Abstract
Hyaluronan (HA), a major extracellular matrix glycosaminoglycan, is a biomarker for cirrhosis. However, little is known about the regulatory and downstream mechanisms of HA overproduction in liver fibrosis. Hepatic HA and HA synthase 2 (HAS2) expression was elevated in both human and murine liver fibrosis. HA production and liver fibrosis were reduced in mice lacking HAS2 in hepatic stellate cells (HSCs), whereas mice overexpressing HAS2 had exacerbated liver fibrosis. HAS2 was transcriptionally up-regulated by transforming growth factor–β through Wilms tumor 1 to promote fibrogenic, proliferative, and invasive properties of HSCs via CD44, Toll-like receptor 4 (TLR4), and newly identified downstream effector Notch1. Inhibition of HA synthesis by 4-methylumbelliferone reduced HSC activation and liver fibrosis in mice. Our study provides evidence that HAS2 actively synthesizes HA in HSCs and that it promotes HSC activation and liver fibrosis through Notch1. Targeted HA inhibition may have potential to be an effective therapy for liver fibrosis.
INTRODUCTION
Liver fibrosis is a consequence of chronic liver diseases, including chronic hepatitis B and C viral infections, and alcoholic and non-alcoholic steatohepatitis (NASH) (1, 2). Cirrhosis, the end stage of liver fibrosis, is the 12th leading cause of death in the United States, and about 32,000 people die annually (3). Although two antifibrotic agents have been approved for treating idiopathic pulmonary fibrosis (4), currently there are no Food and Drug Administration– approved antifibrotics for cirrhosis. The curative therapy for cirrhosis remains liver transplantation unless the underlying disease can be successfully treated. Furthermore, coincident with increased obesity, the prevalence of NASH is rapidly rising (5). To date, NASH-mediated fibrosis has become the second leading cause of liver transplantation (6). The unmet medical needs for liver fibrosis, particularly as mediated by NASH, are remarkable.
The production and deposition of extracellular matrix (ECM) components, such as collagen, that replace functional liver tissues feature prominently in liver fibrosis (1, 2). Hyaluronan (HA), another major ECM component, consists of repeating disaccharides D-glucuronic acid and N-acetyl-d-glucosamine. HA is synthesized as high–molecular weight (HMW) forms (MW, >1000 kDa) through HA synthase 1 (HAS1), HAS2, and HAS3 (7). During tissue injury and inflammation, HMW-HA is degraded into lower–molecular weight (LMW; MW, ~100 to 300 kDa) species through hyaluronidases or nonenzymatically by reactive oxygen species (7). Accumulated LMW-HA induces tissue injury and inflammation through the HA receptors CD44 and Toll-like receptor 4 (TLR4) (7, 8). LMW-HA is further degraded into HA oligomers (<100 kDa) and then eliminated from the body.
HA accumulates in the ECM in the context of tissue remodeling and drives inflammation, fibrosis, and cancer development through activation of TLR4 and CD44 (7, 8). The mechanistic roles of TLR4 and CD44 have been reported in lung injury and fibrosis (7–9). HA-TLR4 interaction mediates chemokine production in lung macrophages and initiates a regenerative response in type 2 alveolar epithelial cells (AEC2) after injury (8, 10). CD44 promotes an HA-mediated invasive phenotype in lung fibroblasts and ECM production in lung fibrosis (9). In the liver, TLR4 activates hepatic stellate cells (HSCs), which then promote liver fibrosis. Gut-derived lipopolysaccharide (LPS) has been identified as a TLR4 ligand involved in the progression of liver fibrosis (11). CD44 is required for macrophage and liver sinusoidal endothelial cell (LSEC) clearance of HA from sites of inflammation (12, 13). However, the role of HA-mediated TLR4 and CD44 signaling in HSCs in the development of liver fibrosis is unknown.
Notch signaling is involved in cellular processes including myofibroblast differentiation and promotes lung, kidney, and skin fibrosis (14). In the liver, Notch1 is overexpressed in patients with hepatitis B or C infections and hepatocellular carcinoma (15). Jagged-1, a Notch ligand, is up-regulated in biliary fibrosis and non-alcoholic fatty liver disease (NAFLD) (16, 17). Previous studies using nonselective Notch inhibitors have suggested that Notch signaling plays a role in HSC activation and liver fibrosis (18, 19). However, the role of Notch1 in HSC activation and liver fibrosis, and the relationship between HA and Notch1 in HSCs, has not been explored.
In healthy participants, serum HA concentrations are low because they are exquisitely fine-tuned through the rapid clearance by LSECs. In cirrhosis, dysfunctional LSECs reduce the uptake of HA because of the decreased expression of CD44, and HA accumulates in the blood (12, 13). Measurement of blood HA concentration has been used as a noninvasive biomarker for cirrhosis (20). Here, we provide evidence that HA overproduction through dysregulated HAS2 expression contributes to the development of liver fibrosis. We show that HAS2 is transcriptionally regulated by Wilms tumor 1 (WT1) in HSCs and that HA promotes fibrogenic, proliferative, and invasive phenotypes of HSCs through activation of CD44 and TLR4 in an autocrine manner. We found that Notch1 contributes to HA-mediated HSC activation and fibrosis, and we demonstrated that inhibiting HA synthesis has potential for the treatment of liver fibrosis.
RESULTS
HAS2 and HA are overexpressed in human and murine liver fibrosis
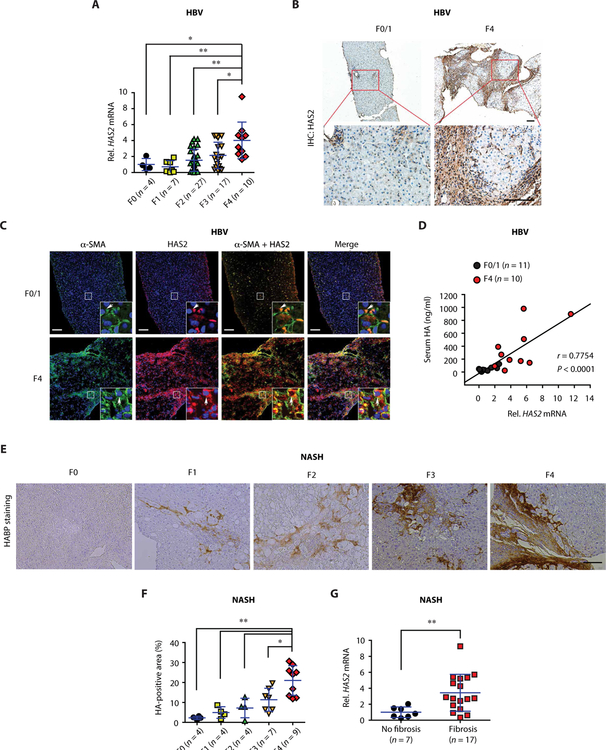
To determine the roles of HA synthases in human liver fibrosis, we examined liver specimens from three different etiologies: hepatitis B–, hepatitis C–, and NASH-induced liver fibrosis. In both hepatitis B– and hepatitis C–induced liver fibrosis, we observed increased hepatic expression of HAS2 in advanced fibrosis (F4) compared to earlier stages (Fig. 1A and fig. S1A). In contrast, the hepatic expression of HAS1 and HAS3 was unchanged between advanced and early fibrosis (fig. S1, A and B). The analysis of Gene Expression Omnibus database (GSE84044) for hepatitis B–induced fibrosis corroborated our results (fig. S1C). HAS2 protein expression was also elevated in fibrotic patient livers, as compared to those from patients with early-stage disease (Fig. 1B). HAS2 was expressed in activated HSCs, as identified by increased α–smooth muscle actin (α-SMA) expression (Fig. 1C). Furthermore, serum HA correlated with hepatic HAS2 mRNA (Fig. 1D) but not with HAS1 or HAS3 (fig. S1D). NASH is the most common etiology of liver fibrosis in recent years. Similar to our observations in viral hepatitis-induced fibrosis, NASH-induced fibrosis showed increased HA accumulation and HAS2 expression in the liver (Fig. 1, E to G).
Fig. 1. Expression of HAS2 and HA is increased in human liver fibrosis.
(A) Hepatic HAS2 mRNA expression in patients with fibrosis and chronic hepatitis B virus (HBV). (B) Representative immunohistochemical (IHC) staining images of HAS2 in liver sections of patients with HBV with F0/1 or F4 stages. Boxed regions are shown at higher magnification in the bottom row. Scale bar, 100 μm. (C) Colocalization of α-SMA and HAS2 in the liver of patients with HBV. Representative immunofluorescence staining images are shown (F0/1, n = 10; F4, n = 10). Boxed regions are shown at higher magnification. Blue, 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 100 μm. (D) Pearson correlation coefficient analysis of HAS2 mRNA expression and serum HA concentration of patients with HBV. (E and F) Representative images and quantification of HA staining in liver sections of patients with NASH-mediated liver fibrosis. HABP, HA-binding protein. Scale bar, 100 μm. (G) Hepatic HAS2 mRNA expression in patients with NAFLD. F0, no fibrosis; F1 to F4, fibrosis. In (A), (F), and (G), data are means ± SD. One-way analysis of variance (ANOVA) with Tukey’s post hoc analysis (A and F) and two-tailed Student’s t test (G). *P < 0.05 and **P < 0.01.
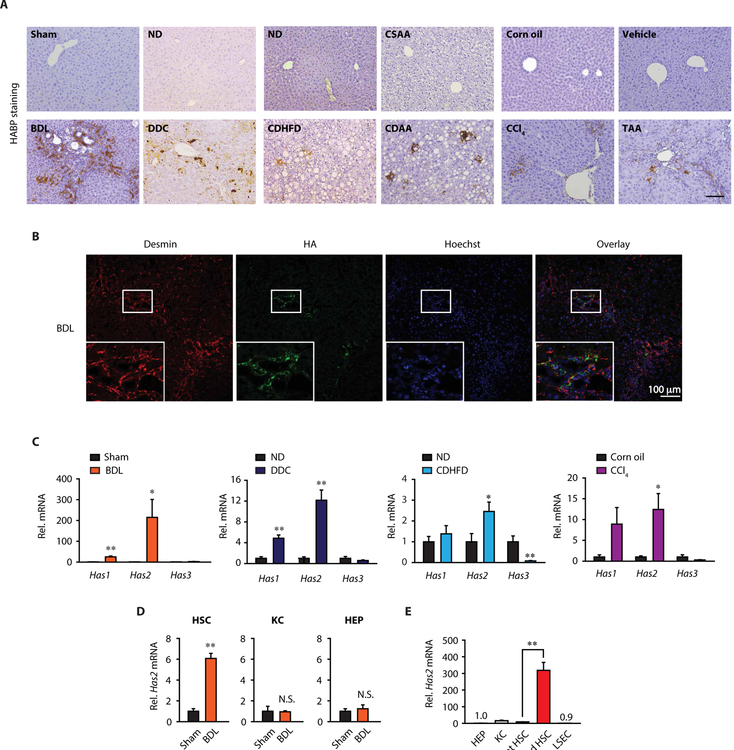
To explore the role of HAS2 in liver fibrosis, we examined the expression of the HAS2 product HA in mouse liver fibrosis models, including cholestasis-induced [bile duct ligation (BDL) or 3,5-diethylcarbonyl-1,4-dihydrocollidine (DDC) diet], NASH-induced [choline-deficient high-fat diet (CDHFD) or choline-deficient amino acid–defined (CDAA) diet], and toxin-induced [carbon tetrachloride (CCl4) or thioacetamide (TAA)] fibrosis models. HA was present in nonparenchymal cells in fibrotic livers (Fig. 2A). We costained for HA and cell-specific markers in BDL-induced fibrotic livers and found that HA was mainly expressed in Desmin+ HSCs but not in HNF4α+ hepatocytes, F4/80+ Kupffer cells, or CD31+ LSECs (Fig. 2B and fig. S2A). Next, we assessed HAS expression in the liver. Has1 and Has2 expression was up-regulated in the fibrotic liver, whereas Has3 was not (Fig. 2C). The up-regulation of Has2 was much greater than that of Has1 (Fig. 2C). We then examined the expression of Has2 in HSCs, Kupffer cells, and hepatocytes from fibrotic livers. Has2 expression was up-regulated in in vivo–activated HSCs from BDL mice, whereas it was not up-regulated in Kupffer cells and hepatocytes after BDL (Fig. 2D). We further compared Has2 mRNA expression among hepatocytes, Kupffer cells, HSCs, and LSECs. Has2 expression was low in all cell populations examined, except for activated HSCs (Fig. 2E). Activated HSCs showed high expression of Has2, which was 20-fold higher than in Kupffer cells (Fig. 2E). In contrast, Has1 and Has3 mRNA expression was lower in activated HSCs than in quiescent HSCs (fig. S2B). Our results suggest that HAS2 is the critical HA synthase in HSC activation and liver fibrosis.
Fig. 2. HAS2 and HA are overexpressed in murine liver fibrosis.
(A) Representative images of HA staining in mouse liver fibrosis. ND, normal diet; CSAA, choline-sufficient amino acid–defined. Scale bar, 100 μm. (B) Representative double immunofluorescence staining images of desmin and HA. Boxed regions are shown at higher magnification. (C) Hepatic Has mRNA expression in mouse liver fibrosis (sham, n = 10 or BDL, n = 12; ND, n = 5 or DDC, n = 9; ND, n = 5 or CDHFD diet, n = 7; corn oil, n = 5 or CCl4, n = 8). Mice underwent BDL for 3 weeks. *P < 0.05 and **P < 0.01 versus relative control. (D) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) for Has2 mRNA in primary HSCs, Kupffer cells (KC), and hepatocytes (HEP) isolated from sham- or BDL-operated mice 5 days after surgery (n = 3 per group). **P < 0.01; N.S., not significant. (E) qRT-PCR assays for Has2 mRNA in mouse liver cells (n = 3 per group). Activated HSC: 7-day culture-activated HSC. **P < 0.01. Data are means ± SEM. Two-tailed Student’s t test (C and D) and one-way ANOVA with Tukey’s post hoc analysis (E).
HSC-derived HAS2 is the critical enzyme for HA production and liver fibrosis in mice
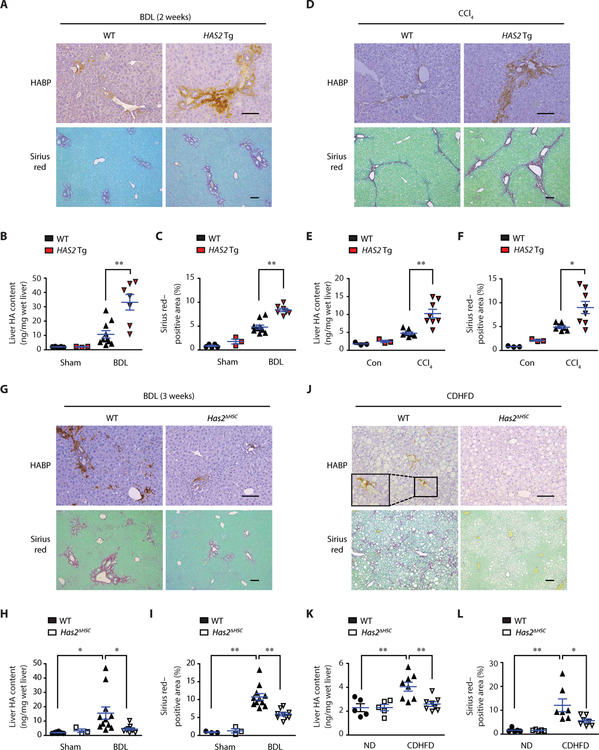
To investigate the role of HAS2 in liver fibrosis, we performed a gain-of-function study using ASMA-HAS2 transgenic (Tg) mice (9). These mice overexpress HAS2 in α-SMA–expressing cells (including HSCs) under control of the Acta2 promoter. Mice were subjected to BDL- or CCl4-induced liver fibrosis, which increased the liver HA content in ASMA-HAS2 Tg mice (Fig. 3, A, B, D, and E). ASMA-HAS2 Tg mice showed augmented collagen deposition along with increased HSC activation, as shown by Sirius red staining, α-SMA staining, and Acta2 and Timp1 mRNA expression (Fig. 3, A, C, D, and F, and fig. S3). These findings demonstrate that overproduction of HA by overexpressing HAS2 in liver myofibroblasts promotes HSC activation and liver fibrosis.
Fig. 3. HSC-derived HAS2 promotes liver fibrosis.
(A) Representative images of HA and Sirius red staining of sections of mouse liver. Mice underwent BDL for 2 weeks (Sham-WT, n = 5; Sham-Tg, n = 3; BDL-WT, n = 10; BDL-Tg, n = 7). (B) Liver HA content by enzyme-linked immunosorbent assay (ELISA). (C) Sirius red–positive area (collagen) from (A). (D) HA and Sirius red staining of sections of mouse liver. Mice were injected with corn oil (Con) or CCl4 intraperitoneally twice a week for 5 weeks (Con, n = 3 per group; CCl4-WT, n = 7; CCl4–ASMA-HAS2 Tg, n = 8). (E) Liver HA content by ELISA. (F) Sirius red–positive area from (D). (G) Representative images of HA and Sirius red staining of sections of mouse liver (Sham-WT, n = 8; Sham-Has2ΔHSC, n = 3; BDL-WT, n = 11; BDL-Has2ΔHSC, n = 9). (H) Liver HA content by ELISA. (I) Quantification of Sirius red–positive area from (G) (Sham groups, n = 3, each). (J) HA and Sirius red staining of sections of mouse liver. Mice were fed with ND or CDHFD for 12 weeks. (K) Liver HA content by ELISA (ND, n = 5 per group; CDHFD, n = 8 per group). (L) Sirius red–positive area from (J) (ND, n = 5 per group; CDHFD, n = 7 per group). Data are means ± SEM. *P < 0.05 and **P < 0.01. One-way ANOVA with Tukey’s post hoc analysis. Scale bars, 100 μm.
HSCs are the primary cells that constitute the myofibroblast pool in liver fibrosis (21). To characterize the role of HSC-derived HAS2 in liver fibrosis, we generated HSC-specific Has2 knockout (Has2ΔHSC) mice, in which specific deletion of Has2 in HSCs was confirmed (fig. S4). These mice were subjected to BDL and DDC diet–induced liver fibrosis models. In parallel with HA content, liver fibrosis induced by BDL and DDC diet in wild-type (WT) mice was diminished in Has2ΔHSC mice (Fig. 3, G to I, and fig. S5, A to C). These findings demonstrate that HAS2 expressed in HSCs plays a crucial role in hepatic HA production and liver fibrosis.
NASH-induced liver fibrosis was developed by CDHFD feeding, which closely recapitulates the liver pathology of human NASH (22). CDHFD feeding increased the hepatic HA content and liver fibrosis in WT mice, but these increases were reduced in Has2ΔHSC mice (Fig. 3, J to L). These results indicate that HSC-produced HAS2 plays a major role in NASH-mediated fibrosis. Has2 deletion in HSCs did not affect CDHFD-induced weight gain, hepatic lipid accumulation, or glucose intolerance (fig. S6).
HAS2 regulates profibrogenic phenotypes of HSCs
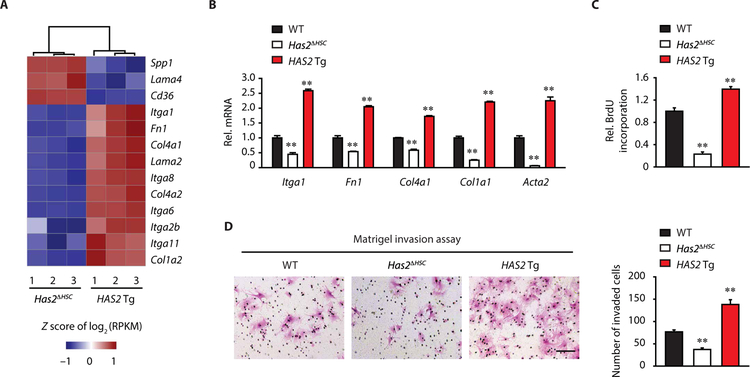
Using HSCs with Has2 ablation and HAS2 overexpression, we performed RNA sequencing (RNA-seq) to examine the expression of genes encoding ECM proteins and ECM-associated pathways. ECM-encoded genes and integrins were up-regulated in HAS2-overexpressing HSCs compared to Has2-deleted HSCs (Fig. 4A). The RNA-seq data were then validated by qPCR. Compared to WT HSCs, the mRNA expression of Itga1, Fn1, Col4a1, Col1a1, and Acta2 was reduced in Has2-ablated HSCs but increased in HAS2-overexpressing HSCs (Fig. 4B). Proliferative and invasive capacities of HSCs were reduced by Has2 deletion but enhanced by HAS2 overexpression (Fig. 4, C and D). These results suggest that HAS2 regulates HSC functions including ECM production, proliferation, and invasion, likely via autocrine production of HA.
Fig. 4. HSC-derived HAS2 regulates HSC function.
(A) A heat map of genes related to ECM-receptor interaction. RNA-seq was performed using quiescent Has2ΔHSC HSCs or HAS2 Tg HSCs (n = 3 per group). RPKM, reads per kilobase million. (B) qRT-PCR assays for Itga1, Fn1, Col4a1, Col1a1, and Acta2 mRNA expression in quiescent WT, Has2ΔHSC, or HAS2 Tg HSCs (n = 3 to 6 per group). (C) BrdU (5-bromo-2′-deoxyuridine) incorporation assay. Proliferation of WT, Has2ΔHSC, or HAS2 Tg HSCs was assessed by BrdU incorporation for 24 hours (n = 3 per group). (D) Microscopic images of the Matrigel-invading capacity of HSCs isolated from WT, Has2ΔHSC, or HAS2 Tg mice. Representative images are shown. Scale bar, 100 μm (n = 4 per group). Data are means ± SEM. **P < 0.01 versus WT HSC. One-way ANOVA with Tukey’s post hoc analysis.
LMW-HA mediates proinflammatory and profibrogenic HSC functions through CD44 and TLR4
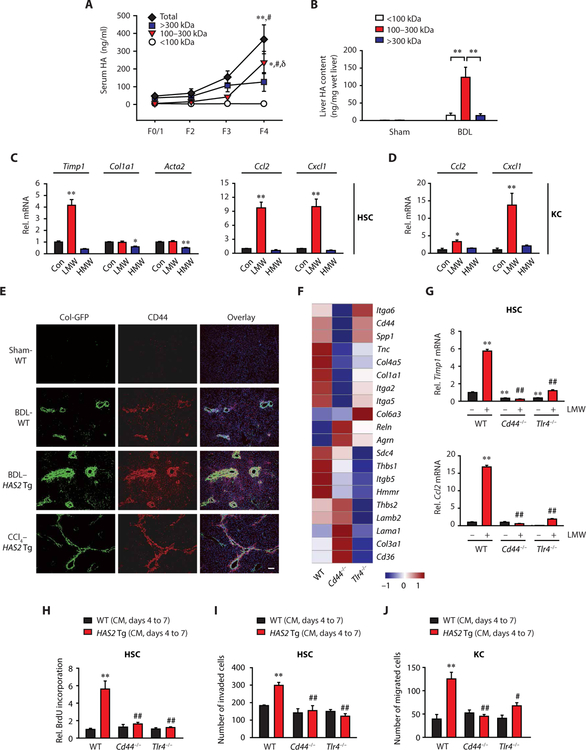
HA production and degradation is a dynamic process. HA generated as HMW forms can be degraded into LMW-HA in the presence of inflammation (7). We detected total and HMW forms of HA in sera of individuals without fibrosis, whereas LMW-HA (100 to 300 kDa) forms were negligible (Fig. 5A). During the progression of liver fibrosis, total HA concentration increased (Fig. 5A), and LMW forms of HA were increased in sera from patients with fibrosis as compared to individuals without fibrosis (Fig. 5A). In mouse fibrotic livers, the 100- to 300-kDa LMW-HA was the dominant form of HA (Fig. 5B). To examine the effect of size of HA, HSCs were treated with LMW- and HMW-HA. The treatment of HSCs with LMW-HA increased Timp1, Ccl2, and Cxcl1 mRNA expression, whereas HMW-HA treatment did not (Fig. 5C). Although continuously exposing HSCs to endogenous, HAS2 overexpression–produced HA enhanced ECM production (Fig. 4, A and B), Col1a1 and Acta2 mRNA expression was not up-regulated by a short LMW-HA treatment (Fig. 5C). Conversely, HMW-HA suppressed expression (Fig. 5C). Similarly, the treatment of Kupffer cells with LMW-HA up-regulated Ccl2 and Cxcl1, whereas HMW-HA had no effect (Fig. 5D). Our results suggest that LMW-HA, but not HMW forms, generated in injured tissues augments inflammatory and fibrogenic responses in HSCs and Kupffer cells.
Fig. 5. CD44 and TLR4 mediate HA-induced HSC profibrogenic phenotypic change.
(A) Total and fractionated serum HA content in patients with fibrosis and NAFLD (F0/1, F2, and F3, n = 5 per group; F4, n = 9). *P < 0.05 and **P < 0.01 versus F0/1; #P < 0.05 versus F2; δP < 0.05 versus F3. (B) Fractionated HA content in murine liver (sham-operated, n = 7; BDL-operated, n = 10). (C) Profibrogenic and proinflammatory gene expression in mouse HSCs and (D) in mouse Kupffer cells (n = 3 to 5 per group). Cells were treated with vehicle (Con), LMW-HA, or HMW-HA. *P < 0.05 and **P < 0.01 versus Con. (E) Representative images of immunofluorescence staining of sections of mouse liver. Blue, DAPI. Scale bar, 100 μm. (F) A heat map of genes related to ECM-receptor interaction. Each lane repre sents pooled WT, Cd44−/−, or Tlr4−/− HSCs from three mice. (G) qRT-PCR assay (n = 3 per group). Mouse HSCs were treated with or without LMW-HA for 12 hours. (H) BrdU incorporation assay. HSCs were treated with conditioned medium (CM, days 4 to 7) (n = 6 per group). (I) Matrigel invasion assay (n = 4 per group). WT, Cd44−/−, or Tlr4−/− HSCs were plated onto the upper chamber, and conditioned medium was placed in the lower chamber. (J) Migration of WT, Cd44−/−, or Tlr4−/− Kupffer cells toward HSC-conditioned medium (n = 3 to 6 per group). In (G) to (J), **P < 0.01 versus vehicle (or WT CM)–treated WT; #P < 0.05 and ##P < 0.01 versus LMW-HA (or HAS2 Tg CM)–treated WT. Data are means ± SEM. One-way ANOVA with Tukey’s post hoc analysis.
TLR4 and CD44 are major HA receptors (8). We previously reported that TLR4 expressed in HSCs promotes liver fibrosis (11). In normal liver, CD44 is weakly expressed by lymphocytes and Kupffer cells but not hepatocytes (13). Fibrotic insults by BDL or CCl4 increased the CD44 expression, especially in collagen-producing cells. CD44 expression was further augmented in mice with HAS2 overexpression (Fig. 5E). Cd44 and Tlr4 mRNA expression was increased in HSCs from BDL mice compared to controls (fig. S7), underscoring that fibrotic HSCs up-regulate CD44 and TLR4 expression. To dissect the role of CD44 and TLR4 in HSCs, we used RNA-seq analysis on WT, Cd44−/−, and Tlr4−/− HSCs after in vitro culturing in which HSCs should be affected by endogenously produced HA (fig. S7). Col1a1, Itga2, and Itga5 were predominantly regulated by CD44, whereas Sdc4 and Thbs1 were TLR4 dominant. In contrast, Tnc, Col4a5, Itgb5, and Hmmr were regulated by both TLR4 and CD44 (Fig. 5F). Both CD44 and TLR4 regulated LMW-HA–induced Timp1 and Ccl2 expression (Fig. 5G). We also found that WT HSCs treated with HA-enriched conditioned medium showed enhanced HSC proliferation and invasion, whereas Cd44−/− or Tlr4−/− HSCs did not (Fig. 5, H and I, and fig. S8). Furthermore, HA-enriched conditioned medium also enhanced Kupffer cell migration, which was abolished by Cd44 or Tlr4 deficiency (Fig. 5J). These results suggest that the biological effects of HA on HSCs and Kupffer cells are mediated through CD44 and TLR4.
HAS2 mediates HSC activation and liver fibrosis through Notch1
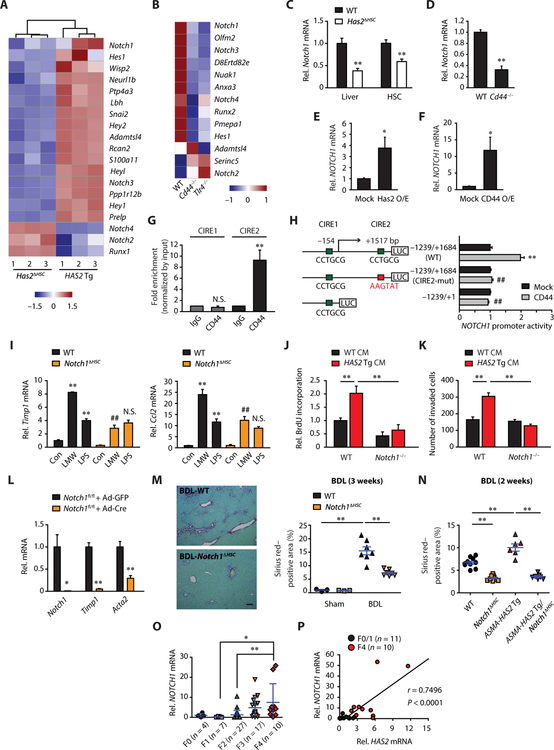
To seek downstream effectors of HAS2, we analyzed the RNA-seq data of HAS2 Tg versus Has2ΔHSC HSCs and WT versus Cd44−/− or Tlr4−/− HSCs. We found that Notch-associated genes were regulated by HAS2, CD44, and TLR4 (Fig. 6, A and B). HSCs are the primary cells expressing Notch1 and its target Hes1 (fig. S9A). Hes1 expression was up-regulated in activated HSCs (fig. S9A), suggesting that Notch signaling is activated during HSC activation. Consistent with the RNA-seq results, liver tissue and HSCs from Has2ΔHSC and Cd44−/− mice had reduced expression of Notch1 and Hes1 (Fig. 6, C and D, and fig. S9B). These findings prompted us to examine whether HAS2-mediated HA promotes HSC activation and liver fibrosis through Notch1. Has2 and CD44 overexpression increased NOTCH1 mRNA expression (Fig. 6, E and F), suggesting the transcriptional regulation of NOTCH1 by HA-CD44. To elucidate the underlying mechanism of HA-CD44–mediated transcriptional regulation of NOTCH1, we searched for the putative CD44 intracellular domain (ICD) response element (CIRE) “CCTGCG” on the NOTCH1 promoter/enhancer region. We found two putative CIREs located in the proximal NOTCH1 promoter region (CIRE1) and intron 1 (CIRE2). Chromatin immunoprecipitation (ChIP) analysis revealed that CD44 interacted with the CIRE2 (Fig. 6G). CD44 overexpression increased NOTCH1 promoter/enhancer activity with the WT full-length reporter construct, whereas the reporter constructs with mutated CIRE2 or lacking both exon 1 and intron 1 abrogated the ability of CD44 to increase NOTCH1 promoter/enhancer activity (Fig. 6H). Together, HA-CD44 signaling transcriptionally regulates Notch1 in HSCs.
Fig. 6. Notch1, as a downstream molecule of HAS2-HA-CD44 signaling, promotes liver fibrosis.
(A and B) Heat maps of genes related to Notch pathway. (A) RNA-seq for HSCs from Has2ΔHSC or HAS2 Tg mice. (B) RNA-seq for HSCs from WT, Cd44−/−, or Tlr4−/− mice. (C to F) Effect of Has2 or CD44 modulation on Notch1. (C) Liver (n = 8 per group), (C and D) HSCs (n = 3 to 4 per group), and (E and F) human HSC line LX-2 (n = 5 to 6). (G) ChIP assay (n = 4). IgG, immunoglobulin G. (H) NOTCH1 promoter activity in human embryonic kidney 293 A (HEK293A) (three replicates in each of four experiments). **P < 0.01 versus −1239/+1684 (WT) and mock-transfected; ##P < 0.01 versus −1239/+1684 (WT) and CD44-transfected. (I) qRT-PCR for Timp1 and Ccl2 (n = 3). Mouse HSCs were treated with vehicle (Con), LMW-HA, or LPS for 12 hours. **P < 0.01 versus WT-Con; ##P < 0.01 versus WT-LMW; N.S. versus WT-LPS. (J) BrdU incorporation assay (n = 9). (K) Matrigel invasion assay (n = 3). (L) Effect of Notch1 depletion on expression of fibrogenic mRNAs in HSCs (n = 3). (M) Sirius red staining and quantification of collagen deposition in liver sections from mice with HSC-specific Notch1 deletion 3 weeks after BDL (sham-operated, n = 3 per group; BDL-operated, n = 7 per group). Scale bar, 100 μm. (N) Sirius red staining in liver sections 2 weeks after BDL (WT, n = 10; Notch1ΔHSC, n = 9; ASMA-HAS2 Tg, n = 6; ASMA-HAS2 Tg/Notch1ΔHSC, n = 7). (C to N) Data are means ± SEM. (O) qRT-PCR (means ± SD) and (P) Pearson correlation coefficient analysis in liver tissue from patients with fibrosis and chronic hepatitis B. Two-tailed Student’s t test (C to F and L) and one-way ANOVA with Tukey’s post hoc analysis (G to K and M to O). *P < 0.05 and **P < 0.01.
To investigate the role of Notch1 in liver fibrosis, we generated mice lacking Notch1 in HSCs (Notch1ΔHSC). LMW-HA–induced Timp1 and Ccl2 mRNA up-regulation was reduced in Notch1-ablated HSCs, whereas LPS-induced expression was unaffected (Fig. 6I). HSC proliferation and invasion induced by HA-enriched conditioned media from HAS2 Tg HSCs were also abolished by Notch1 ablation (Fig. 6, J and K), demonstrating that Notch1 is required for HA-mediated profibrogenic phenotypes of HSCs. Notch1-ablated HSCs showed decreased Notch1 expression, along with reduced Timp1 and Acta2 mRNA expression (Fig. 6L). Hepatic Notch1 and Hes1 expression was increased in BDL-induced liver fibrosis in WT mice (fig. S9C). Moreover, Notch1ΔHSC mice had reduced liver fibrosis compared with WT mice (Fig. 6M). In ASMA-HAS2 Tg mice that show increased liver fibrosis, additional deletion of HSC Notch1 reduced liver fibrosis, of which the fibrosis degree was similar to mice with single Notch1 deletion (Fig. 6N), demonstrating the causal link of HAS2, Notch1 signaling, and liver fibrosis. In addition, NOTCH1 expression was up-regulated in human fibrotic livers and strongly correlated with HAS2 expression (Fig. 6, O and P). These results suggest that Notch1, as a downstream effector of the HAS2-HA-CD44 signaling, contributes to the development of liver fibrosis. Furthermore, the Notch1 ligand Jagged-1 was highly expressed in Kupffer cells and HSCs in comparison to hepatocytes (fig. S10). This further suggests that Kupffer cell– and/or HSC-derived Jagged-1 may engage Notch1 in HSCs through juxtacrine interaction because of their close proximity, thereby activating profibrogenic Notch1 signaling.
HAS2 expression is transcriptionally regulated by WT1 in HSCs
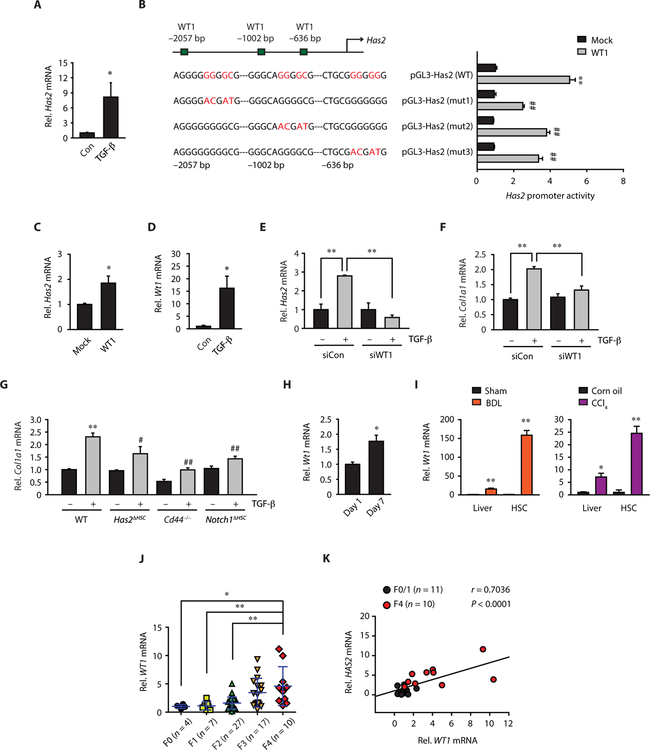
Next, we investigated the transcriptional regulation of HAS2 expression in HSCs. Although transforming growth factor–β (TGF-β) up-regulated Has2 mRNA expression in HSCs (Fig. 7A), the HAS2 promoter region does not have putative SMAD-binding sequences; rather, it contains three putative WT1-binding sequences (Fig. 7B) identified through the PROMO analysis program (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). WT1 overexpression increased Has2 promoter activity and mRNA (Fig. 7, B and C). The site-directed mutagenesis analysis of WT1-binding sites in the Has2 promoter revealed that all three WT1-binding sites are required for the Has2 promoter activation (Fig. 7B). Because TGF-β treatment up-regulated Has2 and Wt1 expression (Fig. 7, A and D), we investigated the contribution of WT1 to TGF-β–mediated Has2 expression and fibrogenic responses. Silencing Wt1 diminished TGF- –induced Has2 and Col1a1 up-regulation (Fig. 7, E and F). We then examined the contribution of HAS2-CD44-Notch1 to TGF-β–mediated fibrogenic response. Deficiency of Has2, Cd44, or Notch1 decreased TGF-β– induced Col1a1 mRNA expression (Fig. 7G). These results suggest that the WT1-HAS2-HA-Notch1 pathway is a key contributor to TGF-β–induced profibrogenic activity. In addition to TGF-β treatment, in vitro culture and in vivo–activated HSCs showed up-regulated Wt1 mRNA expression (Fig. 7, H and I). Hepatic WT1 expression was also up-regulated in fibrotic mouse livers and human cirrhosis (Fig. 7, I and J). There is a positive correlation between WT1 and HAS2 expression in human liver fibrosis (Fig. 7K). Our results demonstrate that WT1 is an upstream regulator of HAS2 induction in HSCs.
Fig. 7. WT1 transcriptionally regulates HAS2 in HSCs.
(A) qRT-PCR assay for Has2 after TGF-β treatment in mouse HSCs (n = 5). (B) Site-directed mutagenesis analysis of WT1-binding sites in the Has2 promoter (three replicates in each of five experiments). **P < 0.01 versus pGL3-Has2 (WT) and mock-transfected cells; ##P < 0.01 versus pGL3-Has2 (WT) and WT1-transfected cells. (C) Effect of WT1 overexpression on Has2 mRNA expression (n = 3). (D) qRT-PCR assays for Wt1 after TGF-β treatment (n = 5). (E and F) Effect of WT1 knockdown on TGF-β–induced Has2 and Col1a1 mRNA (n = 3 to 7). (G) Col1a1 mRNA expression (n = 7 to 8). Mouse HSCs were treated with or without TGF-β. (H) Wt1 mRNA expression in quiescent (day 1) or activated (day 7) HSCs (n = 3 per group) and (I) in the liver (sham, n = 10; BDL, n = 12; Con, n = 8; CCl4, n = 24) and in vivo–activated HSCs (n = 3 per group). (J) WT1 mRNA expression in patients with fibrosis and chronic hepatitis B. (K) Pearson correlation coefficient analysis of WT1 and HAS2. (A to I) Data are means ± SEM and (J) SD. *P < 0.05 and **P < 0.01 versus relative control; #P < 0.05 and ##P < 0.01 versus WT–TGF-β. Two-tailed Student’s t test (A, C and D, and H and I) and one-way ANOVA with Tukey’s post hoc analysis (B, E to G, and J).
Pharmacological inhibition of HA synthesis suppresses liver fibrosis progression
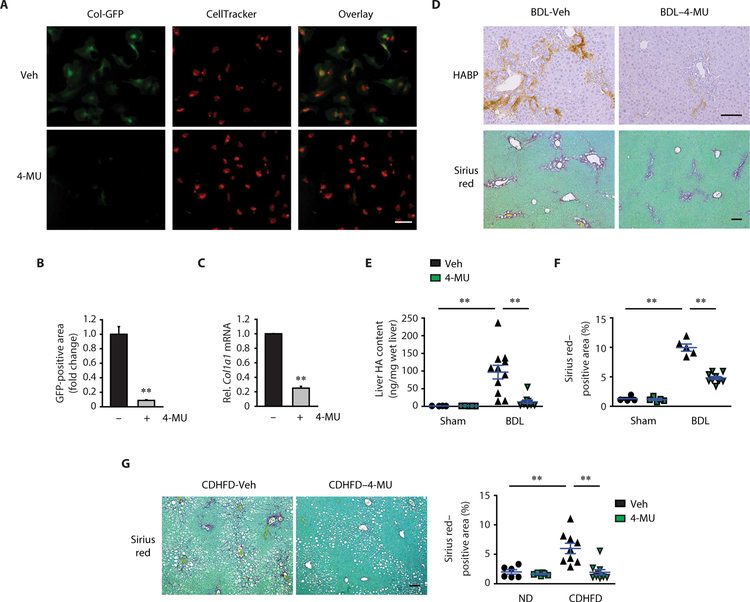
Last, we investigated whether inhibiting HA production could suppress the development of liver fibrosis using the HA synthesis inhibitor 4-methylumbelliferone (4-MU). 4-MU, also known as hymecromone, is an approved drug in Europe and Asia for treating biliary spasm and a dietary supplement for improving liver function (23). We used HSCs isolated from Col1a1 promoter-driven green fluorescent protein (GFP) Tg mice, in which cells express GFP upon increased Col1a1 promoter activity. 4-MU treatment reduced Col1a1 promoter activity and mRNA expression in HSCs (Fig. 8, A to C). To examine the therapeutic potential of 4-MU on liver fibrosis, 4-MU or vehicle was given orally daily to mice with BDL. The treatment of 4-MU began 1 week after BDL surgery and continued until the end of the experiments. Serum alanine aminotransferase concentration was similar between vehicle and 4-MU treatment groups, suggesting no remarkable hepatotoxicity by 4-MU (fig. S11A). 4-MU treatment suppressed liver HA content, Notch1 mRNA expression, HSC activation, macrophage infiltration, and mitigated liver fibrosis (Fig. 8, D to F, and fig. S11, B to D). To determine whether the antifibrotic effect of 4-MU was mediated by inhibiting signaling pathways activated by HAS2 or Notch1, we treated Has2ΔHSC and Notch1ΔHSC mice with 4-MU. 4-MU had no further antifibrotic effect beyond Has2 or Notch1 deficiency (fig. S11E). Furthermore, we tested the efficacy of 4-MU on NASH-mediated liver fibrosis. 4-MU efficiently attenuated CDHFD-induced collagen deposition (Fig. 8G). These results suggest that pharmacological inhibition of HA synthesis could be pursued as a therapeutic approach for liver fibrosis.
Fig. 8. 4-MU treatment prevents liver fibrosis progression.
(A) Representative immunofluorescence staining images. HSCs were isolated from Col-GFP mice. Two days after seeding, cells were treated with either vehicle (Veh) or 0.5 mM 4-MU for 6 days. (B) Quantification of GFP-positive area in (A) by ImageJ (n = 3). (C) Col1a1 mRNA expression (n = 3). (D) Representative images of HA and Sirius red staining in mouse liver. One week after BDL, mice were orally gavaged with either Veh or 4-MU (450 mg/kg) once daily for 2 weeks. (E) Liver HA content after 4-MU treatment (Sham-Veh, n = 4; Sham–4-MU, n = 5; BDL-Veh, n = 11; BDL–4-MU, n = 10). (F) Quantification of Sirius red staining in (D) (Sham-Veh, n = 4; Sham–4-MU, n = 5; BDL-Veh, n = 5; BDL–4-MU, n = 10). (G) Sirius red staining in mouse liver. At 11 weeks of CDHFD feeding, mice were orally gavaged daily with Veh or 4-MU (450 mg/kg) for 4 weeks (ND, n = 6 per group; CDHFD-Veh, n = 9; CDHFD–4-MU, n = 10). Data are means ± SEM. **P < 0.01. Two-tailed Student’s t test (B and C) and one-way ANOVA with Tukey’s post hoc analysis (E to G). Scale bars, 100 μm.
DISCUSSION
Currently, there are no effective antifibrotic agents for liver fibrosis. Many of the known pathways, such as TGF-β, cannot be easily targeted because of side effects (24). Our current study establishes that HA—to date, only known as a noninvasive serum biomarker for liver fibrosis—is actively synthesized in the liver and contributes to the development of liver fibrosis. Among the three major HAS enzymes, hepatic HAS2 expression correlates well with fibrosis stages and serum HA concentrations (Fig. 1). Has2 was markedly up-regulated in activated HSCs and in murine and human liver fibrosis. Targeted deletion of Has2 in HSCs greatly abrogated HA production and reduced liver fibrosis induced by various etiologies, indicating that HSCs are the dominant source of HAS2 and HA in the mouse models of liver fibrosis that we examined. Hence, liver fibrosis is associated with not only increased serum HA due to loss of LSEC-mediated HA clearance (12, 13) but also active HA synthesis by HAS2 within the liver. We identified that Notch1 is a downstream effector of HA and plays a crucial role in HSC activation and liver fibrosis. We also demonstrated the interventional potential of targeting HA synthesis for the treatment of liver fibrosis. We have provided evidence that, in NASH-mediated fibrosis, HAS2 plays a critical role in the progression of fibrosis. Because the prevalence of NASH-mediated liver fibrosis is increasing and no effective treatment for NASH currently exists, our study provides new insight into targeting HAS2 and HA synthesis as attractive interventions for NASH-induced fibrosis.
In a physiological condition, circulating and hepatic HA concentrations are regulated by the appropriate balance between production and clearance. The half-life of plasma HA is 2.5 to 5.5 min, and one-third of the total body HA is turned over daily (25, 26). LSECs play a prominent role in the elimination of circulating HA in the liver, contributing to the rapid clearance and turnover of HA. This could be associated with a few or no deposition of HA in the normal liver (Figs. 2A and 5B). Tracing of circulating HA labeled by an in vivo injection of fluorescently conjugated HA-binding protein using intravital microscopy demonstrated that the labeled circulating HA was bound to LSECs (27). This finding suggests that LSECs could capture and eliminate HA from the circulation.
We previously reported the pathological role of HAS2 overexpression and HA production in lung injury and fibrosis in mice and patients with idiopathic pulmonary fibrosis (9). In the lung, CD44 expressed in lung fibroblasts contributes to HAS2-mediated fibrosis (9). We recently demonstrated that HA expressed in AEC2s plays a role in renewal of these lung progenitor cells (10). In cancer, HA participates in cancer stem cell maintenance through CD44. It appears that HAS2-generated HA has distinct roles, depending on the site of expression, context of disease, and the size of HA (7). HMW-HA is protective against injury and inflammation (8). However, LMW-HA and HA oligomers are proinflammatory, except for the 35-kDa HA that has been shown to prevent alcoholic liver disease (28). During the progression of liver fibrosis, LMW-HA markedly increases in blood and liver tissues and is almost absent in normal conditions. This suggests that LMW-HA may be a sensitive biomarker for evaluating early liver fibrosis and/or regression of fibrosis after treatment.
Our experiment using Has2ΔHSC mice determined that HSCs are the responsible cells expressing HAS2 for HA production in liver fibrosis. We examined the regulatory mechanisms driving HAS2 expression in HSCs. TGF- β is a robust fibrotic mediator that up-regulates Has2 expression in HSCs. We identified WT1-binding elements in the HAS2 promoter. In idiopathic pulmonary fibrosis, WT1 is overexpressed in mesothelial and mesenchymal cells and promotes pulmonary fibroblast activation and ECM production (29), further corroborating our data showing that WT1 plays a crucial role in TGF-β–mediated HAS2 induction and HSC activation. It is conceivable that WT1 plays a fibrogenic role in general populations of HSCs, which may not be limited in mesothelial cell–derived HSCs.
We found that HAS2-produced HA can mediate HSC transdifferentiation to ECM-producing myofibroblasts in an autocrine manner. HAS2-generated HA induces production of ECM through CD44 or TLR4. HAS2-generated HA also promotes invasive phenotypes of HSCs, which corroborates our previous study showing HAS2-mediated invasive phenotypes of lung myofibroblasts through CD44 (9). Activated myofibroblasts migrate toward the fibrillary matrix by acquiring an invasive capacity to destroy the basement membrane in fibrotic disease. In liver fibrosis, activated HSCs migrate to inflammatory and fibrotic sites and must navigate the basement membrane and ECM in the space of Disse, activities that further perpetuate HSC activation and enhance inflammation and fibrosis (30, 31). Previous studies suggest that matrix metalloproteinases (MMPs) and integrins α1β1 and α2β1 mediate the degradation of surrounding ECM and promote HSC migration in response to ECM (30, 31).
Notch1 signaling requires sequential proteolytic cleavage of Notch receptors to release the Notch-ICD, which translocates to the nucleus and binds to RBPj to induce Notch target genes, such as Hes1 and Hey1. Patients with Alagille syndrome, who have mutations in the NOTCH ligand JAGGED1, display hypoplastic intrahepatic bile ducts and slow fibrosis progression with limited ECM deposition, suggesting Notch as a driver for liver fibrosis (32). Notch transcripts are up-regulated in patients with biliary fibrosis (33), and Notch inhibitors suppress TGF-β–induced HSC activation and liver fibrosis (18). Jagged-1 and Hes1 induce fibrogenic phenotypes of HSCs (14, 32). Hes1 can bind to COL1A1 and COL1A2 promoters and induce these genes (34). These findings corroborate our data showing that Notch1 promotes ECM production in HSCs. Notch1 expression could be transcriptionally regulated by HAS2 and CD44. Similar to the Notch1 receptor, CD44 activation induces CD44 cleavage by metalloproteases and β-secretase, releasing the CD44-ICD into the cytoplasm (35). CD44-ICD is then translocated into the nucleus and where it regulates expression of specific genes, such as MMP-9 (36). Our data show that CD44-ICD promotes NOTCH1 transcription. Furthermore, TLR4 regulation of Notch1 has been reported (37, 38). Conversely, Notch signaling can augment or inhibit TLR4-triggered inflammatory responses, suggesting that Notch1 regulation of TLR4 activation is context dependent (39, 40).
While HMW-HA has tissue protective effects, HMW-HA is degraded into LMW-HA in the setting of inflammation (7), which alters the homeostatic environment. Thus, the administration of HMW-HA may not be an ideal therapy for liver fibrosis. A phase 3 clinical trial is in progress investigating cancer treatment with hyaluronidase(s) to accelerate HA degradation (41). However, this strategy could produce intermediate LMW-HA products that accumulate because of dysfunctional LSECs in cirrhosis, causing detrimental effects. 4-MU can reduce fibroblast HA production by depleting cellular uridine diphosphate glucuronic acid (23). The pharmacological efficacy of 4-MU was widely tested in cancers (23). 4-MU is a safe and well-tolerated agent, and no serious adverse effects were reported (23). Our data show that 4-MU treatment inhibited HSC activation, suggesting that 4-MU inhibits autocrine production of HA and subsequent HSC activation. Moreover, our study revealed that inhibiting endogenous HA synthesis by 4-MU reduced liver fibrosis. Thus, repurposing 4-MU could be a safe interventional strategy for treating liver fibrosis.
There are limitations regarding HA-mediated Notch1 activation in our study. We have shown the underlying mechanism of HA-CD44– mediated Notch1 up-regulation. However, Notch1 receptor activation requires engagement by its ligand, and its exact activation mechanism in liver fibrosis remains elusive. HA is known to capture various ligands, shuttling them to cell surface receptors (42). The soluble form of Jagged-1 may be shuttled by HA to Notch1. Unlike membrane-bound Jagged-1, the soluble forms of Jagged-1 generally antagonize Notch1 signaling (43). The engagement of Notch1 by the membrane-bound Jagged-1 could be crucial for Notch1 activation. Because Kupffer cells and HSCs express Jagged-1, it is conceivable that HSC-derived HA and chemokines, such as CCL2, promote the recruitment of Jagged-1–expressing Kupffer cells and HSCs to Notch1-expressing HSCs. Membrane-bound Jagged-1 could then bind to Notch1 in HSCs through cell-cell interactions, thereby activating Notch1. However, the hypothesis that HA-mediated Jagged-1 and Notch1 interact in HSCs needs further investigation. Nevertheless, our study provides new insight into the critical role of overexpressed HAS2 and HA in mediating HSC activation and liver fibrosis (fig. S11F). The profibrogenic role of Notch1 and a relationship between Notch1 and HAS2 in regulating liver fibrosis were also identified. Our study suggests that targeting HAS2 and inhibiting HA production through repurposing 4-MU may be an effective strategy to treat or prevent liver fibrosis. This strategy is likely to avoid the many known side effects associated with TGF-β inhibition (24).
MATERIALS AND METHODS
Study design
This study was designed to determine the role of HAS2-mediated HA production in liver fibrosis and to investigate the therapeutic potential of HA inhibition. All human sample collection and study protocol were approved by Clinical Ethics Committee of Putuo Hospital of Shanghai University of Traditional Chinese Medicine, Cedars-Sinai Medical Center, California Liver Research Institute, and the Princess Alexandra Hospital. For in vivo studies, study protocols were approved by Cedars-Sinai Medical Center Institutional Animal Care and Use Committee. The ARRIVE (Animal Research: Reporting of In Vivo Experiments) guideline was followed. Sample size for animal studies was based on statistical analysis of variance and on previous experience with similar in vivo studies. On the basis of pre-established methodology in our laboratory, we rarely exclude values except for technical reasons such as inability to obtain enough serum/tissue or animals reported to be in distress by Comparative Medicine staff at Cedars-Sinai Medical Center. Animals were randomly assigned to experimental groups. Experimentalists were not blinded to different treatments. However, analysis of samples was performed blindly. There was no blinding in cell culture experiments. Most experiments were independently replicated three or more times. A sample size (n) for each experimental group/condition is indicated in the figure legends. Individual subject-level values are reported in data file S1.
Human samples
The study used deidentified liver tissues that were obtained from 65 patients with chronic HBV infection with clinically diagnosed liver fibrosis who underwent liver biopsy from February 2013 to July 2016 in the Putuo Hospital, Shanghai University of Traditional Chinese Medicine (Shanghai, China). Before biopsy, the patients had not received antiviral or other drugs. The stage of liver fibrosis was determined by Department of Pathology, Putuo Hospital. Exclusion criteria included renal and/or hepatic failure, acute coronary syndromes, valvular heart diseases, autoimmune thyroid diseases, or systematic inflammatory diseases. The liver biopsy samples of 55 patients with chronic hepatitis C were collected between 2000 and 2015 at the Princess Alexandra Hospital (Brisbane, Australia). Details of this cohort are detailed in (17). Liver biopsy samples and serum samples of patients with NAFLD were collected from April 2016 to February 2018 in Cedars-Sinai Medical Center (Los Angeles, CA) and California Liver Research Institute (Pasadena, CA). Pathologists reviewed the specimens and determined the degrees of fat, lobular/portal inflammation, hepatocyte ballooning, and fibrosis stage. Controls (F0) were obtained from patients with NAFLD who have found no fibrosis on biopsies. Deidentified samples were used. Written informed consent was obtained from all patients.
Mice
Lrat-Cre Tg, Has2fl/fl, ASMA-HAS2 Tg, Collagen α1(I)–GFP Tg, and Cd44−/− mice, described previously (21, 44–47), were used at 8 to 12 weeks of age in C57BL/6 background. Tlr4−/− mice were provided by S. Akira (Osaka University, Japan) (48). Notch1fl/fl mice were purchased from the Jackson Laboratory. Mice carrying the floxed allele of Has2 or Notch1 were crossed with Lrat-Cre Tg mice to generate Has2ΔHSC or Notch1ΔHSC, respectively. To generate Notch1ΔHSC/ ASMA-HAS2 Tg mice, ASMA-HAS2 Tg mice were crossed with Notch1ΔHSC mice. Collagen α1(I)–GFP Tg mice were crossed with ASMA-HAS2 Tg. Mice were back-crossed at least 10 generations onto the C57BL/6 background. All studies were performed in accordance with National Institutes of Health (NIH) recommendations outlined in the Guide for the Care and Use of Laboratory Animals.
Liver fibrosis models
For the BDL model, mice were anesthetized with the injection of ketamine and xylazine. After midline laparotomy, the common bile duct was ligated twice with 6–0 silk sutures, and the abdomen was closed. Sham operation was performed similarly, except that the bile duct was not ligated. For DDC model, mice were fed with 0.1% DDC-supplemented diet (catalog no. S4643, Bio-Serv) for 4 weeks. For diet-induced NASH fibrosis model, mice were fed with CDHFD (60% of calorie from fat; catalog no. D05010403, Research Diets) for 12 weeks or CDAA diet (catalog no. 518753, Dyets Inc.) for 22 weeks. Rodent standard chow was used as a control diet. For the CCl4 model, mice were injected with CCl4 (0.5 μl CCl4/g mouse body weight, 1:4 dilution with corn oil) (catalog no. 319961, Sigma-Aldrich) intraperitoneally every 3 days for a total of 10 injections. Control mice received corn oil. For TAA model, TAA (catalog no. 163678, Sigma-Aldrich) was administered at a dose of 300 mg/liter in drinking water for 20 weeks.
Histologic examination
For α-SMA staining in mouse liver tissues, mouse anti–α-SMA monoclonal antibody (1:200; catalog no. M0851, Dako) and the M.O.M. Kit (catalog no. MKB-2225, Vector Laboratories) were applied according to the manufacturer’s instruction. For F4/80 staining, monoclonal antibody to F4/80 (clone BM8; catalog no. 14–4801, eBioscience) was applied. To detect HA in livers of DDC diet–fed mice (fig. S4A), Avidin, AP conjugate (catalog no. 434422, Invitrogen), and ImmPACT Vector Red Alkaline Phosphatase substrate (catalog no. SK-5105, Vector Laboratories) were used to distinguish HA from endogenous bile. Neutral lipids were analyzed by Oil Red O staining. Oil Red O–positive area was evaluated from randomly selected 10 fields of ×200 magnification per slide by ImageJ. HA staining was performed as previously described (8). After routine deparaffinization, hydration, and blockage of endogenous peroxidase, tissues were incubated with biotin-labeled HA-binding protein (4 μg/ml) [rhAggrecan aa20–675/His (NSO/7), biotin, R&D Systems] for 2.5 hours and then incubated and developed using a VECTASTAIN Elite ABC kit and DAB Peroxidase Substrate kit (catalog nos. PK-6100 and SK-4100, Vector Laboratories).
HA ELISA and HA fragmentation
We measured amounts of HA in the serum or the liver homogenates or the culture supernatants with the commercial Hyaluronan DuoSet ELISA Kit (catalog no. DY3614, R&D Systems). Samples were run in duplicate. A size-specific HA fraction in human serum and mouse liver homogenates was determined as previously described (47), with modification. Liver homogenates were digested with Pronase at 60°C for 4 hours and then boiled for 30 min. The samples were fractionated on 100- and 300-kDa molecular weight cutoff columns (catalog nos. 13269-E and 13279-E, Centrisart, Sartorius). The filtered HA with molecular weights less than 100 kDa or less than 300 kDa were collected, and the amount of HA in the unfiltered and filtered serum or liver homogenate samples was quantified by the Hyaluronan DuoSet ELISA Kit.
RNA-seq sample preparation and sequencing
Total RNA was extracted from primary HSCs using TRIzol and purified using the NucleoSpin RNA Kit, according to the manufacturer’s instructions. RNA purity, concentration, and integrity were evaluated with an Agilent 2100 BioAnalyzer (Agilent Technologies). Beijing Genomic Institute performed the RNA-seq. RNA libraries were prepared for sequencing via an Illumina HiSeq 4000 using standard BGISEQ-500 protocols.
RNA-seq data analysis
TopHat2 (version 2.0.12) was applied to align sequencing reads to the reference mouse genome (GRCm38/mm10). The relative gene expression RPKM and read counts were estimated using SAMMate (version 2.7.4) and HTSeq (version 0.6.1) with Ensembl database (Mus Musculus.GRCm38.82). DESeq2 was applied to conduct differential gene expression analysis using read counts. The P values of multiple tests were adjusted using Benjamini-Hochberg’s method, and the significance level was designated as false discovery rate (FDR) < 0.01 and |log2 FC| > = 1. Pathway and Gene Ontology term enrichment analysis of differentially expressed genes was performed using DAVID. For analysis of HAS1, HAS2, and HAS3 expression in the hepatitis C virus (HCV) patient cohort, normalized RNA-seq read counts (counts per million) on unfiltered data were analyzed and presented as previously described (17).
Quantitative real-time polymerase chain reaction
Total RNA from snap-frozen liver tissues were extracted using the NucleoSpin RNA kit (catalog no. 740955.50, Macherey-Nagel), and the total RNA from cells were extracted using TRIzol RNA isolation reagents (catalog no. 15596–018, Invitrogen). It was reverse-transcribed to complementary DNA (cDNA) with the PrimeScript RT Reagent Kit (catalog no. RR037A, Takara) or High-Capacity cDNA Reverse Transcription Kit (catalog no. 4368814, Applied Biosystems). qPCR was performed using SYBR Green (catalog no. RR420A, Takara) or iTaq Univer SYBR Green Supermix 1000 (catalog no. 1725122, Bio-Rad) by ABI ViiA 7Dx real-time PCR system (Applied Biosystems) or CFX96 real-time PCR system (Bio-Rad). The qPCR primer sequences used in this study are listed in table S1. 18S ribosomal RNA was used as an internal control for the normalization.
In vitro treatment
The HSCs were isolated and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) for 3 days. The cells were deprived of serum overnight and subsequently exposed to LMW-HA (100 μg/ml; 135 kDa) in the presence of polymyxin B (10 μg/ml; catalog no. 5291, Sigma-Aldrich) (8), HMW-HA (600 μg/ml; Healon, Kabi Pharmacia Ophthalmics Inc.), LPS (100 ng/ml; catalog no. L9764, Sigma-Aldrich), or TGF-β (5 ng/ml; catalog no. 580704, BioLegend) for 12 hours. Kupffer cells were maintained in RPMI medium with 10% FBS for 6 hours and then serum deprived for overnight. The next day, cells were treated with LMW-HA (100 μg/ml) in the presence of polymyxin B (10 μg/ml) or HMW-HA (600 μg/ml) for 4 hours. To deplete Notch1 in HSCs, cells were isolated from Notch1fl/fl mice and cultured in DMEM with 10% FBS for 2 days and then infected with recombinant adenovirus (Ad) expressing GFP or Cre recombinase. Cells were collected at day 10. The infection efficiency was confirmed by GFP visualization, and the efficiency of Notch1 deletion by Ad-Cre transduction was monitored by qPCR.
Promoter luciferase assay
The HEK293A cells were maintained in DMEM containing 10% FBS, penicillin (50 U/ml), and streptomycin (50 μg/ml) at 37°C in a humidified atmosphere with 5% CO2. The human NOTCH1 promoter (−1239/+1) and customized human NOTCH1 promoter (−1239/+1684) vectors were purchased from GeneCopoeia (catalog no. HPRM48887-PG04). The NOTCH1 promoter luciferase mutant exhibiting point mutations in the CIRE2 at position +1517 (WT: CCTGCG; mut: AAGTAT) from NOTCH1 promoter (−1239/+1684) vector was generated by GENEWIZ Inc. The 2880–base pair (bp) Has2 promoter luciferase reporter plasmid was provided by C. Chiang (Vanderbilt University Medical Center) (49). Three different Has2 promoter luciferase mutants exhibiting point mutations in the WT1-binding motifs at positions −2057 bp (WT: AGGGGGGGGCG; mut1: AGGGGacGatG), −1002 bp (WT: GGGCAGGGGCG; mut2: GGGCAacGatG), and −636 bp (WT: CTGCGGGGGGG; mut3: CTGCGacGatG) from the transcription start site (TSS) were generated from the original 2880-bp Has2 promoter reporter. We performed mutagenesis of pGL3-Has2 starting construct via 5′ Pvull and 3′ BspHI to generate pGL3-Has2-mut1 or pGL3-Has2-mut2 vectors, with sequence verification across the region of interest (1373 bp). Mutagenesis starting construct via 5′ BspHI and 3′ Spel was done to generate pGL3-Has2-mut3, with sequence verification (160 bp) by sequencing (GENEWIZ Inc.). Luciferase assays were performed using the Secrete-Pair Dual Luminescence Assay Kit (catalog no. LF032, GeneCopoeia) or Dual-Glo Luciferase Assay System (catalog no. E2920, Promega), in accordance with the manufacturers’ instruction. HEK293 cells were cotransfected with promoter constructs and CD44- or WT1-expressing plasmid (catalog no. MR225851, Origene) for 24 to 48 hours before the luciferase assay.
4-MU in vivo treatment
4-MU or vehicle control (2% sucrose) was given via oral gavage of 450 mg/kg once daily. WT or Has2ΔHSC or Notch1ΔHSC mice were treated with 4-MU or vehicle control starting from days 7 to 21 after BDL. In a separate set of experiments, after 11 weeks of CDHFD feeding, C57BL/6 mice were orally gavaged daily with vehicle or 4-MU for 4 weeks under continuous feeding of CDHFD.
Statistical analysis
Statistical significance was assessed by using GraphPad Prism 5.01 software (GraphPad Software Inc). Differences between the two groups were compared using a two-tailed unpaired Student’s t test. Differences between multiple groups were compared using one-way ANOVA, followed by Tukey’s post hoc analysis. Multiple linear regression analysis was performed to find the correlation of hepatic HAS2 expression with serum HA contents or genes of interests. P values of <0.05 were considered significant.
Supplementary Material
Fig. S1. Expression of HAS in patients with fibrosis.
Fig. S2. Expression of HA and Has expression in mouse liver cells.
Fig. S3. Increased susceptibility to BDL- or CCl4-induced liver fibrosis in ASMA-HAS2 Tg mice.
Fig. S4. Generation of HSC-specific Has2 knockout mice.
Fig. S5. Effect of HSC-specific Has2 deficiency on DDC-induced liver fibrosis.
Fig. S6. No effect of HSC-specific Has2 deletion on CDHFD-mediated fat accumulation or glucose intolerance.
Fig. S7. Cd44 and Tlr4 mRNA expression in in vivo–activated HSCs.
Fig. S8. HA concentration in the supernatant of WT HSCs or HAS2 Tg HSCs.
Fig. S9. Notch1 and its target gene Hes1 mRNA expression.
Fig. S10. Jagged-1 expression.
Fig. S11. Inhibition of BDL-induced HSC activation and macrophage infiltration by 4-MU treatment.
Table S1. Sequence of primers used for qPCR.
Data file S1. Individual subject-level data.
Acknowledgments:
We thank L. Rodríguez-Rodríguez and K. E. Miletti-González for pcDNA/ CD44 plasmid vector and F. Miao for technical support.
Funding: This work is supported by the NIH (R01DK085252 to E.S. and M.N., R21AA025841 to E.S. and S.C.L., R01AA027036 to E.S., R01DK107288 to S.C.L., R01HL122068 to D.J. and P.W.N., R01AI052201 to P.W.N., 1T32HL134637–01 to Y.M.Y. and P.W.N., P01HL108793 to P.W.N.), the American Liver Foundation Postdoctoral Fellowship (to Y.M.Y. and Y.S.R.), AASLD The Leonard B. Seeff Young Investigator Award (to Y.M.Y. and Y.S.R.), Winnick Research award from Cedars-Sinai Medical Center (to E.S.), Basic Science Research Program of the Ministry of Education (NRF-2014R1A6A3A01054056 to Y.M.Y.), the National Natural Science Foundation of China (no. 81673788 to C.L.), Peak Discipline of Colleges in Shanghai (no. A-U151902 to C.L.), the IMB Centre for Inflammation and Disease Research at the University of Queensland and the National Health and Medical Research Council of Australia (NHMRC Senior Research Fellowship to M.J.S., ID: 1107914), and the National Research Foundation of Korea (2017R1A5A2015541 and 2017R1C1B2004423 to Y.S.R.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Lrat-Cre Tg mice are available from R.F.S. under a material transfer agreement with Columbia University. RNA-seq data have been uploaded to the GEO (accession nos. GSE108593 and GSE109200). HCV RNA-seq data have been submitted to ArrayExpress (www.ebi.ac.uk/arrayexpress/) under accession no. E-MTAB-6863.
stm.sciencemag.org/cgi/content/full/11/496/eaat9284/DC1
Materials and Methods
REFERENCES AND NOTES
- 1.Bataller R, Brenner DA, Liver fibrosis. J. Clin. Invest 115, 209–218 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuchida T, Friedman SL, Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol 14, 397–411 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Murphy SL, Xu J, Kochanek KD, Deaths: Final data for 2010. Natl. Vital Stat. Rep 61, 1–117 (2013). [PubMed] [Google Scholar]
- 4.Sgalla G, Cocconcelli E, Tonelli R, Richeldi L, Novel drug targets for idiopathic pulmonary fibrosis. Expert Rev. Respir. Med, 1–13 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M, Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Chedid MF, Nonalcoholic steatohepatitis: The second leading indication for liver transplantation in the USA. Dig. Dis. Sci 62, 2621–2622 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Jiang D, Liang J, Noble PW, Hyaluronan as an immune regulator in human diseases. Physiol. Rev 91, 221–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, Homer RJ, Goldstein DR, Bucala R, Lee PJ, Medzhitov R, Noble PW, Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med 11, 1173–1179 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Jiang D, Liang J, Meltzer EB, Gray A, Miura R, Wogensen L, Yamaguchi Y, Noble PW, Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J. Exp. Med 208, 1459–1471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang J, Zhang Y, Xie T, Liu N, Chen H, Geng Y, Kurkciyan A, Mena JM, Stripp BR, Jiang D, Noble PW, Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat. Med 22, 1285–1293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seki E, De Minicis S, Österreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF, TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med 13, 1324–1332 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Tamaki S, Ueno T, Torimura T, Sata M, Tanikawa K, Evaluation of hyaluronic acid binding ability of hepatic sinusoidal endothelial cells in rats with liver cirrhosis. Gastroenterology 111, 1049–1057 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Satoh T, Ichida T, Matsuda Y, Sugiyama M, Yonekura K, Ishikawa T, Asakura H, Interaction between hyaluronan and CD44 in the development of dimethylnitrosamine-induced liver cirrhosis. J. Gastroenterol. Hepatol 15, 402–411 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Hu B, Phan SH, Notch in fibrosis and as a target of anti-fibrotic therapy. Pharmacol. Res 108, 57–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Xia Y, Chen K, Zheng Y, Wang J, Lu W, Yin Q, Wang F, Zhou Y, Guo C, Oncogenic role of the Notch pathway in primary liver cancer. Oncol. Lett 12, 3–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nijjar SS, Wallace L, Crosby HA, Hubscher SG, Strain AJ, Altered Notch ligand expression in human liver disease: Further evidence for a role of the Notch signaling pathway in hepatic neovascularization and biliary ductular defects: Further evidence for a role of the Notch signaling pathway in hepatic neovascularization and biliary ductular defects. Am. J. Pathol 160, 1695–1703 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramnath D, Irvine KM, Lukowski SW, Horsfall LU, Loh Z, Clouston AD, Patel PJ, Fagan KJ, Iyer A, Lampe G, Stow JL, Schroder K, Fairlie DP, Powell JE, Powell EE, Sweet MJ, Hepatic expression profiling identifies steatosis-independent and steatosis-driven advanced fibrosis genes. JCI Insight 3, 120274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal R, van Baarlen J, Storm G, Prakash J, The interplay of the Notch signaling in hepatic stellate cells and macrophages determines the fate of liver fibrogenesis. Sci. Rep 5, 18272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu C, Kim K, Wang X, Bartolome A, Salomao M, Dongiovanni P, Meroni M, Graham MJ, Yates KP, Diehl AM, Schwabe RF, Tabas I, Valenti L, Lavine JE, Pajvani UB, Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med 10, eaat0344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuman MG, Cohen LB, Nanau RM, Hyaluronic acid as a non-invasive biomarker of liver fibrosis. Clin. Biochem 49, 302–315 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF, Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun 4, 2823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf MJ, Adili A, Piotrowitz K, Abdullah Z, Boege Y, Stemmer K, Ringelhan M, Simonavicius N, Egger M, Wohlleber D, Lorentzen A, Einer C, Schulz S, Clavel T, Protzer U, Thiele C, Zischka H, Moch H, Tschöp M, Tumanov AV, Haller D, Unger K, Karin M, Kopf M, Knolle P, Weber A, Heikenwalder M, Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell 26, 549–564 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Nagy N, Kuipers HF, Frymoyer AR, Ishak HD, Bollyky JB, Wight TN, Bollyky PL, 4-methylumbelliferone treatment and hyaluronan inhibition as a therapeutic strategy in inflammation, autoimmunity, and cancer. Front. Immunol 6, 123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walton KL, Johnson KE, Harrison CA, Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol 8, 461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraser JRE, Laurent TC, Engström-Laurent A, Laurent UGB, Elimination of hyaluronic acid from the blood stream in the human. Clin. Exp. Pharmacol. Physiol 11, 17–25 (1984). [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi Y, Yamamoto H, Tobisawa Y, Irie F, TMEM2: A missing link in hyaluronan catabolism identified? Matrix Biol 78–79, 139–146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald B, McAvoy EF, Lam F, Gill V, de la Motte C, Savani RC, Kubes P, Interaction of CD44 and hyaluronan is the dominant mechanism for neutrophil sequestration in inflamed liver sinusoids. J. Exp. Med 205, 915–927 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saikia P, Bellos D, McMullen MR, Pollard KA, de la Motte C, Nagy LE, MicroRNA 181b-3p and its target importin α5 regulate toll-like receptor 4 signaling in Kupffer cells and liver injury in mice in response to ethanol. Hepatology 66, 602–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sontake V, Kasam RK, Sinner D, Korfhagen TR, Reddy GB, White ES, Jegga AG, Madala SK, Wilms’ tumor 1 drives fibroproliferation and myofibroblast transformation in severe fibrotic lung disease. JCI Insight 3, 121252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C, Zeisberg M, Mosterman B, Sudhakar A, Yerramalla U, Holthaus K, Xu L, Eng F, Afdhal N, Kalluri R, Liver fibrosis: Insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology 124, 147–159 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Olaso E, Ikeda K, Eng FJ, Xu L, Wang L-H, Lin HC, Friedman SL, DDR2 receptor promotes MMP-2-mediated proliferation and invasion by hepatic stellate cells. J. Clin. Invest 108, 1369–1378 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisler F, Strazzabosco M, Emerging roles of Notch signaling in liver disease. Hepatology 61, 382–392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka A, Leung PSC, Kenny TP, Au-Young J, Prindiville T, Coppel RL, Ansari AA, Gershwin ME, Genomic analysis of differentially expressed genes in liver and biliary epithelial cells of patients with primary biliary cirrhosis. J. Autoimmun 17, 89–98 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Hu M, Ou-Yang H-F, Wu C-G, Qu S-Y, Xu X-T, Wang P, Notch signaling regulates col1α1 and col1 α2 expression in airway fibroblasts. Exp. Biol. Med 239, 1589–1596 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC, Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 14, 357–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miletti-González KE, Murphy K, Kumaran MN, Ravindranath AK, Wernyj RP, Kaur S, Miles GD, Lim E, Chan R, Chekmareva M, Heller DS, Foran D, Chen W, Reiss M, Bandera EV, Scotto K, Rodríguez-Rodríguez L, Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem 287, 18995–19007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, Osborne BA, Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol 38, 174–183 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-García A, López-López S, García-Ramírez JJ, Baladrón V, Ruiz-Hidalgo MJ, López-Sanz L, Ballesteros A, Laborda J, Monsalve EM, Díaz-Guerra MJM, The Tetraspanin TSPAN33 Controls TLR-Triggered Macrophage Activation through Modulation of NOTCH Signaling. J. Immunol 197, 3371–3381 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Zeng Q, Jin C, Ao L, Cleveland JC Jr., Song R, Xu D, Fullerton DA, Meng X, Cross-talk between the Toll-like receptor 4 and Notch1 pathways augments the inflammatory response in the interstitial cells of stenotic human aortic valves. Circulation 126, S222–S230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Wang C, Liu Z, Liu X, Han C, Cao X, Li N, Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J. Biol. Chem 287, 6208–6217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doherty GJ, Tempero M, Corrie PG, HALO-109–301: A Phase III trial of PEGPH20 (with gemcitabine and nab-paclitaxel) in hyaluronic acid-high stage IV pancreatic cancer. Future Oncol 14, 13–22 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Turley EA, Wood DK, McCarthy JB, Carcinoma cell hyaluronan as a “portable” cancerized prometastatic microenvironment. Cancer Res 76, 2507–2512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klose R, Berger C, Moll I, Adam MG, Schwarz F, Mohr K, Augustin HG, Fischer A, Soluble Notch ligand and receptor peptides act antagonistically during angiogenesis. Cardiovasc. Res 107, 153–163 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, Dealy CN, Toole BP, Takeda J, Yamaguchi Y, Kosher RA, Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development 136, 2825–2835 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai S, Chai Q, Danielsen CC, Hjorth P, Nyengaard JR, Ledet T, Yamaguchi Y, Rasmussen LM, Wogensen L, Overexpression of hyaluronan in the tunica media promotes the development of atherosclerosis. Circ. Res 96, 583–591 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, Rippe RA, DNase I-hypersensitive sites enhance α1(I) collagen gene expression in hepatic stellate cells. Hepatology 37, 267–276 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Schmits R, Filmus J, Gerwin N, Senaldi G, Kiefer F, Kundig T, Wakeham A, Shahinian A, Catzavelos C, Rak J, Furlonger C, Zakarian A, Simard JJL, Ohashi PS, Paige CJ, Gutierrez-Ramos JC, Mak TW, CD44 regulates hematopoietic progenitor distribution, granuloma formation, and tumorigenicity, and tumorigenicity. Blood 90, 2217–2233 (1997). [PubMed] [Google Scholar]
- 48.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S, Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol 162, 3749–3752 (1999). [PubMed] [Google Scholar]
- 49.Liu J, Li Q, Kuehn MR, Litingtung Y, Vokes SA, Chiang C, Sonic hedgehog signaling directly targets Hyaluronic Acid Synthase 2, an essential regulator of phalangeal joint patterning. Dev. Biol 375, 160–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of HAS in patients with fibrosis.
Fig. S2. Expression of HA and Has expression in mouse liver cells.
Fig. S3. Increased susceptibility to BDL- or CCl4-induced liver fibrosis in ASMA-HAS2 Tg mice.
Fig. S4. Generation of HSC-specific Has2 knockout mice.
Fig. S5. Effect of HSC-specific Has2 deficiency on DDC-induced liver fibrosis.
Fig. S6. No effect of HSC-specific Has2 deletion on CDHFD-mediated fat accumulation or glucose intolerance.
Fig. S7. Cd44 and Tlr4 mRNA expression in in vivo–activated HSCs.
Fig. S8. HA concentration in the supernatant of WT HSCs or HAS2 Tg HSCs.
Fig. S9. Notch1 and its target gene Hes1 mRNA expression.
Fig. S10. Jagged-1 expression.
Fig. S11. Inhibition of BDL-induced HSC activation and macrophage infiltration by 4-MU treatment.
Table S1. Sequence of primers used for qPCR.
Data file S1. Individual subject-level data.