Abstract
Transcription elongation through the nucleosome is a precisely coordinated activity to ensure timely production of RNA and accurate regulation of co-transcriptional histone modifications. Nucleosomes actively participate in transcription regulation at various levels and impose physical barriers to RNA polymerase II (RNAPII) during transcription elongation. Despite its high significance, the detailed dynamics of how RNAPII translocates along nucleosomal DNA during transcription elongation and how the nucleosome structure dynamically conforms to the changes necessary for RNAPII progression remain poorly understood. Transcription elongation through the nucleosome is a complex process and investigating the changes of the nucleosome structure during this process by ensemble measurements is daunting. This is because it is nearly impossible to synchronize elongation complexes within a nucleosome or a sub-nucleosome to a designated location at a high enough efficiency for desired sample homogeneity. Here we review our recently developed single-molecule FRET experimental system and method that has fulfilled this deficiency. With our method, one can follow the changes in the structure of individual nucleosomes during transcription elongation. We demonstrated that this method enables the detailed measurements of the kinetics of transcription elongation through the nucleosome and its regulation by a transcription factor, which can be easily extended to investigations of the roles of environmental variables and histone post-translational modifications in regulating transcription elongation.
Keywords: Single-molecule FRET, Transcription elongation, RNA polymerase II, Nucleosome
1. Introduction
Nucleosomes impose physical barriers to many vital nuclear processes such as transcription during which RNA polymerase II (RNAPII) must contend with the tight wrapping of DNA around the histone core [1–5]. Histone-DNA contacts need to be broken during transcription elongation by RNAPII [2, 6–11], and subsequently reformed after its passage to maintain the structural integrity of the genome [3, 12, 13]. Understanding how this complicated task is achieved at such a high efficiency requires the fundamental knowledge on the detailed dynamics of histone-DNA interactions during transcription elongation.
Nucleosome-induced RNAPII pausing [3, 10] that may lead to arrests [3, 14] is arguably the most important determinant of the overall efficiency of transcription elongation through the nucleosome [7, 15–40]. Paused RNAPII can resume spontaneously while arrested RNAPII requires additional factors to be rescued. Based on the pause/arrest points revealed by in vitro transcript assays and the recent cryo-EM structures of RNAPII-nucleosome complexes, several key static structural intermediates of the nucleosome during transcription have been suggested [3, 11, 41, 42]. The results indicate that the interaction between DNA and histone dimer or tetramer induces 4 major pauses and that the dimer and tetramer must dissociate from the DNA at least once during RNAPII passage. In a small population, the histones return to the nucleosome or a sub-nucleosome intermediate upon RNAPII passage. These changes are relevant to moderately transcribed genes as heavily transcribed genes may have the nucleosomes completely disassembled because of frequent RNAPII invasion [43]. Most genes are moderately transcribed, and therefore, a method to investigate the complex dynamics of transcription elongation through the nucleosome is essential to investigating roles of the inherent nucleosome structure, post-translational modifications to histone, environmental variables, and transcription factors in regulating the overall efficiency and rate of transcription elongation.
The difficulty associated with investigating the details of a process as long and complex as transcription elongation through the nucleosome arises from the facts that a signal from an ensemble is convolved with heterogeneous and unsynchronized signals and that synchronizing an ensemble to a single state is often impossible. Single-molecule methods provide a powerful means to interrogate such a complex process in real-time in a time-resolved manner. This review is to focus on our single-molecule experimental systems and methods that we reported to investigate nucleosome dynamics [44–57]. In particular, we recently reported an experimental system and method with which we can investigate the real-time dynamics of RNAPII translocation along nucleosomal DNA [56]. The frequency and duration of pauses and the rate of elongation are two direct outcomes of such monitoring. The FRET-based method described here enables the monitoring of RNAPII progression through a nucleosome particle without any perturbation such as a mechanical force. Previous studies based on single molecule force measurements reported various aspects of the nucleosomal barriers during transcription elongation [58, 59]. They reported that histone-DNA contacts play essential roles in regulating elongation kinetics and that transcription factor IIS (TFIIS) inhibits pauses while IIF (TFIIF) helps escape from pauses. These studies rely on the measurements of the length of stretched DNA by optical tweezers. The applied force would facilitate overcoming the nucleosomal barriers and subsequently accelerate transcription elongation, and more importantly, abolish or significantly attenuate smaller barriers. This limitation results in coarse nucleosome barrier mapping which does not allow for probing individual histone-DNA barriers. Our method that is based on single-molecule FRET (smFRET) can be employed to monitor the elongation kinetics through various histone-DNA contacts within a nucleosome. The smFRET method described here is generally applicable to studies of protein-DNA interactions that depend on or result in structural changes of the DNA.
2. Methods and Materials
2.1. Transcription template preparation
The transcription template is constructed by ligating a fragment that will allow for end-initiated transcription to a high affinity nucleosome-positioning sequence [56]. We use the 601 nucleosome positioning sequence [60] due to its high affinity and because many studies have used it to map RNAPII arrest sites (Fig. 1). Labeled nucleosomal DNA (~147bp ds-DNA) is produced by ligating 5 overlapping fragments with a length of 40 ~ 75 nucleotides [56]. The amount of DNA obtained by this method is typically on the order of 1 nmole which is far too small for ensemble kinetics measurements but enough for many single molecule measurements (1 nmole can be used for 20 to 200 thousand measurements in our setup that require 20 ~ 40 μL × 0.1 ~ 1 nM nucleosome). This is one of the most distinct benefits of a single-molecule method.
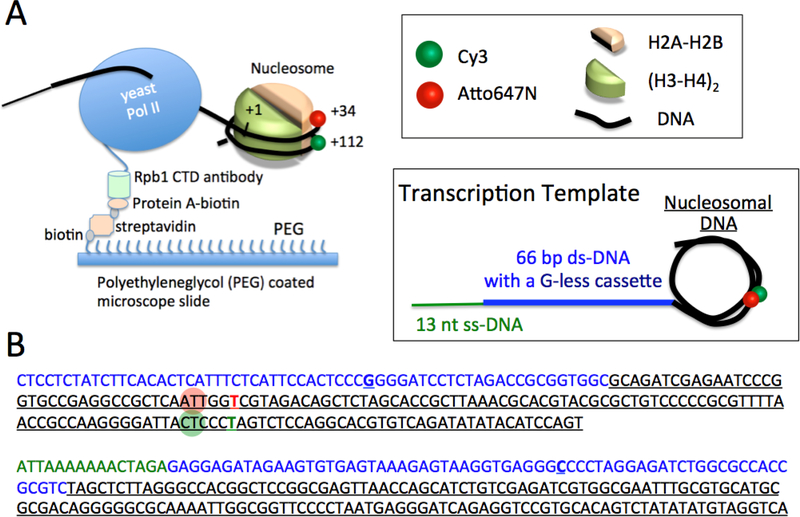
Figure 1. The schematics of the single-molecule FRET system to investigate the dynamics of the nucleosome structure during transcription elongation.
(A) Nucleosome assembled on the shown template is complexed with RNAPII (yeast Pol II). Rpb1 CTD antibody is immobilized on a microscope slide surface via biotinylated Protein A that is conjugated to streptavidin coated on the slide. A FRET pair (Cy3 and Atto647N) is labeled at the +34th and +112th nucleotide. This FRET pair location is sensitive to the nucleosomal dynamics at the proximal-dimer/DNA contact region. (B) The sequence of the upper and lower strands of the transcription template illustrated in A.
Typically two DNA fragments are labeled with a fluorophore to form a FRET pair in the nucleosome. Fluorophore labeling positions can be designed according to the histone-DNA contact points of interest. We prefer using Thymine nucleotide modified at the base with additional C6 linker terminated with an amine group (IDT Inc., Coralville IA). This strategy for fluorophore labeling is one of the most efficient and non-intrusive ways for nucleosome labeling. As the 601 sequence has many Thymine bases, the nucleosome can be labeled at virtually any region of DNA-histone contacts of interest. For instance, DNA unwrapping and rewrapping around a histone H2A-H2B dimer-DNA contact region can be investigated with a nucleosome labeled at the +34th nucleotide (Fig. 1, Thymine base at the +34th position from the entry site of the nucleosome) with a FRET acceptor (e.g. Atto647N or Cy5) and at the +112th nucleotide with a FRET donor (e.g. Cy3). Our fluorophore labeling protocol follows a standard amine conjugation reaction with an NHS-ester functionalized fluorophore.
2.1.1. Template purification
We purify the FRET-labeled nucleosomal DNA construct with a PCR purification kit (QIAquick PCR Purification Kit, Qiagen) to remove ligase. The ligation reaction is nearly complete as shown in figure S1. We add the flanking EC42 DNA (blue/green-colored in Fig. 1B) with a sticky end to the nucleosomal DNA by ligation. Two-step ligation makes it efficient and economical to alter the linker part of the template once the nucleosome portion of the template is prepared. The EC42 DNA can be synthesized easily. The near complete ligation of the nucleosomal DNA can be achieved because the overlapping reagion between fragments have Tm >60 °C. Unlike the nucleosomal DNA ligation, EC42 ligation to the nucleosomal DNA relies on hybridization of a short overhang (4 bp), and therefore, its yield is low. We purify the final product by native PAGE. This DNA construct is used for nucleosome reconstitution. This template does not result in any noticeable heterogeneity in the initial FRET signal due to nucleosome positioning heterogeneity. This is expected from the strong and homogeneous nucleosome positioning property of the 601 sequence as was previously reported with a flanking 70 bp linker [61].
EC42 Template Strand:
5’P−CTGCGCCACCGCGGTCTAGAGGATCCCCGGGAGTGGAATGAGAAATGAGTGTGAAGATAGAGGAGAGATCAAAAAAATTA 3’
EC42 Non-template Strand:
5’CTCCTCTATCTTCACACTCATTTCTCATTCCACTCCCGGGGATCCTCTAGACCGCGGTGGC 3’
Top strand (labeled with Cy5 at +35 and with Cy3 at +112)
GCAGATCGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCAGT
Bottom strand
ACTGGATGTATATATCTGACACGTGCCTGGAGACTAGGGAGTAATCCCCTTGGCGGTTAAAACGCGGGGGACAGCGCGTACGTGCGTTTAAGCGGTGCTAGAGCTGTCTACGACCAATTGAGCGGCCTCGGCACCGGGATTCTCGAT
An equal molar amount of template oligo is added and the two strands are annealed by incubating at 95°C for 5 minutes followed by gradual cooling in 10°C steps for 5 minutes at each step in a thermal cycler (MJ Mini™ Personal Cycler, Part # PTC1148EDU, Bio-Rad Laboratories, Inc.). The annealed EC42 template is then purified on a 12% native PAGE in 0.5X TBE by a crush and soak method. The purified DNA is re-suspended in a volume of 20 μL 1/10th TE. This is done to maintain high concentrations of both DNA’s prior to ligation.
A two fold molar excess of annealed EC42 (5 – 10 μg) with phosphorylated template strand is mixed with fluorescently labeled 601 nucleosomal DNA construct and ligated overnight in 2.5 μL ligation buffer and 20 Units of T4 DNA ligase (Promega) at 15°C.
The template is gel purified on a 2% agarose gel in 1.5X TAE using a gel purification kit (Qiagen). The transcription template is eluted in TE 8.0 (20 – 30 μL) and quantified by nanodrop.
2.1.2. Nucleosome reconstitution
The nucleosome template is reconstituted using the histone chaperone Nap1 as follows.
Recombinant yeast histone octamer (6 pmole, purified and folded by published protocols [62]) is mixed with 36 pmole of Nap1 (purified by a published protocol [63]) in 10 mM Tris-HCl (pH 7.8) with 50 mM NaCl, 0.5 mM MgCl2 and 3 % glycerol to make a total volume of 10 μL.
The mixture is incubated for 2 hours at 25 °C and then a stoichiometric amount of the transcription template is added.
The reaction is incubated for 4 hours at 25 °C and then stored at 4 °C.
The nucleosome template upon reconstitution is subject to quality check by native gel electrophoresis (4 % PAGE at 0.2x TBE buffer, figure S2).
2.2. Microscope slide preparation
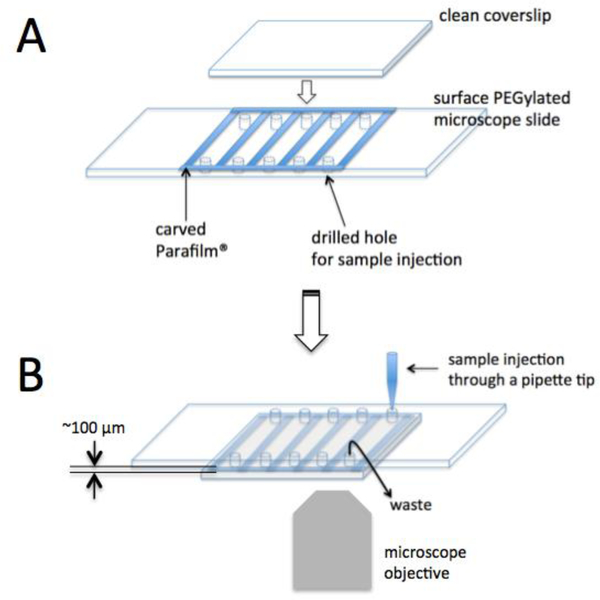
Our single-molecule FRET measurements are performed on the surface of a quartz microscope slide that is a part of a 100 μm thick fluidic channel (Fig. 2). In order to make a slide with five fluidic channels, we first drill holes on the slide that will be used for sample injection. A typical batch size is 5 slides and 10 coverslips. We use a diamond drill bit (Starlite Industries, Rosemont PA, Part # 115005) on a benchtop drill press (Cameron Micro Drill Press, Sonora, CA, Model # 163) to drill 10 holes (5 inlets and 5 outlets, see figure 2) per slide. We subsequently clean the microscope slides and glass coverslips as follows.
A set of 5 new quartz slides and 10 coverslips is sequentially sonicated in acetone, dichloromethane, and methanol for 15 min each, followed by thorough rinsing with deionized distilled water (dd-water from Nanopure™, ThermoFisher Scientific).
A propane torch is used to flame the surfaces of the slides and coverslips. In case of the coverslips, expose the surfaces only briefly (<1 sec) to the flame because prolonged exposure can warp the surface. Place the flamed slides into a glass staining rack and coverslips to a porcelain or glass staining rack (e.g. Thomas Scientific, Swedesboro, NJ, product no. 8542E40). Put the coverslip containing rack to a glass beaker.
Prepare a glass cleaning solution by mixing half a pack of Nochromix® (Sigma-Aldrich) into 500 mL 100 % sulfuric acid.
Carefully pour the cleaning solution to the slide staining rack and the beaker containing the porcelain staining rack and leave them overnight.
Thoroughly wash the slides, coverslips, and the slide staining rack with dd-water. Use filtered 99.99 % nitrogen gas to blow water drops of the slides, if necessary.
Store the coverslips in a desiccator kept under vacuum.
Figure 2. A schematic description of reaction chamber fabrication.
(A) A carved Parafilm® is placed on a microscope slide with 10 holes for sample injection and washing. A clean coverslip is placed on top of the film layer and the sandwich is subsequently heated at 110 °C for 90 seconds to fabricate 5 reaction chambers. (B) The slide reaction chamber is place upside down on an inverted microscope to monitor the surface immobilized elongation complexes.
Next, we coat the slides with Polyethyleneglycol (PEG) as follows.
Place 100 mg Silane-PEG (Mw 5000, Laysan Bio Inc., Arab AL) and 1 mg Silane-PEG-Biotin (Mw 5000, Laysan Bio Inc., Arab AL) into a 500 mL beaker.
Add 100 mL anhydrous acetonitrile to the beaker to dissolve PEG as quickly as possible and pour the solution into the slide rack promptly.
Sonicate the slides in the PEG solution for 30 minute in a bath sonicator (e.g. Bransonic® CPX-952–318R, Branson).
Sonicate the slides in acetonitrile twice for 5 minute each.
Rinse the slides thoroughly with dd-water. Use filtered 99.99 % nitrogen gas to blow water drops from the slide, if necessary.
Store the slides in a desiccator kept under vacuum.
We construct fluidic channels on a PEG coated slide right before measurements. We use a carved double-sided tape or Parafilm® sheet (Sigma-Aldrich) to construct the walls for the channels (see figure 2B for the shape of the carved Parafilm® sheet). The top ceiling of the channel is one of the clean glass coverslips. In case of using double-sided tape for the channel walls, a glass coverslip is gently pushed against the tape walls to complete channel construction. In case of Parafilm®, we heat the slide/coverslip pair with a carved Parafilm® sheet sandwiched between them at 110 °C for 90 seconds.
2.3. Microscope setup
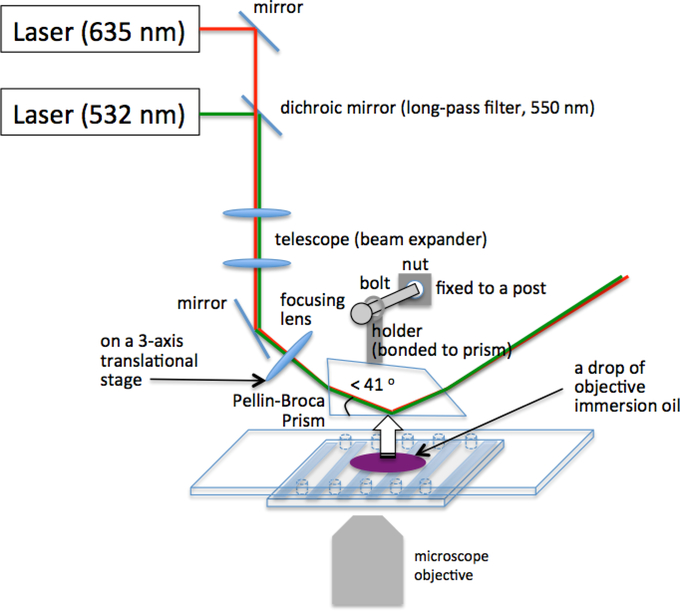
For smFRET measurements, we utilize a prism-coupled total internal reflection fluorescence (TIRF) microscopy geometry for fluorophore excitation. We have an excitation laser beam expanded with a telescope (Fig. 3) and re-focused with a focusing lens right next to the microscope stage (Fig. 3) so that the size and location of the illumination area can be easily changed when necessary. Figure 3 shows the schematics of our instrumental setup. Our setup is on a commercial microscope (Nikon TE2000, Japan) with two lasers at wavelengths 532 nm (150 mW, CrystaLaser, Reno, NV) and 635 nm (100 mW, Cube, Coherent Inc., Santa Clara CA). We use a telescope lens pair to expand a laser beam and re-focus to a target illumination area on the slide that is optically coupled with a Pellin-Broca prism made of quartz (CVI Laser Optics, PLBC-10.0–77.2, 10 mm thickness, Albuquerque, NM) by a thin layer of fluorescence-free objective immersion oil (Cargille Labs, Cedar Grove, NJ, type FF). The lenses and mirrors can be purchased from Thorlabs Inc. (Newton, NJ) and the dichroic mirrors and microscope fluorescence filters can be purchased from Chroma Technology (Bellows Falls, VT). A quartz prism provides far less scattering and fluorescence background than a glass prism does. The prism is mounted via a holder (Fig. 3) that is mechanically separate from the microscope stage that can be moved freely to locate the illumination area to a desired sample spot on the slide.
Figure 3. Fluorescence excitation source, optics, and their alignment.
Detailed description can be found in the main text. The objective must have a long enough working distance to ensure optical access to the elongation complexes immobilized upside down on the slide surface. Our setup is equipped with Nikon CFI Plan APO VC 60x water immersion objective (NA = 1.2, working distance 0.28 – 0.31 mm).
Alignment of the illumination beam to the center of the microscope field of view is achieved via following steps.
Separate the telescope lens pair by the sum of their focal lengths. The ratio of the two focal lengths determines the magnification of the laser beam. Our lasers needed 4~6 times of magnification to expand their beam sizes to 1~1.5 cm diameter. For this, we use one lens with a 2.5 cm focal length (part # LA1951-A, Thorlabs Inc., Newton, NJ) and the other with a 12.5 cm focal length (part # LA1384-A, Thorlabs Inc., Newton, NJ). The lenses have anti-reflection coating for the visible wavelength of light.
Place the focusing lens (Fig. 3) (e.g. part # AC508–150-A, Thorlabs Inc., Newton, NJ) at an approximate focal length from the left rim of the objective lens top and at an angle slightly below the TIRF critical angle (~41 degrees from the microscope stage, Fig. 3).
Make a fluorescent bead slide. To a flow channel of a biotinylated sample slide, inject a 0.01 % solution of fluorescent beads coated with streptavidin (ThermoFisher Scientific, F8777). Flush with dd-water.
Mount the bead slide on the microscope and add a drop of the immersion oil on its top. Put the prism on top of the oil and fasten the bolt to fix its position.
Locate the laser beam at the center of the objective lens as precisely as possible via visual inspection through the prism by moving the 3-axis translational stage where the focusing lens is mounted.
Look in to the microscope eyepiece and see if any fluorescence emission is observed. By randomly moving the focusing lens up, down, left, and right, locate the illumination spot at the center of the field of view.
By moving the focusing lens further or closer to the slide along the optical axis (i.e. the laser beam path), adjust the size of TIR illumination area.
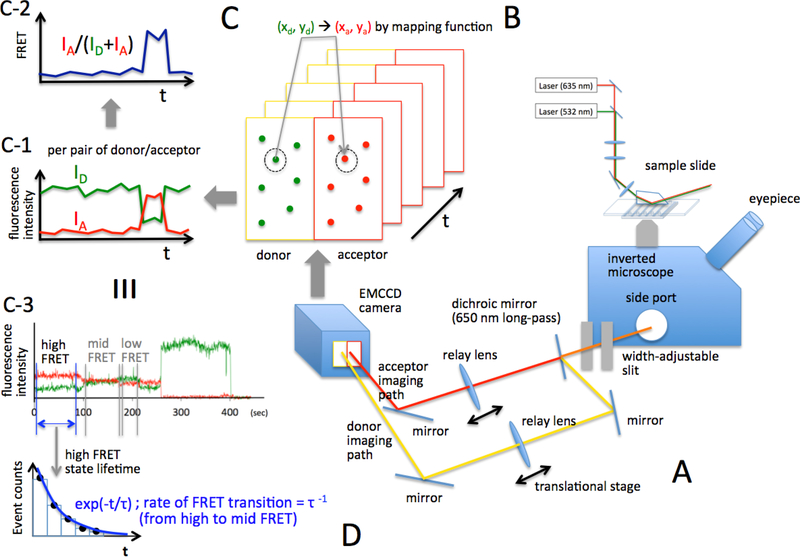
The next step is to align the imaging optics. We currently have two- and three-color [49, 50] smFRET imaging setups. In the two-color smFRET setup (Fig. 4A), the imaging optic system is composed of a width-adjustable slit, a fixed dichroic mirror, three mirrors mounted on a kinematic mount (part # KM100, Thorlabs Inc., Newton, NJ), two relay-lenses (part # AC508–100-A, Thorlabs Inc., Newton, NJ) mounted on a single-axis translational stage (Thorlabs Inc., XR25C, Newton, NJ) (Fig. 4A). Imaging optic alignment can be achieved via following steps.
Position the tube lenses at a 2x focal length from the imaging plane of the side port of the microscope.
Place the charge-coupled-device (CCD) area of an electron-multiplying CCD (EMCCD) camera (Photometrics, Tucson, AZ, Cascade II:512, or Andor Technology, Belfast Ireland, iXonEM+897(back-illuminated)) at a 2x focal length from the tube lens.
Image the fluorescent beads on the camera.
Roughly point the image in the donor spectral range to the left side of the camera chip by adjusting the angles of the kinematic mirror mount. Adjust the slit size and location to make the image fill the left half of the chip.
Focus the image by moving the corresponding tube lens back and forth and adjust the mirror mount and the slit width again to make the image fill the left half of the chip.
Roughly point the image in the acceptor spectral range on the right side of the camera chip by adjusting the angles of the kinematic mirror mount.
Focus the image by moving the corresponding tube lens back and forth.
If the donor image size is approximately the same as the acceptor image size, alignment is done. Otherwise, move the camera slightly and repeat steps 4 to 7. If the donor image is smaller than the acceptor image, move the camera slightly closer to the microscope.
Figure 4. Imaging optics alignment and the process of data collection and analysis.
(A) Imaging optics alignment. A fluorescence image out of the side port of an inverted microscope is passed through a width-adjustable slit. The image is split into two colors (Cy3 and Atto647N) with a 650 nm dichroic mirror. Each image is reformed by a relay lens on another conjugate image plane that is located at the CCD camera. (B) The reformed two-color images at the CCD camera are spatially separate (left: Cy3, right: Atto647N). A typical image is shown in figure S3. A time series of the images are taken and recorded as a movie. (C) On each movie frame, all pairs of Cy3 and Atto647N spots are identified. Each FRET pair at each movie frame gives the fluorescence intensities of the donor and the acceptor. A series of the fluorescence intensities plotted along time makes fluorescence intensity time trajectories for the donor and acceptor (C-1), which is used to generate a FRET time trajectory (C-2). C-3 shows a computer-simulated example of how data looks. (D) An illustration to show how a FRET state lifetime is measured. Detailed descriptions can be found in the main text.
The principles of aligning the three-color setup are the same as the above with additional spectral range. Two images from two separate spectral ranges may have a different size from each other. The following steps are to map the donor coordinates to the acceptor coordinates (Fig. 4C).
Take an image of the fluorescent beads.
Pick one bead on the donor side and determine its center precisely by fitting the intensity pattern to a 2D Gaussian distribution or to a centroid function.
Identify the same bead on the acceptor side and determine its center precisely by fitting the intensity pattern to a 2D Gaussian distribution or to a centroid function.
Pick another bead and repeat steps 3 to 4. Iterate this process as many beads as possible.
With the set of donor coordinates mapped to the acceptor coordinates, determine the function to map a donor coordinate pair (xd, yd) to the acceptor coordinate pair (xa, ya), such that xa = a1xd + a2yd + a3xdyd and ya = a4xd + a5yd + a6xdyd. For this, at least three beads whose centers have been determined precisely in the donor and the acceptor sides are required (i.e. at least three pairs of (xd, yd) mapped precisely to (xa, ya)). With the three sets of mapped coordinates, solve the above two equations for coefficients ai (i = 1 ~ 6). If more than three beads have been mapped, use least squares estimation to determine coefficients.
The principles of mapping coordinates between two spectral ranges in the three-color setup are the same as the above with additional spectral range. Two independent mapping functions need to be determined. One is to map the donor coordinates to the first acceptor coordinates, and the other is to map the first acceptor coordinates to the second acceptor coordinates.
2.4. Elongation Complex Formation and Data collection
The data we collect out of this system is time-resolved donor/acceptor fluorescence (or FRET) intensities during transcription elongation from individual nucleosomes. To collect data exclusively from an active elongation complex, we immobilize the elongation complexes (ECs) via CTD of the Rpb1 subunit of RNAPII on the microscope slide (Fig. 1A). RNAPII is purified from source as described in the companion manuscript and another publication [64, 65].
2.4.1. Preparation of slides for RNAPII immobilization
PEG-biotin coated slides prepared as described in section 2.2 are coated with streptavidin by injecting 50 μL 0.1 mg/ml streptavidin in buffer A (10 mM Tris-HCl at pH 7.8 containing 10 mM NaCl) into a sample chamber followed by washing with 50 μL blank buffer A.
20 μL biotin-protein A at 0.1 mg/ml in buffer A is injected and incubated for 15 min followed by washing with 40 μL blank buffer A.
Purified 8WG16 IgG (Biolegend) is diluted in buffer A to 0.1 mg/ml and 50 μL is injected and incubated for 2 hrs, followed by washing with 50 μL blank buffer A.
2.4.2. Elongation complex formation
RNAPII and the nucleosome template prepared as described in section 2.1 are mixed and diluted to 10 nM and 9.5 nM, respectively, using transcription buffer (50 mM HEPES at pH 7.5 containing 150 mM KCl, 1 mM MnCl2, 0.5 mM DTT, 10% glycerol, 0.01 mM UpG primer, 0.1 ug/μL BSA, and 1 unit/μL RNAsin) to make the total volume of 13 μL.
The mixture is incubated for 5 min at 23°C.
Transcription is initiated by adding 5 μL of a nucleotide mix containing 0.4 mM ATP, CTP, and UTP. The mixture is incubated for 30 min at 23°C.
The EC complexes are diluted 3-fold in transcription buffer and then injected into the chamber and incubated for 5 min to capture RNAPII through the immobilized 8WG16. The free templates are washed out with transcription buffer.
2.4.3. Collecting FRET data
A 40 μL solution containing a complete NTP mix at 0.1 mM, TFIIS at 30 nM, PCA (protocatechuic acid, Sigma-Aldrich) at 4 mM, PCD (protocatechuate dioxygenase, Sigma-Aldrich) at 0.4 u/ml, and Trolox (Sigma-Aldrich) at 1 mM in transcription buffer is then injected. TFIIS is added to rescue the significant fraction of ECs that backtracked during surface immobilization. TFIIS increases the number of elongation complexes released from the G-induced stop and those whose dynamics can be observed within the nucleosome. PCD/PCA/Trolox is added to reduce fluorophore photobleaching and blinking.
FRET signals are recorded as a series of fluorescence images in a movie. In the presence of PCD, PCA, and Trolox, DNA-labeled fluorophores can last for more than 3 minutes up to 5 minutes under an excitation power yielding a minimum 5.0 signal-to-noise ratio at 100 ms imaging duration.
FRET data is collected for 20 minutes. Within the 20 min, 4 consecutive stacks of images are taken from 4 different sets of ECs on 4 different surface regions each of which generates a 5 min FRET movie containing 10 ~ 50 actively transcribing ECs. At the end of each 5 min data collection period, the surface is illuminated with a 635 nm laser in order to examine whether an extinguished FRET acceptor is due to completely unwrapped DNA or photobleaching (Fig. 5A).
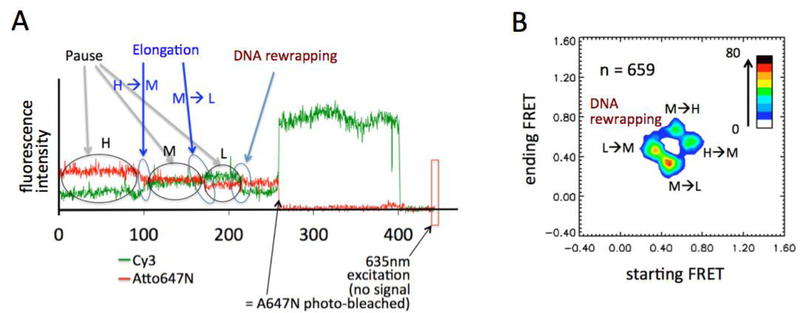
Figure 5. A typical smFRET time trajectory and a FRET transition histogram during transcription elongation.
(A) A typical smFRET time trajectory reveals pauses and elongation between pauses. Detailed description can be found in the main text. (B) FRET transition histogram obtained from 659 FRET transitions. “H”, “M”, and “L” FRET states represents high-, mid-, and low-FRET states, respectively. Further details can be found in the main text.
3. Data processing, analysis and interpretation
3.1. Data processing
In order to construct a FRET time trajectory from an EC, we compute the FRET donor and acceptor intensities from individual ECs in a sequence of frames from the captured movie. On the first movie frame, we identify a donor fluorophore. The pixel values from a 9 × 9 pixel matrix (or 5 × 5 matrix if 3 × 3 binned) centering at the identified donor are sorted and a set of 5 pixel values centering around the median value is averaged and used as the background fluorescence. Each pixel value in the matrix is subtracted with the background fluorescence. The 15 (or 5 in case of 3 × 3 binned images) brightest pixel values within the matrix are summed up to give the donor intensity at the specific time point when the movie frame is taken. We identify the corresponding FRET acceptor spot according to the mapping function as described in 2.3. Using the same method we compute the acceptor intensity. These two intensities result in a FRET efficiency according to the formula IA / (ID + IA), where I is the fluorescence intensity. One can try to be more accurate with the distance measurement by correcting this ratio with the quantum efficiency ratio such as in IA / (γID + IA), where is γ is the quantum efficiency ratio between the donor and the acceptor. In our setup, it is unnecessary to perform this correction, as accurate FRET distance measurement is not pursued or needed. Moreover, the fluorophores are conjugated through a linker and the quantum efficiencies depend often significantly on the fluorophore environment, which makes this correction and consequently accurate distance measurements nearly impossible. This FRET efficiency measurement is repeated at each time point. The time series of the FRET efficiencies measured from the time course of movie frames make an smFRET time trajectory from one EC (Fig. 4C–1). This process is repeated for the entire set of ECs identified on the first movie frame. This process is illustrated in figure 4C.
Another correction we make before we compute the FRET efficiencies of an EC is an inter-channel leakage correction. Donor emission spectrum cannot be completely separated from acceptor emission with a single dichroic mirror. One can easily calibrate this inter-channel leakage by measuring the fluorescence intensities in the donor and acceptor channels from a DNA fragment labeled only with the donor. In our setup, 4% of donor fluorescence leaks to the acceptor channel. Therefore, the equations we use to compute the FRET efficiency at a given time point from a donor-acceptor pair is (IA - δID) / (ID + IA), where δ is the inter-channel leakage correction factor, or 0.04 in our setup.
Finally, we correct the residual background fluorescence by measuring the donor and acceptor intensities after they photobleach. An average of 5~50 intensities after photobleaching is subtracted from the entire time trajectories of the donor and acceptor intensities before FRET efficiencies are computed at each time point. The constructed time series of FRET efficiencies per donor/acceptor pair, or their FRET efficiency time trajectory (Fig. 4C–2), reveals how the distance between the FRET pair is changing over time. An example of real FRET time trajectory data is shown in figure 4C–3.
3.2. Data analysis for the kinetics of transcription elongation
Upon obtaining FRET time trajectories from individual ECs, we extract the kinetic rate constants of transcription elongation relevant to the changes taking place near the FRET location. We use FRET state lifetime analysis or hidden Markov model (HMM) analysis depending on the level of noise and complexity of the kinetic scheme.
For simple kinetic scheme comprising two or three states where each state shows a unique FRET efficiency, we cut FRET time trajectories based on their FRET efficiencies and measure their average lifetimes (Fig. 4C–3). We do not include a region with a gradually changing FRET to any specific state. There is always an error involved in somewhat arbitrary cuts of state durations. In the end, we bin the durations and as such the error is dominated by coarse binning of the histogram, not by the precision of the individual lifetime estimates. We typically fit the lifetime histogram of each state with a single exponential decay function. A good fit confirms that a given FRET state corresponds to a single physical state. We then count the number of transitions from a given state to another. One can use the following formula to compute the kinetic rate constant for each transition.
, where k is the rate constant for transition form state i to state f, τ is the lifetime of state i, and ni→j is the number of transitions from state i to state j.
For example, if one state (state 1) has a lifetime of 2 sec and it transitions 60 times during observation among which 15 times goes to another FRET state (state 2) and 45 times to a different FRET state (state 3), the kinetic rate constant for state 1 to 2 transition, and that for state 1 to 3 transition, .
This lifetime analysis takes a long time. It works well only when there are two or three states in the system so that it is easy to visually inspect a FRET time trajectory and assign a period with a constant FRET efficiency to a FRET state (Fig. 4C–3). If the amount of data is large and the number of FRET states is more than 2, we can use a hidden Markov model (HMM) analysis [54, 56, 66]. The benefits of HMM are that the analysis can be applied to a large amount of data and that the result does not depend on visual inspection. Regardless of the choice of the method between the lifetime analysis and HMM analysis, we need a kinetic model to analyze the data, which is not an issue with hypothesis-driven investigations. HMM analysis results in the kinetic rate constants of all possible transitions among FRET states whose number should be given as a model parameter. Details of the analysis principle and method have been published, which is what we used for transcription elongation kinetics analysis [54, 56].
In both lifetime analysis and HMM analysis, one can identify the time points of state transitions from a data set. At these transition points, we can identify the FRET efficiencies before and after the transitions that can be used to build a 2D transition histogram from an entire data set. A 2D FRET transition is a heat map that shows the frequency of FRET transitions between any given pair of FRET efficiencies. It is often necessary to show the transition dynamics observed in the entire data set. For this purpose, we use a 2D FRET transition histogram (Fig. 5B).
3.3. Data Interpretation
A sample data set that we obtained from the above described setup and method is shown in figure 5. The flat regions in the time trajectories (Fig. 5A) are where the elongation complex paused. Between pauses, gradually changing fluorescence intensities are observed, suggesting that the DNA is unwrapping as the elongation complex translocates. Gradual dissociation of H2A-H2B with no change in DNA is another possibility, which is extremely unlikely considering the structure of the nucleosome and according to our published data even at an elevated salt concentration [49]. If it is still desired to rule out this possibility, a control experiment can be carried out with a FRET pair labeled at both gyres of DNA near the dimer-DNA contact region. The FRET trajectories should not show any dynamics if the nucleosomal change were dimer dissociation only. In some cases, unwrapped DNA rewraps to show increasing FRET (at the ~215 sec point in figure 5A). In the shown trajectory, we named the apparent three FRET states as high- (H), mid- (M), and low- (L) FRET states, respectively, according to their relative FRET efficiencies. A 2D FRET transition histogram constructed from a set of fluorescence trajectories is shown in figure 5B, verifying that the system has only these three FRET states. FRET efficiency alone cannot be used to assign an elongation complex to a unique physical state. Instead, the fluorescence time trajectories of the FRET donor and acceptor from various parallel and control experiments in conjunction with other data (e.g. cryo-EM structure) are often necessary. For example, the apparent FRET efficiencies of the high-FRET states in the beginning (0 – 90 sec) and at the end (215 – 250 sec) of the time trajectory are nearly identical while the absolute intensities of the Cy3 and Atto647N are not. This result suggests that before and after the DNA rewrapping the nucleosome may not be in the same physical state, which can be investigated further. As it is most often impossible to analytically explore the enormous parameter space of such a complex system, hypotheses are necessary to restrict it. In the above example, two viable hypotheses would be that upon RNAPII passage DNA rewrapping around the histone core is non-canonical or that the histone core is damaged. One can test these hypotheses based on nucleosomal changes monitored at various FRET locations that are designed specifically for these hypotheses.
4. Conclusions and Outlook
As evident from our published results, our system can provide unprecedentedly detailed and clear information on the kinetics of transcription elongation through a nucleosome particle. The fact that we can label fluorophores at virtually any location within a nucleosome enables us to investigate the roles of any given specific DNA-histone interaction in regulating transcription elongation. These interactions are the most important determinants for inducing pauses and arrests that essentially govern the overall efficiency of transcription. Our system will greatly facilitate investigating the mechanisms of how the nucleosome structure during transcription elongation and consequently the pauses and arrests are regulated by various transcription factors, environmental variables, and histone modifications.
Supplementary Material
Highlights.
The method enables investigation of transcription dynamics through the nucleosome.
The method is based on single-molecule FRET in a highly refined in vitro system.
The method measures RNAPII nucleosomal pause frequencies and durations.
The method measures RNAPII elongation rates within the nucleosome.
Acknowledgements
This publication was supported by the National Institutes of Health [grant number R01GM123164].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bunch H, TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release, Nat Struct Mol Biol, 21 (2014) 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD, Architecture of RNA polymerase II and implications for the transcription mechanism, Science (New York, N.Y.), 288 (2000) 640–649. [DOI] [PubMed] [Google Scholar]
- [3].Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M, Nature of the nucleosomal barrier to RNA polymerase II, Mol Cell, 18 (2005) 97–108. [DOI] [PubMed] [Google Scholar]
- [4].Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P, Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA, Mol Cell, 34 (2009) 710–721. [DOI] [PubMed] [Google Scholar]
- [5].Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C, DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC, Mol Cell, 24 (2006) 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Krumm A, Hickey LB, Groudine M, Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation, Genes Dev, 9 (1995) 559–572. [DOI] [PubMed] [Google Scholar]
- [7].Adelman K, Lis J, Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans, Nature Rev. Genet, 13 (2012) 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C, Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner, Nature, 446 (2007) 820–823. [DOI] [PubMed] [Google Scholar]
- [9].Brueckner F, Armache KJ, Cheung A, Damsma GE, Kettenberger H, Lehmann E, Sydow J, Cramer P, Structure-function studies of the RNA polymerase II elongation complex, Acta crystallographica. Section D, Biological crystallography, 65 (2009) 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II, Science (New York, N.Y.), 325 (2009) 626–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM, Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II, Nat Struct Mol Biol, 16 (2009) 1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kristjuhan A, Svejstrup JQ, Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo, EMBO J, 23 (2004) 4243–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chang HW, Kulaeva OI, Shaytan AK, Kibanov M, Kuznedelov K, Severinov KV, Kirpichnikov MP, Clark DJ, Studitsky VM, Analysis of the mechanism of nucleosome survival during transcription, Nucleic acids research, 42 (2014) 1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gaykalova DA, Kulaeva OI, Volokh O, Shaytan AK, Hsieh FK, Kirpichnikov MP, Sokolova OS, Studitsky VM, Structural analysis of nucleosomal barrier to transcription, Proc Natl Acad Sci U S A, 112 (2015) E5787–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kwak H, Lis JT, Control of transcriptional elongation, Annu. Rev. Genet, 47 (2013) 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jonkers I, Lis JT, Getting up to speed with transcription elongation by RNA polymerase II, Nature reviews. Mol Cell biology, 16 (2015) 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Veloso A, Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications, Genome Res, 24 (2014) 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou Q, Li T, Price DH, RNA polymerase II elongation control, Annu. Rev. Biochem, 81 (2012) 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alexander R, Innocente S, Barrass J, Beggs J, Splicing-dependent RNA polymerase pausing in yeast, Mol. Cell, 40 (2010) 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ardehali MB, Spt6 enhances the elongation rate of RNA polymerase II in vivo, EMBO J, 28 (2009) 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C, The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes, Nat Struct Mol Biol, 18 (2011) 1394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Buckley MS, Kwak H, Zipfel WR, Lis JT, Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation, Genes Dev, 28 (2014) 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Danko C, Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells, Mol. Cell, 50 (2013) 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mata M, l. de, A slow RNA polymerase II affects alternative splicing in vivo, Mol. Cell, 12 (2003) 525–532. [DOI] [PubMed] [Google Scholar]
- [25].Moehle EA, Braberg H, Krogan NJ, Guthrie C, Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate, RNA Biol, 11 (2014) 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Min IM, Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells, Genes Dev, 25 (2011) 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gardini A, Integrator regulates transcriptional initiation and pause release following activation, Mol. Cell, 56 (2014) 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gilchrist D, Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation, Cell, 143 (2010) 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gilchrist D, Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks, Genes Dev, 26 (2012) 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hazelbaker D, Marquardt S, Wlotzka W, Buratowski S, Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination, Mol. Cell, 49 (2013) 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ip J, Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation, Genome Res, 21 (2011) 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jishage M, Transcriptional regulation by Pol II(G.) Involving mediator and competitive interactions of Gdown1 and TFIIF with pol II, Mol. Cell, 45 (2012) 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Galburt EA, Grill SW, Bustamante C, Single molecule transcription elongation, Methods (San Diego, Calif.), 48 (2009) 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nudler E, RNA polymerase active center: the molecular engine of transcription, Annu Rev Biochem, 78 (2009) 335–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Finkelstein IJ, Greene EC, Molecular traffic jams on DNA, Annual review of biophysics, 42 (2013) 241–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu X, Bushnell DA, Kornberg RD, RNA polymerase II transcription: structure and mechanism, Biochimica et biophysica acta, 1829 (2013) 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reese JC, The control of elongation by the yeast Ccr4-not complex, Biochimica et biophysica acta, 1829 (2013) 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou J, Schweikhard V, Block SM, Single-molecule studies of RNAPII elongation, Biochimica et biophysica acta, 1829 (2013) 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C, Molecular mechanisms of transcription through single-molecule experiments, Chemical reviews, 114 (2014) 3203–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cheung AC, Cramer P, Structural basis of RNA polymerase II backtracking, arrest and reactivation, Nature, 471 (2011) 249–253. [DOI] [PubMed] [Google Scholar]
- [41].Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM, Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription, Mol Cell, 9 (2002) 541–552. [DOI] [PubMed] [Google Scholar]
- [42].Kujirai T, Ehara H, Fujino Y, Shirouzu M, S.-i. Sekine, H. Kurumizaka, Structural basis of the nucleosome transition during RNA polymerase II passage, Science (New York, N.Y.), (2018). [DOI] [PubMed] [Google Scholar]
- [43].Kulaeva OI, Hsieh FK, Studitsky VM, RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones, Proc Natl Acad Sci U S A, 107 (2010) 11325–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Choy JS, Lee TH, Structural dynamics of nucleosomes at single-molecule resolution, Trends in biochemical sciences, 37 (2012) 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH, DNA methylation increases nucleosome compaction and rigidity, J Am Chem Soc, 132 (2010) 1782–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dhall A, Wei S, Fierz B, Woodcock CL, Lee TH, Chatterjee C, Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions, J Biol Chem, 289 (2014) 33827–33837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim J, Lee J, Lee TH, Lysine Acetylation Facilitates Spontaneous DNA Dynamics in the Nucleosome, The journal of physical chemistry. B, 119 (2015) 15001–15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kim J, Wei S, Lee J, Yue H, Lee TH, Single-Molecule Observation Reveals Spontaneous Protein Dynamics in the Nucleosome, The journal of physical chemistry. B, 120 (2016) 8925–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee J, Lee TH, Single-molecule investigations on histone H2A-H2B dynamics in the nucleosome, Biochemistry, 56 (2017) 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lee JY, Lee J, Yue H, Lee TH, Dynamics of nucleosome assembly and effects of DNA methylation, J Biol Chem, 290 (2015) 4291–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lee JY, Lee TH, Effects of histone acetylation and CpG methylation on the structure of nucleosomes, Biochimica et biophysica acta, 1824 (2012) 974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee JY, Lee TH, Effects of DNA methylation on the structure of nucleosomes, J Am Chem Soc, 134 (2012) 173–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lee JY, Wei S, Lee TH, Effects of histone acetylation by Piccolo NuA4 on the structure of a nucleosome and the interactions between two nucleosomes, J Biol Chem, 286 (2011) 11099–11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lee T-H, Extracting kinetics information from single molecule fluorescence resonance energy transfer data using hidden Markov models, J. Phys. Chem. B, 113 (2009) 11535–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wei S, Falk SJ, Black BE, Lee TH, A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome, Nucleic acids research, 43 (2015) e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Crickard JB, Lee J, Lee TH, Reese JC, The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome, Nucleic acids research, 45 (2017) 6362–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yue H, Fang H, Wei S, Hayes JJ, Lee TH, Single-Molecule Studies of the Linker Histone H1 Binding to DNA and the Nucleosome, Biochemistry, 55 (2016) 2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal elements that control the topography of the barrier to transcription, Cell, 151 (2012) 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, Bustamante C, Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics, Proc Natl Acad Sci U S A, 111 (2014) 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lowary PT, Widom J, New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning, J Mol Biol, 276 (1998) 19–42. [DOI] [PubMed] [Google Scholar]
- [61].Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee T-H, Bartholomew B, INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers, Nature communications, 8 (2017) 15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Luger K, Rechsteiner TJ, Richmond TJ, Expression and purification of recombinant histones and nucleosome reconstitution, Methods Mol. Biol, 119 (1999) 1. [DOI] [PubMed] [Google Scholar]
- [63].Toth KF, Mazurkiewicz J, Rippe K, Association states of nucleosome assembly protein 1 and its complexes with histones, J Biol Chem, 280 (2005) 15690–15699. [DOI] [PubMed] [Google Scholar]
- [64].Crickard JB, Reese JC, Methods (San Diego, Calif.), (in this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jiang H, Reese JC, (submitted). [Google Scholar]
- [66].Kumar R, Nashine VC, Mishra PP, Benkovic SJ, Lee TH, Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies, Proc Natl Acad Sci U S A, 107 (2010) 19736–19741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.