases of the DNA-RNA hybrid upstream and 18–19 bases of the double-stranded DNA downstream, whereas the full-bubble scaffold contains an intact transcription bubble (matched or mismatched) with a preformed short DNA-RNA hybrid (see Table S1 for examples). Both systems are widely used for biochemical and structural studies. Here, we will describe the assembly of both systems.
Figure 1. Schematic representation of investigating the effect of DNA lesions by transcription assay.
To investigate the effect of DNA lesion or modification (black diamond) on transcription, mini-scaffold or full-bubble scaffold is used. Mini-scaffold consists of short template strand, non-template strand and RNA. This miniature transcription bubble is a useful tool in understanding how elongation complex handles obstacles during transcription. Full-bubble scaffold has longer template strand DNA and non-template strand DNA. Additional steps are required for the preparation of elongation complex (EC).
To prepare the mini-scaffold, a ratio of 1:3:3 of P32-labeled RNA, DNA-lesion containing DNA template strand, and non-template strand DNA are mixed in the elongation buffer. 5’-P32 labeled RNA oligo is generated using T4 PNK (NEB). Final concentration of the labeled RNA is 200 nM. The reaction mixture is annealed by incubating at 95 °C in a thermocycler or heat block for 2 min and slowly cooling down to room temperature (23 °C). (Note: If the DNA lesion is heat sensitive, lower temperature and shorter incubation time are needed for denaturation and annealing. For DNA lesions that are light-sensitive, the tubes are wrapped in aluminum foil).
The full-bubble scaffold requires a two-step assembly. In the first step, 1:1 molar ratio of labeled RNA and DNA-lesion containing DNA template strand are mixed and annealed as described, with a final concentration of 200 nM each. The non-template DNA strand is added after the incubation with Pol II (see section 2.4 step a).
2.4. In vitro transcription assay
For the transcription assay with mini-scaffold, a minimum of 2- to 5-fold excess amount of Pol II is incubated with pre-assembled mini-scaffold in elongation buffer (EB, See Table 1) at room temperature for at least 10 min to ensure the formation of a stable Pol II elongation complex. For the transcription assay with full-bubble scaffold, we use a slightly modified annealing method (Fig. 1). First, 1:1 molar ratio of P32-labeled RNA and template strand DNA are annealed. This DNA-RNA hybrid is mixed with 2.5 molar excess of 12-subunit Pol II and incubated for 10 min at room temperature, followed by 2 min at 37 °C. Subsequently, after the addition of 5’-biotinylated non-template strand DNA, the reaction mixture is incubated for 5 min at 37 °C and then for 20 min at room temperature. Final concentrations are 100 nM of RNA and template strand DNA, and 250 nM of Pol II and non-template strand DNA. To remove unbound RNA and Pol II, the elongation complex is incubated with streptavidin magnetic beads (NEB) for 30 min at room temperature and washed successively with EB, EB with 0.3 M NaCl, EB with 1 M NaCl, EB with 0.3 M NaCl, and EB. The assembled Pol II elongation complex is ready for transcription assay.
To start the transcription reaction, an equal volume of Pol II elongation complex in (a) is mixed with various concentration of rNTPs or single nucleotide. Final reaction condition is 100 nM of Pol II, 20 nM of scaffold, and varying concentration of NTPs in elongation buffer. For the RNA cleavage assay, TFIIS is incubated with/without NTPs and mixed with elongation complex when the reaction starts, because pre-incubation of TFIIS and elongation complex without NTPs can lead to the degradation of labeled RNA, especially for the damaged scaffold. For reaction with Rad26, 3 mM of dATP is also included with NTPs to facilitate the ATPase activity (Fig. 2B).
For each designated time point, four volumes of the quench-loading buffer (see Table 1) is added to the reaction mixture to stop the reaction. Time point zero can be prepared by adding quench-loading buffer to the elongation complex without NTPs. The KinTek RQF-3 Quench-Flow instrument is needed for rapid time points in pre-steady state kinetic study. After quenching, the samples are incubated at 95 °C for at least 5 min to denature the protein complex and scaffold. After denaturation, the RNA transcripts samples are ready for denaturing urea/PAGE gel analysis.
The quenched products (RNA transcripts) are analyzed by denaturing urea/PAGE gel in 1× TBE at 200 V for 2.5–3.5 h. 8–16% of acrylamide can be used depending on the predicted size of RNA product. RNA transcripts are visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad).
For intrinsic transcript cleavage assays, the Pol II complex is assembled as described above in a 20 mM Tris-HCl (pH 7.5) buffer without Mg2+. Intrinsic cleavage is initiated after the addition of Mg2+. Final concentrations for intrinsic cleavage are 20 mM Tris-HCl (pH 9.0), 100 nM Pol II, 25 nM scaffold, and 50 mM MgCl2. The reaction is stopped by adding four volumes of quench loading buffer at various time points and analyzed by denaturing PAGE. For TFIIS-stimulated transcript cleavage assays, Pol II complex is assembled as aforementioned in a 20 mM Tris-HCl (pH 7.5) buffer without Mg2+. The solution is then mixed with an equal volume of solution containing TFIIS and MgCl2 in elongation buffer. The final reaction solution contains 100 nM Pol II, 25 nM scaffold, 1 μM TFIIS, and 5 mM MgCl2. The reactions are quenched at various time points by the addition of an equal volume of quench loading buffer. The quenched products are again analyzed by denaturing urea/PAGE and visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad).
For detailed single-turnover kinetic analysis, non-linear regression data fitting is performed using Prism 6. The time dependence of product formation is fit to a one-phase association equation to determine the observed rate (kobs). The substrate concentration dependence is fit to a hyperbolic equation to obtain values for the maximum rate of NTP incorporation (kpol) and apparent Kd (Kd,app) governing NTP binding as previously described. The specificity constant is determined by kpol/Kd,app. These kinetic parameters are used for measuring the impacts of DNA lesion on nucleotide incorporation and transcriptional fidelity in a quantitative manner.
Troubleshooting: If a high level of unextended RNA is detected, check the Pol II activity with undamaged DNA template as a control. If Pol II is active, increase Pol II concentration or preincubation time to make sure all scaffolds formed Pol II elongation complex. If the bands are smeared, increase denaturation time. Please also use RNase-free water to prepare buffers. The same method can be used with other eukaryotic and prokaryotic RNA polymerases.
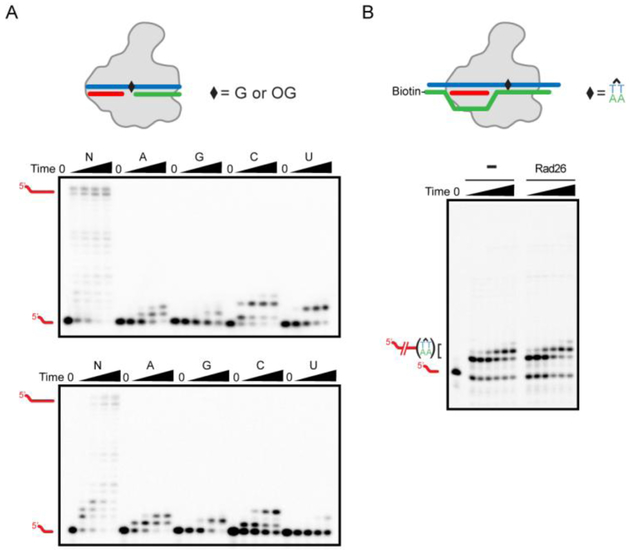
Figure 2. Transcription assay results using mini-scaffold or full-bubble scaffold.
(A) An example of transcription assay is performed by using mini-scaffold harboring non-damaged G (upper panel) or 8-oxoguanine (8OG, bottom panel) in its template strand DNA. Reaction is initiated by adding 50 μM of NTPs, ATP, GTP, CTP, or UTP. Time points are: 0 (without NTPs), 10 sec, 3 min, 30 min, and 2 hr. Note that 8OG can be bypassed by adding A or C, producing full-length transcripts after 3 min, as described in previous paper [13]. (B) Example of full-bubble transcription assay, using the same scaffold in our previous paper [24]. Time points are: 0, 1, 3, 10, 30, and 60 min, respectively. In this experiment, cyclobutane pyrimidine dimer (CPD) is tested to see whether presence of Rad26 can rescue arrested elongation complex (EC). As previously reported [24], Rad26 fails to rescue CPD induced EC arrest. Gel image was processed by Image Lab (Bio-rad). DNA and RNA oligos used in this study are summarized in Table S1.
3. Structural analysis of DNA-lesion arrested Pol II complexes
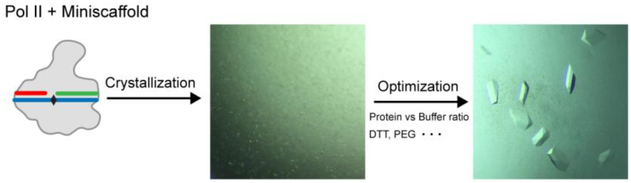
Crystallization methods are essentially the same as previously reported [9, 15, 26]. However, optimization is needed to determine the best crystallization conditions for different types of DNA lesion-arrested Pol II complex (Fig. 3). The optimization procedures include designing different lengths and sequences of DNA and RNA scaffold for structural studies, changing initial protein concentration, crystallization buffer concentration, ratios of protein:crystallization buffer, and variation of pH, salt, detergents, and temperature. Note: If DNA lesion is light-sensitive, the trays are covered with aluminum foil. If DNA lesion is sensitive to redox, special consideration is needed for choosing compatible reducing agents in the buffer.
Figure 3. Schematic overview of crystal optimization.
Transcription assay results can be used to design mini-scaffold for crystallization. Crystals of 10-subunit Pol II crystallized with 8OG containing mini-scaffold are shown. Initial crystals are produced by mixing same volume of protein and crystallization buffer (see Table 1) with 10 % PEG 6,000 and without DTT. Optimized crystals were grown in 5 mM DTT buffer with 2:1 protein to buffer ratio.
3.1. Preparation of DNA-lesion arrested Pol II complexes
For complexes the mini-scaffold, mix 1:2:2 molar ratio of DNA-lesion containing template DNA, non-template DNA, and RNA oligonucleotides in elongation buffer. Final concentration of template DNA can vary but is usually adjusted to 0.1 mM. Annealing is performed by incubating at 65 °C for 5 min and slowly cooling down to 25 °C.
Centrifuge ammonium sulfate precipitated Pol II to remove residual supernatant and dissolve in elongation buffer to final concentration of 5 μM.
Add five molar equivalents of prepared mini-scaffold to purified Pol II in elongation buffer. Final concentrations of Pol II and template DNA are 2.5 μM and 12.5 μM, respectively. Incubate elongation complex at room temperature for 30 min, followed by at least 30 min on ice.
Ultrafiltration is performed to change buffer and exclude excess scaffold, by using 100 kDa molecular cut-off centrifuge filters (Merck). We add two equal volumes of exchange buffer to elongation complex and centrifuge to concentrate. This process is repeated 3 times. Final concentration is set to 6–8 mg/ml by Bradford assay (Bio-rad). Prepared elongation complex can be directly used for crystallization, or flash-frozen and stored in −80 °C for crystallization repeat and optimization.
3.2. Crystallization of elongation complex and ligand soaking
The hanging drop crystallization plate (Qiagen) is set up by mixing protein and crystallization reservoir solution at 1:1 to 2:1 ratio. 5 mM of fresh reducing agents (such as DTT) is added to the reservoir right before crystallization to prevent the oxidation of Pol II. Crystals are incubated at 22 °C for 4 days to 2 weeks.
The size of crystal is one of the most important factors correlated to diffraction quality. Optimization of crystallization buffer and ratio of protein:reservoir can be done to obtain higher quality crystals. An example of crystal optimization is shown in Figure 3. Ten-subunit Pol II is crystallized with mini-scaffold containing a site-specific 8-oxo-G lesion at the template strand DNA (same scaffold as in Fig. 2A, bottom panel). Initial crystallization buffer is 10 % PEG 6,000 without DTT (Table 1), and 1:1 volume ratio is used. Well-diffracting crystals are obtained by changing the ratio of protein and crystallization buffer to 2:1 and adding additional 5 mM fresh DTT to the crystallization buffer to prevent oxidation.
For the data collection, selected crystals are transferred from crystallization drop to cryo buffer in a step-wise manner. The intermediate buffers are prepared by mixing crystallization buffer and cryo buffer in a ratio of 2:1 and 1:2. The transferred crystals are then incubated overnight at 4 °C and then flash-frozen and stored in liquid nitrogen for remote data collection. In the case of soaking experiments, various ligands are added to the final cryo buffer. If the ligand is nucleotide triphosphate or its analogs, the same or higher concentration of MgCl2 should also be added to prevent Mg2+ depletion in the crystal.
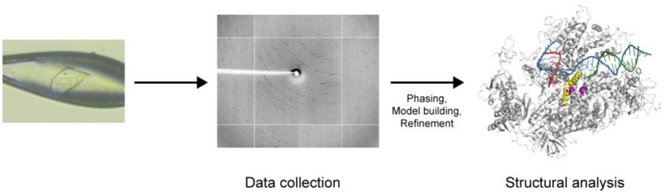
Collected datasets can be processed by using iMosflm [41]. CC1/2 around 0.5 is usually used to determine the resolution of each dataset [42]. Initial structure can be determined with PHENIX molecular replacement by using unmodified Pol II elongation complex as a search model [9]. Manual model building and refinement by COOT and PHENIX is necessary to get the final model [43, 44]. Summary of structure determination is depicted in Figure 4.
Figure 4. Structure determination of Pol II elongation complex.
An example of crystal and diffraction pattern of Pol II elongation complex is shown. X-ray diffraction image was collected at synchrotron (beamline 8.2.1, Advanced Light Source, Lawrence Berkeley National Laboratory). An example of structural representation of Pol II elongation complex with mini-scaffold was prepared by Pymol [9]. Color codes are the same as in schematic representation (Fig. 1). Bridge helix and trigger loop are colored as yellow and purple, respectively.
4. Conclusion
Investigation of transcriptional dynamics is critical for us to fully understand transcription regulation as well as its interplay with other DNA processes, such as DNA repair and replication. Indeed, the causes of transcriptional pausing and arrest are exceptionally diverse: Pol II complex can pause/arrest through specific interactions with transcription factors (such as promoter proximal-pause), or naturally occurring sequences (such as repetitive sequence or non-B form DNA), or at DNA lesions and modifications [45, 46]. The interplays between Pol II transcription machinery and DNA lesions and modifications dictate distinct transcriptional outcomes. Some of the DNA lesions cause transient transcriptional pausing, which can be bypassed by Pol II itself or with the help from other elongation factors [31], whereas bulky DNA lesions can cause prolonged transcriptional pausing and arrest, which provides a strong signal for transcription coupled repair. In addition, prolonged transcriptional arrest also leads to the degradation of complex or apoptosis [47].
In this mini-review, we aimed to summarize the current methods of transcription assay and crystallization of DNA lesion-arrested yeast RNA polymerase II. Transcription assay can be used to investigate the transcriptional consequences of modification or DNA binding ligands. For example, the ratio of run-off product, the severity of transcription pausing, pausing position, nucleotide preference, calculating kinetic values, and TFIIS induced cleavage of RNA transcript can be studied by transcription assay. Structural analysis of Pol II and damaged scaffold provides structural snapshots of lesion-arrested Pol II. These combined methods provide novel mechanistic insights into the understanding of how RNA Pol II transcription machinery recognizes different types of lesions and dictates translesion synthesis or transcription coupled repair.
Supplementary Material
Table S1. The sequences of DNA and RNA oligonucleotides used in this study.
Highlights.
Description of a combined approach including biochemical transcription assays and structural analysis to investigate the mechanism of DNA lesion arrested RNA Pol II.
Method for in vitro transcription biochemical assay with RNA Pol II on a DNA lesion-containing template
Crystallization method for DNA lesion-arrested RNA Pol II complex
Acknowledgements
This work was supported by the National Institutes of Health (R01 GM102362 to D.W.). This research used resources of the Advanced Light Source, which is a DOE Office of Science User Facility under contract no. DE-AC02–05CH11231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN, MicroRNA genes are transcribed by RNA polymerase II, EMBO J. 23(20) (2004) 4051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hernandez N, Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription, J. Biol. Chem 276(29) (2001) 26733–6. [DOI] [PubMed] [Google Scholar]
- [3].Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell 153(6) (2013) 1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].De Bont R, van Larebeke N, Endogenous DNA damage in humans: a review of quantitative data, Mutagenesis 19(3) (2004) 169–85. [DOI] [PubMed] [Google Scholar]
- [5].Cramer P, Bushnell DA, Kornberg RD, Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution, Science 292(5523) (2001) 1863–76. [DOI] [PubMed] [Google Scholar]
- [6].Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD, Architecture of RNA polymerase II and implications for the transcription mechanism, Science 288(5466) (2000) 640–9. [DOI] [PubMed] [Google Scholar]
- [7].Armache KJ, Kettenberger H, Cramer P, Architecture of initiation-competent 12-subunit RNA polymerase II, Proc. Natl. Acad. Sci. U. S. A 100(12) (2003) 6964–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bushnell DA, Kornberg RD, Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription, Proc. Natl. Acad. Sci. U. S. A 100(12) (2003) 6969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD, Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis, Cell 127(5) (2006) 941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brueckner F, Hennecke U, Carell T, Cramer P, CPD damage recognition by transcribing RNA polymerase II, Science 315(5813) (2007) 859–62. [DOI] [PubMed] [Google Scholar]
- [11].Damsma GE, Alt A, Brueckner F, Carell T, Cramer P, Mechanism of transcriptional stalling at cisplatin-damaged DNA, Nat. Struct. Mol. Biol 14(12) (2007) 1127–33. [DOI] [PubMed] [Google Scholar]
- [12].Brueckner F, Cramer P, Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation, Nat. Struct. Mol. Biol 15(8) (2008) 811–8. [DOI] [PubMed] [Google Scholar]
- [13].Damsma GE, Cramer P, Molecular basis of transcriptional mutagenesis at 8-oxoguanine, J. Biol. Chem 284(46) (2009) 31658–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P, RNA polymerase II-TFIIB structure and mechanism of transcription initiation, Nature 462(7271) (2009) 323–30. [DOI] [PubMed] [Google Scholar]
- [15].Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD, Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution, Science 324(5931) (2009) 1203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang D, Zhu G, Huang X, Lippard SJ, X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct, Proc. Natl. Acad. Sci. U. S. A 107(21) (2010) 9584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D, 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription, Nat. Struct. Mol. Biol 19(8) (2012) 831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kellinger MW, Park GY, Chong J, Lippard SJ, Wang D, Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis, J. Am. Chem. Soc 135(35) (2013) 13054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu L, Da L, Plouffe SW, Chong J, Kool E, Wang D, Molecular basis of transcriptional fidelity and DNA lesion-induced transcriptional mutagenesis, DNA Repair (Amst) 19 (2014) 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Walmacq C, Wang L, Chong J, Scibelli K, Lubkowska L, Gnatt A, Brooks PJ, Wang D, Kashlev M, Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions, Proc. Natl. Acad. Sci. U. S. A 112(5) (2015) E410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, Chong J, Li H, He C, Fu XD, Wang D, Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex, Nature 523(7562) (2015) 621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P, Transcription initiation complex structures elucidate DNA opening, Nature 533(7603) (2016) 353–8. [DOI] [PubMed] [Google Scholar]
- [23].Xu L, Wang W, Gotte D, Yang F, Hare AA, Welch TR, Li BC, Shin JH, Chong J, Strathern JN, Dervan PB, Wang D, RNA polymerase II senses obstruction in the DNA minor groove via a conserved sensor motif, Proc. Natl. Acad. Sci. U. S. A 113(44) (2016) 12426–12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu J, Lahiri I, Wang W, Wier A, Cianfrocco MA, Chong J, Hare AA, Dervan PB, DiMaio F, Leschziner AE, Wang D, Structural basis for the initiation of eukaryotic transcription-coupled DNA repair, Nature 551(7682) (2017) 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xu L, Wang W, Wu J, Shin JH, Wang P, Unarta IC, Chong J, Wang Y, Wang D, Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis, Proc. Natl. Acad. Sci. U. S. A 114(34) (2017) E7082–E7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang W, Walmacq C, Chong J, Kashlev M, Wang D, Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II, Proc. Natl. Acad. Sci. U. S. A 115(11) (2018) E2538–E2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P, Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA, Mol. Cell 34(6) (2009) 710–21. [DOI] [PubMed] [Google Scholar]
- [28].Cheung AC, Cramer P, Structural basis of RNA polymerase II backtracking, arrest and reactivation, Nature 471(7337) (2011) 249–53. [DOI] [PubMed] [Google Scholar]
- [29].Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM, FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration, Nature 533(7603) (2016) 407–10. [DOI] [PubMed] [Google Scholar]
- [30].Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P, Structure of transcribing mammalian RNA polymerase II, Nature 529(7587) (2016) 551–4. [DOI] [PubMed] [Google Scholar]
- [31].Hanawalt PC, Spivak G, Transcription-coupled DNA repair: two decades of progress and surprises, Nat. Rev. Mol. Cell Biol 9(12) (2008) 958–70. [DOI] [PubMed] [Google Scholar]
- [32].Walmacq C, Cheung AC, Kireeva ML, Lubkowska L, Ye C, Gotte D, Strathern JN, Carell T, Cramer P, Kashlev M, Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage, Mol. Cell 46(1) (2012) 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wilson MD, Harreman M, Svejstrup JQ, Ubiquitylation and degradation of elongating RNA polymerase II: the last resort, Biochim. Biophys. Acta 1829(1) (2013) 151–7. [DOI] [PubMed] [Google Scholar]
- [34].Kim N, Jinks-Robertson S, Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair, Mol. Cell. Biol 30(13) (2010) 3206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Scicchitano DA, Olesnicky EC, Dimitri A, Transcription and DNA adducts: what happens when the message gets cut off?, DNA Repair (Amst) 3(12) (2004) 1537–48. [DOI] [PubMed] [Google Scholar]
- [36].Da LT, Pardo-Avila F, Xu L, Silva DA, Zhang L, Gao X, Wang D, Huang X, Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue, Nat Commun 7 (2016) 11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Silva DA, Weiss DR, Pardo Avila F, Da LT, Levitt M, Wang D, Huang X, Millisecond dynamics of RNA polymerase II translocation at atomic resolution, Proc. Natl. Acad. Sci. U. S. A 111(21) (2014) 7665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shin JH, Xu L, Wang D, RNA polymerase II acts as a selective sensor for DNA lesions and endogenous DNA modifications, Transcription 7(3) (2016) 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kireeva ML, Lubkowska L, Komissarova N, Kashlev M, Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae, Methods Enzymol. 370 (2003) 138–55. [DOI] [PubMed] [Google Scholar]
- [40].Kaplan CD, Larsson KM, Kornberg RD, The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin, Mol. Cell 30(5) (2008) 547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG, iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM, Acta Crystallogr. D Biol. Crystallogr 67(Pt 4) (2011) 271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Karplus PA, Diederichs K, Linking crystallographic model and data quality, Science 336(6084) (2012) 1030–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. D Biol. Crystallogr 66(Pt 2) (2010) 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr. D Biol. Crystallogr 66(Pt 4) (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT, Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells, Genes Dev. 25(7) (2011) 742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jonkers I, Lis JT, Getting up to speed with transcription elongation by RNA polymerase II, Nat. Rev. Mol. Cell Biol 16(3) (2015) 167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Todd RC, Lippard SJ, Inhibition of transcription by platinum antitumor compounds, Metallomics 1(4) (2009) 280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The sequences of DNA and RNA oligonucleotides used in this study.