Abstract
Detecting conformational heterogeneity in biological macromolecules is a key for the understanding of their biological function. We here provide a comparison between two independent approaches to assess conformational heterogeneity: molecular dynamics simulations, performed without inclusion of any experimental data, and maximum occurrence (MaxOcc) distribution over the topologically available conformational space. The latter only reflects the extent of the averaging and identifies regions which are most compliant with the experimentally measured NMR Residual Dipolar Couplings (RDCs). The analysis was performed for the HIV-1 TAR RNA, consisting of two helical domains connected by a flexible bulge junction, for which four sets of RDCs were available as well as an 8.2 μs all-atom molecular dynamics simulation. A sample and select approach was previously applied to extract from the molecular dynamics trajectory conformational ensembles in agreement with the four sets of RDCs. The MaxOcc analysis performed here identifies the most likely sampled region in the conformational space of the system which, strikingly, overlaps well with the structures independently sampled in the molecular dynamics calculations and even better with the RDC selected ensemble.
Introduction
The fundamental importance of extensive conformational dynamics for allowing non-coding RNAs to carry out a wide variety of regulatory functions is well recognised.1–4 RNA secondary structure consists of stable A-form helical domains that are connected by bulges, internal loops, and higher order junctions. Such helix–junction–helix (HJH) motifs play essential roles in the folding and biological function of non-coding RNAs. They are often points of significant flexibility that guide large adaptive changes in the orientation of helical domains and RNA global structure during folding, ribonucleoprotein assembly, and catalysis. HJH motifs also serve as binding sites for proteins, small molecules, and metal ions. Characterizing the extent and nature of inter-helical flexibility across HJH motifs is of primary importance for understanding the physical principles underlying RNA folding and recognition.5 However, due to the biophysical properties of RNA it remains a major challenge. First collecting rich NMR datasets such as residual dipolar couplings (RDCs) is limited by the difficulties of obtaining significantly independent alignment.6 Then, the presence of large internal motions, couples the internal dynamics to the overall diffusive or alignment properties of the RNA, complicating the interpretation of NMR spin relaxation7–10 or RDC.11–17 Finally, due to the potentially complex conformational dynamics, recovering an ensemble from experimental data remains an under-determined problem.4,15,16,18–23
The transactivation response element (TAR) RNA from the HIV-1 virus is a well-studied RNA drug target that plays essential roles during viral replication.17,24,25 TAR consists of two A-form helical domains connected by a flexible three residue bulge linker. In previous work, each of the two TAR helices were independently elongated as a means of decoupling internal and overall motions.7,17 This made it possible to interpret RDCs in terms of inter-helical motions since the elongated helix dominates the overall alignment. In particular, the measured RDCs could be interpreted in terms of motions of the short helix relative to the elongated one. The RDCs measured on two independently elongated TAR samples made it possible to characterize inter-helical motions with 3D orientation sensitivity.15,17,26
More recently, we showed the feasibility of using a shape-based prediction27–30 of the alignment tensor approach for treating couplings between internal and overall motions.31 This made it possible to integrate additional RDCs measured in partially elongated TAR samples in the determination of atomic resolution ensembles. Such ensembles were composed of conformations selected from a conformational pool obtained using an 8.2 μs molecular dynamics calculation31 computed using the CHARMM36 force-field.32–34 From this long MD trajectory conformational ensembles in agreement with the experimental RDC data were selected.31 This approach permitted to extract from the whole pool of structures determined by MD calculation, the conformations which may better represent the conformational variability of the system. The selected structures clustered into three distinct states, separated by large transitions in inter-helical orientations, coupled to local melting of base-pairs near the junction. The RDC-selected ensemble included conformations that bear strong resemblance to the ligand bound conformations of TAR.
We here apply a different approach for the analysis of the averaged experimental RDCs, based on the compliance of each and any sterically-allowed conformation with respect to the average experimental data. The method, called maximum occurrence (MaxOcc),35,36 aims at identifying conformations that can exist for a large share of the time; this is done by assigning to each conformation the maximum time that it can exist and be in agreement with the experimental observation,37–39 when taken together with an arbitrary number of other conformations. Thus it is possible to identify the conformations, which must necessarily have a negligibly small weight and those which may have a large weight, whatever the real ensemble of conformations experienced by the RNA is. We have previously demonstrated by synthetic tests that the conformations used to construct synthetic ensembles are found to have a high MaxOcc.35,40,41 The analysis was performed without taking advantage of the MD calculations, i.e. without restricting the possible RNA conformations to the pool of structures sampled by the MD trajectory. Strikingly, the RNA structures with large MaxOcc define a conformational region in substantial overlap with the structures sampled in the MD calculations, indicating good convergence between the MD results and the MaxOcc analysis. Furthermore, the previously determined structural ensembles selected from the MD trajectory31 is on average even closer to the most likely region of the MaxOcc landscape.
Materials and methods
Experimental RDC datasets
The experimental RDC data measured using the Pf1 phage alignment medium for four constructs of HIV-1 TAR RNA (non-elongated, with the first helix elongated by 3 base pairs, with either the first or the second helix elongated by 22 base pairs) were previously published.11,17,31,42 The helix elongation causes a strong modulation of the alignment of the RNA strand, leading to a high degree of independence of the different sets of RDCs.
In the study we analyzed the one bond couplings measured between the sugar C1′–H1′, C2′–H2′, C3′–H3′, and C4′–H4′ and base C2–H2, C5–H5, C6–H6, C8–H8, C5–C6, N1–H1, and N3–H3 pairs of atoms for nucleotides in both helical regions. The data measured for the A22–U40 base pair was omitted in the current analysis due to previously reported conformational flexibility of this base pair.31
Generation of the pool of conformers and prediction of RDCs
The MaxOcc analysis of HIV-1TAR was performed using the broadest possible topologically allowed conformational space obtained through exhaustive sampling of inter-helical Euler angles43 in increments of 5°, excluding the orientations violating loose sterical and stereochemical restraints.17 The two separately well-folded regions were assumed to adopt idealized A-form helical structures and the bulge nucleotides were not explicitly modelled in this study. For each conformer, the 4 sets of RDCs were predicted using the PALES software.27 A steric description was used based on the cylindrical wall model with an effective low concentration (0.022 g mL−1) as no significant improvement of the alignment tensor prediction was observed for nucleic acids when the electrostatic model is used.44 To model the alignment of constructs that feature elongation of one of the helices, the proper number of base pairs was added to the initial structure assuming idealized A-form geometry. The helix II is capped by a UUCG apical loop corresponding to the sequence of the experimentally used TAR constructs.
Euler angle definition
The Euler angles were defined as previously described.43 In this definition αh, γh and βh varies from −180 to 180. Other common Euler angle conventions may have βh restricted to only positive values. The degeneracy introduced by this broader definition of βh is lifted by choosing the solution that minimizes .
MaxOcc calculations
The calculation of MaxOcc of each selected conformer is performed by finding optimized ensembles that yield the best agreement with experimental observables, while containing the selected conformer with a given weight. The calculation is repeated for a different weight of the same conformer. As this weight is increased, the agreement with the experimental data may start to deteriorate. The weight at which the quality of the fit reaches a fixed threshold corresponds to the MaxOcc of that conformer, i.e. to the highest weight that it could have in any ensemble that explains the experimental data. The target function used in the fit has the form of the quality factor Q.45 The best fit obtainable without applying any restraint to the weight of the conformers had a Q of 0.22 (corresponding to χ2 ≈ 1.55). A fit was considered good if the corresponding Q was below a threshold defined 20% higher than the lowest Q of 0.22, that is0.264 (this corresponds to a χ2 close to 2.0; as it is only Q, not χ2, that is optimized, the latter rises slightly faster).
When external alignment RDCs are used as experimental observables, the problem of finding an optimized ensemble with one structure (labelled j) present at a fixed weight (xMO) can be expressed as
| (1) |
where x is the vector of the weights of the N structures composing the pool, y is the vector of M experimentally observed RDC values, normalized by their norm, c1,…ck are the scaling factors between the experimental and back-calculated RDC for each of the k constructs (required because the magnitude of alignment induced by the anisotropic solution is not known exactly, and may differ from the one assumed in the PALES calculation), λ is a weighting factor, and A(c1,…ck) is the M × N matrix whose columns contain the RDC values back-calculated for each of the conformers, again normalized by the norm of the experimental RDC data. The A(c1,…ck) matrix is created by stacking the sub-matrices An containing back-calculated RDCs of single constructs multiplied by the appropriate scaling factors cn:
The y vector and the A matrix were normalized in such a way that the ‖Ax − y‖22 term corresponds to the square of the Q factor between the experimental and back-calculated data. The value of λ, set to 10 in the present calculations, is found with the L-curve method, as a compromise between a good fit of the experimental observables and the proximity of the sum of the weights to 1.41
The problem as expressed in eqn (1) would require a nonlinear minimization due to the presence of the unknown c factor. It becomes linear if the scaling factor c is fixed to a constant value. The optimal value of c for a given back-calculated data vector ycalc = Ax (arising either from a single structure or an ensemble) can be readily calculated as . A minimization. procedure was thus applied which involved making an initial guess of the value of scaling factor c, solving eqn (1) for x (with fixed c) using a non-negative least squares method (a frugal coordinate descent algorithm,46 combined with random coordinate search47), then calculating the optimal c for the present x vector, and finally using it as the fixed scaling factor in the next iteration of non-negative least squares minimization, in an iterative fashion, until convergence of the c value was reached.
MaxOR calculations
The MaxOcc analysis of interdomain mobility can yield additional insights into the details of the sampled conformational subspace if it is supplemented by maximum occurrence of regions (MaxOR) calculations.40 This method, which is the natural extension of the MaxOcc approach for single conformations to conformational regions, aims at determining the maximum amount of time for which a group of conformers can collectively exist in agreement with the averaged experimental data. To achieve this goal the algorithm described above is somewhat modified, according to eqn (2). Instead of fixing the weight of one conformer to the desired value xMO, it is the sum of the weights of all conformers composing the chosen group that is fixed to xMO:
| (2) |
where C and D indicate the structures within and outside the selected group, respectively.
Results and discussion
Maximum occurrence of single conformers – a comparison with extensive MD
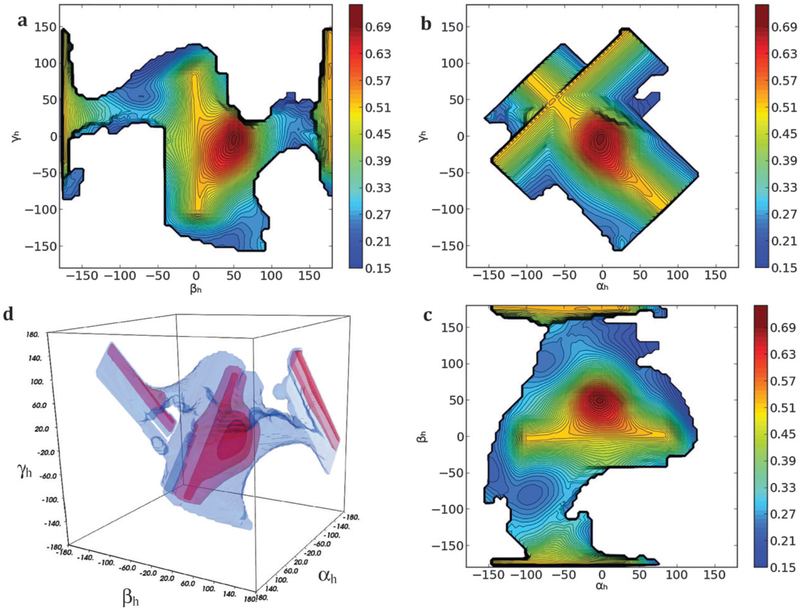
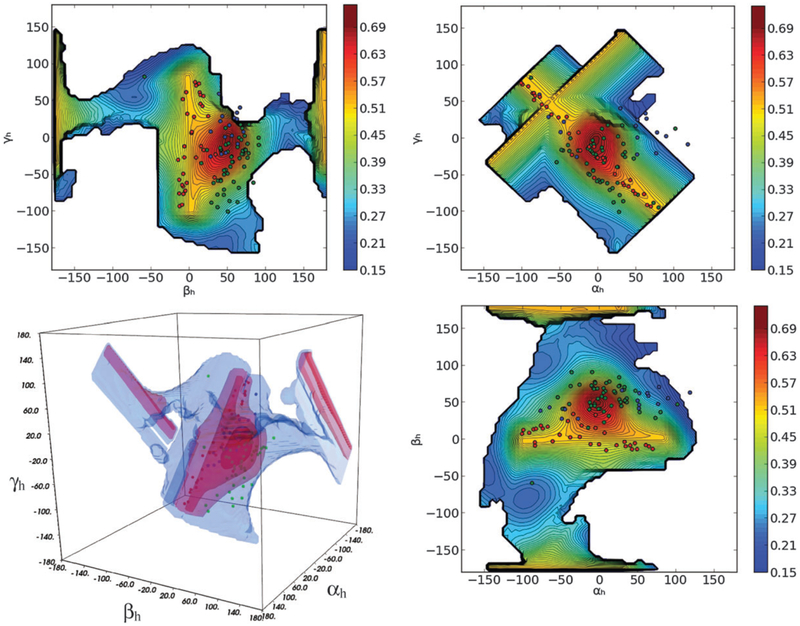
A pool containing all sterically-allowed RNA structures was generated by sampling all topologically allowed combinations of the inter-helical Euler angles αh, βh and γh, defining the inter-domain orientation of the two RNA domains, in steps of 5° for each angle separately. The three angles (see Fig. 1) represent the twisting of the first and second helices around their respective axes (αh and γh) and the inter-helical bending (βh).43 For each of the conformations in the generated pool (37 005 structures) the MaxOcc value was calculated using the implementation of the maximum occurrence method described in Materials and methods section. The obtained MaxOcc values show a considerable spread over the pool of conformations (from 17% to 70%) indicating that indeed specific structures are much more compliant with the experimental data than others. The fine sampling of the conformational space permits to observe that MaxOcc is a smooth function of the three inter-helical Euler angles. Fig. 2 and 4a show the 2D projections of the MaxOcc function on different pairs of inter-helical angles. It can be easily appreciated that the structures with the highest MaxOcc are grouped into a single well-defined conformational region, with a peak at around −10 < αh < 5°, 45 < βh < 55°, −15 < γh < 5°, centered at αh = −5°, βh = 50° and γh = −5°. To ease the understanding of the 3D shape of the high MaxOcc region, a 3D representation is given in Fig. 2d. Additional structures with intermediate-high MaxOcc values (up to 50%) appear at close to βh = 180°. They correspond most likely to a non-physical solution,48 whose high MaxOcc value arises from inherent degeneracy of the RDC data.16,49–51
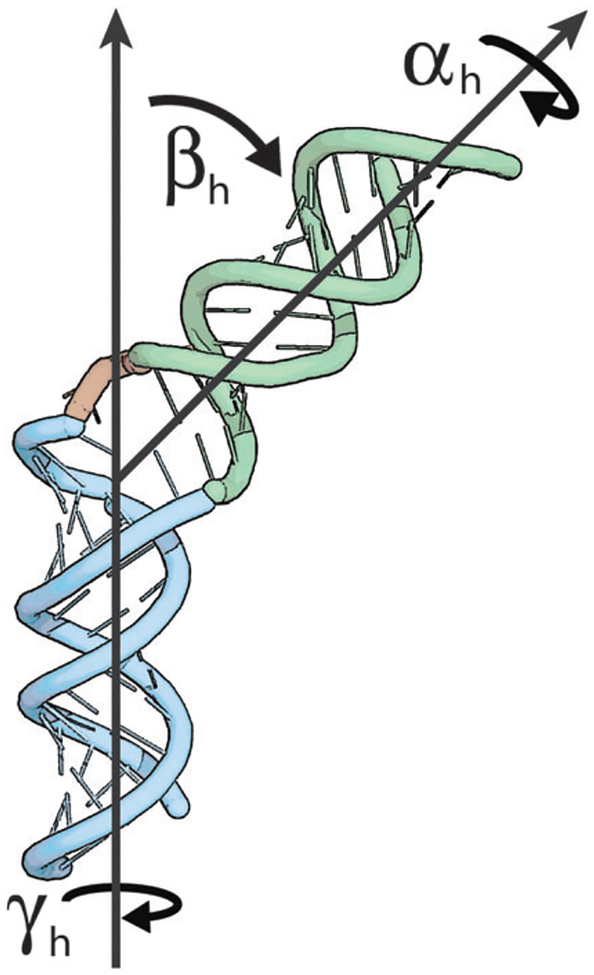
Fig. 1.
Angles (αh, βh, γh) inter-helical Euler angles defining the inter-domain orientation of the two RNA domains: αh and γh report on the twisting of the first and second helices around their respective axis, respectively, and βh on the inter-helical bending.
Fig. 2.
(a–c) The MaxOcc landscape (MaxOcc values color coded) as a function of αh, βh and γh angles (2D projections). White areas correspond to not sampled regions. (d) 3D representation of the full sampled space (blue) and of the area which encompasses high MaxOcc conformations (outer red surface, MaxOcc > 0.4; middle red surface, MaxOcc > 0.5; inner red surface, MaxOcc > 0.6).
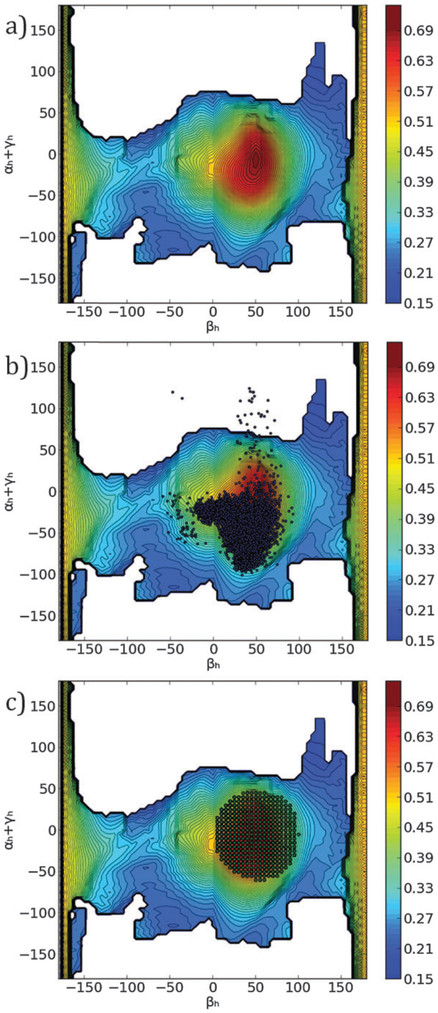
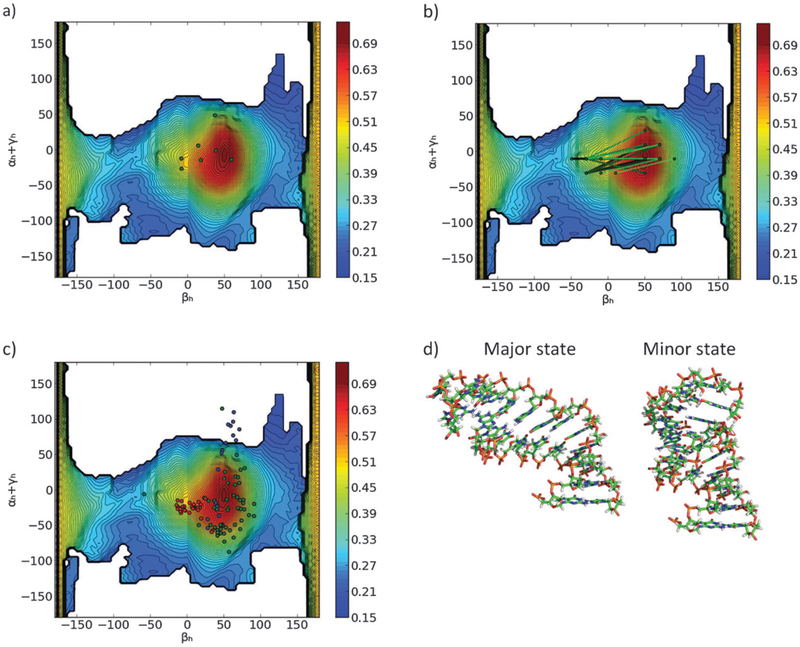
Fig. 4.
(a) The MaxOcc landscape (MaxOcc values color coded) as a function of the βh and αh + γh coordinates. White areas correspond to not sampled regions. (b) Superimposition of the MD trajectory (dark dots) to the MaxOcc landscape. (c) The smallest region centered at the MaxOcc peak with MaxOR = 1 (green dotted area) superimposed to the MaxOcc landscape (color coded) in the (βh, αh + γh) coordinates.
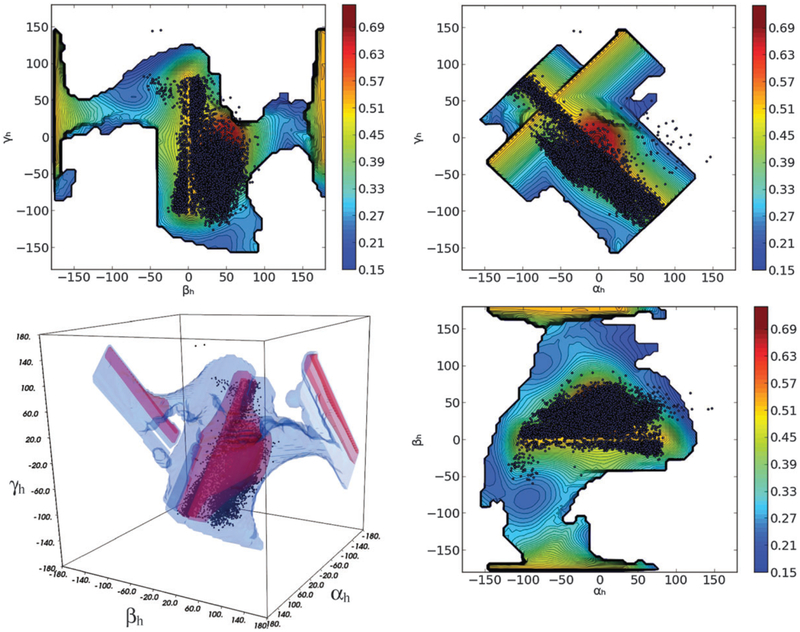
In a previous work31 the HIV-1 TAR RNA was studied by means of an 8.2 μs MD simulation. Interestingly when the coordinates of the structures constituting the MD are superimposed to the MaxOcc profile it appears that practically the entire MD trajectory falls inside the identified high MaxOcc region (Fig. 3 and 4b). It is a very encouraging result that two completely independent approaches suggest similar conformational sampling for the system in question.
Fig. 3.
Superimposition of MD trajectory (dark dots) to the MaxOcc landscape (color coded) as a function of αh, βh and γh angles (2D projections and 3D representation). White areas correspond to not sampled regions.
Even though the geometric center of the MD trajectory (the averaged Euler angles over the whole MD simulation are αh = −22°, βh = 32° and γh = −57) is somewhat shifted with respect to the peak of the MaxOcc profile, one has to keep in mind that the MD trajectory taken as such fits the experimental RDC data rather poorly (χ2 = 6.03). It is actually possible that, despite the overall sampling of conformations is correctly reconstructed by the MD, the populations of the specific conformational regions are not correctly represented, as already pointed out,4,31 owing to a lack of convergence or to imperfection in the applied force field. It is worth noting that the MD trajectory treats both local and global degrees of freedom, while the approach proposed here only considers the conformational dynamics of the bulge. The possibility of imperfect weighing of the MD trajectory was already explored using a sample and select (SAS) approach4,26,31 to properly reweight different regions of the MD trajectory. Small ensembles that fit well the experimental RDCs were repeatedly selected from the original trajectory and then combined to provide the ‘RDC reweighted ensemble’. Interestingly, the geometric center of this reweighted trajectory is located much closer to the MaxOcc peak (the average values of the Euler angles for the SAS ensemble are αh = −15°, βh = 52° and γh = −28°) than the original MD trajectory (Fig. 5). The improved agreement between the MaxOcc analysis and the MD sampling when the latter is adjusted using experimental information may not seem surprising, yet it should not to be taken for granted due to the under-determination of the recovery problem, the differences in the assumptions used in the two approaches, and the different physical meaning of the conformations selected by the two approaches. The fact that the MaxOcc and SAS methods actually favor a similar region of the conformational space can be considered an additional cross-validation of the ensemble previously extracted from the MD31 and further suggests that indeed the structures located in this part of the conformational space are crucial for explaining the HIV-1 TAR conformational sampling in solution.
Fig. 5.
Ensemble selected from the MD trajectory by SAS (‘RDC reweighted trajectory’), divided into three clusters after the original paper (cluster 1 in green, cluster 2 in red and cluster 3 in blue), superimposed to the MaxOcc landscape (color coded) as a function of αh, βh and γh angles (2D projections and 3D representation). White areas correspond to not sampled regions.
Seven distinct structures of HIV-1 TAR RNA bound to different small molecule ligands are available in the PDB.17 When their coordinates are superimposed to the MaxOcc profile (Fig. 6a and Fig. S1, ESI†) it appears that also these structures are located either close to the peak of the MaxOcc function or on its shoulder towards lower values of βh. This finding may suggest that ligand binding occurs by taking advantage of pre-existing conformations of HIV-1 TAR RNA, which are already highly populated in the conformational ensemble of the free nucleic acid.
Fig. 6.
Superposition of the MaxOcc landscape (MaxOcc values color coded), as a function of the βh and αh + γh coordinates, and (a) the ligand bound structures available in the PDB (green dots), (b) the set of pairs of 20°·20° regions with MaxOR > 95% (depicted as dots located in the centers of the regions, connected by a line; pairs including the ‘ghost’ of the minor state are omitted for clarity), (c) the ensemble selected from the MD trajectory by SAS (‘RDC reweighted trajectory’) divided into three clusters as in the original paper (cluster 1 in green, cluster 2 in red and cluster 3 in blue). White areas correspond to not sampled regions. (d) Representative RNA conformations of the two compact regions, one located at the peak of the MaxOcc profile (major state) and another at αh = −15, βh = −40, γh = 0° (minor state), able to fit the experimental data.
Maximum occurrence of conformational regions
The MaxOcc analysis identified the part of the conformational space which contains the single structures that can explain the largest share of the experimental observables by themselves. However, even the structures with the highest MaxOcc can contribute only up to 70% to the conformational ensemble sampled by HIV-1 TAR RNA. The next question to ask is what is the smallest compact ensemble or the simplest mobility scheme which can account for the experimental observables. One of the simplest mobility schemes that one can conceive consists of a motion around a single center. The MaxOR approach was applied to quantify the smallest amount of conformational heterogeneity that has to occur around the peak of the MaxOcc profile in order to obtain an ensemble which fully reproduces the experimental data. For this purpose, several regions were built around the conformation with the highest MaxOcc comprising all structures that can be obtained from the central conformation by changing the inter-helical orientation through a single axis rotation in any direction by less than a fixed angle (the quaternion representation of rotations was used at this step, because the Euler angle representation is not the best way to define distances between two structures). By increasing the maximum allowed rotation in steps of 10° and calculating the MaxOR of the corresponding regions, it was found that rotations up to 50° from the central conformer have to occur in order to obtain a MaxOR of 1 (i.e.: full agreement with the experimental data) (Fig. 4c). Thus if mobility in a symmetric region around a single center is assumed, inter-helical motions of high amplitude (the most distant conformations are 100° of rotation apart from one another) have to be considered to explain the experimental RDC values, in good agreement with initial studies of TAR dynamics.42
The size of the conformational space to be sampled by the system can likely be reduced if instead of an isotropic distribution around a single center, two or more separated, yet compact, regions are allowed to be explored.40,41,52 In order to identify other compact regions in the conformational space that can best complement the MaxOcc peak, a broad series of MaxOR calculations was performed. In each calculation the considered region was composed of two parts: the structures composing the peak of the MaxOcc profile (−10 < αh < 5°, 45 < βh < 55°, −15 < γh < 5°) and another group of structures constituting a 5°·5° square in the (βh, αh + γh) 2D projection of the conformational space. The second part of the region was changed in the different calculations in a systematic way in order to cover the whole (βh, αh + γh) space. The results of the whole procedure, shown in Fig. S2 (ESI†), demonstrate that there exist only two compact areas in the (βh, αh + γh) space which, when added to the MaxOcc peak, lead to a considerable increase of MaxOR. These two areas are located around βh = −40, αh + γh = −15, and βh = 165, αh + γh = −20. Because these regions are separated by an almost 180° rotation, it is probable that one of them arises from the inherent degeneracy of the RDC data (i.e. it is just a ‘ghost’51 of the other). As the region with high values of βh is located close to the edge of the available conformational space, possibly hardly sterically allowed if a more physically accurate modelling of the bulge was applied,48 it is quite safe to assume that this region is indeed a ‘ghost’ of the other region. Thus the MaxOR analysis shows that conformers situated around βh = −40°, αh + γh = −15° are the best suitable to complement the structures located close to the peak of MaxOcc, and when the two are taken together they are nearly enough to explain the whole experimental observables (MaxOR of the pair is 99%).
The size of the complementing region is, as said above, a 5°·5° square in the (βh, αh + γh) 2D projection, yet it has the shape of a long rod in the whole (αh, βh, γh) 3D conformational space. In order to locate more precisely the actual structures responsible for the high MaxOR, such rod can be thus further subdivided into 5°·5°·5° cubes in the full 3D Euler angle space with the centers at αh = x, βh = −40, γh = −x −15°, where x runs over all the values of αh sterically allowed at this point of space, in steps of 5°. Fig. S3 (ESI†) presents the MaxOR values of each cube as a function of the αh angle. The MaxOR function has a single maximum at αh = −15° (and γh = 0°) and its value at this point is only slightly lower (MaxOR = 97%) than when the whole rod is considered. The volume occupied by these regions is much smaller than the volume occupied by the single region with MaxOR equal to 1 identified before. Therefore, we have identified a pair of compact regions in the Euler angle space, one located at the peak of the MaxOcc profile and another at αh = −15, βh = −40 γh = 0° (Fig. 6d), that constitute a compact conformational sampling able to fit the experimental data (MaxOR of 100% is easily obtainable with this pair by slightly increasing the size of either region).
The possible existence of other, clearly distinct, two-centered ensembles not containing a region close to the peak of the MaxOcc profile was examined by performing a series of additional calculations over all pairs of 2D regions of size of 20°·20° (Fig. 6b). Interestingly, all the two-centered ensembles with the highest maxOR (>95%) are composed of a region located in proximity of the MaxOcc peak (with the coordinates of their centers in the range 50 < βh < 90 and −30 < αh + γh < 30) and of another region very close either to the identified minor state (−50 < βh < 10 and −30 < αh + γh < −10) or to its ghost solution described above. Therefore, although the positions of the two states may be subject to some uncertainty, yet the existence of any other distinct two-centered ensemble with high maxOR value can be excluded.
Comparison of MaxOR results and previous results
Having identified such a two-region scheme as the most compact ensemble capable of explaining the experimental averaged RDCs, one can re-examine Fig. 6a, where the positions of the ligand bound structures are shown. It can be noted that these structures (all except one) are either located within the regions defined by the two-center MaxOR calculations or in the conformational space between them.
In ref. 31, the conformations that were selected by the SAS algorithm from the MD trajectory could be clustered into three main states, on the basis of the bending angle and the inter-nucleotide distance between A22, the last nucleotide in helix 1, and U23, the first nucleotide in the bulge. Whereas the present pool lacks the information about the inter-nucleotide distance, we could compare the location of the three clusters in the Euler angles space. The results of such a comparison are shown in Fig. 5 and 6c. Although the clusters selected from the MD are more spread than the MaxOcc peak, there is a clear similarity between the SAS cluster 1 (in green in Fig. 5 and 6c) and the main state identified by MaxOcc/MaxOR, and between the SAS cluster 2 (in red in Fig. 5 and 6c) and the minor state found by MaxOR. This correspondence is particularly striking if we extend the comparison to the generalized positions of the MaxOR regions shown in Fig. 6b. Also our qualitative identification of the major and minor states is in line with the relative importance of the clusters found by SAS, as cluster 1 was sampled for 66% of time and cluster 2 for 19%. The third cluster, representing 15% of weight in the SAS ensemble and located approximately in between the two others states, does not find its counterpart in the current analysis. A possible explanation can be found from the analysis of the structural details of the conformers composing this third cluster. The latter cluster features the melting of the A22–U40 base pair (the last base pair of the first helix), which allows them to sample inter-helical angles which are sterically disallowed when the helices are modelled as rigid bodies, like in the current MaxOcc analysis. Furthermore, the SAS ensemble actively incorporates experimental data within the bulge, potentially requiring a more complex model of motion to be adequately explained. A glance at Fig. 6c reveals that a significant fraction of the structures from the SAS cluster 3 is indeed located outside of what was considered the sterically allowed space for the Max-Occ analysis, while the remaining part is practically within the ranges of the Euler angles of the other two identified states.
Finally, we note that if only conformations in the first or second half of the MD trajectory were considered, either the conformations in the MaxOcc peak or in the minor state are scarcely sampled, thus suggesting that significantly shorter MDs would not be able to capture the structural variability detected by the RDC data. This is in line with the previous observation that the quality of the RDC fit deteriorates considerably when applying SAS to a shorter 80 ns MD trajectory.31
Conclusions
We have applied the MaxOcc and MaxOR approaches to analyse the RDC datasets previously acquired by some of us for the HIV-1 TAR RNA strand. Our analysis shows that all conformations which can provide the highest contributions to the experimental averaged data are clustered into one broad but well-defined peak in the conformational space defined by the three Euler angles providing the inter-domain orientation of the two RNA strands. Very interestingly many of the known ligand bound structures of HIV-1 TAR RNA turn out to be very similar to the conformers with the highest MaxOcc suggesting that known ligands may actually bind to a HIV-1 TAR conformation that is already highly present in the free RNA ensemble. A comparison of the present analysis with the MD simulation previously performed for this system shows that the MD sampling largely covers the medium-high MaxOcc regions. It is intriguing to observe how two completely different approaches tend to converge to a common result: molecular dynamics is in fact only based on the driving force of a general force field, whereas the MaxOcc results only reflect the regions of the conformational space which mostly comply with the experimental data. Moreover the agreement between the two approaches is significantly improved when the MD trajectory is reweighted based on averaged experimental RDCs, suggesting the validity of the SAS approach used for that purpose.31
Finally, another compact region of conformations, apart from the MaxOcc peak, was identified, which is the best suitable to complement the latter in a two centered conformational ensemble. We have also shown that this pair of regions constitutes the simplest conformational ensemble capable of reproducing the experimental RDC values and that they resemble the two principal states determined by selecting conformational ensembles from the MD trajectory.
Supplementary Material
Acknowledgements
This work was supported by Ente Cassa di Risparmio di Fire-nze, MIUR PRIN 2012SK7ASN, the European FP7 ITN contract pNMR No. 317127, and Instruct, part of the European Strategy Forum on Research Infrastructures (ESFRI). HMA acknowledges support from the US National Institutes of Health (R01AI066975 and PO1GM0066275).
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5cp03993b
References
- 1.Dethoff EA, Chugh J, Mustoe AM and Al Hashimi HM, Nature, 2012, 482, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustoe AM, Brooks CL and Al Hashimi HM, Annu. Rev. Biochem, 2014, 83, 441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinnenthal J, Buck J, Ferner J, Wacker A, Furtig B and Schwalbe H, Acc. Chem. Res, 2011, 44, 1292–1301. [DOI] [PubMed] [Google Scholar]
- 4.Salmon L, Yang S and Al Hashimi HM, Annu. Rev. Phys. Chem, 2014, 65, 293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailor MH, Sun X and Al Hashimi HM, Science, 2010, 327, 202–206. [DOI] [PubMed] [Google Scholar]
- 6.Latham MP, Hanson P, Brown DJ and Pardi A, J. Biomol. NMR, 2008, 40, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Sun X, Watt ED and Al Hashimi HM, Science, 2006, 311, 653–656. [DOI] [PubMed] [Google Scholar]
- 8.Hansen AL and Al-Hashimi HM, J. Am. Chem. Soc, 2007, 129, 16072–16082. [DOI] [PubMed] [Google Scholar]
- 9.Ryabov YE and Fushman D, Magn. Reson. Chem, 2006, 44, S143–S151. [DOI] [PubMed] [Google Scholar]
- 10.Ryabov YE and Fushman D, J. Am. Chem. Soc, 2007, 129, 3315–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dethoff EA, Hansen AL, Zhang Q and Al Hashimi HM, J. Magn. Reson, 2010, 202, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipari G and Szabo A, J. Am. Chem. Soc, 1982, 104, 4546–4559. [Google Scholar]
- 13.Brüschweiler R, Roux B, Blackledge M, Griesinger C, Karplus M and Ernst RR, J. Am. Chem. Soc, 1992, 114, 2289–2302. [Google Scholar]
- 14.Iwahara J and Clore GM, J. Am. Chem. Soc, 2010, 132, 13346–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravera E, Salmon L, Fragai M, Parigi G, Al-Hashimi HM and Luchinat C, Acc. Chem. Res, 2014, 47, 3118–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fragai M, Luchinat C, Parigi G and Ravera E, Coord. Chem. Rev, 2013, 257, 2652–2667. [Google Scholar]
- 17.Zhang Q, Stelzer AC, Fisher CK and Al-Hashimi HM, Nature, 2007, 450, 1263–1267. [DOI] [PubMed] [Google Scholar]
- 18.Salmon L, Nodet G, Ozenne V, Yin G, Jensen MR, Zweckstetter M and Blackledge M, J. Am. Chem. Soc, 2010, 132, 8407–8418. [DOI] [PubMed] [Google Scholar]
- 19.Guerry P, Salmon L, Mollica L, Ortega Roldan JL, Markwick P, van Nuland NA, McCammon JA and Blackledge M, Angew. Chem., Int. Ed. Engl, 2013, 52, 3181–3185. [DOI] [PubMed] [Google Scholar]
- 20.Cavalli A, Camilloni C and Vendruscolo M, J. Chem. Phys, 2013, 138, 094112. [DOI] [PubMed] [Google Scholar]
- 21.Lindorff-Larsen K, Kristjansdottir K, Teilum K, Fieber W, Dobson CM, Poulsen FM and Vendruscolo M, J. Am. Chem. Soc, 2004, 126, 3291–3299. [DOI] [PubMed] [Google Scholar]
- 22.Boomsma W, Ferkinghoff-Borg J and Lindorff-Larsen K, PloS Comput. Biol, 2014, 10, e1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernadò P, Mylonas E, Petoukhov MV, Blackledge M and Svergun DI, J. Am. Chem. Soc, 2007, 129, 5656–5664. [DOI] [PubMed] [Google Scholar]
- 24.Musiani F, Rossetti G, Capece L, Gerger TM, Micheletti C, Varani G and Carloni P, J. Am. Chem. Soc, 2014, 136, 15631–15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD and Krogan NJ, Nature, 2012, 481, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank AT, Stelzer AC, Al-Hashimi HM and Andricioaei I, Nucleic Acids Res, 2009, 37, 3670–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zweckstetter M and Bax A, J. Am. Chem. Soc, 2000, 122, 3791–3792. [Google Scholar]
- 28.Zweckstetter M and Bax A, J. Biomol. NMR, 2001, 20, 365–377. [DOI] [PubMed] [Google Scholar]
- 29.Zweckstetter M, Nat. Protoc, 2008, 3, 679–690. [DOI] [PubMed] [Google Scholar]
- 30.Berlin K, O’Leary DP and Fushman D, J. Magn. Reson, 2009, 201, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon L, Bascom G, Andricioaei I and Al Hashimi HM, J. Am. Chem. Soc, 2013, 135, 5457–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denning EJ and Mackerell AD Jr., J. Am. Chem. Soc, 2011, 133, 5770–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foloppe N and Mackerell AD Jr., J. Comput. Chem, 2000, 21, 86–104. [Google Scholar]
- 34.Mackerell AD Jr., Banavali N and Foloppe N, Biopolymers, 2000, 56, 257–265. [DOI] [PubMed] [Google Scholar]
- 35.Bertini I, Giachetti A, Luchinat C, Parigi G, Petoukhov MV, Pierattelli R, Ravera E and Svergun DI, J. Am. Chem. Soc, 2010, 132, 13553–13558. [DOI] [PubMed] [Google Scholar]
- 36.Bertini I, Ferella L, Luchinat C, Parigi G, Petoukhov MV, Ravera E, Rosato A and Svergun DI, J. Biomol. NMR, 2012, 53, 271–280. [DOI] [PubMed] [Google Scholar]
- 37.Gardner RJ, Longinetti M and Sgheri L, Inv. Probl, 2005, 21, 879–898. [Google Scholar]
- 38.Longinetti M, Luchinat C, Parigi G and Sgheri L, Inv. Probl, 2006, 22, 1485–1502. [Google Scholar]
- 39.Bertini I, Gupta YK, Luchinat C, Parigi G, Peana M, Sgheri L and Yuan J, J. Am. Chem. Soc, 2007, 129, 12786–12794. [DOI] [PubMed] [Google Scholar]
- 40.Andralojc W, Luchinat C, Parigi G and Ravera E, J. Phys. Chem. B, 2014, 118, 10576–10587. [DOI] [PubMed] [Google Scholar]
- 41.Andralojc W, Berlin K, Fushman D, Luchinat C, Parigi G, Ravera E and Sgheri L, J. Biomol. NMR, 2015, DOI: 10.1007/s10858-015-9951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A and Patel DJ, J. Mol. Biol, 2012, 315, 95–102. [DOI] [PubMed] [Google Scholar]
- 43.Bailor MH, Mustoe AM, Brooks CL III and Al Hashimi HM, Nat. Protoc, 2011, 6, 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zweckstetter M, Hummer G and Bax A, Biophys. J, 2004, 86, 3444–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornilescu G, Marquardt J, Ottiger M and Bax A, J. Am. Chem. Soc, 1998, 120, 6836–6837. [Google Scholar]
- 46.Potluru VK, Frugal Coordinate Descent for Large-Scale NNLS, 2012, Ref Type: Conference Proceeding. [Google Scholar]
- 47.Nesterov Y, SIAM J Control, 2012, 22, 341–362. [Google Scholar]
- 48.Mustoe AM, Al Hashimi HM and Brooks CL III, J. Phys. Chem. B, 2014, 118, 2615–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK and Prestegard JH, J. Magn. Reson, 2000, 143, 402–406. [DOI] [PubMed] [Google Scholar]
- 50.Bertini I, Longinetti M, Luchinat C, Parigi G and Sgheri L, J. Biomol. NMR, 2002, 22, 123–136. [DOI] [PubMed] [Google Scholar]
- 51.Longinetti M, Parigi G and Sgheri L, J. Phys. A: Math. Gen, 2002, 35, 8153–8169. [Google Scholar]
- 52.Tolman JR, Al-Hashimi HM, Kay LE and Prestegard JH, J. Am. Chem. Soc, 2001, 123, 1416–1424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.