Abstract
The Carboxy-terminal Domain (CTD) of RNA polymerase II (Pol II) plays essential roles in regulating gene expression in eukaryotes. Here, we describe multiple genetic approaches for studying the CTD in Drosophila that complement pre-existing molecular analyses of the Pol II CTD in other experimental models. These approaches will allow one to assess the effects of any CTD mutations in a developmentally complex organism. The approaches discussed in this work can in principle, be applied to analyze other transcription components in eukaryotes.
Keywords: Drosophila, Genetics, Pol II CTD, GAL4/UAS, CRISPR/Cas9
1. Introduction
The Pol II CTD is an essential binding platform for numerous factors regulating gene expression [1,2]. It is at the C-terminus of Rpb1, the largest subunit of Pol II, and consists primarily of repetitive arrays of seven amino acids (“heptads”) in many organisms. For example, the CTDs of budding yeast (S. cerevisiae) and fission yeast (S. pombe) have 26 and 29 repeats respectively and most of these repeats match the seven amino acid consensus sequence: Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (YSPTSPS). Over three decades of mutational analyses of the yeast CTDs have established the significance of residues at each heptad position and that the functional unit of the CTD likely spans across two consecutive heptad repeats [1,2]. Subsequent biochemical and genomic analyses reveal that different patterns of phosphorylation constitute a CTD code that enable or exclude certain CTD:factor interactions as Pol II transcribes across genes [3].
In contrast to the yeast CTDs, the CTDs of higher eukaryotes are longer and have evolved numerous divergent motifs. For example, all mammalian CTDs have 52 repeats and approximately half the repeats are different from YSPTSPS in at least one position. Biochemical and genomic analyses of the mammalian CTD suggest that the divergent motifs serve functions essential in developmentally-complex organisms: the conserved 7th position arginine in the distal portion of the CTD can be methylated [4,5] or citrullinated [6] while the 7th position lysines can be methylated [7] or acetylated [8]. Mutating these residues to alanine, which prevents post-translational modification, led to changes in gene expression in mammalian cultured cells [4,5,6,8]. In addition, the arginine methylation marks are bound by TDRD3 and SMN, factors present in higher eukaryotes [4,5]. One model is that the post-translational marks on the divergent motifs expand the CTD code available in yeast, allowing more complex networks of CTD:factor interactions to occur in higher eukaryotes [3,9]. However, systematic mutational analyses of the mammalian CTD has not been performed in an animal model and most residue substitution mutations in the divergent residues of the CTD do not affect viability of mammalian cultured cells [4,7,8,10]. Therefore, the significance of these CTD residues in animal development and viability remains unclear.
We recently established two genetic approaches in Drosophila that have allowed us to mutate the CTD and test whether the presence of divergent motifs are required for development of the fly [11–13]. Unlike the yeast CTDs which are dominated by the consensus heptads, only 2 of 42 repeats of the Drosophila CTD match the consensus. In addition, the Drosophila CTD contains a 7th position arginine and 7th position lysines in its distal portion similar to what is present in mammals [4,7]. The well-established GAL4/UAS system and the more recently developed CRISPR/Cas9 technology make Drosophila an attractive model to perform genetic analyses of the CTD. Using these approaches, we have made the unexpected discovery that all divergent motifs can be replaced with consensus heptads without affecting the viability of Drosophila, although the number of consensus heptads is critically important for function [13]. Here, we describe the methods that we developed to perform a molecular genetic analysis of the RNA polymerase II CTD in Drosophila. These methods serve as a guide for altering the CTD and provide a framework for analyzing functional domains of other transcription components in Drosophila.
2. Functional analyses of the Drosophila CTD using GAL4/UAS
2.1. Design of an RNAi based approach
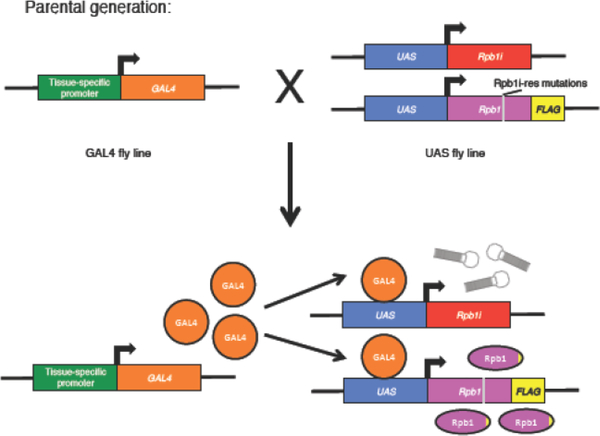
Since mutations in the CTD can lead to lethality, one way to perform a genetic analysis on the CTD is to conditionally inactivate the endogenous Rpb1 while simultaneously expressing an Rpb1 derivative harboring mutations in the CTD. In mammalian cells, this is achieved by inhibiting the endogenous Rpb1 with α-amanitin after transfecting into cells an expression construct encoding an α-amanitin resistant version of Rpb1 [4,5,7,8,10]. A conceptually similar experiment outlined in this section can be performed in Drosophila using the binary GAL4/UAS expression system [14]. As illustrated in Fig. 1, in this system, two UAS-driven transgenes are present in one fly line. One UAS transgene encodes a short hairpin designed to target a 21-nucleotide coding sequence in the body of the Rpb1 mRNA [15], which knocks down expression of the endogenous Rpb1. Ubiquitous expression of this Rpb1 RNAi (Rpb1i) alone in the fly causes lethality [11,12]. The other UAS transgene encodes an RNAi-resistant derivative of Rpb1 that is rendered resistant to the RNAi with synonymous changes in the region targeted by the RNAi (Fig. 1 and 2, Rpb1i-res mutations). Neither transgene is expressed until the flies are mated to another fly line expressing GAL4. This system allows the simultaneous activation of both UAS transgenes in tissues where GAL4 is expressed. The Drosophila community has generated hundreds of GAL4 stocks that can be used to drive expression in various tissues. This allows for functional tests of CTD mutations in specific tissue or throughout the entire fly. Table 1 provides a list of GAL4 stocks that we have used to analyze the effects of CTD mutations. The GAL4 lines were obtained from the Bloomington Drosophila Stock Center (BDSC).
Fig. 1.
Schematic of GAL4/UAS based Rpb1 RNAi rescue approach. Parental GAL4 and UAS bearing fly lines (upper) are mated to generate offspring expressing GAL4 that activates expression of the UAS-target genes (lower).
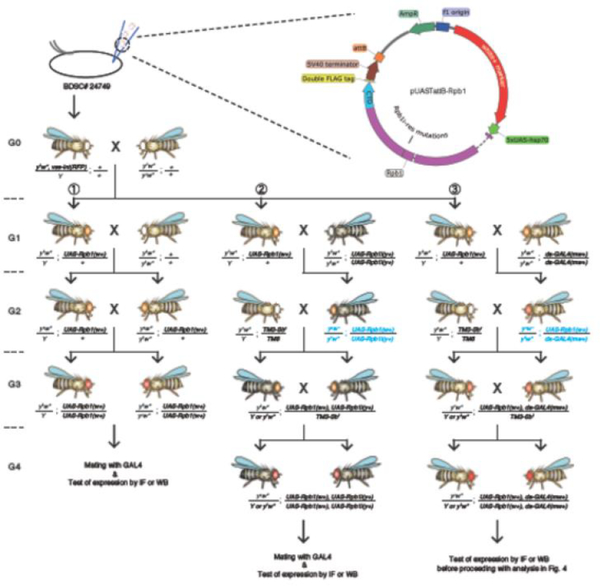
Fig. 2.
Design of pUASTattB-Rpb1 and cross schemes to generate stable fly lines expressing various GAL4 and UAS transgenes. Blue genotypes designate steps where meiotic recombination events occur. In Drosophila, meiotic recombination is limited to females. Offspring with TM3 third chromosome balancer can be distinguished by their short bristles (Sb1). Primers for verifying the presence of pUASTattB-Rpb1 in the fly genome: fw: 5’-CGCCTTCGGCTGCATCGG-3’ and rev: 5’-ACAAAGATCCTCTAGAGGTACCCTCGAGC-3’. Primers for detecting UAS-Rpb1i transgene: fw: 5’-GGTGATAGAGCCTGAACCAG-3’ and rev: 5’-TAATCGTGTGTGATGCCTACC-3’. Primers for detecting da-GAL4 transgene: fw: 5’-GGATGCTCTTCATGGATTTG-3’ and 5’-CAACATCATTAGCGTCGGTGAG-3’.
Table 1.
GAL4 lines useful for functional analysis of the CTD.
| Name | Symbol | Chr. | BDSC# | Expression | References |
|---|---|---|---|---|---|
| Act-GAL4 | P{w[+mC]=Act5CGAL4}25FO1 | 2 | 4414 | Ubiquitous | [16] |
| da-GAL4 | P{w[+mW.hs]=GAL4da.G32}UH1 | 3 | 55850 | Ubiquitous | [17] |
| ms1096GAL4 | P{w[+mW.hs]=GawB}Bx[MS1 096] | X | 8860 | Wing, robust expression | [18] |
| C5-GAL4 | P{w[+mW.hs]=GawB}cvc[C5] | 3 | 30839 | Wing, relatively low expression | [19] |
| Ab1- GAL4 | P{w[+mW.hs]=GawB}AB1 | 3 | 1824 | Salivary glands | [20] |
| Sgs3- GAL4 | P{w[+mC]=Sgs3GAL4.PD}TP1 | 3 | 6870 | Salivary glands, activated at mid third-instar | [21] |
| CG-GAL4 | P{w[+mC]=Cg-GAL4.A}2 | 2 | 7011 | Fat body | [22] |
GAL4 drivers that are ubiquitously expressed serve to determine if CTD mutations affect development. The effects of a CTD mutation can be assess by monitoring the developmental stage at which individuals die when expression is driven by da-GAL4. A quantitative analysis can be readily obtained by using flies with a GAL4 transgene kept over a marked balancer chromosome. The rescue efficiency can be presented as the ratio of progeny with versus without the balancer marker. We have previously tested a series of CTD mutants using Act-GAL4 (BDSC# 4414), which is ubiquitously expressed. The Act-GAL4 transgene is maintained over a balancer marked by curly wings (CyO). In cases where an Rpb1 derivative is fully rescued, approximately equal numbers of adult progeny with normal and curly wings will be produced whereas this ratio is skewed if the Rpb1 derivative is defective [11–13].
For Rpb1 derivatives with mutations that provide no rescue when ubiquitously expressed, a follow-up analysis with a tissue-specific GAL4 driver can be performed to determine if a mutant is completely inactive. A test with wing-specific expression works well because wings are non-essential organs for flies and a range of mutant phenotypes are readily observed. Alternatively, tissue-specific expression provides the possibility to perform molecular characterization on severe mutants that fail to develop any analyzable tissue when expressed ubiquitously. For example, the Sgs3-GAL4 driver is not activated in the salivary glands till mid third-instar larval stage after the salivary glands are well-developed [21].
It is important to note that although in general, the level of expression driven by GAL4/UAS tends to be higher than the endogenous level, the ubiquitous expression of wild-type Rpb1 alone does not produce any phenotype. Moreover, co-expression of the Rpb1i-resistant wild-type Rpb1 was sufficient to rescue defects of Rpb1i in all cases examined, indicating that there is no evidence of off-target effects of the Rpb1 RNAi. In addition, unlike its Rpb1i-resistant counterpart, the Rpb1i sensitive form of Rpb1 was unable to rescue Rpb1i, showing that the elevated level of Rpb1 mRNA alone is not overwhelming the capacity of the Rpb1i [12]. In cases where the co-expression of a mutant Rpb1 with Rpb1i is unable to fully rescue, a control mating of the same GAL4 driver to UAS-driven mutant Rpb1 alone should be done to determine if the expression of the mutant alone produces any dominant-negative phenotype. An example of this is provided in Fig. 3.
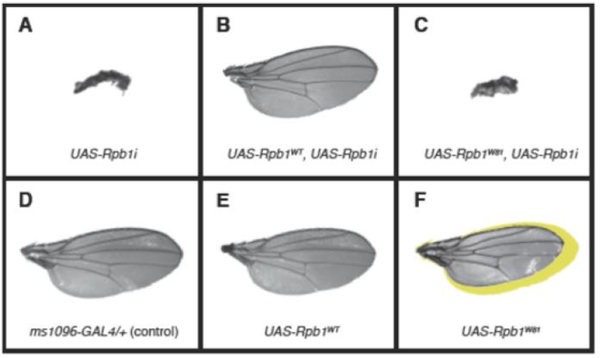
Fig. 3.
Testing the functionality of Rpb1W81 [12,32,33] in Drosophila wings. Wing-specific expression was driven by ms1096-GAL4. Mating to y1w* serves as a control (D). The expression of Rpb1i alone gave rise to miniature-sized wings (A) that were rescued by the co-expression of wild-type Rpb1 (Rpb1WT, B), but not by the co-expression of Rpb1W81 (C). The expression of Rpb1WT alone did not cause any defect (E) whereas the expression of Rpb1W81 alone led to a reduction in wing size (F). Yellow shadow in (F) corresponds to the size of the control wing shown in (D).
2.2. Test of mutant CTD function using an Rpb1 embryonic lethal allele
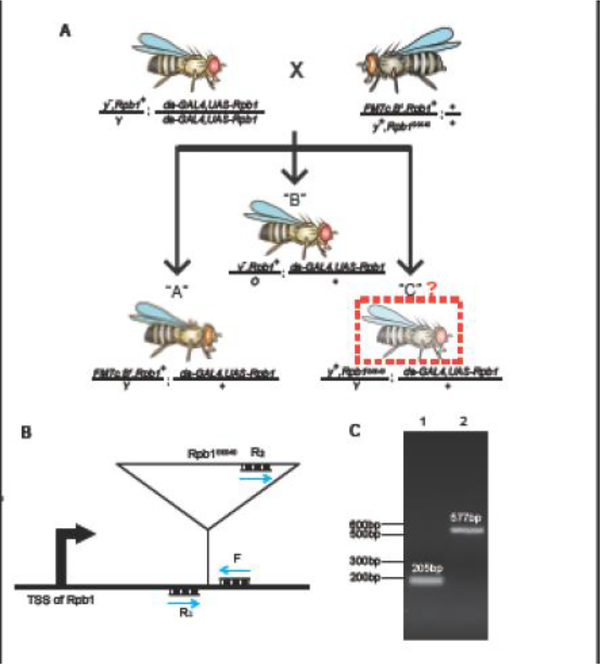
An orthogonal approach to the RNAi rescue assay is to test if the ubiquitous expression of an Rpb1 derivative rescues an embryonic lethal Rpb1 allele. This allele of Rpb1, dubbed Rpb1G0040 (Kyoto #111768), has a P-element inserted into the 5’ UTR of Rpb1 and is maintained over the X balancer chromosome FM7c [12]. In this assay, females carrying the Rpb1G0040 allele (Rpb1G0040/FM7c) are mated to males ubiquitously expressing an Rpb1 derivative (da-GAL4, UAS-Rpb1). As illustrated in Fig. 4A, this mating will result in at least two types of male progeny: “A” progeny, marked by narrow eyes (B1), survive because of the wild-type Rpb1 allele on the FM7c balancer; “B” progeny, marked by yellow body color (y−) and normal eyes, are the products of nondisjunction. Additionally, if the GAL4-mediated expression of the Rpb1 derivative fully rescues Rpb1G0040, “C” male progeny with wild-type body color (y+) and normal eyes will be present at a comparable number as “A” males. The presence of the P-element insertion in the Rpb1G0040 allele can be validated by PCR (Fig. 4B and C). As this rescue approach relies on the generation of a stable fly line ubiquitously expressing an Rpb1 derivative, it is limited to testing mutants that do not have severe dominant negative effects.
Fig. 4.
Testing the functionality of UAS-Rpb1 derivatives using the Rpb1G0040 allele. (A) Schematic of the Rpb1G0040 rescue assay. “C” type males have normal body color due to y+ while “B” type males have yellow body color due to y−. (B) Schematic for the PCR evaluation of the endogenous Rpb1 allele located on the X chromosome in progeny from the Rpb1G0040 rescue assay. Transcription start site (TSS) of Rpb1 is shown. Blue arrows represent primers. PCR amplification of the wild-type endogenous allele with primers F and R1 produces a 205 bp DNA. PCR amplification with primers F and R2 of the Rpb1G0040 allele produces a 577 bp DNA. Sequences of primers: F: 5’-AGAAGGCTGGGTAAACAATCAC-3’, R1: 5’-CTGCTACAACGACCGCAATA-3’ and R2: 5’-AATGAACAGGACCTAACGCACA-3’. (C) PCR results for the endogenous Rpb1 allele following the scheme shown in (B). Genomic DNA was isolated from one male fly [34] and amplified with a mixture of primers F, R1 and R2. Lane 1 shows the product from one male containing the endogenous wild-type allele but not the Rpb1G0040 allele. Consequently, only the 205 bp DNA is produced. Lane 2 shows the product from a “C” male. The presence of a 577 bp product confirms the presence of Rpb1G0040 while the absence of the 205 bp product indicates the absence of the endogenous wild-type allele.
2.3. Design and generation of pUAST plasmids expressing Rpb1 derivatives
We use Clontech In-Fusion Cloning to insert fragments encoding various Rpb1 derivatives into pUASTattB plasmid (DGRC #1419). Most fragments can be obtained directly by amplification from genomic DNA or subclones of the amplified DNA, although we occasionally order 200 bp to 1 kb synthetic DNA fragments (Integrated DNA Technologies or Genscript) for sequences that are difficult to obtain by PCR. Each final pUASTattB-Rpb1 vector (Fig. 2) contains: (1) a 5XUAS-hsp70 promoter [14]; (2) the entire Rpb1 coding region with the first two introns; (3) synonymous mutations changing the Rpb1 RNAi target sequences from 5′-AACGGTGAAACTGTCGAACAA-3′ to 5′-AACCGTCAAGTTGAGCAACAA-3’; (4) white+ (w+) eye color marker for identifying transformants; (5) a double FLAG tag appending to the C-terminus of Rpb1 that distinguishes the transgenic Rpb1 from the endogenous counterpart; (6) an attB site for genome integration. The entire coding region of Rpb1 on the plasmid, including the double FLAG tag, is sequenced before submitting for microinjection services (Rainbow Transgenic Flies, Inc.).
2.4. PhiC31 integrase-mediated transformation and molecular validation
The Rpb1-containing pUASTattB plasmids are microinjected into embryos of a fly line with an attP site at a known genomic locus on its third chromosome (86Fb, BDSC# 24749). This fly line also carries a transgene on its X chromosome that expresses the PhiC31 integrase in its germline [23]. The integrase transgene can be removed by selecting orange-eyed G1 males in the second mating (Fig. 2). Alternatively, because the integrase is marked by the expression of eye-specific EGFP, it can also be removed by selecting against progeny with green fluorescent eyes during subsequent matings. PhiC31 integrase causes recombination between attB on the plasmid and attP in the fly genome and results in incorporation of the entire plasmid into the genome. The injected individuals (G0) are then outcrossed to y1w*. As both parents carry w* mutation (a recessive allele that exhibits the white eye phenotype), the presence of progeny with the white+ (orange eyes when only one copy is present in the w* background) marker identifies the desired recombinants. Homozygous fly lines are established by selecting for red eye progeny (having two copies of white+) resulting from mating of orange eye siblings (Fig. 2, scheme 1). As the expression of the UAS transgenes is not induced in the absence of GAL4, such red eye flies can be maintained as stable fly lines regardless of whether the Rpb1 derivative encoded in the pUAST plasmid is functional.
The PhiC31 transformation efficiency is rather high - we typically obtain tens to hundreds of orange-eyed G1 transformants per 100 injected embryos. To verify that the sequence encoding a CTD mutant is present, the region encompassing the transgenic CTD is PCR amplified from genomic DNA isolated from the orange eye recombinants and the resulting PCR product is sequenced. One PCR primer is complementary to sequence just upstream of the CTD while the other PCR primer is complementary to a sequence specific to the pUAST vector (see legend of Fig. 2 for sequences of primers). DNA isolated from y1w* flies is used as a control for non-specific amplification. PCR products that match predicted sizes are gel purified and sequenced to confirm that the transformed sequence is correct.
The GAL4-mediated expression of UAS-driven Rpb1 derivatives can be assessed by immunofluorescence or by western blotting tissue lysates using anti-FLAG antibody. Progeny arising from a mating of y1w* to the same GAL4 driver serves as a negative control for the expression analyses. We have used salivary glands (Act-GAL4, da-GAL4, Ab1-GAL4 or Sgs3-GAL4) and fat body (CG-GAL4) for immunofluorescence analyses and employed indirect immunofluorescence to detect FLAG-tagged Rpb1 on spread polytene chromosomes [24] and in fixed intact tissues. We have had success detecting the expression of FLAG-Rpb1 derivatives by western blot using lysate prepared from embryos (da-GAL4), larval fat body (CG-GAL4), whole third-instar larvae (da-GAL4), whole early pupae (da-GAL4), whole late pupae (Act-GAL4) and adult heads (da-GAL4). Western blot analysis using lysate prepared from salivary glands is problematic because something migrates just above the wild-type Drosophila Rpb1 (>200 kD) that interferes with the western blot analysis. We routinely use home-made 7% Tris-acetate gels [25] or commercial 3–8% Tris-acetate gradient gels (Fisher Scientific) because we find that the Drosophila Rpb1 does not migrate well on a SDS-PAGE gel [26].
2.5. Establishment of stable fly lines containing two transgenes
The UAS-Rpb1 transgene needs to be meiotically recombined with UAS-Rpb1i (BDSC# 36830) for the functional analyses in section 2.1 and with da-GAL4 (BDSC# 55850) for the functional analyses in section 2.2. These three transgenes are all located on the third chromosome. The original UAS-Rpb1i deposited at the Bloomington Drosophila Stock Center is maintained over a third chromosome balancer, likely due to a spontaneous recessive lethal mutation on the same chromosome that UAS-Rpb1i resides. We have established a stable fly line homozygous for UAS-Rpb1i by outcrossing the original UAS-Rpb1i fly line to y1w*. The y1 allele is a recessive allele that gives rise to yellowish body color. The UAS-Rpb1i transgene is marked by a dominant y+ allele that reverts the yellowish body color of y1 flies back to the normal darker color. Three sequential matings are performed to produce the final fly line homozygous for both UAS-Rpb1i and UAS-Rpb1 transgenes (Fig. 2, scheme 2). First, UAS-Rpb1i is mated to UAS-Rpb1/+ to generate progeny heterozygous for both transgenes. Second, the female offspring from the cross are mated to y1w* males containing third chromosome balancers. Third, normal body color (=UAS-Rpb1i present), orange-eyed (=UAS-Rpb1 present) siblings from the second cross are mated to each other and the resulting progeny lacking the balancer marker are kept as stable homozygous stocks.
The da-GAL4 transgene is marked by mini-white+ (mw+), which gives rise to orange eyes (yellowish eye when only one copy is present). An approach similar to the one described above is applied to generate flies homozygous for both da-GAL4 and UAS-Rpb1 except that in the third step, light red eye progeny (one copy of white+ and one copy of mini-white+) are selected instead of normal body color (y1) (Fig. 2, scheme 3).
The presence of UAS-Rpb1i and da-GAL4 can be confirmed using the same PCR approach described above but with primers specific to either transgene. In addition, the co-existence of da-GAL4 and UAS-Rpb1 can be validated by immunofluorescence or western blot using anti-FLAG antibody as described in section 2.4.
2.6. Potential pitfalls
While Rpb1 derivatives that fail to rescue Rpb1i or Rpb1G0040 are defective, those Rpb1 derivatives that provide rescue have two caveats. First, expression driven by GAL4/UAS tends to be high which could compensate for the potential loss of function of a Rpb1 derivative. Second, residual endogenous Rpb1 could be present either due to incomplete depletion or maternal loading of Rpb1, although we have recently determined that the level of knockdown driven by Act-GAL4 is efficient both at the transcript [12] and protein levels [13] (both are at least ten-fold less than wild-type). To definitively assess if the mutants that function like wild-type are sufficient to support development of Drosophila, one can evaluate their performance following the CRISPR/Cas9 scheme described in section 3. Besides, the promoter in the pUAST vector is hardly activated in the female germline [27]. Therefore, rescue analyses in the female germline and assessment of female fertility should be interpreted with caution.
3. Functional analyses of the Drosophila CTD using CRISPR/Cas9
3.1. Design of a CRISPR/Cas9 based approach
CRISPR/Cas9 allows the functional test of an Rpb1 derivative under conditions where expression is driven by the endogenous Rpb1 promoter and the endogenous Rpb1 is absent if a viable fly line homozygous for the mutant Rpb1 allele can be established. In this assay, the endogenous genomic sequence encoding the CTD is replaced with sequence encoding a desired CTD mutation via CRISPR/Cas9-mediated genome editing. A DNA mixture containing three plasmids is injected into embryos that stably express Cas9 [28]. Two plasmids express sgRNAs that direct CRISPR/Cas9 cutting to genomic regions upstream and downstream of the CTD respectively. The third “donor” plasmid serves two purposes: (1) it contains two sequences that are homologous to outside sequences immediately adjacent to cut sites, which serve as templates for homologous recombination mediated repair; (2) it encodes the CTD mutations to be introduced, which is placed in between the two homology sequences. Successful CRISPR-ed recombinants can be identified as described in section 3.4. Subsequent functional analyses are described in section 3.5.
3.2. Design and generation of the two plasmids expressing sgRNAs
We have identified two CRISPR target sequences with zero predicted off-target sites using flyCRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). These sites were selected with “NGG PAM sequence only” and “Maximum Stringency” search filters so that they should have higher cleavage efficiency and are less likely to be off-target. In addition, the two target sequences flank the CTD so that various CTD mutations can be introduced with the same targeting plasmids and homology sequences. One of the target sites is immediately upstream of the coding region of the CTD while the other is in the coding region of the downstream neighboring gene PGRP-SA. In hindsight, this genome editing scheme is not ideal as it disrupts the coding sequence of PGRP-SA. However, this has not had any detectable impact on our studies.
Regions encompassing the sequences of the guides are PCR amplified from the w1118 strain, which has the same X chromosome as the injected fly strain, and sequenced to confirm that the CRISPR target sequences match the sequence of the fly line that is being edited. Unphosphorylated, complementary strands encoding the sgRNAs are used (Integrated DNA Technologies): Rpb1_fw1: 5’- CTTCGAGAAACACTCGGCGAGGCT-3’ and Rpb1_rev1: 5’-AAACAGCCTCGCCGAGTGTTTCTC-3’ direct cleavage adjacent to the start of the Drosophila CTD; Rpb1_fw2: 5’-CTTCGTAGGGATTTGAGAGCCAGTG-3’ and Rpb1_rev2: 5’-AAACCACTGGCTCTCAAATCCCTAC-3’ direct cleavage downstream of the 3’ UTR of Rpb1. They are annealed, phosphorylated by T4 polynucleotide kinase and cloned into the BbsI site of pU6-BbsI-chiRNA (Addgene #45946) following the protocol developed by flyCRISPR (http://flycrispr.molbio.wisc.edu/protocols/gRNA).
3.3. Design and generation of the donor plasmid
Similar to the generation of pUASTattB-Rpb1 plasmids, we also employ Clontech In-Fusion Cloning to insert various fragments into pHD-ScarlessDsRed plasmid (DGRC # 1364). Most fragments can be obtained via PCR amplification from w1118 genomic DNA. Sequences encoding mutant CTDs can also be either amplified from pre-existing pUASTattB-Rpb1 plasmids or synthesized as DNA fragments (Integrated DNA Technologies or Genscript). Each final pHD-ScarlessDsRed-Rpb1 vector (Fig. 5) contains: (1) two ~1 kb homology arms; (2) sequence encoding a mutant CTD followed by the entire 3’ UTR of Rpb1; (3) a 3XP3 driven DsRed marker for identifying CRISPR-ed candidates; (4) a double FLAG tag appended to the C-terminus of Rpb1 that distinguishes the modified Rpb1 from the unmodified counterpart; (5) synonymous mutations in one of the target sequences that render the plasmid sequence resistant to CRISPR cutting (this CRISPR targeted sequence was changed from 5′-GAGAAACACTCGGCGAGGCT −3′ to 5′-GACTAACTGATGGGCTAGCT-3’; the other target sequence is separated from the PAM site by the DsRed cassette). The region spanning Homology Arm I, the CTD, the double FLAG tag and the 3’ UTR is cloned into the SapI sites while Homology Arm II is cloned into the AarI sites. Both inserts are sequenced before submitting for microinjection services (Bestgene Inc).
Fig. 5.
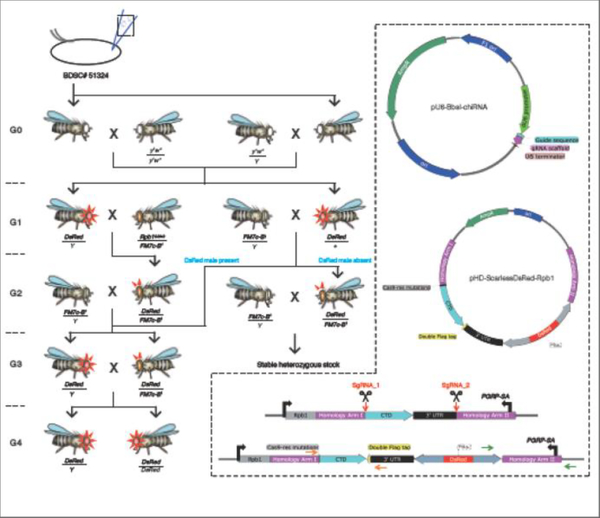
Design of CRISPR/Cas9 plasmids and cross schemes to generate stable fly lines with modified CTDs. Rpb1G0040/FM7c was used as a convenient source of FM7c. Red triangles surrounding the eyes designate DsRed flies with bright red fluorescent eyes and ocelli. Red triangles above the fly head designate DsRed flies with bright red fluorescence in ocelli but not in eyes because fluorescence in eyes is largely masked by the eye pigment. Sequences of orange primers: fw: 5’-GGCGATCGAGCGTAGTCGGTACTT-3’ and rev: 5’-CCAGGACCTTCGATGTCGCCGTATTT-3’. Sequences of green primers: fw: 5’-CGGCATGCGAGCCTTCTATATCTT-3’ and rev: 5’-ACGTACGTCACAATATGATTATCTTTCTAGGGTT-3’.
3.4. CRISPR microinjection, subsequent crosses and molecular validation
The above DNA plasmids are mixed with guide plasmids at 100 ng/ul each and the donor plasmid at 500 ng/ul, and microinjected into embryos that stably express Cas9 in the germline (BDSC# 51324). The Cas9 transgene is marked by 3XP3-GFP, which can be eliminated in subsequent crosses by selecting against progeny with green fluorescent eyes or ocelli. CRISPR/Cas9-mediated genome editing occurs in the gametes produced by the injected individuals. The injected individuals (referred to as G0) are then mated to w1118 flies and CRISPR-ed candidates among the G1 offspring will be marked with bright red fluorescent eyes (3XP3 driven DsRed) (Fig. 5). Integration of the DsRed cassette into the genome can occur in 3 ways: (1) successful modification of the endogenous Rpb1 locus as desired; (2) integration of the entire donor plasmid into one of the CRISPR target sites as a result of a single crossover; (3) integration of the entire donor plasmid elsewhere in the genome. Therefore, unlike the generation of UAS-Rpb1 fly lines where numerous G1 transformants can be pooled in subsequent crosses, all G1 DsRed candidates should be outcrossed individually and maintained as separate lines even if they are derived from the same parent. G1 DsRed males are mated to Rpb1G0040/FM7c females whereas G1 DsRed females are mated to FM7c/Y males. DsRed-Rpb1/FM7c G2 females, which are marked by red fluorescent ocelli and light orange nicked eyes, will be mated to FM7c/Y males. If hemizygous DsRed-Rpb1/Y males are present and fertile, stable fly lines homozygous for DsRed-Rpb1 can be generated by selecting against the FM7c balancer. Otherwise, DsRed-Rpb1/FM7c females and FM7c/Y males are maintained as final stocks.
The DsRed recombination rate is relatively low compared to PhiC31-mediated transformation - we typically obtain only a few to tens of DsRed G1 transformants per 100 injected embryos. In our experience, the percentage of desired CRISPR-ed recombinants among all DsRed candidates varies from 20% to 50% for mutants that were previously shown to support development into adulthood when tested in the ubiquitous GAL4/UAS assay and 0 to 5% for mutants that do not support development to adulthood in the GAL4/UAS assay. A single crossover event that occurs in the homology arm upstream of the CTD coding region also results in the replacement of the endogenous CTD so these single crossover candidates can serve as backups in cases where double crossover candidates cannot be obtained. Therefore, we first perform a western blot on G2 DsRed late pupae to test if the FLAG-tagged Rpb1 derivative is expressed. Lysate prepared from w1118 serves as a negative control. Candidates that express FLAG-Rpb1 are subjected to PCR analysis using their genomic DNA as templates with the following primers: (1) one primer pair that contains a primer that anneals to genomic sequence outside of the homology arm and the other anneals to sequence in the DsRed cassette (Fig. 5, green arrows); (2) a primer pair flanking the CTD coding region (Fig. 5, orange arrows). Genomic DNA isolated from w1118 can be used as a negative control template. The expression of the FLAG-Rpb1 derivative indicates that a crossover has occurred in the region that corresponds to Homology Arm I. As illustrated in Fig. 5, PCR validation using the green primer pair will determine if a crossover has also occurred within Homology Arm II. If a crossover has occurred in both homology arms, an amplicon containing Homology Arm II will be produced.
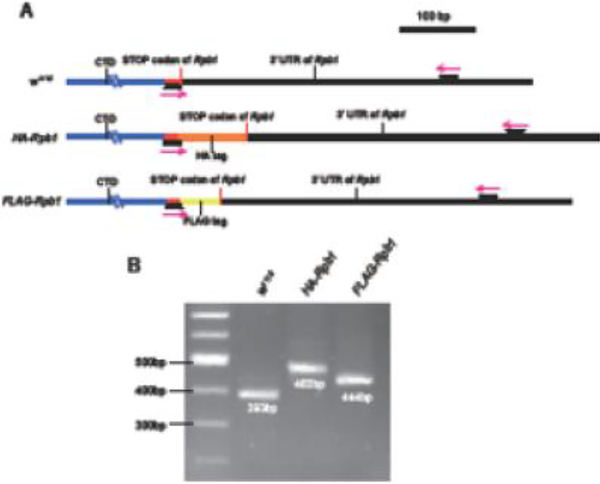
Once a DsRed male is available, an alternative approach is to amplify DNA from the male with the magenta primers shown in Fig. 6A. One primer anneals to sequence near the end of the coding sequence of Rpb1 and the other anneals to sequence in the 3’ UTR. Desired candidates with the additional C-terminal tag can be distinguished from the unmodified counterparts based on differences in the sizes of their amplicons. Examples of detecting HA-tagged and FLAG-tagged Rpb1 using this approach are provided in Fig. 6B.
Fig. 6.
Alternative detection of the modified endogenous Rpb1 locus by PCR. (A) Schematic of PCR validation of the tagged CTD fly lines. Magenta arrows represent primers. One of the primers hybridizes to the acidic tip (red) that is retained in CTD mutants. The other primer hybridizes to the 3’ UTR of Rpb1. Sequences of primers: fw: 5’-TCATACAGTGGGTTGTGCAAAGAA-3’ and rev: 5’-ACGTTCGAGGAGAGCGAAGAC −3’. (B) PCR reactions with primers annotated in (A). Genomic DNA was isolated from one male fly [34]. The increased sizes of the PCR products with HA-Rpb1 and FLAG-Rpb1 compared with w1118 suggested that the desired tags were introduced.
3.5. Survival analyses
To analyze the performance of Rpb1 mutants that cannot be maintained as stable homozygous stocks, we monitor how long males hemizygous for mutant Rpb1 survive over the course of development. This can be achieved by maintaining the DsRed-Rpb1 allele over a GFP labeled X chromosome balancer (FM7i, P{w[+mC]=ActGFP}JMR3, BDSC# 4559) instead of FM7c. GFP positive individuals can be distinguished from the GFP negative counterpart beginning at the onset of larval stage. The effects of of various CTD mutations on development can be determined by monitoring the stage at which hemizygous DsRed-Rpb1 males (GFP negative) disappear from the population.
To examine if Rpb1 mutants that can be maintained as stable homozygous stocks have defects, survival rates (embryonic hatch rates, percentages that develop into adults, life span, etc.) can be compared to a FLAG-tagged wild-type Rpb1 line that is generated via the same CRISPR/Cas9 editing scheme. In yeast, some CTD variants exhibit growth phenotypes under cold stress while supporting growth under normal culture conditions [29,30]. To further examine whether the Rpb1 mutants that function like wild-type under normal growth conditions (24°C) also function normally under temperature stresses, survival rates can be measured at 18°C or 30°C.
3.6. Potential pitfalls and discussion
Compared to GAL4/UAS, CRISPR/Cas9 has several limitations. First, as described in section 3.4, it is much more difficult to obtain a correct CRISPR-ed transformant containing an Rpb1 derivative that is defective in the GAL4/UAS assay. Second, CRISPR/Cas9 assay does not allow the assessment of function in a tissue-specific manner. Therefore, potential partial function of an Rpb1 derivative could be missed if a mutant Rpb1 allele fails to support development past the embryonic stage. Third, Rpb1 mutants that fail to develop at an early stage will not provide analyzable material. This might make it challenging to perform certain molecular analyses on these mutants in the absence of the endogenous Rpb1.
Considering these limitations, CRISPR/Cas9 assay should be viewed as a complementary approach to GAL4/UAS assays. Because the success rate of generating a CRISPR-ed Rpb1 allele tends to be much lower than that of integrating a UAS-Rpb1 transgene into the genome, it is best to first perform the GAL4/UAS assays so that one has a rough estimate of how many DsRed G1 transformants are needed to obtain one desired CRISPR-ed candidate.
4. Future perspective
Using the approaches described in this work, we have found a handful of CTD mutants that exhibit varying levels of dysfunction [11–13]. More defective mutants could be identified using the same methods. We envision that follow-up molecular characterization of these defective mutants will shed light on many questions. For example, various genome-wide sequencing analyses will determine if a mutant CTD is defective in transcription initiation, elongation, termination and RNA processing. If so, are these effects gene-specific? Affinity tags can be appended to the CTD following the same mutagenesis schemes to assist in purifying mutant Pol II so that CTD:factor interactions that are perturbed by these mutations can be identified. In addition, suppressors or enhancers of mutant CTD phenotypes can be identified following modifier screens in stable fly lines co-expressing Rpb1i and a CTD mutant specific in the fly wings, or with CRISPR-ed mutants that appear by themselves to function normally.
Highlights.
Drosophila provides a multicellular system for analyzing the effects of CTD mutations
How to design, generate and validate UAS-Rpb1 fly lines harboring CTD mutations
How to modify the endogenous CTD of Drosophila using CRISPR/Cas9
How to assess the functionality of CTD mutations using GAL4/UAS and CRISPR/Cas9
Acknowledgments
We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the transgenic Rpb1 RNAi fly stock used in this study. This work was funded by NIH GM047477 (to D.S.G.). Illustrations of genetic mating schemes in all figures were generated using Genotype Builder [31].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Eick D, Geyer M, The RNA polymerase II carboxy-terminal domain (CTD) code, Chem. Rev 113 (2013) 8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- [2].Corden JL, RNA polymerase II C-terminal domain: Tethering transcription to transcript and template, Chem. Rev 113 (2013) 8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zaborowska J, Egloff S, Murphy S, The pol II CTD: new twists in the tail, Nat. Struct. Mol. Biol 23 (2016) 771–777. doi: 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- [4].Sims RJ 3rd, Rojas LA, Beck DB, Bonasio R, Schüller R, Drury WJ 3rd, Eick D, Reinberg D, The C-terminal domain of RNA polymerase II is modified by site-specific methylation, Science. 332 (2011) 99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, Vedadi M, Min J, Pawson TJ, Blencowe BJ, Greenblatt JF, SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination, Nature. 529 (2016) 48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- [6].Sharma P, Lioutas A, Fernandez-Fuentes N, Wright RHG, Vona CD, Le Dily F, Schüller R, Eick D, Oliva B, Beato M, Arginine Citrullination at the C-Terminal Domain Controls RNA Polymerase II Transcription[J]. Molecular cell, (2019) 84–96. doi: 10.1101/216143. [DOI] [PubMed] [Google Scholar]
- [7].Dias JD, Rito T, Torlai Triglia E, Kukalev A, Ferrai C, Chotalia M, Brookes E, Kimura H, Pombo A, Methylation of RNA polymerase II non-consensus Lysine residues marks early transcription in mammalian cells, Elife. 4 (2015). doi: 10.7554/eLife.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schröder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, Schnölzer M, Cole PA, Geyer M, Bruneau BG, Adelman K, Ott M, Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells, Mol. Cell 52 (2013) 314–324. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Egloff S, Dienstbier M, Murphy S, Updating the RNA polymerase CTD code: adding gene-specific layers, Trends Genet. 28 (2012) 333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- [10].Chapman RD, Conrad M, Eick D, Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation, Mol. Cell. Biol 25 (2005) 7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gibbs EB, Lu F, Portz B, Fisher MJ, Medellin BP, Laremore TN, Zhang YJ, Gilmour DS, Showalter SA, Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain, Nat. Commun 8 (2017) 15233. doi: 10.1038/ncomms15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Portz B, Lu F, Gibbs EB, Mayfield JE, Rachel Mehaffey M, Zhang YJ, Brodbelt JS, Showalter SA, Gilmour DS, Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain, Nat. Commun 8 (2017) 15231. doi: 10.1038/ncomms15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu F, Portz B, Gilmour DS, Interplay between consensus and divergent RNA polymerase II C-terminal domain repeats in viability and targeting, bioRxiv. (2018). https://www.biorxiv.org/content/early/2018/10/21/448506.abstract.
- [14].Brand AH, Perrimon N, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes, Development. 118 (1993) 401–415. https://www.ncbi.nlm.nih.gov/pubmed/8223268. [DOI] [PubMed] [Google Scholar]
- [15].Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, Yang-Zhou D, Shim H-S, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N, A genome-scale shRNA resource for transgenic RNAi in Drosophila, Nat. Methods 8 (2011) 405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D, The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells, Development. 124 (1997) 761–771. https://www.ncbi.nlm.nih.gov/pubmed/9043058. [DOI] [PubMed] [Google Scholar]
- [17].Wodarz A, Hinz U, Engelbert M, Knust E, Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila, Cell. 82 (1995) 67–76. https://www.ncbi.nlm.nih.gov/pubmed/7606787. [DOI] [PubMed] [Google Scholar]
- [18].Capdevila J, Guerrero I, Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings, EMBO J. 13 (1994) 4459–4468. https://www.ncbi.nlm.nih.gov/pubmed/7925288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yeh E, Gustafson K, Boulianne GL, Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila, Proc. Natl. Acad. Sci. U. S. A 92 (1995) 7036–7040. https://www.ncbi.nlm.nih.gov/pubmed/7624365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Munro S, Freeman M, The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD, Curr. Biol 10 (2000) 813–820. https://www.ncbi.nlm.nih.gov/pubmed/10899003. [DOI] [PubMed] [Google Scholar]
- [21].Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P, EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue, Development. 130 (2003) 271–284. https://www.ncbi.nlm.nih.gov/pubmed/12466195. [DOI] [PubMed] [Google Scholar]
- [22].Asha H, Nagy I, Kovacs G, Stetson D, Ando I, Dearolf CR, Analysis of Ras-induced overproliferation in Drosophila hemocytes, Genetics. 163 (2003) 203–215. https://www.ncbi.nlm.nih.gov/pubmed/12586708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bischof J, Maeda RK, Hediger M, Karch F, Basler K, An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schwartz BE, Werner JK, Lis JT, Indirect immunofluorescent labeling of Drosophila polytene chromosomes: visualizing protein interactions with chromatin in vivo, Methods Enzymol. 376 (2004) 393–404. doi: 10.1016/S0076-6879(03)76026-1. [DOI] [PubMed] [Google Scholar]
- [25].Cubillos-Rojas M, Amair-Pinedo F, Tato I, Bartrons R, Ventura F, Rosa JL, Tris-acetate polyacrylamide gradient gels for the simultaneous electrophoretic analysis of proteins of very high and low molecular mass, Methods Mol. Biol 869 (2012) 205–213. doi: 10.1007/978-1-61779-821-4_17. [DOI] [PubMed] [Google Scholar]
- [26].Schägger H, Tricine–SDS-PAGE, Nat. Protoc 1 (2006) 16. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- [27].Rørth P, Gal4 in the Drosophila female germline, Mech. Dev 78 (1998) 113–118. https://www.ncbi.nlm.nih.gov/pubmed/9858703. [DOI] [PubMed] [Google Scholar]
- [28].Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM, Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila, Genetics. 196 (2014) 961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nonet M, Sweetser D, Young RA, Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II, Cell. 50 (1987) 909–915. https://www.ncbi.nlm.nih.gov/pubmed/3304659. [DOI] [PubMed] [Google Scholar]
- [30].West ML, Corden JL, Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations, Genetics. 140 (1995) 1223–1233. https://www.ncbi.nlm.nih.gov/pubmed/7498765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Roote J, Prokop A, How to design a genetic mating scheme: a basic training package for Drosophila genetics, G3. 3 (2013) 353–358. doi: 10.1534/g3.112.004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL, The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro, Proc. Natl. Acad. Sci. U. S. A 85 (1988) 3698–3702. https://www.ncbi.nlm.nih.gov/pubmed/3131761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brickey WJ, Greenleaf AL, Functional studies of the carboxy-terminal repeat domain of Drosophila RNA polymerase II in vivo, Genetics. 140 (1995) 599–613. https://www.ncbi.nlm.nih.gov/pubmed/7498740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR, Type I repressors of P element mobility, Genetics. 135 (1993) 81–95. https://www.ncbi.nlm.nih.gov/pubmed/8224830. [DOI] [PMC free article] [PubMed] [Google Scholar]